Summary
Aquaporin-4 (AQP4) is a water channel expressed in astrocytes, skeletal muscle and epithelial cells that forms supramolecular aggregates in plasma membranes called orthogonal arrays of particles (OAPs). AQP4 is expressed as a short isoform (M23) that forms large OAPs, and a long isoform (M1) that does not form OAPs by itself but can mingle with M23 to form relatively small OAPs. AQP4 OAPs were imaged with ∼20 nm spatial precision by photoactivation localization microscopy (PALM) in cells expressing chimeras of M1- or M23-AQP4 with photoactivatable fluorescent proteins. Native AQP4 was imaged by direct stochastic optical reconstruction microscopy (dSTORM) using a primary anti-AQP4 antibody and fluorescent secondary antibodies. We found that OAP area increased from 1878±747 to 3647±958 nm2 with decreasing M1:M23 ratio from 1:1 to 1:3, and became elongated. Two-color dSTORM indicated that M1 and M23 co-assemble in OAPs with a M1-enriched periphery surrounding a M23-enriched core. Native AQP4 in astrocytes formed OAPs with an area of 2142±829 nm2, which increased to 5137±1119 nm2 with 2-bromopalmitate. PALM of AQP4 OAPs in live cells showed slow diffusion (average ∼10−12 cm2/s) and reorganization. OAP area was not altered by anti-AQP4 IgG autoantibodies (NMO-IgG) that cause the neurological disease neuromyelitis optica. Super-resolution imaging allowed elucidation of novel nanoscale structural and dynamic features of OAPs.
Key words: AQP4, OAP, PALM, Water channel, Neuromyelitis optica, Supramolecular assembly
Introduction
Aquaporin-4 (AQP4) is a water-selective channel expressed at the plasma membrane in astrocytes in brain, spinal cord and sensory tissues, in skeletal muscle, and in epithelial cells in kidney, stomach, airways and exocrine glands (Frigeri et al., 1995). Studies in AQP4 knockout mice have implicated the involvement of AQP4 in brain water balance (Manley et al., 2000; Papadopoulos et al., 2005) neuroexcitation (Padmawar et al., 2005), astrocyte migration (Saadoun et al., 2005) and neuroinflammation (Li et al., 2011). AQP4 is the target antigen in the neuroinflammatory demyelinating disease neuromyelitis optica (NMO), where pathogenic anti-AQP4 autoantibodies (NMO-IgG) cause astrocyte cytotoxicity, leading to paralysis and blindness (Lennon et al., 2005).
AQP4 forms supramolecular aggregates in cell plasma membranes called orthogonal arrays of particles (OAPs), which are regular square arrays of particles that were originally visualized by freeze-fracture electron microscopy (FFEM) (Landis and Reese, 1974; Wolburg et al., 2011). Our lab discovered that AQP4 is the OAP-forming protein, demonstrating OAPs in AQP4-transfected cells (Yang et al., 1996) and their absence in AQP4 knockout mice (Verbavatz et al., 1997). Speculated, though unproven, roles of OAPs include enhanced water permeability (Silberstein et al., 2004; Fenton et al., 2010), cell–cell adhesion (Hiroaki et al., 2006; Zhang and Verkman 2008; Ho et al., 2009) and AQP4 polarization to astrocyte end-feet (Frydenlund et al., 2006; Noell et al., 2009). OAP formation by AQP4 has been reported to be altered in some neuromuscular diseases (Wakayama et al., 2002; Frigeri et al., 2002) and appears to be regulated by various signaling events and post-translational modifications such as N-terminus palmitoylation (Suzuki et al., 2008; Crane et al., 2009).
AQP4 is expressed in two isoforms: a long (M1) isoform with translation initiation at Met-1, and a short (M23) isoform with translation initiation at Met-23 (Jung et al., 1994; Yang et al., 1995; Lu et al., 1996) (Fig. 1A). Like other AQPs, AQP4 is present as tetramers in cell membranes. The M1 and M23 isoforms of AQP4 form heterotetramers, which further assemble into OAPs (Neely et al., 1999; Crane et al., 2009; Tajima et al., 2010; Rossi et al., 2011). M1-AQP4 is present mainly as individual tetramers, whereas M23-AQP4 forms large aggregates containing up to 600 M23-AQP4 tetramers (Rash et al., 2004). Co-expression of M1- and M23-AQP4 produces OAPs that are smaller than those formed by M23-AQP4 alone (Furman et al., 2003; Silberstein et al., 2004). We have extensively studied the determinants and dynamics of OAP formation utilizing biophysical methods, including quantum dot single particle tracking, single molecule photobleaching, and total internal reflection fluorescence microscopy (TIRFM) (Crane et al., 2008; Tajima et al., 2010; Rossi et al., 2011). We found that OAP formation by M23 involves N-terminal interactions between M23 molecules by residues just downstream of Met-23, and that the inability of M1 to form OAPs involves prevention of the N-terminus interaction by residues just upstream of Met-23 (Crane and Verkman, 2009). Recently, we developed a mathematical model of AQP4 OAP formation based on M23–M23 association and M1–M23 heterotetramer formation (Jin et al., 2011). Though some predictions of the model were verified with available freeze-fracture, biophysical and biochemical (native gel) data, the limited resolution of conventional microscopy methods to visualize OAPs in cell membranes precluded direct testing of key predictions.
Fig. 1.
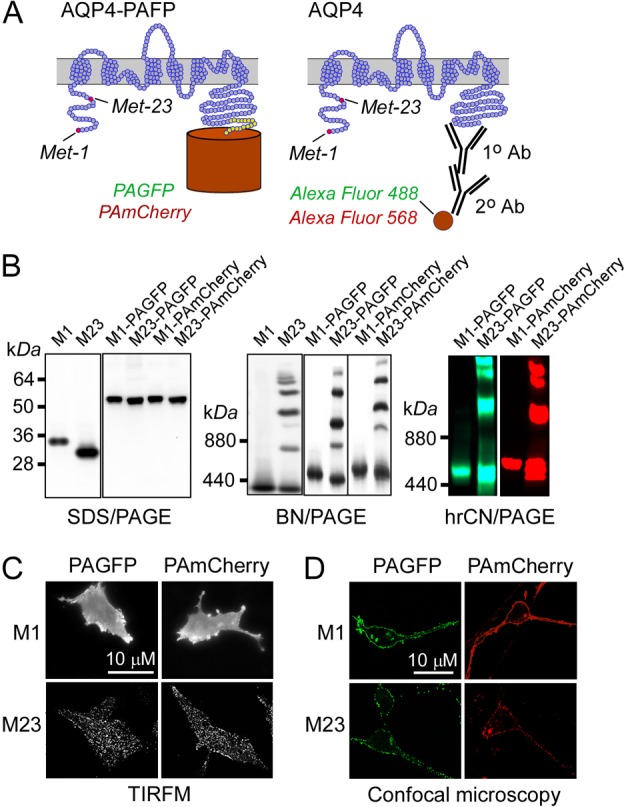
Characterization of cells expressing chimeras of AQP4 and photoactivatable fluorescent proteins. (A) Membrane topography of AQP4 showing C-terminus chimeras with photoactivatable proteins (left) and native AQP4 labeled with an anti-C-terminus AQP4 primary antibody and fluorescent secondary antibodies (right). M1 and M23 translation initiation sites are shown. (B) SDS-PAGE (left) and BN-PAGE (center) immunoblots of total homogenates of U87MG cells expressing native M1- and M23-AQP4 (labeled ‘M1’ and ‘M23’) and M1- and M23-AQP4 containing C-terminus PAGFP or PAmCherry. (Right) hrCN-PAGE showing PAGFP and PAmCherry fluorescence. (C,D) TIRFM (C) and confocal microscopy (D) of indicated AQP4–fluorescent protein chimeras.
Here, we imaged AQP4 OAPs in cell membranes using super-resolution microscopy, including PALM (Betzig et al., 2006) and dSTORM (Heilemann et al., 2009; Löschberger et al., 2012). Based on our prior characterization of AQP4–GFP chimeras showing similar targeting and properties to those of native AQP4 (Tajima et al., 2010; Rossi et al., 2012a), for PALM we generated chimeras of M1- and M23-AQP4 with photoactivatable and photoswitchable fluorescent proteins. Native AQP4 in astrocytes was imaged by dSTORM using an anti-C-terminus AQP4 antibody and fluorescent secondary antibodies. Super-resolution imaging allowed the determination of OAP size, shape and composition in transfected cells and primary astrocyte cultures, elucidating novel structural features of OAPs and testing of a recent hypothesis linking NMO-IgG binding to AQP4 to NMO neuropathology.
Results
Characterization of cells expressing AQP4–fluorescent protein chimeras
PALM was done on AQP4 chimeras containing photoactivatable fluorescent proteins, and dSTORM using Alexa-fluorophore-conjugated secondary antibodies against an anti-C-terminus AQP4 antibody (Fig. 1A). Cells expressing AQP4–fluorescent protein chimeras were characterized to ensure that C-terminus fluorescent protein addition did not interfere with AQP4 plasma membrane targeting or OAP formation. Fig. 1B (left) shows the expected molecular sizes, as determined by SDS-PAGE and AQP4 immunoblot, of the untagged M1 and M23 isoforms and the fluorescent protein chimeras containing photoactivatable (PA)GFP or PAmCherry. OAP formation by untagged M23 and M23–fluorescent protein conjugates was found by blue native polyacrylamide gel electrophoresis (BN-PAGE; Fig. 1B, center) and high resolution colorless native polyacrylamide gel electrophoresis (hrCN-PAGE; Fig. 1B, right), in which multiple bands were seen, as compared to single major bands for untagged M1 or M1–fluorescent protein chimeras. TIRFM confirmed OAP formation in the plasma membrane of cells expressing M23– but not M1–fluorescent protein chimeras (Fig. 1C). Confocal imaging indicated plasma membrane targeting of chimeras containing AQP4 (Fig. 1D), with M1 chimeras showing a smooth pattern of fluorescence and M23 chimeras a punctate pattern, reflecting their different oligomeric states on the plasma membrane. Other photoactivatable or photoconvertible chimeras were tested but not used because some (e.g. AQP4–PATagRFP) were not efficiently targeted to the plasma membrane, while others (e.g. AQP4–Dendra2 and AQP4–EOSFP) showed bright-green fluorescence prior to photoconversion, precluding their use in two-color imaging.
Super-resolution imaging of AQP4 OAPs
The localization precision of our system was characterized by repeated localization of individual spots in coverglass-immobilized lysates of cells expressing AQP1–PAmCherry; AQP1 is a water channel with homology to AQP4 that does not form OAPs. Fig. 2A shows a localization precision of ∼21 nm, as deduced from the full width at half maximum of a normal distribution of spot x,y-positions on repeated measurements. As determined similarly, the localization precision of PAGFP (in AQP1–PAGFP lysates) by PALM was ∼20 nm, and of Alexa Fluor 488 and 568 (on labeled antibodies) by dSTORM were ∼18 nm. Our system was also tested by imaging the F-actin cytoskeletal network in fixed cells using Alexa-Fluor-488-conjugated phalloidin. Fig. 2B (top) shows the substantially improved resolution of the cytoskeleton by dSTORM compared to TIRFM, allowing visualization of individual fibers. Fig. 2B (bottom) shows a smooth pattern of AQP1 fluorescence (labeled with Alexa-Fluor-488-conjugated secondary antibody against an anti-C-terminus AQP1 antibody) in transfected HeLa cells by TIRFM, but well-demarcated individual spots by dSTORM. The average spot diameter was 27±8 nm, providing a measure of the system resolution for dSTORM of labeled cells.
Fig. 2.
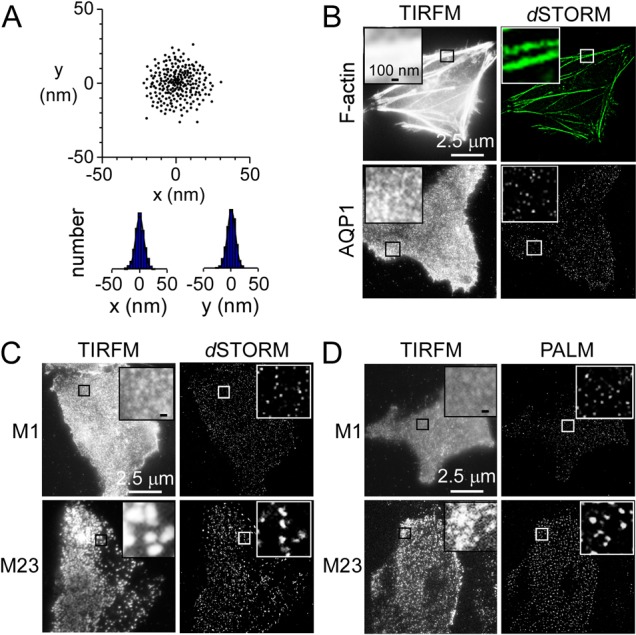
PALM and dSTORM of cells expressing M1- and M23-AQP4. (A) The localization precision of a single fluorophore was determined by localization of immobilized single AQP1–PAmCherry tetramers. Centers-of-mass and deduced spatial histograms with Gaussian fits are shown. (B) Immunofluorescence (TIRFM) and dSTORM of F-actin (top) and AQP1 (bottom) in HeLa cells. Insets show magnified views of boxed regions. (C,D) Immunofluorescence (TIRFM) of M1- and M23-AQP4 in U87MG cells and corresponding dSTORM (C) and PALM (D) reconstructions.
TIRFM and super-resolution imaging (dSTORM, Fig. 2C; PALM, Fig. 2D) of U87MG cells expressing M1- and M23-AQP4 were compared. dSTORM was done labeling an anti-C-terminus AQP4 antibody with an Alexa-Fluor-488-conjugated antibody, and PALM using PAmCherry. TIRFM showed a punctate pattern of M23-AQP4 fluorescence, with the minimal diameter of individual spots >300 nm, representing the diffraction limit. By super-resolution imaging, average spot diameters were 110±31 nm (dSTORM, M23), 121±37 nm (PALM, M23), 26±8 nm (dSTORM, M1), and 28±9 nm (PALM, M1). The similar small spot diameters for M1-AQP4 and AQP1, which are at the system resolution, supports the conclusion that the long AQP4 isoform does not form aggregates in cell membranes.
PALM was done to investigate how relative M1-to-M23 AQP4 expression affects OAP size and shape. U87MG cells were transfected with M1-AQP4–PAmCherry and M23-AQP4–PAmCherry at different ratios. PALM of cells expressing M23-AQP4–PAmCherry alone showed large spots with considerable irregularity and occasional defects (Fig. 3A,C), with average diameter 127±41 nm. However, in cells expressing both AQP4 isoforms at an M1:M23 ratio of 1:3, OAPs were quite elongated, as predicted by our prior mathematical modeling (Jin et al., 2011). At an M1:M23 ratio of 1:1, OAPs were smaller, with average diameter 52±26 nm, and more rounded in shape. Cells expressing M1 alone showed mostly small spots of diameter 25±8 nm, the approximate system resolution. Fig. 3B shows number histograms of the OAP size distribution. M23-AQP4-expressing cells formed OAPs of average area 7853±1243 nm2, whereas cells expressing M1-AQP4 or AQP1 showed small spots of areas 394±41 nm2 and 384±31 nm2, respectively. Co-expression of both isoforms gave OAPs of areas 3647±958 nm2 at an M1:M23 ratio of 1:3 and 1878±747 nm2 at an M1:M23 ratio of 1:1. Fig. 3C summarizes elongation ratios (long-to-short diameter of fitted ellipses).
Fig. 3.
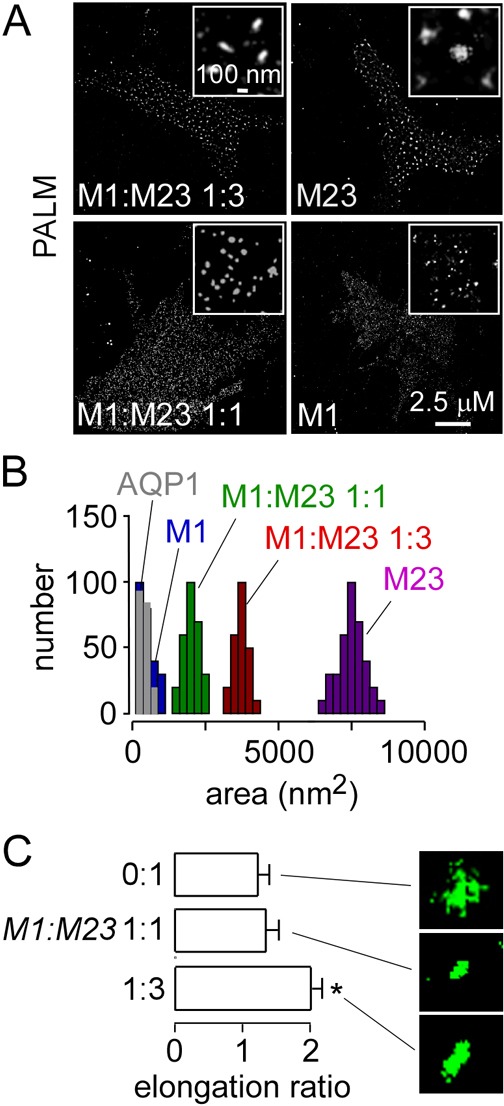
Dependence of OAP size on the ratio of M1- to M23-AQP4. (A) PALM of U87MG cells transfected with M1- and M23-AQP4-PAmCherry at the indicated ratios. Insets show magnified views. (B) Cluster size distributions (numbers of clusters analyzed: M1 = 245, M1:M23 1:1 = 250, M1:M23 1:3 = 220, M23 = 335). (C) OAP elongation ratios (mean ± s.e.m., n = 5 cells; *P<0.05; numbers of clusters analyzed: M23 = 145, M1:M23 1:1 = 115, M1:M23 1:3 = 160).
A major prediction of our model of OAP organization is that co-expression of M1- and M23-AQP4 should produce mixed OAPs in which M1-AQP4 is polarized to the OAP periphery and M23-AQP4 to the core (Jin et al., 2011). To test this prediction, U87MG cells were co-transfected with M1-AQP4-PAGFP and (untagged) M23-AQP4 at an M1:M23 ratio of 1:4 to generate relatively large OAPs. PALM showed many large spots of diameter ∼100 nm in which M1-AQP4–PAGFP fluorescence was concentrated in an annular pattern surrounding a relatively dark interior (Fig. 4A). Two-color dSTORM was done in U87MG cells co-expressing M1-AQP4–myc and M23-AQP4–HA (M1:M23 ratio of 1:4), labeled respectively with Alexa-Fluor-568- and -488-conjugated secondary antibodies against anti-myc and anti-HA antibodies. dSTORM showed red M1-AQP4 fluorescence at the periphery surrounding green M23-AQP4 fluorescence (Fig. 4B).
Fig. 4.
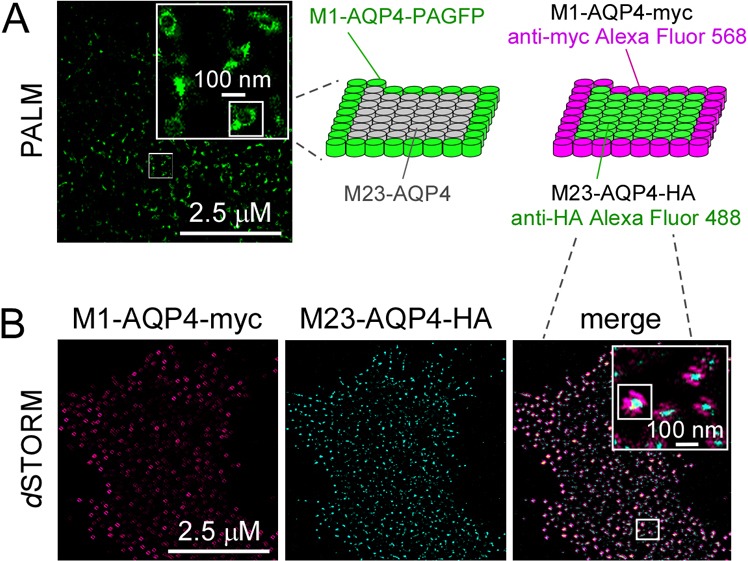
Polarized distribution of M1- and M23-AQP4 in OAPs. (A) PALM of U87MG cells co-expressing M1-AQP4–PAGFP and unlabeled M23-AQP4 in a 1:4 ratio. A diagram of polarized AQP4 distribution is shown on the right. (B) Two-color dSTORM of U87MG cells co-expressing M1-AQP4–myc and M23-AQP4–HA, labeled with fluorescent anti-myc (red) and anti-HA antibodies (green).
Super-resolution imaging of native AQP4 in astrocytes
Although expression of AQP4 in cell models is useful in elucidating the structural features and molecular determinants of OAP assembly, analysis of native AQP4 in astrocytes, which contain both M1- and M23-AQP4 in a native environment, provides information about OAP regulation and involvement in human disease (NMO disease pathogenesis). Two labeling approaches were used for dSTORM of AQP4 in primary astrocyte cultures: labeling of cells with an anti-C-terminus AQP4 primary antibody or a recombinant monoclonal NMO-IgG against an extracellular epitope. Fig. 5A shows the substantially better resolution of AQP4 OAPs by dSTORM versus TIRFM. OAPs with average area 2142±829 nm2 were seen, similar of those observed in astrocytes by FFEM (Furman et al., 2003). Two-color dSTORM (‘merge’ image at the right) showed co-localization of the C-terminus and external epitope-targeted AQP4 antibodies.
Fig. 5.
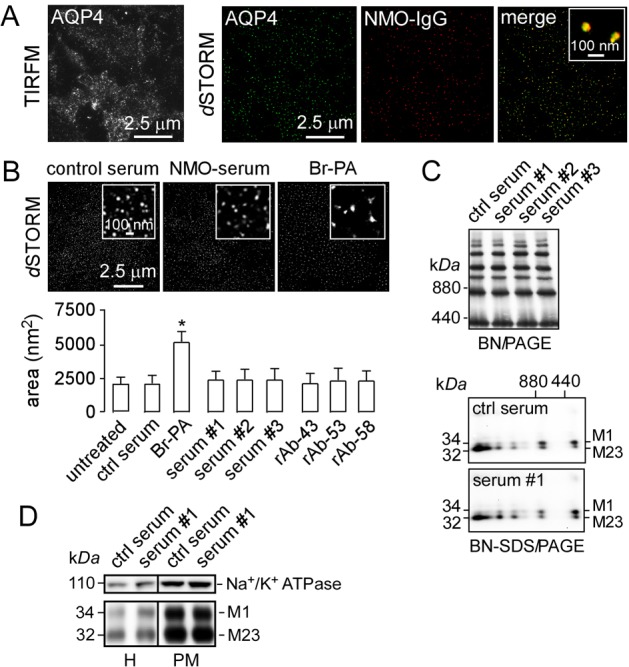
OAP imaging in astrocytes. (A) Primary, well-differentiated cultures of mouse astrocytes were fixed and stained with NMO-IgG (red) and C-terminus anti-AQP4 antibody (green) and imaged using TIRFM and two-color dSTORM. (B) dSTORM of astrocytes exposed to control or NMO serum, or treated with 2-bromopalmitate. Average OAP areas summarized at the bottom (mean ± s.e.m., n = 6, P<0.05). (C) BN-PAGE (top) and two-dimensional BN-SDS-PAGE (bottom) of whole-astrocyte homogenates after incubation with control or NMO sera as in B. (D) SDS-PAGE of astrocyte homogenate (H) and plasma membrane fraction (PM) after incubation with control or NMO serum, probed with antibodies against Na+/K+ ATPase (top) and AQP4 (bottom).
Autoantibodies against AQP4 on astrocytes are thought to be pathogenic in NMO (Papadopoulos and Verkman, 2012). A recent paper suggested a novel mechanism of NMO pathogenesis involving preferential NMO-IgG-induced internalization of M1-AQP4 in astrocytes, resulting in relative M23-AQP4 enrichment at the plasma membrane and consequent increased OAP size and complement-dependent cytotoxicity (Hinson et al., 2012).
We examined whether NMO-IgG affects AQP4 supramolecular assembly in mouse astrocyte cultures using dSTORM, BN-PAGE and BN-SDS-PAGE. Fig. 5B shows dSTORM of AQP4 in astrocytes labeled with a C-terminus AQP4 antibody. The average OAP area of 2163±803 nm2 was not significantly affected by exposure to a high concentration of NMO serum. Sera from three different NMO patients (each 1:5 dilution of original serum) and three NMO recombinant antibodies (rAbs; each 50 µg/ml) did not significantly alter OAP area (Fig. 5B, bottom). As a positive control, astrocyte cultures were incubated for 16 h with 2-bromopalmitate (Br-PA), which inhibits M1-AQP4 N-terminus palmitoylation and increases OAP size (Suzuki et al., 2008; Crane et al., 2009). Br-PA increased average OAP area significantly from 2163±803 nm2 to 5137±1119 nm2.
Biochemical studies on cell homogenates provided independent support for conclusion that NMO-IgG does not alter AQP4 supramolecular assembly. BN-PAGE showed no effect of NMO sera (Fig. 5C, top) or NMO-rAbs (not shown) on the OAP band pattern, which is a very sensitive measure of OAP assembly (Rossi et al., 2011). Two-dimensional BN-SDS-PAGE also showed no effect of NMO serum on the detailed pattern of M1- and M23-AQP4 in different OAP bands (Fig. 5C, bottom). SDS-PAGE of plasma membrane-enriched fractions of mouse astrocyte cultures exposed to control and NMO serum showed little or no internalization of native M1- or M23-AQP4 (Fig. 5D). Enrichment of the plasma membrane fraction was verified by Na+/K+ ATPase immunoblot. These results thus challenge the conclusion of Hinson et al. (Hinson et al., 2012) that NMO-IgG by itself causes OAP clustering.
To investigate potential reasons for the different results from Hinson et al. we tested the hypotheses that complement C1q binding causes OAP clustering in the presence of NMO-IgG, and that OAP clustering might be a non-selective consequence of astrocyte cytotoxicity (Hinson et al., 2012). Fig. 6A shows that C1q binds to astrocytes when NMO-IgG is present, as expected from our recent work showing that complement-dependent cytototoxity requires AQP4 OAP formation (Phuan et al., 2012). However, C1q did not cause OAP clustering, as quantified by OAP area (Fig. 6A,C). However, incubation with NMO-IgG in the presence of fresh human complement (Fig. 6B), which causes progressive cytotoxicity, greatly increased average OAP area to 10,834±3189 nm2. Such apparent OAP clustering appeared to be a non-specific effect of cell toxicity, as incubation with thapsigargin also increased OAP area.
Fig. 6.
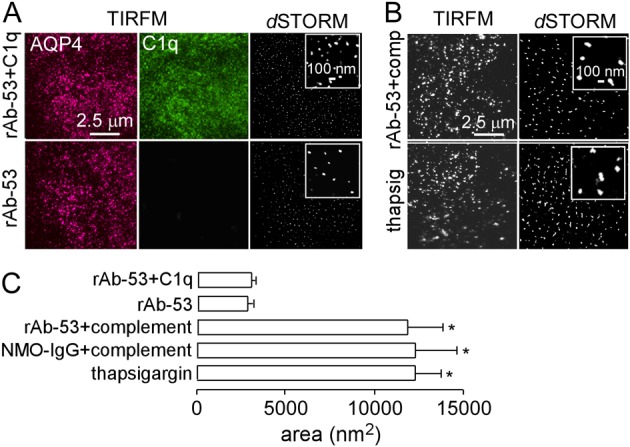
OAP clustering as a non-specific response to cytotoxicity. (A) TIRFM of primary cultures of mouse astrocytes incubated with rAb-53, with or without C1q, fixed, and stained with AQP4 (red) and C1q (green) antibodies. Right panels show dSTORM of AQP4, labeled with Alexa-Fluor-488-conjugated C-terminus anti-AQP4 antibody. (B) TIRFM and dSTORM of mouse astrocytes incubated with rAb-53 and 20% fresh human complement or thapsigargin, stained as in right panels of A. (C) OAP areas (mean ± s.e.m., n = 3, P<0.01).
Live-cell super-resolution imaging of AQP4 OAPs
The high sensitivity of PALM and the ability of PAGFP-labeled M23 to form OAPs allowed measurement of the nanoscale dynamics of individual OAP in live cells. For these studies, the relatively photon-tolerant CHO cells were transfected with M23-AQP4-PAGFP and imaged at 37°C for 45 min. PALM in live cells showed large, distinct spots (Fig. 7A), representing OAPs, similar to those seen on fixed cells. Similar images were obtained at 0 and 45 min, though several subtleties were seen including random diffusion of individual OAPs and occasional OAP fusion and fission events (Fig. 7B). Analysis of OAP diffusion showed approximately linear mean square displacement with time over 45 min, and a wide distribution of diffusion coefficients with mean ∼10−12 cm2/s (Fig. 7C).
Fig. 7.
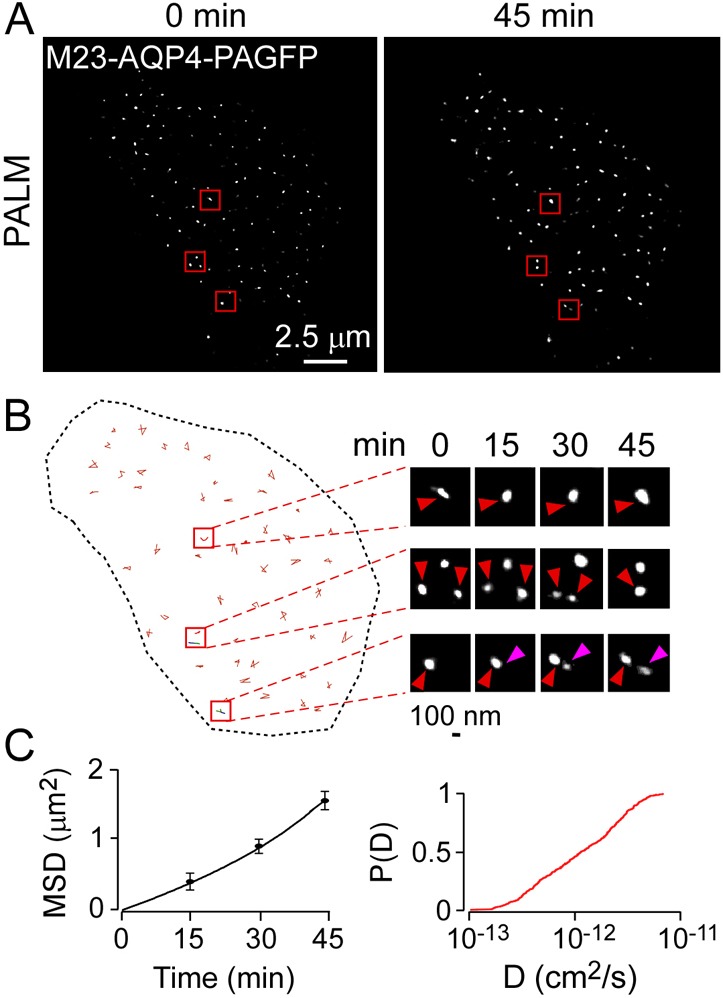
Live-cell PALM of CHO cells expressing M23-AQP4. PALM images (4-min acquisitions) were obtained every 15 min. (A) PALM of M23-AQP4–PAGFP measured at 0 and 45 min. (B) Deduced OAP trajectories over 45 min, with examples of OAP diffusion, fusion and fission shown at the right. Purple arrowhead indicates nascent OAP. (C) Mean squared displacement (MSD); and cumulative probability distribution [P(D)] of diffusion coefficients deduced from single OAP trajectories.
Discussion
Super-resolution imaging provides a unique tool to study OAPs, which complements the classical approach of FFEM, the biochemical approach of native gel electrophoresis, and the imaging approaches of single particle tracking and TIRFM. PALM and dSTORM allowed resolution of OAP size and structure to 20–30 nm spatial resolution in AQP4-transfected cells and astrocytes, obviating the concerns about sample preparation and sampling bias in FFEM. PALM allowed imaging of OAPs in live cells, providing information about their nanometer-scale dynamics. We focused attention on questions about OAPs that could be addressed effectively by super-resolution imaging, including AQP4 isoform-specific effects on OAP size and shape, spatial heterogeneity in isoform composition, and nanoscale effects of NMO-IgG binding.
An important finding was that OAPs in cells expressing M1- and M23-AQP4 consist of a M23-enriched core surrounded by a M1-enriched periphery. This finding supports our prediction, based on modeling (Jin et al., 2011) and mutagenesis (Crane and Verkman, 2009), that intermolecular M23–M23 interactions provide the adhesive energy to stabilize OAPs, whereas M1 prevents the adhesive interaction and limits OAP growth by ‘coating’ their periphery. The finding that increasing M1:M23 ratio reduces OAP size supports prior FFEM, biochemical and imaging data (Furman et al., 2003; Crane et al., 2009; Rossi et al., 2011), and the oblong OAP shape at intermediate M1:M23 ratio supports predictions from mathematical modeling (Jin et al., 2011).
OAP formation by AQP4 is of central importance in the pathogenesis of the multiple sclerosis-like disease NMO, where binding of autoantibodies to AQP4 on astrocytes in the central nervous system causes cytotoxicity, inflammation, myelin loss and neurological deficit (Papadopoulos and Verkman, 2012). We found previously that immunoglobulin G autoantibodies against AQP4, NMO-IgG, generally bind with much greater affinity to array-assembled AQP4 than to AQP4 tetramers (Crane et al., 2011). We recently reported a second, major effect of AQP4 OAP assembly in NMO involving complement-dependent cytotoxicity, in which binding of the multivalent complement protein C1q is greatly enhanced when NMO-IgG is clustered on AQP4 OAPs (Phuan et al., 2012). OAPs are thus of central importance in NMO pathogenesis.
Here, we tested a recent hypothesis about NMO pathogenesis in which NMO-IgG binding to AQP4 on astrocytes causes preferential internalization of M1-AQP4 leading to OAP clustering and resulting in enhanced complement-dependent cytotoxicity (Hinson et al., 2012). By super-resolution imaging, we found, using multiple different NMO sera and recombinant monoclonal antibodies, that exposure of astrocytes to NMO-IgG did not change OAP size. We also found that exposure of astrocyte cultures to NMO-IgG did not induce internalization of M1- or M23-AQP4, which adds to prior results from our laboratory (Ratelade et al., 2011; Rossi et al., 2012b). In trying to account for the different conclusion of Hinson et al. (2012), we found that apparent OAP clustering could be seen following different toxic insults to astrocytes, including complement-dependent cytotoxicity and thapsigargin. Though the mechanism of OAP clustering in dying cells is unclear, it appears to be a general phenomenon that has been reported in astrocytes in vitro and in vivo (Anders and Brightman, 1982).
In summary, super-resolution imaging resolves OAP geometry and dynamics with high spatial resolution in fixed and live cells, allowing the investigation of questions that are not easily addressed by convention electron microscopy, biochemical or light microscopy methods. Though a substantial body of data now exists on OAP geometry, dynamics and molecular interactions, and the role of OAPs in NMO, major questions remain about the significance of AQP4 OAPs in the normal functioning of astrocytes, skeletal myocytes and epithelial cells where they are found.
Materials and Methods
DNA constructs
cDNAs encoding human and rat AQP1, M1- and M23-AQP4 were PCR-amplified using whole-brain cDNA as template. AQP4 (M1 and M23) C-terminal chimeras with EGFP were generated as described (Tajima et al., 2010). AQP4 C-terminus chimeras with PAmCherry, PAGFP, EOSFP, PAtagRFP and Dendra2 were generated by PCR amplification of the respective coding sequences as NotI and AgeI fragments, and swapping with EGFP in M1- and M23-AQP4 pEGFP-N1 (Clontech, Mountain View, CA). All constructs were verified by sequencing. cDNAs encoding human M1- and M23-AQP4, each containing a 10-residue Myc (NH2-EQKLISEEDL-COOH) or hemagglutinin (HA) (NH2-YPYDVPDYA-COOH) epitope in the second extracellular loop, were generated as described (Crane et al., 2009).
NMO sera and recombinant AQP4 autoantibodies
NMO serum was obtained from three NMO-IgG-seropositive individuals who met the revised diagnostic criteria for clinical disease (Wingerchuk et al., 2006). Control (non-NMO) human serum was purchased from the UCSF cell culture facility. Purified human monoclonal recombinant NMO-IgGs rAb-10, rAb-53, rAb-58, and control (non-NMO) recombinant human IgG were generated as described (Bennett et al., 2009). For some studies, IgG was purified from NMO or control sera using a Melon Gel IgG Purification Kit (Thermo Fisher Scientific, Rockford, IL) and concentrated using Amicon Ultra Centrifugal Filter Units (Millipore, Billerica, MA).
Cell culture and transfections
U87MG (human glioblastoma-astrocytoma, ATCC HTB-14), HeLa (ATCC CCL-2) and CHO (ATCC CCL-61) cell cultures were maintained at 37°C in 5% CO2, 95% air in appropriate medium (without phenol red) containing 10% fetal bovine serum, 100 units/ml penicillin and 100 µg/ml streptomycin. Cells were transfected in glass bottom culture dishes (MatTek, Ashland, MA) using X-tremeGENE HP (Roche, Basel, Switzerland) one day prior to measurements in antibiotic-free medium. Primary astrocyte cultures were generated from cortex of neonatal wild-type and AQP4-null mice, as described (Li et al., 2011). After 8–10 days in culture, cells were treated for two weeks with 0.25 mM dibutyryl cAMP (Sigma-Aldrich, St. Louis, MO) to induce differentiation. Greater than 95% of cells were positive for the astrocyte marker, glial fibrillary acidic protein (GFAP). In some experiments, cells were incubated for 16 h with 100 µM Br-PA or for 36 h with 5 µg/ml thapsigargin (Sigma-Aldrich). In some experiments cells were incubated for 24 h at 37°C with 20% NMO or control sera, 50 µg/ml NMO or control rAb, or 2 mg/ml IgG purified from NMO or control sera plus fresh or heat-inactivated human complement (20%, pooled human complement serum; Innovative Research, Novi, MI). C1q binding was measured as described (Phuan et al., 2012) in which cells were incubated with 50 µg/ml NMO-rAb and 150 µg/ml recombinant human C1q protein for up to 24 h at 37°C. C1q immunofluorescence was detected using a fluorescein-conjugated rabbit polyclonal anti-C1q antibody (Abcam, Cambridge, MA).
Cell fixation and labeling for super-resolution imaging
For PALM, cells were fixed for 20 min in PBS containing 3% paraformaldehyde and 0.1% glutaraldehyde, and then washed in PBS. For dSTORM, cells were fixed as above, permeabilized for 15 min in PBS containing 0.3% v/v Triton X-100, incubated for 1 h with rabbit anti-C-terminus AQP4 antibody or rabbit anti-AQP1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and then for 1 h with goat secondary anti-rabbit antibody conjugated with Alexa Fluor 488 or 568 (Invitrogen, Carlsbad, CA) (10−6 M in PBS). Cells were washed three times in PBS containing 0.1% v/v Tween 20. F-actin was stained using Alexa-Fluor-488-conjugated phalloidin (Invitrogen; 10−6 M in PBS) for 20 min and washed three times in PBS containing 0.1% v/v Tween 20. The degree of labeling was determined by absorption spectroscopy to be 1–2 fluorophores/antibody. Alexa Fluor dyes were switched reversibly between on and off states by incubation of fixed cells with 100 mM β-mercaptoethylamine (MEA; Sigma-Aldrich) in PBS (Heilemann et al., 2009).
Super-resolution imaging
Super-resolution imaging was performed using a custom-built instrument. The outputs of four continuous-wave diode lasers (CLAS2-405-100C, BSR, Milpitas, CA; Cube 488-50C, Coherent, Santa Clara, CA; Cobolt Jive 50, Market Tech, Scotts Valley, CA; 56 RCS 009/HS, Melles Griot, Albuquerque, NM) were each spectrally cleaned with band-pass filters (FF01-405/10-25, LL01-488-12.5, FF01-561/4-25, FF01-642/10-25; Semrock, Rochester, NY), controlled by an acousto-optic tunable filter (AOTFnC-400.650-TN and MDSnC, AA Opto-Electronic, Orsay, France), and fiber-coupled into a total internal reflection fluorescence attachment (TIRF2, Nikon, Melville, NY) of a motorized inverted microscope (TE2000-E, Nikon) equipped with infrared autofocus. Cells were viewed using a high-numerical aperture (NA) TIRF objective (Apo TIRF 100×, oil, NA 1.49, Nikon). Fluorescence was imaged using an EM-CCD camera (QuantEM∶512SC, Photometrics, Tucson, AZ) using quad-band filter cube (LF405/488/561/635-A-NTE, Semrock) and appropriate emission filters (FF02-520/28-25, FF01-600/37-25, Semrock) mounted on a Lambda 10-3 optical filter changer (Sutter Instruments, Novato, CA). The AOTF and microscope were controlled with open source software Micro-Manager 1.4 (Edelstein et al., 2010) and Laser Control for QuickPALM (Henriques et al., 2010).
Data acquisition and analysis
Super resolution images were reconstructed from a series of TIRF images, acquired with Micro-Manager. Typically, 20,000–50,000 frames (TIRF images) were processed with the ImageJ plug-in QuickPALM (Henriques et al., 2010). The acquisition time for one TIRF image was 50 ms (frame rate 20 Hz), the laser power for fluorescence excitation was ∼9 mW (cw, 561 nm for PAmCherry) at the back of the microscope objective, and the activation laser (405 nm) power was 1–5 µW. For PALM the activation laser was pulsed at 1–20 Hz with pulse duration 1–10 ms, which was adjusted empirically to provide optimal stochastic activation of fluorophores. Dual-color dSTORM with Alexa Fluor 488 and Alexa Fluor 568 was carried out in PBS containing 100 mM MEA. Fluorophores were excited using moderate laser power at 488 nm (Alexa Fluor 488; 3 kW/cm2) and 568 nm (Alexa Fluor 568; 3 kW/cm2). Typically, 20,000 images were recorded at 20 Hz frame rate. Between 500–3000 photons were used to compute fluorophore localizations. For imaging, Alexa Fluor 488 and Alexa Fluor 568 were sequentially imaged and the images were aligned by using multi-fluorescent beads and gold nanoparticles, as described previously (Löschberger et al., 2012).
Live-cell PALM was performed as described previously (Shroff et al., 2008). Images were recorded at 20 Hz frame rate. CHO cells transfected with M23-AQP4-PAGFP or M23-AQP4-PAmCherry were exposed (for 4 min every 15 min) simultaneously to light at 405 nm (0.05 kW/cm2) to activate single PA molecules and at 520 nm or 600 nm (1 kW/cm2) to excite fluorescence and cause bleaching. Both lasers were shuttered between image acquisitions. More than 103 molecules per µm2 were recorded. Analysis of OAP diffusion was done as described for live cell PALM imaging and quantum dot single particle tracking (Shroff et al., 2008; Crane et al., 2009).
Individual fluorophores were localized using QuickPALM, generally specifying a minimum signal-to-noise ratio of 10, a maximum full-width at half-maximum of 6 pixels, and an image plane pixel size of 106 nm (related to the optical configuration of the microscope). Asymmetry and photon thresholds were also applied, excluding spots appearing too wide or too elliptical, as well as spots with less than 300 or more than 2000 photons per spot per frame. All acquisition frames were corrected for any mechanical or thermal drift during the image acquisition, chromatic aberration and frame shifts, which was done using immobile 100-nm gold nanoparticles (Corpuscular, Cold Spring, NY). The pixel size of rendered (reconstructed) super-resolution images was 21 nm. The image processing package Fiji was used to analyze the size of the OAPs in reconstructed images as follows. The cell outline was traced with the polygon selection tool and the area outside the cell was cropped to exclude extracellular noise. Images were converted from 16-bit to 8-bit gray scale and transformed into binary images by threshold adjustment. The particle analyzer plug-in was then used to determine the size (area) of connected pixels by fitting to ellipses. Elongation ratios were determined as the long-to-short diameter of fitted ellipses using the FracLac plug-in for ImageJ.
Subcellular fractionation
An enriched plasma membrane fraction from mouse astrocyte cultures was obtained by differential centrifugation, as described (Rossi et al., 2012a). Cells were homogenized by 20 strokes in a glass Dounce homogenizer, centrifuged at 4000 g for 15 min and the pellet discarded. A plasma membrane-enriched fraction was obtained by centrifugation at 17,000 g for 45 min. Immunoblot with Na+/K+ ATPase antibody (Abcam, Cambridge, MA) was used to assess plasma membrane enrichment.
Electrophoresis and immunoblotting
For blue-native PAGE (BN-PAGE) cells were lysed in native buffer (500 mM ε-aminocaproic acid, 50 mM imidazole (pH 7.0), 12 mM NaCl, 10% glycerol, 1% Triton X-100, protease inhibitor cocktail), incubated for 30 min on ice, centrifuged at 22,000 g for 30 min and the supernatants were supplemented with Coomassie Blue G-250 as described (Rossi et al., 2012a). The running buffers were: 25 mM imidazole, pH 7 (anode buffer) and 50 mM tricine, 7.5 mM imidazole, 0.02% Coomassie Blue G-250, pH 7 (cathode buffer). Proteins were blotted onto polyvinylidene difluoride (PVDF) membranes using a native transfer buffer (50 mM tricine, 7.5 mM imidazole). Laemmli SDS-PAGE gels consisted of a 12% running gel and 3% stacking gel. hrCN-PAGE was performed as described (Rossi et al., 2012a). For immunoblot, membranes were blocked with 3% BSA and incubated at 4°C overnight with goat or rabbit anti-AQP4 antibody (Santa Cruz Biotechnology). Membranes were then rinsed, incubated for 1 h with horseradish peroxidase-conjugated goat anti-rabbit IgG or donkey anti-goat IgG (Santa Cruz Biotechnology), rinsed extensively, and labeled proteins were detected using the ECL Plus enzymatic chemiluminescence kit (Amersham Biosciences, Pittsburgh, PA). The Laemmli SDS-PAGE gels consisted of a 12% running gel and 4% stacking gel.
Acknowledgments
We thank Bo Huang (PAtagRFP), Matthew F. Krummel (EOSFP), Jennifer Lippincott-Schwartz (PAmCherry C1) and Dianne Cox (Dendra2 N1) for providing cDNAs encoding fluorescent proteins, and Jeff Bennett for providing recombinant monoclonal NMO antibodies.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant numbers EY13574, EB00415, DK35124, HL73856, DK86125, DK72517, all to A.S.V.]; and the Guthy-Jackson Charitable Foundation [to A.S.V.]. Deposited in PMC for release after 12 months.
References
- Anders J. J., Brightman M. W.1982). Particle assemblies in astrocytic plasma membranes are rearranged by various agents in vitro and cold injury in vivo. J. Neurocytol. 11, 1009–1029 [DOI] [PubMed] [Google Scholar]
- Bennett J. L., Lam C., Kalluri S. R., Saikali P., Bautista K., Dupree C., Glogowska M., Case D., Antel J. P., Owens G. P., et al. (2009). Intrathecal pathogenic anti-aquaporin-4 antibodies in early neuromyelitis optica. Ann. Neurol. 66, 617–629 10.1002/ana.21802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott–Schwartz J., Hess H. F. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Crane J. M., Verkman A. S.2009). Determinants of aquaporin-4 assembly in orthogonal arrays revealed by live-cell single-molecule fluorescence imaging. J. Cell Sci. 122, 813–821 10.1242/jcs.042341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. M., Bennett J L., Verkman A. S.2009). Live cell analysis of aquaporin-4 M1/M23 interactions and regulated orthogonal array assembly in glial cells. J. Biol. Chem. 284, 35850–35860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. M., Van Hoek A. N., Skach W. R., Verkman A. S. (2008). Aquaporin-4 dynamics in orthogonal arrays in live cells visualized by quantum dot single particle tracking. Mol. Biol. Cell 19, 3369–3378 10.1091/mbc.E08-03-0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane J. M., Lam C., Rossi A., Gupta T., Bennett J. L., Verkman A. S. (2011). Binding affinity and specificity of neuromyelitis optica autoantibodies to aquaporin-4 M1/M23 isoforms and orthogonal arrays. J. Biol. Chem. 286, 16516–16524 10.1074/jbc.M111.227298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. (2010). Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chap. 14, unit 14 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton R. A., Moeller H. B., Zelenina M., Snaebjornsson M. T., Holen T., MacAulay N.2010). Differential water permeability and regulation of three aquaporin 4 isoforms. Cell. Mol. Life Sci. 67, 829–840 10.1007/s00018-009-0218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigeri A., Gropper M. A., Turck C. W., Verkman A. S. (1995). Immunolocalization of the mercurial-insensitive water channel and glycerol intrinsic protein in epithelial cell plasma membranes. Proc. Natl. Acad. Sci. USA 92, 4328–4331 10.1073/pnas.92.10.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigeri A., Nicchia G. P., Repetto S., Bado M., Minetti C., Svelto M. (2002). Altered aquaporin-4 expression in human muscular dystrophies: a common feature? FASEB J. 16, 1120–1122 [DOI] [PubMed] [Google Scholar]
- Frydenlund D. S., Bhardwaj A., Otsuka T., Mylonakou M. N., Yasumura T., Davidson K. G. V., Zeynalov E., Skare O., Laake P., Haug F. M., et al. (2006). Temporary loss of perivascular aquaporin-4 in neocortex after transient middle cerebral artery occlusion in mice. Proc. Natl. Acad. Sci. USA 103, 13532–13536 10.1073/pnas.0605796103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman C. S., Gorelick–Feldman D. A., Davidson K. G., Yasumura T., Neely J. D., Agre P., Rash J. E. (2003). Aquaporin-4 square array assembly: opposing actions of M1 and M23 isoforms. Proc. Natl. Acad. Sci. USA 100, 13609–13614 10.1073/pnas.2235843100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M., van de Linde S., Mukherjee A., Sauer M. (2009). Super-resolution imaging with small organic fluorophores. Angew. Chem. Int. Ed. Engl. 48, 6903–6908 10.1002/anie.200902073 [DOI] [PubMed] [Google Scholar]
- Henriques R., Lelek M., Fornasiero E. F., Valtorta F., Zimmer C., Mhlanga M. M. (2010). QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 10.1038/nmeth0510-339 [DOI] [PubMed] [Google Scholar]
- Hinson S. R., Romero M. F., Popescu B. F., Lucchinetti C. F., Fryer J. P., Wolburg H., Fallier–Becker P., Noell S., Lennon V. A. (2012). Molecular outcomes of neuromyelitis optica (NMO)-IgG binding to aquaporin-4 in astrocytes. Proc. Natl. Acad. Sci. USA 109, 1245–1250 10.1073/pnas.1109980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroaki Y., Tani K., Kamegawa A., Gyobu N., Nishikawa K., Suzuki H., Walz T., Sasaki S., Mitsuoka K., Kimura K., et al. (2006). Implications of the aquaporin-4 structure on array formation and cell adhesion. J. Mol. Biol. 355, 628–639 10.1016/j.jmb.2005.10.081 [DOI] [PubMed] [Google Scholar]
- Ho J. D., Yeh R., Sandstrom A., Chorny I., Harries W. E., Robbins R. A., Miercke L. J., Stroud R. M. (2009). Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance. Proc. Natl. Acad. Sci. USA 106, 7437–7442 10.1073/pnas.0902725106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B. J., Rossi A., Verkman A. S. (2011). Model of aquaporin-4 supramolecular assembly in orthogonal arrays based on heterotetrameric association of M1-M23 isoforms. Biophys. J. 100, 2936–2945 10.1016/j.bpj.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. S., Bhat R. V., Preston G. M., Guggino W. B., Baraban J. M., Agre P. (1994). Molecular characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc. Natl. Acad. Sci. USA 91, 13052–13056 10.1073/pnas.91.26.13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis D. M., Reese T. S. (1974). Differences in membrane structure between excitatory and inhibitory synapses in the cerebellar cortex. J. Comp. Neurol. 155, 93–125 10.1002/cne.901550107 [DOI] [PubMed] [Google Scholar]
- Lennon V. A., Kryzer T. J., Pittock S. J., Verkman A. S., Hinson S. R. (2005). IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J. Exp. Med. 202, 473–477 10.1084/jem.20050304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang H., Varrin–Doyer M., Zamvil S. S., Verkman A. S. (2011). Proinflammatory role of aquaporin-4 in autoimmune neuroinflammation. FASEB J. 25, 1556–1566 10.1096/fj.10-177279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löschberger A., van de Linde S., Dabauvalle M. C., Rieger B., Heilemann M., Krohne G., Sauer M. (2012). Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125, 570–575 10.1242/jcs.098822 [DOI] [PubMed] [Google Scholar]
- Lu M., Lee M. D., Smith B. L., Jung J. S., Agre P., Verdijk M. A., Merkx G., Rijss J. P., Deen P. M. (1996). The human AQP4 gene: definition of the locus encoding two water channel polypeptides in brain. Proc. Natl. Acad. Sci. USA 93, 10908–10912 10.1073/pnas.93.20.10908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley G. T., Fujimura M., Ma T., Noshita N., Filiz F., Bollen A. W., Chan P., Verkman A. S. (2000). Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat. Med. 6, 159–163 10.1038/72256 [DOI] [PubMed] [Google Scholar]
- Neely J. D., Christensen B. M., Nielsen S., Agre P. (1999). Heterotetrameric composition of aquaporin-4 water channels. Biochemistry 38, 11156–11163 10.1021/bi990941s [DOI] [PubMed] [Google Scholar]
- Noell S., Fallier–Becker P., Deutsch U., Mack A. F., Wolburg H. (2009). Agrin defines polarized distribution of orthogonal arrays of particles in astrocytes. Cell Tissue Res. 337, 185–195 10.1007/s00441-009-0812-z [DOI] [PubMed] [Google Scholar]
- Padmawar P., Yao X., Bloch O., Manley G. T., Verkman A. S. (2005). K+ waves in brain cortex visualized using a long-wavelength K+-sensing fluorescent indicator. Nat. Methods 2, 825–827 10.1038/nmeth801 [DOI] [PubMed] [Google Scholar]
- Papadopoulos M. C., Verkman A. S. (2012). Aquaporin 4 and neuromyelitis optica. Lancet Neurol. 11, 535–544 10.1016/S1474-4422(12)70133-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos M. C., Binder D. K., Verkman A. S. (2005). Enhanced macromolecular diffusion in brain extracellular space in mouse models of vasogenic edema measured by cortical surface photobleaching. FASEB J. 19, 425–427 [DOI] [PubMed] [Google Scholar]
- Phuan P. W., Ratelade J., Rossi A., Tradtrantip L., Verkman A. S. (2012). Complement-dependent cytotoxicity in neuromyelitis optica requires aquaporin-4 protein assembly in orthogonal arrays. J. Biol. Chem. 287, 13829–13839 10.1074/jbc.M112.344325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash J. E., Davidson K. G. V., Yasumura T., Furman C. S. (2004). Freeze-fracture and immunogold analysis of aquaporin-4 (AQP4) square arrays, with models of AQP4 lattice assembly. Neuroscience 129, 915–934 10.1016/j.neuroscience.2004.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratelade J., Bennett J. L., Verkman A. S. (2011). Evidence against cellular internalization in vivo of NMO-IgG, aquaporin-4, and excitatory amino acid transporter 2 in neuromyelitis optica. J. Biol. Chem. 286, 45156–45164 10.1074/jbc.M111.297275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Crane J. M., Verkman A. S. (2011). Aquaporin-4 Mz isoform: brain expression, supramolecular assembly and neuromyelitis optica antibody binding. Glia 59, 1056–1063 10.1002/glia.21177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Baumgart F., van Hoek A. N., Verkman A. S. (2012a). Post-Golgi supramolecular assembly of aquaporin-4 in orthogonal arrays. Traffic 13, 43–53 10.1111/j.1600-0854.2011.01299.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Ratelade J., Papadopoulos M. C., Bennett J. L., Verkman A. S. (2012b). Consequences of NMO-IgG binding to aquaporin-4 in neuromyelitis optica. Proc. Natl. Acad. Sci. USA 109, E1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadoun S., Papadopoulos M. C., Watanabe H., Yan D., Manley G. T., Verkman A. S. (2005). Involvement of aquaporin-4 in astroglial cell migration and glial scar formation. J. Cell Sci. 118, 5691–5698 10.1242/jcs.02680 [DOI] [PubMed] [Google Scholar]
- Shroff H., Galbraith C. G., Galbraith J. A., Betzig E. (2008). Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 10.1038/nmeth.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein C., Bouley R., Huang Y., Fang P., Pastor–Soler N., Brown D., Van Hoek A. N. (2004). Membrane organization and function of M1 and M23 isoforms of aquaporin-4 in epithelial cells. Am. J. Physiol. 287, F501–F511 10.1152/ajprenal.00439.2003 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nishikawa K., Hiroaki Y., Fujiyoshi Y. (2008). Formation of aquaporin-4 arrays is inhibited by palmitoylation of N-terminal cysteine residues. Biochim. Biophys. Acta 1778, 1181–1189 10.1016/j.bbamem.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Tajima M., Crane J. M., Verkman A. S. (2010). Aquaporin-4 (AQP4) associations and array dynamics probed by photobleaching and single-molecule analysis of green fluorescent protein-AQP4 chimeras. J. Biol. Chem. 285, 8163–8170 10.1074/jbc.M109.093948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbavatz J. M., Ma T., Gobin R., Verkman A. S. (1997). Absence of orthogonal arrays in kidney, brain and muscle from transgenic knockout mice lacking water channel aquaporin-4. J. Cell Sci. 110, 2855–2860 [DOI] [PubMed] [Google Scholar]
- Wakayama Y., Jimi T., Inoue M., Kojima H., Murahashi M., Kumagai T., Yamashita S., Hara H., Shibuya S. (2002). Reduced aquaporin 4 expression in the muscle plasma membrane of patients with Duchenne muscular dystrophy. Arch. Neurol. 59, 431–437 10.1001/archneur.59.3.431 [DOI] [PubMed] [Google Scholar]
- Wingerchuk D. M., Lennon V. A., Pittock S. J., Lucchinetti C. F., Weinshenker B. G. (2006). Revised diagnostic criteria for neuromyelitis optica. Neurology 66, 1485–1489 10.1212/01.wnl.0000216139.44259.74 [DOI] [PubMed] [Google Scholar]
- Wolburg H., Wolburg–Buchholz K., Fallier–Becker P., Noell S., Mack A. F. (2011). Structure and functions of aquaporin-4-based orthogonal arrays of particles. Int. Rev. Cell Mol. Biol. 287, 1–41 10.1016/B978-0-12-386043-9.00001-3 [DOI] [PubMed] [Google Scholar]
- Yang B., Ma T., Verkman A. S. (1995). cDNA cloning, gene organization, and chromosomal localization of a human mercurial insensitive water channel. Evidence for distinct transcriptional units. J. Biol. Chem. 270, 22907–22913 10.1074/jbc.270.39.22907 [DOI] [PubMed] [Google Scholar]
- Yang B., Brown D., Verkman A. S. (1996). The mercurial insensitive water channel (AQP-4) forms orthogonal arrays in stably transfected Chinese hamster ovary cells. J. Biol. Chem. 271, 4577–4580 10.1074/jbc.271.9.4577 [DOI] [PubMed] [Google Scholar]
- Zhang H., Verkman A. S. (2008). Evidence against involvement of aquaporin-4 in cell-cell adhesion. J. Mol. Biol. 382, 1136–1143 10.1016/j.jmb.2008.07.089 [DOI] [PMC free article] [PubMed] [Google Scholar]


