Abstract
The E2F transcription factors are important regulators of the cell cycle whose function is commonly misregulated in cancer. To identify novel regulators of E2F1 activity in vivo, we used Drosophila to conduct genetic screens. For this, we generated transgenic lines that allow the tissue-specific depletion of dE2F1 by RNAi. Expression of these transgenes using Gal4 drivers in the eyes and wings generated reliable and modifiable phenotypes. We then conducted genetic screens testing the capacity of Exelixis deficiencies to modify these E2F1-RNAi phenotypes. From these screens, we identified mutant alleles of Suppressor of zeste 2 [Su(z)2] and multiple Polycomb group genes as strong suppressors of the E2F1-RNA interference phenotypes. In validation of our genetic data, we find that depleting Su(z)2 in cultured Drosophila cells restores the cell-proliferation defects caused by reduction of dE2F1 by elevating the level of dE2f1. Furthermore, analyses of methylation status of histone H3 lysine 27 (H3K27me) from the published modENCODE data sets suggest that the genomic regions harboring dE2f1 gene and certain dE2f1 target genes display H3K27me during development and in several Drosophila cell lines. These in vivo observations suggest that the Polycomb group may regulate cell proliferation by repressing the transcription of dE2f1 and certain dE2F1 target genes. This mechanism may play an important role in coordinating cellular differentiation and proliferation during Drosophila development.
Keywords: cell proliferation, E2F1, Su(z)2, PcG, Drosophila
The E2F family of transcription factors provides temporal control of genes that are necessary for the G1/S-phase transition and are critical for controlling cell proliferation (Burkhart and Sage 2008; van den Heuvel and Dyson 2008). In early G1 phase of the cell cycle, the RB family proteins bind to and inhibit E2F transcriptional activities. In late G1 to S phase, cyclin-dependent kinases (CDKs) phosphorylate the RB family proteins, which then dissociate, resulting in E2F liberation and activation of E2F-dependent transcription (Burkhart and Sage 2008; van den Heuvel and Dyson 2008). E2F-regulated genes are required in dividing cells for proper DNA replication and subsequent mitosis (Müller and Helin 2000; Ren et al. 2002). The basic unit of E2F is a heterodimer composed of an E2F and a DP subunit. Eight E2F genes have been characterized in mammals (Stevaux and Dyson 2002; Trimarchi and Lees 2002; van den Heuvel and Dyson 2008): three activating E2Fs (E2F1∼3), two DP interacting repressive E2Fs (E2F4∼5), and three DP independent repressive E2Fs (E2F6∼8). The E2F family members display partial redundancy as well as antagonizing functions; thus, it is challenging to elucidate the functions of individual mammalian E2Fs. The RB-E2F pathway is streamlined in Drosophila because it contains only two E2Fs, the activator dE2F1 and the repressor dE2F2 (Frolov and Dyson 2004). Therefore, genetic and developmental analyses using Drosophila as a model organism may provide important insights into the mechanisms regulating the RB-E2F pathway during development.
We used a modifiable dE2F1 RNA interference system in Drosophila to identify novel regulators of E2F1 activity. By conducting a dominant modifier genetic screen, we have identified a set of genetic interactions between dE2F1 and members of the Polycomb group (PcG) genes. Several PcG complexes have been characterized, including polycomb repressive complex 1 (PRC1), PRC2, Pho-repressive complex (PhoRC), dRING-associated factors, and the Polycomb repressive deubiquitinase complex [PR-DUB (Levine et al. 2004; Schuettengruber et al. 2007; Schwartz and Pirrotta 2007; Müller and Verrijzer 2009; Margueron and Reinberg 2011)]. Of these complexes, the PRC2 contains the sole histone methyl-transferase, Enhancer of zeste (E(z)), specific for histone 3 lysine 27 (H3K27). Methylation of H3K27 by PRC2 is shown to facilitate the recruitment of the PRC1 complex through direct binding with the chromodomain of Polycomb (Pc) (Cao et al. 2002; Fischle et al. 2003; Min et al. 2003; Cao and Zhang 2004). However, in vivo regulations of these complexes in development are less well understood.
There are several reports linking PcG complexes to the RB-E2F pathway in vertebrates. First, the INK4b-ARF-INK4a tumor suppressor locus is regulated by the PcG complexes (Gil and Peters 2006). The INK4b-ARF-INK4a locus is vertebrate-specific and encodes the INK4 family of inhibitors that target CDK4/6-cyclin D (CycD), which phosphorylate and inactivate pRB family members in mammals (Sherr 2004; Gil and Peters 2006). Second, RB was reported to regulate the G2/M-phase transition by forming an E2F-RB-CtBP-HPC2 complex, thus repressing the expression of cyclin A and Cdc2 in cultured human cells (Dahiya et al. 2001). Third, E2F6, one of the repressive E2F family members in mammals, forms complexes with RYBP, Bmi1, EPC1, and other PcG subunits (Trimarchi et al. 2001; Ogawa et al. 2002; Attwooll et al. 2005) and regulates Hox gene expression and axial skeleton development in mouse (Storre et al. 2002; Courel et al. 2008). Finally, the RB-E2F pathway has been shown to regulate the expression of certain PcG subunits, such as EZH2 and EED (Bracken et al. 2003). Although it is not known whether these mechanisms are conserved in evolution, these studies suggest that the interactions between the RB-E2F pathway and PcG-mediated silencing can occur at multiple levels.
In Drosophila, PcG complexes have been reported to regulate the expression of several cell-cycle regulators. Polycomb responsive elements have been identified in the promoter and coding region of dCycA and dE2f1 (Martinez et al. 2006). Similarly, the PhoRC subunit Pleiohomeotic (Pho) and the PRC1 component Ph are found at the promoters of dCycB, dDp, dE2f1, and Rbf1 in Drosophila embryos (Oktaba et al. 2008). These studies suggest a direct role for multiple PcG complexes in regulating key Rb-E2F pathway components and that PcG complexes may affect cell proliferation by controlling the expression of different cell-cycle regulators in development. The relationships between PcG complexes and cell proliferation in different developmental contexts are important and far from clear, thus further investigations using diverse model systems and approaches are necessary.
We have identified a set of genetic interactions between PcG genes and dE2F1. As summarized in this report, our results suggest that PcG complexes may directly repress the transcription of dE2f1 and certain dE2F1 target genes. Together with the previous reports linking PcG complexes to cell-cycle regulators (Martinez et al. 2006; Oktaba et al. 2008), our genetic analyses provide in vivo evidence that supports a role for different PcG complexes in coordinating cell proliferation and differentiation during Drosophila development by controlling the expression of several key cell-cycle regulators.
Materials and Methods
Generation of UAS-dE2f1-dsRNA (tissue-specific dE2f1-RNAi) transgenic lines
A 650-bp fragment of DNA sequence was amplified by polymerase chain reaction (PCR) using dE2f1 cDNA as the template, and the primer sequences were 5′-TTATTTCAAACGCCCTACCG-3′ and 5′-GAATTGCATCTGCAGTGAGC-3′. This fragment was previously used as the target sequence to generate double-strand RNA (dsRNA) in our microarray analyses for dE2F1 target genes (Dimova et al. 2003). The PCR product was gel purified and subsequently subcloned into the pWIZ vector in an inverted configuration [for the detailed procedure, see (Lee and Carthew 2003)] and verified by sequencing. The final pWIZ-dE2f1-dsRNA vector, as referred to as “UAS-dE2f1-dsRNA” in the text, was injected into early Drosophila embryos (w1118) to generate transgenic flies. Approximately 30 different transgenic lines carrying one or multiple transgenes, as indicated by their eye color because pWIZ carries mini-white as a selection marker, were balanced, crossed, and recombined with different Gal4 lines using standard genetic crosses. Because the dE2f1dsRNA phenotypes in both the eye (w1118; GMR-Gal4, UAS-dE2f1RNAi #10 or #8/+; +/+ at 25°) and the wing (w1118; ptc-Gal4, UASdE2f1dsRNA#3/+; +/+ at 22∼23°) are modifiable by known RB-E2F pathway factors in expected manners and the phenotypes are fully penetrate, these two recombined stocks were used for genetic analyses in this work.
Genetic screen using the Exelixis deficiency (Df) lines
Flies were maintained on standard cornmeal-yeast agar medium. Exelixis Df lines and most of the mutant alleles used in this work were obtained from the Bloomington Drosophila Stock Center. The null allele of Polycomb (Pc3) allele was obtained from Dr. Antonio Garcia-Bellido (Castelli-Gair et al. 1990). For genetic screen using the Exelixis Df lines: approximately 5∼10 female virgins from either w1118; GMR-Gal4, UAS-dE2f1RNAi #10 (or #8)/CyO; +/+, or w1118; ptc-Gal4, UASdE2f1dsRNA#3/CyO; +/+ lines were crossed with 5∼10 males from each Df line on second or third chromosomes, and the crosses were maintained at either 25° (for the eye phenotype) or 22∼23° (for the wing phenotype). As an example for the eye phenotype, the female F1 with the following genotypes were scored for potential modifications: w1118; GMR-Gal4, UAS-dE2f1RNAi #10/Df(2R/2L)Exel#; +, or w1118; GMR-Gal4, UAS-dE2f1RNAi #10/+; Df(3R/3L)Exel#/+. The reverse crosses were performed for Df lines on the X chromosome and F1 female flies with the following genotype were scored: Df(1)Exel#/w1118; GMR-Gal4, UAS-dE2f1RNAi #10/+; +/+.
Scanning electron microscopy and measurement of the L3-L4 intervein region
The F1 female flies were stepwise dehydrated using ethanol, and scanning electron micrographs were taken following standard procedures at the Northeastern University. To measure L3-L4 intervein region, wings are removed, briefly treated with isopropanol and then mounted in Canada Balsam (Sigma-Aldrich, St. Louis, MO). The width of L3-L4 was measured under a Nikon i90 microscope using the Nikon NIS Elements software.
Drosophila RNAi in SL2 cells and the MTT assay
The dsRNAs used in this work were synthesized using the RiboMax Large Scale RNA Production Systems (Promega, Madison, WI) following the manufacturer’s instructions. The following primer sets were used to generate dsRNAs to dE2f1 (F: 5′-CGAGTAAGAAGCAGCAGCAC; R: 5′-CTGCCGGTTCTATCGTGATT), Su(z)2 (F: 5′-TCTGCTACCGGATTCTGCTTTACG; R: 5′-AACTCCCTTTCGATTCGCTGTCTT), Psc (F: 5′-CAACGCCAAGCCGAACATCAAATC; R: 5′-AGCGGCTGGGGCGACTCATAAAC), Pc (F: 5′-TGCCAATGCAATAGATTGTAAA; R: 5′-CGCTTTGAATTGCTGTTTTG), E(Pc) (F: 5′-TCAGCCCTTCTACGATGCCTACTA; R: 5′-CTCGCGTCGCCTCACCATCTCCAG), and white with T7 sequence (F: 5′-CTAATACGACTCACTATAGGGAGGGAAGATGGCTCCG; R: 5′-CTAATACGACTCACTATAGGGAGTTTCGCTCAGCAAATG). Treatment of the Drosophila SL2 cells with 50 μg of dsRNA was performed as described previously (Dimova et al. 2003). The white-dsRNA was used as a control, and it is also used to normalize the total amount of dsRNA in codepletion experiments. The MTT assay was performed as described (Hansen et al. 1989) in 96-well format, and the O.D. at 570nm was measured using a standard plate-reader.
RNA preparation and quantitative reverse-transcription (qRT)-PCR analysis
The total RNA isolation, quantification, reverse transcription, and the subsequent qRT-PCR analyses were performed as described previously (Zhao et al. 2012). The following primers were used for qRT-PCR for data presented in Figure 4: stg (F: 5′-AAACCAGCTGCTCGGCATATT; R: 5′-ATCTCAATTCACCGAACGAGGA), rnrL (F-5′-CGGTTAAGGCTCAATCCCTGT; R: 5′-TGGTTGCTCTTCCTGTTGCA), his2AvD (F: 5′-TCACTCCTCGCCACTTACAGCT; R: 5′-CGACTTGTGTATGTGCGGAATG), Mars (F: 5′-ATCTTGGATCCTCAGCAGACGA; R: 5′-GGCATTCCATTGGATTCGC), Mcm 5 (F: 5′-GAAGCTAAAGAGCCGCTACGTG; R: 5′-TCCAACTGACGCACAGTGATG), PCNA (F: 5′-GAATCGGCTAACCAGGAGAAGG; R: 5′-ACCACGCACGAGAAGTCTGTCT), Nebbish (F: 5′-AGTCGCATTGCCCTTAATCTGA; R: 5′-ATGTCTGTCGCGGTGTATTGC), dE2f1 (F: 5′-CTCTTTCTCCGCGTGTGGATT; R: 5′-GCGACGAAAAGCGAACTGAA), dCycA (F: 5′-AACCACGAACCGCTGAACAA; R: 5′-GGCAGCGTTGGAATTAGTTT), dCycE (F: 5′-ATGTGGCGCATAAGGTGCA; R: 5′-CCCGATCTTTGGCGGATAA), and rp49 gene (F: 5′-ACAGGCCCAAGATCGTGAAGA; R: 5′-CGCACTCTGTTGTCGATACCCT) was used as the internal loading control.
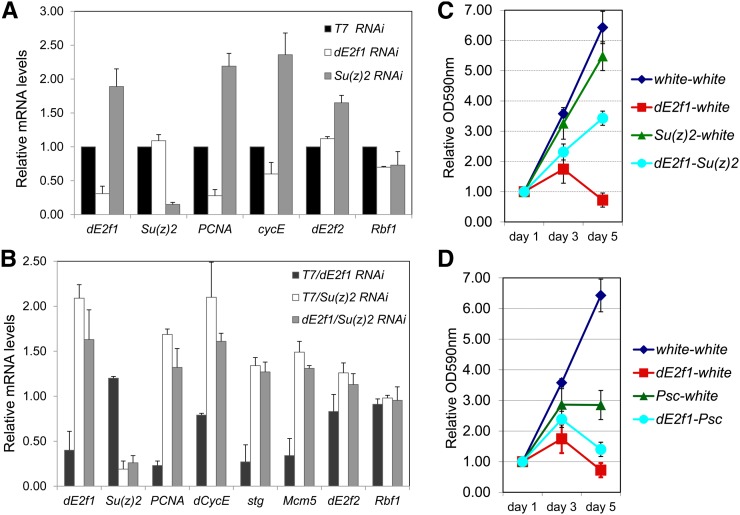
Figure 4 .
Su(z)2 regulates the transcription of dE2f1 and some of the dE2F1 target genes. (A) Knocking down Su(z)2 (gray bars) leads to up-regulation of dE2f1, and some of the dE2F1 target genes, such as PCNA, dCycE, and to a less extent dE2f2 and no effect of Rbf1, based on qRT-PCR assay. The samples treated with dE2f1-dsRNAs (white bars) serve as a positive control, and T7-dsRNA treated samples are negative controls. (B) Codepletion (gray bars) of Su(z)2 and dE2f1 suppresses the effect of dE2f1-dsRNA treatment and leads to increased expression of dE2f1, PCNA, dCycE, stg, and mcm5. The total dsRNAs are normalized with T7-dsRNA. (C and D) Effect of dsRNA treatment of the growth of SL2 cells: knocking down of Su(z)2 (C), but not Psc (D), suppresses the effect of dE2f1-dsRNA treatment at day 5. For each sample, the total amount of dsRNA is normalized with white-dsRNA and cell viability was determined by using the dimethyltriazoldiphenyl tetrazolium-formazan assay after 1, 3 or 5 days of dsRNA treatment.
Results
Tissue-specific knockdown of dE2F1 activity produces modifiable phenotypes
Homozygous dE2f1 mutant animals die during larval development (Duronio et al. 1995); thus, we used a dE2f1-dsRNA expression system based on the pWIZ vector (Lee and Carthew 2003). This system allows the tissue-specific expression of the target dsRNA (Hannon 2002) using the Gal4-UAS system (Brand et al. 1994; Lee and Carthew 2003). We generated multiple transgenic lines that produce a 650-bp dsRNA from the dE2f1 gene under control of the UAS, designated as “UAS-dE2f1-dsRNA” (see Materials and Methods for details). The UAS-dE2f1-dsRNA transgenes were then crossed to multiple tissue-specific Gal4 drivers and the resulting phenotypes were characterized. By driving the expression of UAS-dE2f1-dsRNA using the eye-specific GMR-Gal4 and the wing-specific patched-Gal4 (ptc-Gal4), we observed phenotypes with 100% penetrance and limited variation. Expression of dE2f1-dsRNA under the control of GMR-Gal4 caused a rough eye phenotype characterized by fused ommatidia (Figure 1B, compared with the control in Figure 1A), which we refer to as the “dE2f1-dsRNA eye phenotype” hereafter. Expression of dE2f1-dsRNA under the control of ptc-Gal4 reduces the L3-L4 intervein region in the adult wing (Figure 3B, compared with the control in Figure 3A), which is referred as the “dE2f1-dsRNA wing phenotype.”
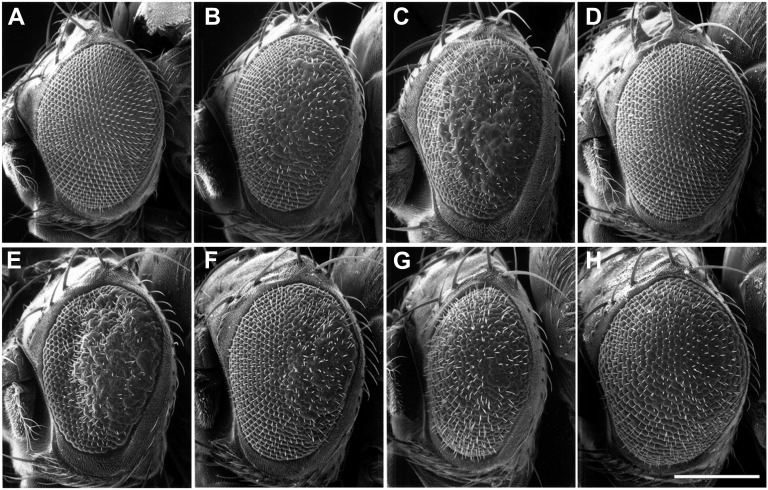
Figure 1 .
Tissue-specific expression of dE2f1-dsRNA generates phenotypes that can be modified by known factors of the dE2F1 pathway. (A) A normal Drosophila eye (w1118; GMR-Gal4/+; +/+). (B) Expressing one copy of the UAS-dE2f1-dsRNA (Line #10) generates a slight rough eye phenotype (w1118; GMR-Gal4, UAS-dE2f1dsRNA#10/+; +/+), which can be enhanced by reducing the endogenous dE2f1 levels, as shown in (C) (w1118; GMR-Gal4, UAS-dE2f1dsRNA#10/+; dE2f1i2/+), and completely rescued by overexpressing wild-type dE2f1, as shown in (D) (w1118; GMR-Gal4, UAS-dE2f1dsRNA#10/+; UAS-dE2f1+/+). A stronger rough eye phenotype is generate when multiple copies of UAS-dE2f1dsRNA (line #8) is expressed, as shown in (E) (w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/+; +/+). This stronger phenotype can be suppressed by overexpressing wild-type dCycA (F: w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/+; UAS-dCycA+/+), wild-type dCycE (G: w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/UAS-dCycE+; +/+), or dCdk4 and dCycD (H: w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/+; UAS-dCdk4+, UAS-dCycD+/+). The scale bar (in H) is 200µm.
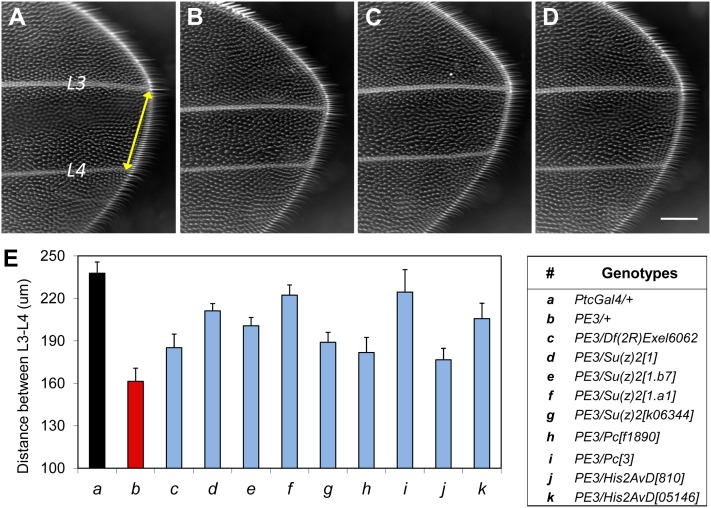
Figure 3 .
Su(z)2 and additional PcG genes are strong suppressors of the dE2f1-dsRNA phenotypes in the wing. (A) Part of L3-L4 intervein region of a control Drosophila wing (ptc-Gal4/+). Ptc-Gal4 is expressed in the L3-L4 intervein region. At 22∼23°, when dE2f1-dsRNA (line #3) is expressed under control of ptc-Gal4, the L3-L4 intervein region is reduced by ∼50%, as shown in (B) (w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; +/+). This wing phenotype can be strongly suppressed by Df(2R)Exel6062 (C: w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Df(2R)Exel6062; +/+), or the Su(z)21.a1 allele (D: w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Su(z)21.a1; +/+). The modification of the wing phenotype can be quantified by measuring the width of L3-L4 intervein region (E), and the genotypes of data presented in (E) are as follows: (a) w1118; ptc-Gal4/+; +; (b) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; +; (c) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Df(2R)Exel6062; +/+; (d) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Su(z)21; +/+; (e) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Su(z)21.b7; +/+; (f) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Su(z)21.a1; +/+; and (g) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/Su(z)2k06344; +/+; (h) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; Pcf01890/+; (i) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; Pc3/+; (j) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; His2AvD810/+; and (k) w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; His2AvD05146/+. At least 15 to 25 wings of each genotype (a∼k) were measured. Each genotype (c∼k) was compared with the control (b: w1118; ptc-Gal4, UAS-dE2f1dsRNA#3/+; +) and each comparison is highly significant (P < 4.9E-06 based on one-tailed t-test). For simplicity, “ptc-Gal4, UAS-dE2f1dsRNA#3” is referred as “PE3” in (E). The scale bar in (D) is 100µm.
To verify the specificity of the dE2f1-dsRNA−induced phenotypes, we recombined different UAS-dE2f1-dsRNA lines to the GMR-Gal4 or ptc-Gal4 drivers on the second chromosome (see Materials and Methods). Using these stocks, we then tested the capacity of components of the Rb-E2F pathway to modifying the phenotypes. We observed that the GMR-driven rough eye phenotypes generated by a weak allele of dE2f1-dsRNA (line #10; Figure 1B) were enhanced by mutant alleles of dE2f1 (Figure 1C). In contrast, the rough-eye phenotypes can be suppressed by introducing a single copy of a UAS-dE2f1+ transgene (Figure 1D). We observed that even the strong effects of dE2f1-dsRNA (line #8; Figure 1E) were suppressed by the overexpression of wild-type dCycA (Figure 1F), dCycE (Figure 1G), or dCdk4-dCycD (Figure 1H). Conversely, mutant alleles of dCdk4, dCycA, or dCycE enhanced the dE2f1-dsRNA phenotypes (data not shown). These genetic analyses show that the dE2f1-dsRNA phenotypes are modified by components of the Rb-E2F pathway in a predictable manner, suggesting that the phenotypes are caused by specific reduction of dE2F1 activity. In support of this, we observed reduced dE2F1 protein levels in both immunostaining and Western blotting experiments when using tissue-specific expression of dE2f1-dsRNA (Morris et al. 2008). We also find that knockdown of dE2F1 in the wing imaginal discs results in reduced expression of a PCNA-GFP reporter, which directly reflects endogenous dE2F1 activity (Thacker et al. 2003; Morris et al. 2008). Taken together, these molecular and genetic analyses suggest that the dE2f1-dsRNA phenotypes result from the specific reduction of dE2F1 activity.
A dominant modifier genetic screen to identify novel regulators of dE2F1 activity
To identify novel regulators of dE2F1 in vivo, we performed a dominant modifier genetic screen based on the dE2f1-dsRNA phenotypes described previously. The initial screen used the Exelixis Df collection (459 lines), which was generated in an isogenic background and all of the breakpoints are molecularly defined (Parks et al. 2004). We conducted a primary screen using the dE2f1-dsRNA eye phenotype because of ease of screening, and only Df lines that were able to modify this eye phenotype were subsequently retested using the dE2f1-dsRNA wing phenotype (Figure 2A). Thus, the Df lines that did not modify the dE2f1-dsRNA eye phenotype (referred to as “no effect” or “NE” in the tables) are excluded from further analysis (referred to as “not determined” or “ND” in the tables). Although this screen strategy may miss the modifiers that only affect the dE2f1-dsRNA wing phenotype, it enabled the identification of general regulators of E2F1 activity rather than tissue-specific modifiers.
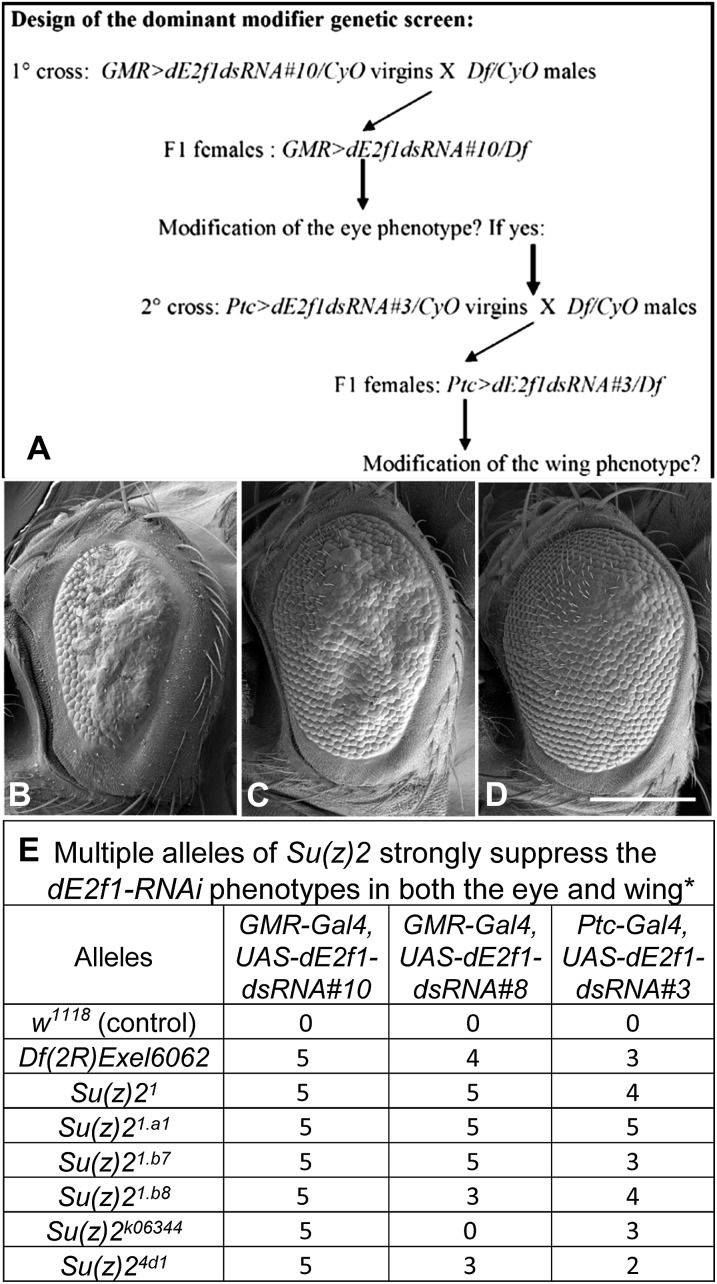
Figure 2 .
Su(z)2 is a strong suppressor of the dE2f1-dsRNA phenotypes in the eye. (A) The design of the dominant modifier genetic screen using deficiency lines. (B-D) shows the modification of the dE2f1-dsRNA eye phenotype by Su(z)2 alleles. The eye phenotype of GMR-Gal4, UAS-dE2f1dsRNA#8/+ (B) flies can be strongly suppressed by the Df(2R)Exel6062 line (C, the genotype is w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/Df(2R)Exel6062; +/+) and a null allele of Su(z)2 (D, the genotype is w1118; GMR-Gal4, UAS-dE2f1dsRNA#8/Su(z)21.b7; +/+). (E) Summary of the genetic interactions between Su(z)2 alleles and dE2f1-dsRNA phenotypes in the eye and wing. The suppressive effect was ranked with scores from 1 to 5, with “1” being the weakest and “5” the strongest. “0” means no genetic interaction. The scale bar in (D) is 200µm.
From these screens, we identified 18 suppressor Df lines (Table 1) and 23 enhancer Df lines (Table 2) that modified both dE2f1-dsRNA phenotypes in the same fashion. The results of all Exelixis Df lines are summarized in Supporting Information, Table S1. Because the dE2f1-dsRNA phenotypes are based on RNAi, we tested the modifier Df lines on the GMR > white-Inverted Repeat (GMR-wIR) line, to identify gene products that change RNAi efficiency rather than the E2F1 directly (Lee et al. 2004). None of the enhancers and suppressors of the dE2f1-dsRNA phenotypes affected the GMR-wIR eye color (Table 1 and Table 2), suggesting that the modifiers identified in our screen are bona fide regulators of dE2F1.
Table 1. Exelixis Df lines that dominantly suppress the dE2f1-dsRNA phenotypes.
| Bloomington Stock No. | Symbol | Breakpoints | GMR-Gal4,UAS-dE2f1-dsRNA#10a,b | ptc-Gal4,UAS-dE2f1-dsRNA#3a,c | GMR-wIR a |
|---|---|---|---|---|---|
| 7699 | Df(1)Exel6221 | 1B4;1B8 | 5 | 2 | NE |
| 7700 | Df(1)Exel6223 | 1C4;1D2 | 5 | 4 | NE |
| 7723 | Df(1)Exel6255 | 20A1;20B1 | 5 | 3 | NE |
| 7772 | Df(2L)Exel7002 | 21B4;21B7 | 5 | 2 | NE |
| 7774 | Df(2L)Exel8003 | 21D1;21D2 | 5 | 2 | NE |
| 7489 | Df(2L)Exel6002 | 21D2;21D3 | 5 | 5 | NE |
| 8000 | Df(2L)Exel6006 | 22B5;22D1 | 5 | 2 | NE |
| 7817 | Df(2L)Exel8024 | 31A2;31B1 | 5 | 2 | ND |
| 7531 | Df(2L)Exel6049 | 40A5;40D3 | 5 | 1 | NE |
| 7540 | Df(2R)Exel6058 | 44C4;44D1 | 5 | 2 | NE |
| 7544 | Df(2R)Exel6062 | 49E6;49F1 | 5 | 4 | NE |
| 7880 | Df(2R)Exel9015 | 51F11;51F12 | 5 | 1 | NE |
| 7883 | Df(2R)Exel7138 | 52D1;52D12 | 5 | 1 | NE |
| 7557 | Df(2R)Exel6077 | 57F10;58A3 | 5 | 4 | NE |
| 7903 | Df(2R)Exel7173 | 58D4;58E5 | 5 | 2 | NE |
| 7921 | Df(3L)Exel9000 | 64A10;64B1 | 5 | 2 | NE |
| 7927 | Df(3L)Exel7210 | 65A1;65A5 | 5 | 2 | NE |
| 7992 | Df(3R)Exel9014 | 95B1;95D1 | 5 | 1 | NE |
The suppressive effect was ranked with scores from 1 to 5, with “1” the weakest and “5” the strongest. ND, not determined (this line is no longer available from the Bloomington stock center); NE, no effect.
These crosses were maintained at 25°.
These crosses were maintained at 22-23°; see Materials and Methods for the detailed genotypes analyzed.
Table 2. Twenty-three enhancers from the Exelixis Df lines.
| Bloomington Stock No. | Symbol | Breakpoints | GMR-Gal4,UAS-dE2f1- dsRNA#10a,b | ptc-Gal4,UAS-dE2f1- dsRNA#3a,c | GMR-wIR a |
|---|---|---|---|---|---|
| 7510 | Df(2L)Exel6027 | 32D2;32D5 | 5 | Lethal | NE |
| 7519 | Df(2L)Exel6036 | 35B1;35B2 | 3 | 5 | NE |
| 7859 | Df(2R)Exel7094 | 44A4;44B4 | 3 | 4 | NE |
| 7538 | Df(2R)Exel6056 | 44A4;44C2 | 4 | 5 | NE |
| 7896 | Df(2R)Exel7162 | 56F11;56F16 | 2 | 3 | NE |
| 7554 | Df(2R)Exel6072 | 57B16;57D4 | 2 | Lethal | NE |
| 7902 | Df(2R)Exel7171 | 58C1;58D2 | 5 | Lethal | NE |
| 7745 | Df(3L)Exel6279 | 66A17;66B5 | 4 | 2 | NE |
| 7602 | Df(3L)Exel6123 | 70D7;70E4 | Pupal lethal | Lethal | NE |
| 7611 | Df(3L)Exel6132 | 74B2;74D2 | 3 | 1 | NE |
| 7614 | Df(3L)Exel6135 | 76B11;76C4 | 5 | Lethal | NE |
| 7624 | Df(3R)Exel6145 | 83C1;83C4 | 5 | Lethal | NE |
| 7627 | Df(3R)Exel6148 | 84F12;85A2 | Pupal lethal | Lethal | NE |
| 7632 | Df(3R)Exel6153 | 85D21;85E1 | 3 | 3 | NE |
| 7633 | Df(3R)Exel6154 | 85E9;85F1 | 4 | 2 | NE |
| 7732 | Df(3R)Exel6265 | 85F10;85F16 | 4 | 2 | NE |
| 7636 | Df(3R)Exel6157 | 86B1;86B3 | 5 | 2 | NE |
| 7641 | Df(3R)Exel6162 | 87A1;87B5 | Pupal lethal | Lethal | NE |
| 7649 | Df(3R)Exel6170 | 87F10;87F14 | 1 | 5 | NE |
| 7742 | Df(3R)Exel6275 | 88D1;88D7 | 5 | Pupal lethal | NE |
| 7659 | Df(3R)Exel6180 | 91B5;91C5 | 3 | 3 | NE |
| 7678 | Df(3R)Exel6199 | 95F8;96A2 | 3 | 5 | NE |
| 7993 | Df(3R)Exel8178 | 95F8;96A6 | 3 | 4 | NE |
The effect of enhancement was ranked with scores from 1 to 5, with “1” the weakest and “5” the strongest. NE, no effect.
These crosses were maintained at 25°.
These crosses were maintained at 22-23°; See Materials and Methods for the detailed genotypes analyzed.
Su(z)2 is a strong suppressor of the dE2f1-dsRNA phenotypes
One of the strongest suppressors (Df(2R)Exel6062) of the dE2f1-dsRNA phenotypes was reported to delete only one characterized gene, Su(z)2 (Parks et al. 2004). Df(2R)Exel6062 suppressed both the eye phenotype (Figure 2C compared to the control Figure 2B) and the wing phenotype (Figure 3C compared to the control Figure 3B). The Df(2R)Exel6062 line deletes a region of ∼54kb between two P-element (XP vector) insertion lines d09185 and d02103 (Parks et al. 2004; Thibault et al. 2004). This deletion starts at 190bp region upstream of the neighboring gene Posterior sex comb (Psc), and includes CG33798 (an uncharacterized gene with unknown function) and the Su(z)2 gene (Parks et al. 2004).
To validate the suppressor gene of the dE2f1-dsRNA phenotypes, we tested the capacity of additional alleles of Su(z)2 from the Bloomington stock center (Su(z)21, Su(z)21.a1, Su(z)21.b7, Su(z)21.b8, Su(z)24d1, Su(z)2k06344) to modify the dE2F1-RNAi phenotypes (mutant alleles of CG33798 are unavailable). These Su(z)2 mutant alleles strongly suppressed the dE2f1-dsRNA (line #10) eye phenotype, and to a less extent with the strong eye phenotype generated by dE2f1-dsRNA (line #8; Figure 2E). Next, we validated these genetic interactions identified in the eye by testing the effect of Su(z)2 mutants on the dE2f1-dsRNA wing phenotype. Reducing Su(z)2 by either Df(2R)Exel6062 (Figure 3C) or Su(z)21.a1 (Figure 3D) increased the L3-L4 intervein region of ptc-Gal4 UAS-dE2f1-dsRNA flies compared with controls (ptc-Gal4 UAS-dE2f1-dsRNA/+, Figure 3B). Measurement of L3-L4 distance demonstrated significant rescue of the intervein distance by these Su(z)2 alleles compared to the control (Figure 3E). Together, these genetic analyses suggest that Su(z)2 is a strong suppressor of dE2f1-dsRNA phenotypes.
We then sought to extend this observation by examining additional Su(z)2 alleles described recently (Emmons et al. 2009). We examined the capacity of Su(z)2 point mutant alleles (Su(z)2s15, Su(z)2s20, Su(z)2s21, Su(z)2s36, Su(z)2s84, Su(z)2s95, and Su(z)2sM) to suppress the dE2f1-dsRNA phenotypes. However, we did not observe any obvious modification of the E2F1-dsRNA phenotypes (data not shown), indicating that these particular Su(z)2 point mutations are insufficient to modify these phenotypes. Similarly, we did not observe any genetic interactions between dE2f1 and multiple alleles of the Su(z)2 paralog, Psc (Pscs14, Psce22, Psch27, PscEY06547, and Psck07804; data not shown), suggesting that Su(z)2 and Psc are not functionally redundant in these genetic analyses.
Multiple PcG and PcG-related genes suppress the dE2f1-dsRNA phenotypes
Having identified Su(z)2 as a suppressor of the dE2f1-dsRNA phenotypes, we tested whether mutants other than Polycomb family members, as well as genes that genetically interact with PcG, such as E(Pc) (Jürgens 1985; Campbell et al. 1995), Mi-2 (Kehle et al. 1998), and His2AvD (Swaminathan et al. 2005), could modify the dE2f1-dsRNA phenotypes. We observed that mutations of Pc, pho, Su(z)12, Scm, and His2AvD suppressed the dE2f1-dsRNA phenotypes, whereas E(Pc) and Mi-2 behaved as enhancers (Table 3, Figure 3E). Importantly, components of three PcG complexes, including PRC 1 complex (Pc, Scm), PRC2 (Esc, Su(z)12), and PhoRC (Pho), were able to suppress the dE2f1-dsRNA phenotypes (Table 3), suggesting that PcG may repress dE2F1 activities. In addition, we observed that mutant alleles of several PcG/TrxG genes, such as ash21, crm7, Dsp1EP355, eff8, lid10424, lidk06801, PclEY08457, Sce1, trx1, trxEY13717, showed variable genetic interactions ranging from suppression to no effect and enhancement of varied degrees (data not shown). These variable interactions might reflect the dynamic and complex interactions in vivo.
Table 3. Some of the PcG and TrxG genes dominantly modify the phenotypes caused by varied dE2F1 and RBF1 in the Drosophila eye and wing.
| Mutant Alleles | GMR-Gal4,UAS-dE2f1-dsRNA#10a,b | ptc-Gal4,UAS-dE2f1-dsRNA#3a,b | Act88F-Gal4,UASdE2fa,b | GMR-Gal4,UASdE2f1, UAS-dDpa,b |
|---|---|---|---|---|
| Suppressors | ||||
| Asx1 | Suppression (5) | Suppression (1) | Enhancement (1) | NE |
| effmer4 | Suppression (5) | Suppression (2) | NE | NE |
| E(Pc)84DET66.1 | Suppression (5) | Suppression (1) | NE | NE |
| esc1 | Suppression (4) | NE | ND | ND |
| esc21 | Suppression (5) | Suppression (4) | ND | ND |
| His2AvD810 | Suppression (5) | Suppression (2) | ND | Enhancement (3) |
| His2AvD05146 | Suppression (5) | Suppression (3) | ND | Enhancement (3) |
| KisBG01657 | Suppression (5) | Suppression (2) | NE | NE |
| Pc3 | Suppression (5) | Suppression (5) | ND | Enhancement (4) |
| Pcf01890 | Suppression (5) | Suppression (2) | ND | Enhancement (1) |
| pho1 | Suppression (5) | Suppression (1) | Enhancement (1) | NE |
| ScmD1 | Suppression (5) | Suppression (4) | ND | ND |
| Su(z)21 | Suppression (5) | Suppression (4) | ND | Enhancement (1) |
| Su(z)21.a1 | Suppression (5) | Suppression (5) | NE | NE |
| Su(z)21.b7 | Suppression (5) | Suppression (3) | Enhancement (1) | NE |
| Su(z)2k06344 | Suppression (5) | Suppression (3) | Enhancement (1) | NE |
| Su(z)123 | Suppression (5) | Suppression (2) | NE | NE |
| tara1 | Suppression (5) | Suppression (1) | ND | NE |
| tou2 | Suppression (5) | Suppression (2) | Enhancement | NE |
| brm2 | Suppression (5) | Suppression (2) | NE | NE |
| trxKG08639 | Suppression (5) | Suppression (1) | NE | NE |
| Enhancers | ||||
| E(Pc)w3 | Enhancement (4) | Enhancement (3) | Suppression (1) | ND |
| E(Pc)D4 | Enhancement (5) | Enhancement (5) | Suppression (1) | ND |
| Mi-2j3D4 | Enhancement (4) | Enhancement (1) | Suppression (4) | NE |
| Mi-2EY08138 | Enhancement (5) | Enhancement (1) | ND | ND |
| Su(z)31 | Enhancement (5) | Enhancement (4) | Suppression (1) | ND |
| taraBG01673 | Pupal lethal | Enhancement (4) | Lethal | Lethal |
The effects of suppression or enhancement were ranked with scores from 1 to 5, with “1” the weakest and “5” the strongest; NE, no effect; ND, not determined.
These crosses were maintained at 25°.
These crosses were maintained at 22-23°; see Materials and Methods for the detailed genotypes analyzed.
Next, we tested whether PcG mutants could modify phenotypes caused by overexpression of dE2f1 alone or together with dDp, as we described previously (Staehling-Hampton et al. 1999; Morris et al. 2008). We found that PcG mutants weakly enhanced phenotypes associated with dE2f1 overexpression (Table 3), which is consistent with the PcG role in repressing dE2F1 activities. Furthermore, to examine whether the PcG genes affect RNAi efficiency, we used the GMR-wIR line and tested several PcG mutants, including E(Pc)w3, Psc1, Psce23, Psce25, Psch28, Su(z)21.b8, Su(z)2k06344, Su(z)24d1, Su(z)2s15, Su(z)2s20, Su(z)2s95, and Su(z)2sM. We did not observe any of these lines affected the light yellow eye color caused by knocking down of white gene (data not shown), suggesting that Psc and Su(z)2 does not affect RNAi process. Taken together, these genetic analyses revealed in vivo regulation of dE2F1 by the PcG complexes, suggesting that several PcG complexes cooperate to restrict dE2F1-dependent cell proliferation.
Su(z)2 represses the expression of dE2f1 and critical proliferation target genes
To examine the role of Su(z)2 in regulating dE2F1 activity, we depleted Su(z)2 in cultured Drosophila SL2 cells and analyzed the expression of dE2f1 and a subset of critical proliferation target genes by qRT-PCR. Depletion of Su(z)2 significantly increased the transcription of dE2f1 and dE2F1 target genes including PCNA and dCycE (Figure 4A). In contrast, reduction of Su(z)2 had little effect on Rbf1 transcription and weakly up-regulates the expression of dE2f2 gene (Figure 4A). These results suggest that Su(z)2 constrains cell proliferation by regulating the expression of dE2f1, PCNA, and dCycE.
To test whether depletion of Su(z)2 could rescue the effect of reduced dE2f1 transcription, we codepleted Su(z)2 and dE2f1 in Drosophila SL2 cells and measured the effect on dE2F target gene expression. As shown in Figure 4B, we observed that compared to knocking down dE2f1 alone, codepletion of Su(z)2 and dE2F1 significantly increased the expression of dE2f1 and several dE2F1 target genes, including PCNA, dCycE, string (stg, encoding Drosophila CDC25 phosphatase), and Mcm5. These results suggest that reduction of Su(z)2 is sufficient to alleviate the effect of dE2f1 depletion in SL2 cells, which is consistent with our observations that Su(z)2 mutants can suppress the dE2f1-dsRNA phenotypes.
Next, to determine the biological consequence of codepleting dE2f1 and Su(z)2, we conducted the dimethyltriazoldiphenyl tetrazolium-formazan cell viability assays, also known as the MTT assay, to analyze the kinetics of cell proliferation in SL2 cells. This assay is based on mitochondrial reduction of a tetrazolium salt to a colored formazan salt, which can be quantified by measuring the absorbance at 570 nm, in living cells (Hansen et al. 1989). Depletion of dE2f1 impairs cellular proliferation, and cells arrest after 5 days of dsRNA treatment (Figure 4C). Reducing Su(z)2 levels alone has little effect on cell proliferation (Figure 4C); however, codepletion of dE2f1 and Su(z)2 significantly rescues the proliferation defects associated with dE2f1 depletion. In contrast, codepletion of dE2f1 and Psc (or E(Pc); data not shown) had no effect in rescue of this defect (Figure 4D), which is consistent with our genetic analyses (Table 3). Interestingly, depleting Psc alone blocked cell proliferation (Figure 4D), consistent with the recently reported role of Psc in regulating the G2-M progression by directly affecting Cyclin B degradation (Mohd-Sarip et al. 2012). In contrast to Psc, depleting Su(z)2 does not affect cell proliferation (Figure 4C), suggesting that unlike Psc, Su(z)2 may not regulate the turnover of CycB and nuclear division. Taken together, these results suggest that Su(z)2 represses the transcription of dE2f1 and certain dE2F1 target genes that are required for cell proliferation.
Discussion
PcG and TrxG complexes play important roles in maintaining the expression of many developmental genes in metazoans, and deregulation of their functions has been linked to human malignancy. Here we identify genetic interactions between multiple components of PcG complexes and a key cell-cycle regulator, E2F1, in Drosophila. We find that mutations compromising the PcG functions suppress the defects caused by dE2F1-RNAi in the Drosophila eye and wing. Our results suggest that PcG complexes may regulate the key cell-cycle regulator dE2F1 and a subset of dE2F1 target genes in Drosophila development. To our knowledge, this is the first work to show functionally that dE2F1 is affected by PcG proteins, especially by Su(z)2.
Mutant alleles of Su(z)2, but not Psc, suppress dE2f1-dsRNA phenotypes
Our dominant modifier genetic screen using Exelixis Df mutants identified Su(z)2 as a strong suppressor of the dE2F1-RNAi phenotypes. By expanding our studies to mutations of other components of the PcG complexes, we found a strong genetic link between PcG and E2F1 activity. However, as summarized in Table 3, not all of mutant alleles of the PcG genes tested modified the dE2f1-dsRNA phenotypes. For example, although Su(z)2 and Psc are paralogs and their functions are partially redundant (Brunk et al. 1991; van Lohuizen et al. 1991; Soto et al. 1995; Wu and Howe 1995; Stankunas et al. 1998), we found that only Su(z)2 could modify the E2F1-RNAi phenotypes. In addition, biochemical analyses suggest that both Psc and Su(z)2 share similar activities in DNA binding, chromatin compacting, and chromatin remodeling inhibition (Lo et al. 2009). However, in multiple analyses, including genetic tests based on phenotypes caused by overexpression or knockdown of dE2F1, and experiments in cultured SL2 cells, we observed a consistent pattern of interaction with Su(z)2 but not Psc (Figure 2, Figure 4, Table 3).
There are several potential explanations to these observations. First, this screen was designed to identify the dominant modifiers and perhaps mutations within some PcG genes remain above a critical threshold during development. Second, the dE2f1-dsRNA phenotypes in both the eye and wing are caused by reduction of dE2F1 protein levels and dE2F1 activity (Morris et al. 2008). Because dE2F1 levels vary during the cell cycle (Shibutani et al. 2008), the dynamic interactions between dE2F1 and PcG gene products may determine whether a phenotypic interaction can be visualized in these adult tissues. Perhaps Su(z)2 has a more important role in the tissues we used to screen for E2F1 modifiers, and our genetic tests alone still cannot rigorously rule out the potential redundant functions of Su(z)2 and Psc. Third, as Psc regulates mitotic progression independently of the transcriptional functions of the canonical PcG complexes, it is likely that Su(z)2 and Psc regulate different sets of targets (Mohd-Sarip et al. 2012). Unlike Psc (Figure 4D), depleting Su(z)2 alone does not affect cell proliferation (Figure 4C), suggesting that Su(z)2 may not have a role in regulating CycB degradation. Nevertheless, the mitotic effects of Psc may mask its role in regulating dE2f1 transcription. Thus, our results are not sufficient to exclude the possibility that Psc might have a redundant role with Su(z)2 in repressing the expression of dE2f1. Additional molecular and biochemical analyses are necessary to further dissect the difference between these two paralog proteins.
The dE2f1 gene is a target repressed by PcG complexes
There are several lines of evidence suggesting that dE2F1 activity is regulated by PcG and TrxG complexes. Mutant alleles of subunits of the SWI/SNF chromatin-remodeling complex (Grimaud et al. 2006), such as brahma (brm) and moira (mor), have been shown to dominantly modify the rough eye phenotype caused by overexpression of dE2f1 and its heterodimeric partner dDp (Staehling-Hampton et al. 1999). Subunits of the Domino chromatin-remodeling complex (PcG-like L3mbt and the related dSfmbt) negatively regulate transcription of an artificial dE2f1 reporter gene (Lu et al. 2007). ChIP assays have identified both Ph and Pho on the promoter and coding regions of the dE2f1 gene in Drosophila embryos (Oktaba et al. 2008).
PcG complexes regulate methylation of H3K27 in Drosophila (Cao and Zhang 2004), we therefore analyzed the status of H3K27 methylation during development or in several Drosophila cell lines using chromatin immunoprecipitation (ChIP) followed by microarray hybridization (ChIP-chip) or high-throughput sequencing (ChIP-Seq) data sets deposited to modENCODE (Celniker et al. 2009) (http://modencode.oicr.on.ca/fgb2/gbrowse/fly/). We found that the genomic loci of dE2f1, dCycE and stg display mono-, di-, or trimethylation of H3K27 (H3K27me1/2/3) during development or in Drosophila cell lines, including SL2, Kc, and BG3 cells (see Figure S1, Figure S2, Figure S3, and Figure S4 for details), suggesting that PcG may directly regulate the expression of these genes. dCycE and stg are critical dE2F1 target genes, which regulate the G1/S-phase and the G2/M-phase transition of the cell cycle, respectively (Edgar and Lehner 1996; Dyson 1998). We did not observe obvious H3K27me modification of other dE2F1 target genes such as PCNA and Mcm5 (data not shown), suggesting that the effect of Su(z)2 on expression of these genes (Figure 4B) is likely indirect through dE2f1. Together, these observations suggest that PcG complexes may repress the expression of dE2f1 and a subset of dE2F1 target genes during development.
These observations are consistent with our genetic studies and suggest the suppressive effect of PcG mutants on dE2f1-dsRNA phenotypes is caused by derepression of dE2f1 and certain dE2F1 target genes, which compensates for the effect of dE2f1-depletion. Together with previous published observations linking PcG complexes to cell-cycle regulators, such as dCycA (Martinez et al. 2006), dCycB (Oktaba et al. 2008), dCycE (Brumby et al. 2002), and dE2f1 (Oktaba et al. 2008), our observations provide further support for the role of PcG in repressing the transcription of cell-cycle genes, including dE2f1, dCycE, and stg (Figure 4 and Figure S4).
Regulation of the key cell-cycle regulators by PcG complexes may present a general mechanism to coordinate cellular differentiation and proliferation during development. Disrupting the coordination between differentiation and proliferation may result in abnormal development and may contribute to tumorigenesis. Consistent with this notion, accumulating evidence shows that the PcG complexes are misregulated in a wide variety of human cancers (Sparmann and van Lohuizen 2006; Ballestar and Esteller 2008; Bracken and Helin 2009). This study, together with previous reports in Drosophila (Staehling-Hampton et al. 1999; Brumby et al. 2002; Grimaud et al. 2006; Martinez et al. 2006; Lu et al. 2007; Oktaba et al. 2008), suggest that mutations compromising PcG activity would elevate E2F activity, thereby providing cells with a strong tumorigenic advantage. Further studies are necessary to elucidate how these two important regulatory mechanisms are coordinated during cellular differentiation and proliferation in development.
Supplementary Material
Acknowledgments
We thank Richard Carthew and Andre Bernard for sharing the pWIZ vector, Fajun Yang for his advice in generating pWIZ-dE2f1 constructs, Doug Renee at the MGH Drosophila transgene core facility for his help in making transgenic flies, Bill Fowle at the Northeastern University for his expertise with scanning electron microscopy, and Rebecca Wang for technical assistant. We appreciate Keith Maggert as well as the anonymous reviewers for critical reading of the manuscript and constructive comments. We are grateful to Richard Carthew, Robert Duronio, Richard Emmons, Antonio García-Bellido, Vincenzo Pirrotta, Chao-Ting Wu, and the Bloomington Drosophila Stock Center for mutant stocks and reagents used in this work. This work is supported in part by a Fund for Medical Discovery Postdoctoral Fellowship from Massachusetts General Hospital, a startup fund from Texas A&M Health Science Center and a grant from the American Heart Association to J.-Y.J., and National Institutes of Health grant (R01GM053203) to N.J.D., the James and Shirley Curvey MGH Research Scholar. The first author would like to dedicate this paper to the memory of Professor Gerold Schubiger for his many years of inspiration and support.
Footnotes
Communicating editor: J. Brill
Literature Cited
- Attwooll C., Oddi S., Cartwright P., Prosperini E., Agger K., et al. , 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J. Biol. Chem. 280: 1199–1208 [DOI] [PubMed] [Google Scholar]
- Ballestar E., Esteller M., 2008. Epigenetic gene regulation in cancer. Adv. Genet. 61: 247–267 [DOI] [PubMed] [Google Scholar]
- Bracken A. P., Helin K., 2009. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9: 773–784 [DOI] [PubMed] [Google Scholar]
- Bracken A. P., Pasini D., Capra M., Prosperini E., Colli E., et al. , 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22: 5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Manoukian A. S., Perrimon N., 1994. Ectopic expression in Drosophila. Methods Cell Biol. 44: 635–654 [DOI] [PubMed] [Google Scholar]
- Brumby A. M., Zraly C. B., Horsfield J. A., Secombe J., Saint R., et al. , 2002. Drosophila cyclin E interacts with components of the Brahma complex. EMBO J. 21: 3377–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk B. P., Martin E. C., Adler P. N., 1991. Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature 353: 351–353 [DOI] [PubMed] [Google Scholar]
- Burkhart D. L., Sage J., 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. B., Sinclair D. A., Couling M., Brock H. W., 1995. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 246: 291–300 [DOI] [PubMed] [Google Scholar]
- Cao R., Zhang Y., 2004. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., et al. , 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J. E., Micol J. L., Garcia-Bellido A., 1990. Transvection in the Drosophila Ultrabithorax gene: a Cbx1 mutant allele induces ectopic expression of a normal allele in trans. Genetics 126: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S., et al. , 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel M., Friesenhahn L., Lees J. A., 2008. E2f6 and Bmi1 cooperate in axial skeletal development. Dev. Dyn. 237: 1232–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya A., Wong S., Gonzalo S., Gavin M., Dean D. C., 2001. Linking the Rb and polycomb pathways. Mol. Cell 8: 557–569 [DOI] [PubMed] [Google Scholar]
- Dimova D. K., Stevaux O., Frolov M. V., Dyson N. J., 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R. J., O’Farrell P. H., Xie J. E., Brook A., Dyson N., 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Dyson N., 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12: 2245–2262 [DOI] [PubMed] [Google Scholar]
- Edgar B. A., Lehner C. F., 1996. Developmental control of cell cycle regulators: a fly’s perspective. Science 274: 1646–1652 [DOI] [PubMed] [Google Scholar]
- Emmons R. B., Genetti H., Filandrinos S., Lokere J., Wu C. T., 2009. Molecular genetic analysis of Suppressor 2 of zeste identifies key functional domains. Genetics 182: 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., et al. , 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov M. V., Dyson N. J., 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117: 2173–2181 [DOI] [PubMed] [Google Scholar]
- Gil J., Peters G., 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell Biol. 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Grimaud C., Negre N., Cavalli G., 2006. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 14: 363–375 [DOI] [PubMed] [Google Scholar]
- Hannon G. J., 2002. RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Hansen M. B., Nielsen S. E., Berg K., 1989. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 119: 203–210 [DOI] [PubMed] [Google Scholar]
- Jürgens G., 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155 [Google Scholar]
- Kehle J., Beuchle D., Treuheit S., Christen B., Kennison J. A., et al. , 1998. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282: 1897–1900 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Carthew R. W., 2003. Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30: 322–329 [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Nakahara K., Pham J. W., Kim K., He Z., et al. , 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Levine S. S., King I. F., Kingston R. E., 2004. Division of labor in polycomb group repression. Trends Biochem. Sci. 29: 478–485 [DOI] [PubMed] [Google Scholar]
- Lo S. M., Ahuja N. K., Francis N. J., 2009. Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol. Cell. Biol. 29: 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Ruhf M. L., Perrimon N., Leder P., 2007. A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression. Proc. Natl. Acad. Sci. USA 104: 9381–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D., 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. M., Colomb S., Dejardin J., Bantignies F., Cavalli G., 2006. Polycomb group-dependent Cyclin A repression in Drosophila. Genes Dev. 20: 501–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Zhang Y., Xu R. M., 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Lagarou A., Doyen C. M., van der Knaap J. A., Aslan U., et al. , 2012. Transcription-independent function of Polycomb group protein PSC in cell cycle control. Science 336: 744–747 [DOI] [PubMed] [Google Scholar]
- Morris E. J., Ji J. Y., Yang F., Di Stefano L., Herr A., et al. , 2008. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature 455: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H., Helin K., 2000. The E2F transcription factors: key regulators of cell proliferation. Biochim. Biophys. Acta 1470: M1–M12 [DOI] [PubMed] [Google Scholar]
- Müller J., Verrijzer P., 2009. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19: 150–158 [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y., 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Oktaba K., Gutierrez L., Gagneur J., Girardot C., Sengupta A. K., et al. , 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., et al. , 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G., 2007. Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Pirrotta V., 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Sherr C. J., 2004. Principles of tumor suppression. Cell 116: 235–246 [DOI] [PubMed] [Google Scholar]
- Shibutani S. T., de la Cruz A. F., Tran V., Turbyfill W. J., 3rd, Reis T., et al. , 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell 15: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto M. C., Chou T. B., Bender W., 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A., van Lohuizen M., 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Ciampa P. J., Brook A., Dyson N., 1999. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics 153: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K., Berger J., Ruse C., Sinclair D. A., Randazzo F., et al. , 1998. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 125: 4055–4066 [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dyson N. J., 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14: 684–691 [DOI] [PubMed] [Google Scholar]
- Storre J., Elsasser H. P., Fuchs M., Ullmann D., Livingston D. M., et al. , 2002. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 3: 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J., Baxter E. M., Corces V. G., 2005. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 19: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker S. A., Bonnette P. C., Duronio R. J., 2003. The contribution of E2F-regulated transcription to Drosophila PCNA gene function. Curr. Biol. 13: 53–58 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Trimarchi J. M., Lees J. A., 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Trimarchi J. M., Fairchild B., Wen J., Lees J. A., 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N. J., 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724 [DOI] [PubMed] [Google Scholar]
- van Lohuizen M., Frasch M., Wientjens E., Berns A., 1991. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 353: 353–355 [DOI] [PubMed] [Google Scholar]
- Wu C. T., Howe M., 1995. A genetic analysis of the Suppressor 2 of zeste complex of Drosophila melanogaster. Genetics 140: 139–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., et al. , 2012. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122: 2417–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.