Abstract
The identification of rare inherited and de novo copy number variations (CNVs) in human subjects has proven a productive approach to highlight risk genes for autism spectrum disorder (ASD). A variety of microarrays are available to detect CNVs, including single-nucleotide polymorphism (SNP) arrays and comparative genomic hybridization (CGH) arrays. Here, we examine a cohort of 696 unrelated ASD cases using a high-resolution one-million feature CGH microarray, the majority of which were previously genotyped with SNP arrays. Our objective was to discover new CNVs in ASD cases that were not detected by SNP microarray analysis and to delineate novel ASD risk loci via combined analysis of CGH and SNP array data sets on the ASD cohort and CGH data on an additional 1000 control samples. Of the 615 ASD cases analyzed on both SNP and CGH arrays, we found that 13,572 of 21,346 (64%) of the CNVs were exclusively detected by the CGH array. Several of the CGH-specific CNVs are rare in population frequency and impact previously reported ASD genes (e.g., NRXN1, GRM8, DPYD), as well as novel ASD candidate genes (e.g., CIB2, DAPP1, SAE1), and all were inherited except for a de novo CNV in the GPHN gene. A functional enrichment test of gene-sets in ASD cases over controls revealed nucleotide metabolism as a potential novel pathway involved in ASD, which includes several candidate genes for follow-up (e.g., DPYD, UPB1, UPP1, TYMP). Finally, this extensively phenotyped and genotyped ASD clinical cohort serves as an invaluable resource for the next step of genome sequencing for complete genetic variation detection.
Keywords: rare variants, gene copy number, chromosomal abnormalities, cytogenetics, molecular pathways
Rare inherited and de novo copy number variations (CNVs) contribute to the genetic vulnerability in autism spectrum disorder (ASD) in as many as 5–10% of idiopathic cases examined (Devlin and Scherer 2012). Smaller intragenic CNVs (often called indels) can be involved, or CNVs can encompass an entire gene, and in some cases they can affect several genes as part of a genomic disorder (Lee and Scherer 2010). Screening for CNVs using microarrays has proven to be a rapid method to identify both large and small genomic imbalances associated with ASD susceptibility (Jacquemont et al. 2006; Sebat et al. 2007; Autism Genome Project Consortium 2007; Christian et al. 2008; Marshall et al. 2008; Pinto et al. 2010; Shen et al. 2010; Sanders et al. 2011).
A trend emerging from recent investigations in autism genetics is that significant heterogeneity and complexity exists. However, some highly penetrant risk genes (e.g., the SHANK, NRXN, and NLGN family members) and CNV loci (e.g., 1q21.1, 15q13.3, and 16p11.2) are now known (Devlin and Scherer 2012). Moreover, there has been some progress in identifying multiple mutations in single individuals (Schaaf et al. 2011), suggesting possible multigenic threshold models for ASD (Cook and Scherer 2008). Scientific designs enabling the discovery of a comprehensive set of genetic variants will be required to fully assess the genetic burden in ASD.
CNV detection in case and control cohorts have been performed using several different microarray platforms, including those utilizing single-nucleotide polymorphisms (SNPs) and others using comparative genomic hybridization (CGH) arrays (Redon et al. 2006; Graubert et al. 2007; Christian et al. 2008; Conrad et al. 2010; Pinto et al. 2011). There are advantages to each microarray platform. SNP-based microarray platforms are typically cost-effective and have the potential to analyze the genomic DNA sample at both the SNP genotype and copy number level, whereas CGH arrays are often used in a clinical diagnostic setting because of the better signal-to-noise-ratio achieved in comparison to SNP arrays (Shinawi and Cheung 2008). Multiple algorithms, some specific to certain array types, are available to detect CNVs, and they can vary considerably in the number of CNV calls made even for an identical array experiment. In recent comprehensive studies, authors have provided performance assessments of CNV detection platforms and methods by examining control DNA samples (Lai et al. 2005; Curtis et al. 2009; Hester et al. 2009; Winchester et al. 2009; Pinto et al. 2011). The consensus is that using multiple microarray technologies and prediction algorithms increases CNV detection rates.
In this study, we used a high-resolution Agilent CGH array comprising 1 million (1M) probes to assay for CNVs in a Canadian cohort of 696 unrelated ASD cases, 615 of which were genotyped previously on SNP arrays, including Illumina 1M single/duo (Pinto et al. 2010), Affymetrix 500K (Marshall et al. 2008), Affymetrix 6.0 (Lionel et al. 2011; and A. C. Lionel, unpublished data), and Illumina Omni 2.5M (A. C. Lionel, unpublished data) arrays. Our high-quality data, interpreted for the first time with CNV control data generated on 1000 individuals using the same 1M CGH array, enabled detection of numerous rare CNVs in each ASD sample examined and identified new potential ASD risk genes. These data allowed us to significantly expand the profile of genetic variants that are potentially causative of ASD and to identify novel molecular pathways affecting ASD vulnerability in this cohort.
Materials and Methods
DNA samples and CNV analysis
ASD case cohort:
The array CGH component of this study included 696 unrelated ASD cases, 39 affected siblings, and 42 parents (17 complete trios) that passed array quality control. All the ASD cases met the criteria for autism on one or both diagnostic measures – Autism Diagnostic Interview-Revised training and Autism Diagnostic Observation Schedule training. The samples came from three Canadian sites: Hospital for Sick Children in Toronto, Ontario; McMaster University, Hamilton, Ontario; and Memorial University of Newfoundland, St. John’s, Newfoundland. For the 696 unrelated ASD cases (571 males and 125 females), CGH experiments were performed on genomic DNA derived from blood for 354 cases, lymphoblastoid cell line DNA was used for 340 cases, and in two instances saliva was the DNA source used.
The SNP microarray component of the study used for comparison included 615 unrelated ASD cases, and the data from 433 of these experiments have been published (Marshall et al. 2008; Pinto et al. 2010). The CNVs detected in these samples were found using previously described stringent calling criteria defined for two different computer algorithms, which yields reliable CNV calls that can be experimentally validated >95% of the time (Redon et al. 2006; Marshall et al. 2008; Conrad et al. 2010; Pinto et al. 2010).
Control cohort:
A control cohort of 1000 DNA samples from reportedly healthy donors (502 males and 498 females) were acquired from BioServe (Beltsville, MD) and are part of a collection of >12,000 control samples originally banked by Genomics Collaborative, Inc. (acquired by BioServe in 2007) for the purpose of large-scale genomic studies (Ardlie et al. 2002). Donors were consented and deidentified via a protocol approved by the institutional review board. The control DNA samples were derived from apparently healthy white donors older than 45 years of age. Health history information, documented at the time of consent, was used to select the samples based on the following attributes: body mass index between 15 and 35, blood glucose level <125 mg/dL, total cholesterol level between 100 and 300, systolic blood pressure between 100 and 150, and no major diseases (e.g., cancer and neurodegenerative diseases) or psychiatric disorders (e.g., alcoholism, mental illness, and depression). The control cohort DNA samples (also called PDx controls) used in this study are proprietary and currently biobanked at Population Diagnostics, Inc. (Melville, NY) for future microarray, sequencing and genotyping validation experiments.
Array CGH
The ASD test and reference samples were labeled with Cy5 and Cy3, respectively using Invitrogen BioPrime CGH labeling kit (Invitrogen, Carlsbad, CA). A pool of 50 sex-matched Caucasian control samples was used as a reference. The PDx control test and reference samples were labeled with Cy3 and Cy5, respectively and one sex-matched sample was used as a reference. All samples have been run on Agilent 1M CGH array according to manufacturer’s protocol (Agilent Technologies, Santa Clara, CA). The Agilent 1M CGH array consists of a total of 974,016 probes providing relatively uniform whole genome coverage. The arrays were scanned using the Agilent microarray scanner, and data were extracted using Feature extraction software version 10.5.1.1. The array experiments for ASD cases and PDx controls were run at The Centre for Applied Genomics (Toronto, Canada) and in the service laboratory of Oxford Gene Technology (Oxford, UK), respectively.
CNV detection and analysis
All array CGH data (both ASD cases and PDx controls) were analyzed in precisely the same manner using two programs—DNA Analytics v.4.0.85 (Agilent Technologies, Santa Clara, CA) and DNAcopy (Venkatraman and Olshen 2007)—to obtain high-confidence calls. At least five consecutive probe sets were used to call a CNV. For DNA Analytics, the aberration algorithm of Aberration Detection Method-2 was used with a threshold of 6.0, a minimum absolute average log2 ratio per region of 0.25, and maximum number of aberrant regions of 10,000 was used to identify all aberrant intervals. A nested filter of two was applied with subsequent removal of nested child calls using a custom script and retaining only the parent aberrations.
The other algorithm used to call CNVs was DNAcopy, from R Bioconductor package, which is a circular binary segmentation algorithm. The default settings and a log2 ratio cutoff of −0.41 and 0.32 for loss and gain, respectively, was used to call CNVs. Any segment with absolute median log2 ratio to median absolute deviation value less than 2 was filtered out. The CNVs detected by DNA Analytics and DNAcopy for each individual were merged using outer probe boundaries. As in our previous SNP microarray studies (Marshall et al. 2008; Pinto et al. 2010; Lionel et al. 2011), we defined a CNV as being ‘stringent’ if it was detected by both algorithms at the sample level. The stringent dataset was utilized for novel rare CNV discovery.
Quality control
Experiments with poor derivative log ratio spread (DLRs > 0.3) were discarded. Any sample that had an excessive number of CNVs detected using either algorithm, measured by mean plus 3 SDs, was identified as an outlier and removed from further analysis. Any experiment that was a gender-mismatch was removed, and we excluded CNVs that were within centromere proximal cytobands. Twenty ASD cases that had CNVs larger than 5 Mb in size were removed from further downstream analyses of overlap with SNP microarrays, global rare CNV burden analysis and gene-set association tests.
Rare ASD CNVs
Stringent CNV calls were classified as rare based on three separate stepwise comparisons with control datasets. First, a subset of stringent CNVs found at a frequency <0.5% for the total sample set (including 676 ASD cases and 1000 PDx controls) was compiled. Second, CNV data from an additional 4139 in-house controls were used to filter CNVs found at ≥0.1% frequency and where 50% by length overlapped with CNVs in the in-house controls. The additional controls consisted of 1782 subjects from the Study on Addiction: Genetics and Environment [SAGE (Bierut et al. 2010)], a total of 1234 unrelated controls from the Ottawa Heart Institute study (Stewart et al. 2009), and 1123 European controls from the PopGen study (Krawczak et al. 2006). The SAGE controls were genotyped with Illumina Human 1M-single BeadChip arrays and a subset of stringent CNVs detected by both iPattern (Pinto et al. 2010) and QuantiSNP (Colella et al. 2007) were used. The Ottawa Heart Institute and PopGen controls were genotyped with Affymetrix Genome-Wide Human SNP 6.0 arrays and the stringent subset consisted of regions that were detected by at least two of the three different CNV calling algorithms, Birdsuite (Korn et al. 2008), iPattern, and Affymetrix Genotyping Console. Third, the list of CNVs that overlapped 50% or more by length with known polymorphic regions in the genome (Conrad et al. 2010; McCarroll et al. 2008) were excluded. The final list of rare CNVs consisted of 1,884 CNVs in ASD cases and 2,299 CNVs in PDx controls.
CNV overlap With SNP microarrays to identify novel ASD CNVs
For each sample for which CNV calls from SNP microarrays were available, stringent CNVs detected using the Agilent 1M array were overlapped with the stringent CNVs detected by corresponding SNP microarray experiments (Marshall et al. 2008; Pinto et al. 2010; A.C. Lionel, unpublished data). The CNVs from the SNP arrays were filtered to include only the regions with five probes or more. The CNVs were considered to be novel when 50% or more by length of the detected call was unique to a platform.
Experimental CNV validation
The experimental validation of novel CNV calls was carried out using SYBR-Green-based (Stratagene) real-time quantitative polymerase chain reaction (qPCR) method by at least two independent PCR assays. Each assay was conducted in triplicate, with one set of primers corresponding to the region of interest and the other mapping to a control region on FOXP2 at 7q31.1 (serving as a negative diploid control). The ratio between the test and control regions were then determined using standard curve method, and a fold-change less than 0.7 was confirmed as deletion and greater than 1.3 was confirmed as duplication. The parents and siblings were also tested for inheritance and segregation of CNVs, respectively.
Selection of European ASD cases
Of the 676 unrelated ASD cases that passed QC, 615 of them had SNP microarray data available [Affymetrix Genome-Wide Human SNP Array 6.0, Affymetrix GeneChip Human Mapping 500K Array Set (Commercial and Early Access), Illumina Human1M and Human1M-Duo, and Illumina HumanOmni2.5-4 BeadChip]. We combined this subset of samples with the HapMap3 data set (International HapMap Consortium 2010), which includes individuals of different ethnicity (e.g., 183 Utah residents with Northern and Western European ancestry, 91 Toscani from Italy, 89 Han Chinese from Beijing, 181 Yorubans from Nigeria). For the ancestry analysis, only a subset of ∼30,000 autosomal, non-MHC SNPs that were common to all the platforms were used. We analyzed this data set using multidimensional scaling (MDS) as implemented in PLINK (Purcell et al. 2007) to identify a putative group of European ASD cases. To avoid confounding ancestry issues, we only used European ASD cases for global rare CNV burden analyses and gene-set association tests.
Global rare CNV burden analyses
CNVs mapping to genes were considered for burden analysis whenever at least one exon was encompassed by or impacted by the CNV. For each subject, we calculated the number of rare CNVs, the log10 of the sum of rare CNV sizes, and the total number of genes harboring a rare CNV. CNV burden differences were assessed by comparing the distribution of these statistics between case and control subjects. Specifically, differences in distribution over case and control subjects were tested using the nonparametric Wilcoxon exact test (as implemented in the R package exactRankTests), whereas the magnitude of the difference was assessed looking at the ratio of case and control means. CNV burden differences were assessed for all rare variants together, as well as for deletions and duplications only.
Gene-set association test
Gene-sets representing pathways, functional annotations and protein domains were tested if they were more frequently affected by rare CNVs in ASD cases compared with controls using the Fisher exact test [FET (Pinto et al. 2010)]. Gene-sets were compiled from Gene Ontology annotations (downloaded from NCBI in April 2011), pathway databases (KEGG, Reactome, bioCarta, NCI in March 2011), and protein domains (PFAM, March 2011). We only tested gene-sets with member genes numbering between 25 and 750: 2456 in total, 1939 from Gene Ontology, 414 from pathways, and 103 from PFAM domains. We found that small gene-sets decrease the statistical power of the analysis, whereas larger gene-sets tend to have a very broad biological scope without much useful biological meaning. For each gene-set, we built a contingency matrix with subjects as sampling units. Subjects were categorized as (1) cases or controls and (2) having at least one gene-set harboring a rare CNV or not. On the basis of this contingency table, we tested higher prevalence of rare CNVs in autism probands vs. controls using a one-tailed FET. Any subject in which a rare CNV affected more than 20 genes was not considered for the gene-set analysis because these individuals may have a broader set of gene functions perturbed by rare variants, which may consequently reduce the quality of gene-set analysis results. The FET nominal p value was corrected for multiple tests using a case/control class permutation procedure to estimate an empirical false discovery rate (FDR). We performed 5000 permutations of case-control labels and for each permutation we tested gene-sets following exactly the same procedure. The p values were ranked from lowest (most significant) to highest (least significant) and for each p value we computed the empirical FDR as the average number of gene-sets with equal or smaller p value over permutations. We have selected 25% as the empirical FDR significance threshold. Significant gene-sets were visualized using the Enrichment Map Cytoscape plugin (Merico et al. 2010).
Results
Detection of CNVs in ASD cases and controls
Of the 696 unrelated ASD cases examined by array CGH, 20 were found to carry CNVs larger than 5 Mb (Supporting Information, Table S1) and excluded from further analysis. Four of these cases were known to have Down syndrome as well as an ASD diagnosis, 14 carried large chromosomal abnormalities previously detected through karyotyping and genotyping with SNP microarrays, and two cases harbored cell line artifacts. The rare stringent CNVs from the remaining 676 unrelated ASD cases were used for gene burden analysis, gene-set association tests and comparison with CNV data from other SNP microarrays to identify novel CNVs (Figure 1, Table 1).
Figure 1 .
CNV analysis workflow. The ASD cases and controls were typed using the Agilent 1M CGH array, and CNVs were identified using two algorithms, DNA Analytics and DNAcopy. CNVs detected by both algorithms were defined as the stringent dataset and were used for novel rare variant discovery. Rare variants were defined as described in the Materials and Methods. The 1884 rare ASD CNVs (as reported in Table S2) were compared with the CNVs obtained from SNP microarray studies and this resulted in identification of 946 rare CNVs that were novel to the Agilent 1M CGH platform. The rare CNVs from only European ASD cases (505 cases) were then used for global rare CNV burden analysis and gene-set association tests by comparison to 1000 PDx controls.
Table 1. Summary statistics of stringent CNVs found in ASD and PDx control data sets.
| Unrelated ASD Cases | PDx Controls | |
|---|---|---|
| No. samples | 676 | 1000 |
| No. males/females | 560/116 | 502/498 |
| No. stringent CNVsa | 23,493 | 49,524 |
| Mean no. CNVs/ sample ± SDb | 34.75 ± 6.40 | 49.52 ± 5.95 |
| Median no. CNVs/sample | 34 | 49 |
| Mean CNV size, kb ± SD | 97.75 ± 229.412 | 87.50 ± 198.59 |
| Median CNV size, kb | 30 | 30 |
| % gain /% loss | 42.36/57.64 | 48.24/51.76 |
| No. CNVs of size >1 Mb (%) | 471 (2) | 942 (1.90) |
| No. CNVs of size from 100 Kb to 1 Mb (%) | 3715 (15.81) | 7982 (16.12) |
| No. CNVs of size from <100 Kb (%) | 19,307 (82.18) | 40,600 (81.98) |
ASD, autism spectrum disorder; CNV, copy number variations; PDx, control cohort DNA samples.
CNVs detected in the same individual using two algorithms, DNA Analytics and DNAcopy, were merged with the outside probes used as boundaries and defined as stringent CNV dataset containing at least five consecutive probes. Samples containing CNVs larger than 5 Mb were excluded (Table S1).
We observed a significant difference in average number of CNVs per sample in PDx controls than in ASD cases (One-tailed exact Wilcoxon test p value < 2.2e-16) possibly due to reference bias.
We also examined the CNV content of the 1000 PDx controls and observed a significantly higher average number of CNVs per sample (p value < 2.2e-16) compared with the ASD cases (Table 1). This finding is likely attributable to the use of a different reference DNA, a single sex-matched individual, in the PDx CGH experiments rather than a pool of 50 sex-matched individuals used as a reference in the ASD CGH experiments. However, when we focused on rare variants (as defined in the materials and methods section), we found a smaller yet significant difference in the opposite direction (p value 0.002287; Table 2). The presence of a significant change in the relation between class (ASD, control) and CNV number after restricting to rare variants was further confirmed using a quasi-Poisson and linear regression model (class variable and rare variant filter variable interaction p value < 2.2e-16). Of the 676 unrelated ASD cases and 1000 PDx controls, 630 ASD cases (93%) and 896 PDx controls (90%) had at least one rare variant detected on the CGH 1M array (Table 2).
Table 2. Summary statistics of rare CNVs found in ASD and PDx control data sets.
| Unrelated ASD Cases | PDx Controls | |
|---|---|---|
| No. samples with ≥1 rare CNV | 630 | 896 |
| No. CNVs | 1884 | 2299 |
| Mean no. CNVs/sample ± SDa | 2.99 ± 1.69 | 2.57 ± 1.44 |
| Median no. CNVs/sample | 3 | 2 |
| Mean CNV size, kb ± SD | 109.79 ± 314.56 | 95.62 ± 215.74 |
| Median CNV size, kb | 28.74 | 30.43 |
| % gain/% loss | 39.81/60.19 | 44.04/55.96 |
| No. CNVs of size >1 Mb (%) | 30 (1.59) | 24 (1.04) |
| No. CNVs of size from 100 Kb to 1 Mb (%) | 362 (19.21) | 498 (21.66) |
| No. CNVs of size from <100 Kb (%) | 1492 (79.19) | 1777 (77.29) |
ASD, autism spectrum disorder; CNV, copy number variations; PDx, control cohort DNA samples.
We observed significant difference in average number of rare CNVs in ASD cases compared with PDx controls (one-tailed exact Wilcoxon test p value 0.002287).
CNV comparison with other microarray platforms
Of the 676 unrelated ASD cases, 615 were genotyped previously with SNP microarrays including 26 cases on Illumina Human Omni 2.5M-quad (2.5 million probes), 234 cases on Affymetrix Genome-Wide Human SNP Array 6.0 (1.8 million probes), 262 cases on Illumina Human 1M single infinium chip (1 million probes), 11 cases on Illumina 1M duo array (1 million probes), and 82 ASD cases were genotyped on lower resolution Affymetrix Mapping 500K chip set (500,000 probes). The other 61 unrelated ASD cases were only run on the Agilent 1M array and thus not included in this CNV platform comparison analysis. For the 615 samples run on both SNP and CGH array platforms, we performed a sample-level 50% one-way overlap of stringent Agilent 1M CNVs with the stringent CNVs from the SNP array platforms. We found that 64% of the Agilent 1M CNVs are novel with respect to the CNVs detected on the SNP microarrays (Figure 2). A more detailed comparison of CNVs detected by the 1M CGH array vs. the various SNP array platforms is shown in Table 3. For example, a comparison between CNVs detected using similar resolution arrays Agilent 1M and Illumina 1M showed that, on average, 24 novel CNVs/sample were detected by the Agilent 1M CGH array whereas only eight novel CNVs/sample were detected by the Illumina 1M SNP array. This platform comparison analysis suggests that use of multiple microarray platforms provides complementary data as every microarray platform detects a unique set of novel CNVs.
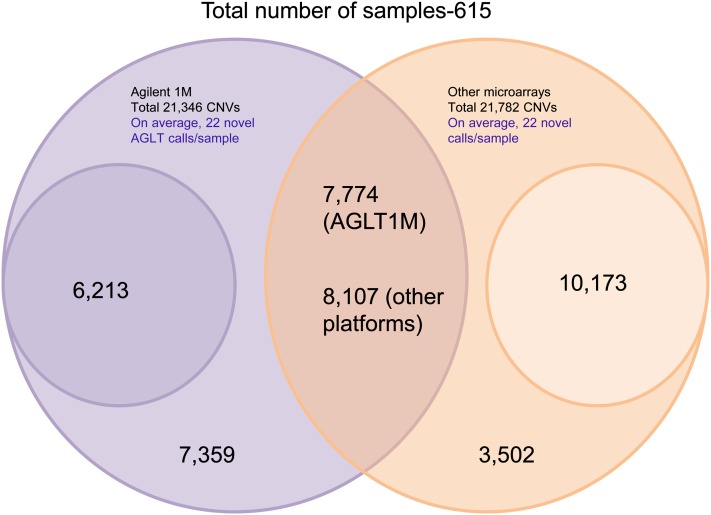
Figure 2 .
A Venn diagram showing comparison of Agilent 1M CNV calls with those detected by other SNP microarray platforms including Illumina 1M single/duo array, Affymetrix500K, Affymetrix6.0, and Illumina Omni 2.5M array for a total of 615 samples. Agilent 1M CGH data yielded 21,346 stringent CNVs and the SNP (other) microarray platforms yielded 21,782 stringent CNVs. A total of 7774 CNVs (36%) detected by the Agilent 1M array (AGLT1M) were detected by other platforms, whereas 8107 CNVs (37%) detected by other microarrays were detected by the Agilent 1M array. A total of 6213 (29%) Agilent 1M CNVs have less than 5 probe coverage in other SNP microarray platforms and 10,173 (47%) CNVs from the other SNP microarray platforms have less than 5 probe coverage in Agilent 1M array and are shown as smaller circles in this figure.
Table 3. Details of platform comparison.
| Arrays | No. Samples | No. Stringent CNVs | Average CNVs/Sample | Average CNV Size, kb | No. Overlapping calls | % Overlap/ Validationa | % CNVs With Insufficient Probes in Other Array | Novel CNVs/Sample | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Agilent 1M | 82 | 2747 | 33.5 ± 6.11 | 107 | 183 | 6 | 67.5 | 31.27 ± 5.38 |
| Affy 500K | 82 | 289 | 3.52 ± 2.28 | 520 | 189 | 65 | 6 | 2.17 ± 1.22 | |
| 2 | Agilent 1M | 262 | 8971 | 34.24 ± 6.45 | 97 | 2725 | 30 | 38 | 23.84 ± 5.30 |
| Ilmn 1M single | 262 | 4779 | 18.24 ± 4.69 | 110 | 2729 | 57 | 59.12 | 7.82 ± 3.11 | |
| 3 | Agilent 1M | 11 | 413 | 37.55 ± 6.76 | 83 | 99 | 24 | 39.70 | 28.55 ± 5.13 |
| Ilmn 1M duo | 11 | 169 | 14.36 ± 4.32 | 84 | 108 | 64 | 67.21 | 5.54 ± 1.81 | |
| 4 | Agilent 1M | 234 | 8270 | 35.49 ± 6.23 | 98 | 4439 | 53 | 5.6 | 16.44 ± 4.27 |
| Affy 6.0 | 234 | 15,315 | 65.45 ± 8.87 | 80 | 4701 | 34 | 77.46 | 45.35 ± 7.39 | |
| 5 | Agilent 1M | 26 | 945 | 36.35 ± 6.27 | 108 | 328 | 35 | 33 | 23.73 ± 4.63 |
| Ilmn Omni 2.5M | 26 | 1230 | 47.31 ± 9.77 | 61 | 380 | 31 | 81.4 | 32.69 ± 8.57 |
ASD, autism spectrum disorder; CNV, copy number variations; PDx, control cohort DNA samples.
% overlap/validation- refers to the percentage of CNVs that were also detected by the other array. For example, for the first platform comparison between Agilent 1M and Affy500K arrays, only 6% of the CNVs detected by Agilent 1M were also detected by Affy500K while 65% of the CNVs detected by Affy500K were also detected or validated by Agilent 1M array.
Rare ASD CNVs that were not detected by SNP microarrays but were detected by the Agilent 1M CGH array were defined as novel rare CNVs (946 of 1884 rare CNVs detected on the CGH 1M array). We examined the size distribution of these 946 novel rare CNVs detected in the ASD cohort and found that approximately 75% of them are <30 kb in size (Figure S1). These smaller CNVs that went undetected by SNP microarrays were missed due to insufficient probes coverage at these loci and because of the lower signal-to-noise often observed for SNP array platforms (e.g., SNP-based arrays are optimized for robust genotyping call rates, which may minimize quantitative probe response to copy number variation). Only 25% of the novel, rare CNVs were >30 kb in size and were mostly missed by previous studies due to insufficient probe coverage, CNV-calling parameters and analysis algorithms chosen to define a CNV as stringent.
Rare ASD-specific CNVs
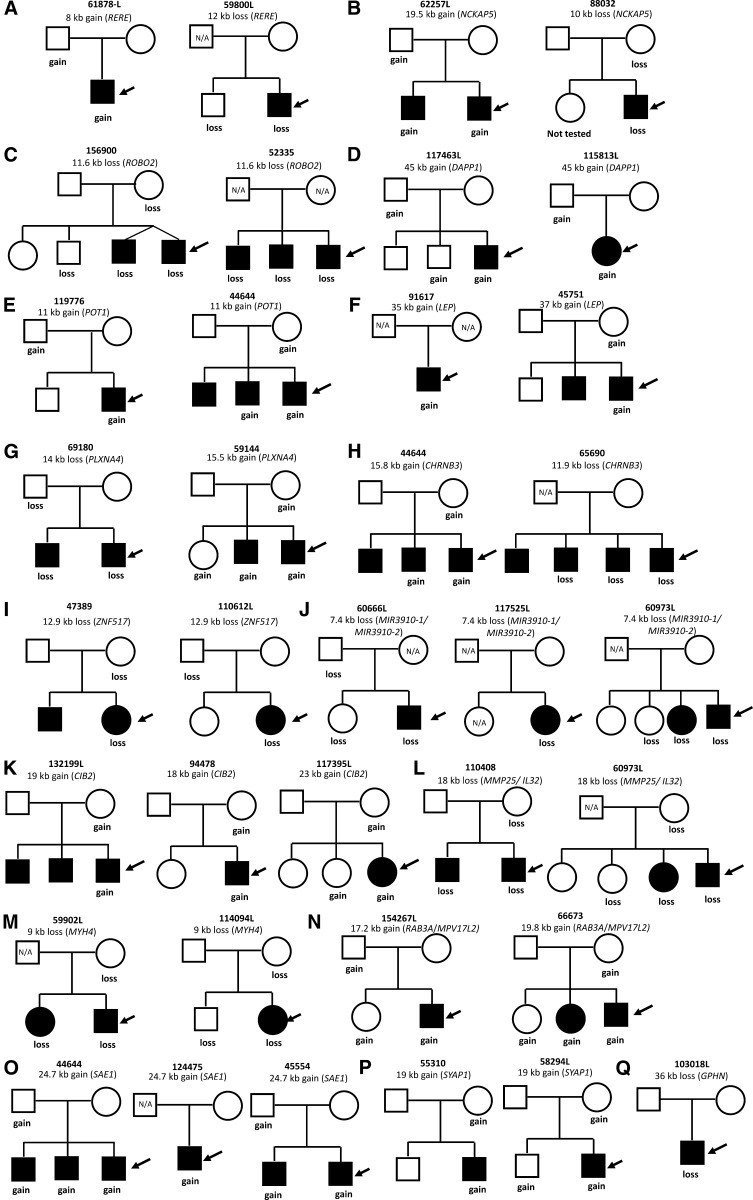
Rare ASD-specific CNVs were defined using a total of 5139 controls (1000 PDx controls summarized in Table 2 and 4139 in-house controls previously reported in Krawczak et al. 2006, Stewart et al. 2009, and Bierut et al. 2010). A total of 1884 rare CNVs were detected in ASD cases, of which 946 of them were novel as compared with previous SNP microarray studies (Table S2). Using qPCR, we validated 117 of 132 (88.6%) novel and rare stringent CNVs that were tested. Of the 946 novel rare ASD CNVs, 57 CNVs are reported in Table 4 that correspond to overlapping CNVs in two or more unrelated ASD cases (32 cases at 14 loci), recurrent CNVs (i.e., same breakpoints) in two or more unrelated ASD cases (24 cases at 11 loci), or are a de novo event [1 case (Table 4)]. Some of the overlapping/recurrent CNVs impacted previously identified ASD genes such as DPYD, RGS7, NRXN1, CNTNAP5, ERBB4, GRM8, NRXN3, YWHAE, and DMD (Serajee et al. 2003; Wu et al. 2005; Autism Genome Project Consortium 2007; Marshall et al. 2008; Bruno et al. 2010; Pagnamenta et al. 2010; Pinto et al. 2010; Carter et al. 2011; Vaags et al. 2012), whereas others were novel, including RERE, NCKAP5, ROBO2, DAPP1, POT1, LEP, PLXNA4, CHRNB3, ZNF517, MIR3910-1/MIR3910-2, CIB2, MMP25/IL32, MYH4, RAB3A/MPV17L2, SAE1, and SYAP1 (Figures 3 and 4).
Table 4. Novel recurrent/overlapping ASD CNVs (≥ 2 cases) and a de novo CNV.
| No. | Chromosome | Sample | Gender | Size, bpa | CNV | Origin | Genesb | Other Rare Variantsc |
|---|---|---|---|---|---|---|---|---|
| 1 | 1p36.23 | 61878-L | M | 8602 | Gain | Paternal | RERE intronic | 7q31.1 (231.3 kb) loss not in any gene |
| 1p36.23 | 59800L | M | 12,682 | Loss | Unknownd | RERE intronic | 2q31.3 (126.4 kb) loss not in any gene;11q14.3 (62.6 kb) loss not in any gene;13q12.13 (9.8 kb) Gain not in any gene;16p13.3 (34 kb) loss LOC342346 | |
| 2 | 1p21.3 | 82302 | M | 11,699 | Loss | Paternal | DPYD intronic | 13q33.2 (25.2 kb) loss not in any gene;18p11.21 (1010.9 kb) gain CXADRP3, ANKRD30B, MC2R, LOC284233, ZNF519, POTEC, MC5R |
| 1p21.3 | 82062L | M | 10,405 | Loss | Maternal | DPYD exonic | 7q31.1 (8.4 kb) Gain not in any gene | |
| 3 | 1q43 | 117370L | F | 13,664 | Loss | Unknownd | RGS7 intronic | 7q21.2 (505.2kb) loss LRRD1, MTERF, AKAP9, CYP51A1;21q21.2 (37.3 kb) gain not in any gene |
| 1q43 | 52401 | M | 13,664 | Loss | Paternal | RGS7 intronic | 15q25.2 (42.8 kb) gain LOC648809;Xp22.33 (25.3kb) loss DHRSX | |
| 4 | 2p16.3 | 87396 | F | 6925 | Loss | Maternal | NRXN1 intronic | 1q32.1 (18.9 kb) Gain not in any gene;2p16.3 (106.6kb) loss not in any gene;5q33.1 (349.1 kb) loss not in any gene |
| 2p16.3 | 78391 | F | 13,027 | Loss | Maternal | NRXN1 exonic | 1q23.1 (21.7 kb) loss OR6K2;5p15.33 (56.2 kb) gain not in any gene;5q34 (35.3 kb) loss not in any gene;11p12 (16.5 kb) loss not in any gene;12q21.32 (13.6 kb) loss MGAT4C;18q12.1 (58.4 kb) loss not in any gene | |
| 2p16.3 | 122686L | M | 8739 | Loss | Paternal | NRXN1 intronic | 1q44 (229 kb) gain ZNF238;3q29 (320.2 kb) loss APOD, ACAP2, PPP1R2;8q24.3 (189.8 kb) gain ZNF7, RPL8, ZNF251, ZNF250, COMMD5, ZNF34, ZNF517 | |
| 2p16.3 | L384 | M | 81,779 | Loss | Paternal | NRXN1 exonic | 1q25.3 (24 kb) loss MR1,STX6;12p12.1 (10.7 kb) loss not in any gene;13q22.1 (11.3 kb) loss KLF12;15q15.3 (14.2 kb) loss CASC4;17p13.1 (18.3 kb) loss KDM6B;19p13.12 (24.8 kb) Gain CYP4F22 | |
| 5 | 2q14.3 | 111520L | M | 17,780 | Loss | Maternal | CNTNAP5 intronic | 3q23 (56.4 kb) loss SPSB4;8p12 (37.6 kb) loss not in any gene |
| 2q14.3 | 129914 | M | 10,249 | Loss | Maternal | CNTNAP5 intronic | 1p36.22 (15 kb) Gain CORT,APITD1-CORT,APITD1;2p13.1 (8.5 kb) loss C2orf65;4q28.1 (51.7 kb) Gain not in any gene;Xq11.1 (18.7 kb) loss not in any gene | |
| 6 | 2q34 | 90188 | F | 6323 | Loss | Maternal | ERBB4 intronic | 2q33.1 (60.5 kb) gain not in any gene;17p13.2 (174.7 kb) loss TEKT1, ALOX12P2, XAF1 ,FBXO39 |
| 2q34 | 138145L | M | 11,613 | Loss | Not qPCR tested | ERBB4 intronic | 4q22.1 (97.3 kb) loss MMRN1; 13q31.3 (22.1 kb) loss GPC5 | |
| 7 | 2q21.2 | 62257L | M | 19,521 | Gain | Paternal | NCKAP5 exonic | 11p14.3 (36k b) gain LUZP2 |
| 2q21.2 | 88032 | M | 10,109 | Loss | Maternal | NCKAP5 intronic | 2q22.1 (23.6 kb) loss THSD7B;3p26.3 (57.6 kb) loss not in any gene;7q32.3 (112 kb) Gain CHCHD3;8p12 (14.1 kb) loss not in any gene;Xq21.1 (108.8 kb) gain not in any gene | |
| 8 | 3p12.3 | 156900 | M | 11,610 | Loss | Maternal | ROBO2 intronic | 15q15.1 (8.7 kb) loss EHD4 |
| 3p12.3 | 52335 | F | 11,610 | Loss | Unknownd | ROBO2 intronic | 8q21.13 (36.3 kb) loss not in any gene;14q23.3 (96 kb) loss not in any gene;17q25.3 (11.5 kb) loss not in any gene | |
| 9 | 4q23 | 115813L | F | 45,723 | Gain | Paternal | DAPP1 exonic | 17q12 (976.7 kb) gain CCL2, ACCN1, CCL13, CCL7, CCL11, CCL8, C17orf102, CCL1 ,TMEM132E |
| 4q23 | 117463L | M | 45,723 | Gain | Paternal | DAPP1 exonic | 1q42.13 (110.3 kb) gain RNF187, HIST3H3, HIST3H2BB, HIST3H2A;6p22.1 (234.7 kb) gain SCAND3, GPX5, GPX6 | |
| 10 | 7q31.33 | 119776 | M | 11,094 | Gain | Paternal | POT1 exonic | 7q34 (15.4 kb) loss not in any gene |
| 7q31.33 | 44644 | M | 11,094 | Gain | Maternal | POT1 exonic | 8p11.21 (15.9 kb) gain CHRNB3;12p13.33 (35.2 kb) loss CACNA2D4;16q22.3 (9.7 kb) loss not in any gene;17p13.2 (12.4kb) loss TM4SF5;19q13.32 (24.7 kb) gain SAE1 | |
| 11 | 7q31.33 | 128860 | M | 34,699 | Gain | Paternal | GRM8 exonic | 7p22.1 (14.8 kb) loss not in any gene;18q22.1 (12.5kb) gain CCDC102B |
| 7q31.33 | 146436L | M | 16,963 | Gain | Maternal | GRM8 intronic | 9q34.3 (38.7 kb) loss PNPLA7 | |
| 7q31.33 | 130293 | F | 21,856 | Loss | Maternal | GRM8 intronic | 13q21.1 (100 kb) loss not in any gene;Xq27.3 (33.2 kb) gain not in any gene | |
| 12 | 7q32.1 | 91617 | M | 35,669 | Gain | Unknownd | LEP exonic | 17q23.2 (9.8 kb) loss BCAS3 |
| 7q32.1 | 45751 | M | 37,923 | Gain | Maternal | LEP exonic | 16q24.2 (12 kb) gain JPH3 | |
| 13 | 7q32.3 | 69180 | M | 14,254 | Loss | Paternal | PLXNA4 exonic | 1p36.32 (40.7 kb) gain not in any gene;13q12.11,13q12.12 (383.8 kb) gain BASP1P1;15q22.2 (21.9 kb) loss not in any gene |
| 7q32.3 | 59144 | M | 15,521 | Gain | Maternal | PLXNA4 UTRs | 2q14.2 (8.5 kb) loss not in any gene;5q35.3 (22.2 kb) gain COL23A1 | |
| 14 | 8p11.21 | 44644 | M | 15,873 | Gain | Maternal | CHRNB3 intronic | 7q31.33 (11.1 kb) gain POT1;12p13.33 (35.2 kb) loss CACNA2D4;16q22.3 (9.7 kb) loss not in any gene;17p13.2 (12.4 kb) loss TM4SF5;19q13.32 (24.7kb) gain SAE1 |
| 8p11.21 | 65690 | M | 11,965 | Loss | Unknownd | CHRNB3 intronic | 4q32.1 (8.8 kb) loss not in any gene;7q11.23 (30.3 kb) Gain not in any gene;15q26.2 (13.8 kb) loss not in any gene;17q12 (25.7 kb) loss not in any gene | |
| 15 | 8q24.3 | 47389 | F | 12,937 | Loss | Maternal | ZNF517 exonic | 8q11.23 (48 kb) loss MRPL15;13q22.1 (68.3kb) loss PIBF1 |
| 8q24.3 | 110612L | F | 12,937 | Loss | Paternal | ZNF517 exonic | 12q13.11 (12.2 kb) loss COL2A1 | |
| 16 | 9q22.31 | 60666L | M | 7480 | Loss | Paternal | MIR3910-1,MIR3910-2 | 6q16.1 (87.9 kb) loss KLHL32;10q23.31 (10.6 kb) loss STAMBPL1;12q21.1 (20.5 kb) loss not in any gene |
| 9q22.31 | 117525L | F | 7480 | Loss | Unknownc | MIR3910-1,MIR3910-2 | 3p26.1 (24.8 kb) loss not in any gene | |
| 9q22.31 | 60973L | M | 7480 | Loss | Maternal | MIR3910-1,MIR3910-2 | 7p21.3 (42.5 kb) gain THSD7A;10p12.33 (23.4 kb) loss not in any gene;16p13.3 (18 kb) loss MMP25,IL32;20p13 (4.3 kb) loss PANK2 | |
| 17 | 14q24.3 | 102350 | M | 14,061 | Loss | Maternal | NRXN3 intronic | 1q42.13 (10.2 kb) loss not in any gene;5q14.3 (96.8 kb) gain not in any gene;6p21.2 (194.5 kb) gain DNAH8,GLP1R |
| 14q31.1 | 95458L | M | 289,711 | Loss | Paternal | NRXN3 exonic | 10q25.3 (135 kb) gain PNLIPRP3 | |
| 18 | 15q25.1 | 117395L | F | 23,541 | Gain | Maternal | CIB2 exonic | 12q21.2 (31.2 kb) loss not in any gene |
| 15q25.1 | 94478 | M | 17,639 | Gain | Paternal | CIB2 exonic | 19p12 (573.7 kb) loss ZNF826P, ZNF737, ZNF682, ZNF486, ZNF90, MIR1270-2, MIR1270-1 | |
| 15q25.1 | 132199L | M | 18,728 | Gain | Maternal | CIB2 exonic | 2p14 (14.9 kb) loss not in any gene;6q12 (411.3 kb) loss EYS;7q35 (12.4 kb) Gain not in any gene;8q23.2 (217.3 kb) loss not in any gene | |
| 19 | 16p13.3 | 110408 | M | 18,045 | Loss | Maternal | MMP25,IL32 | 3q13.2 (21.4 kb) loss not in any gene;4p13 (12.6 kb) loss GRXCR1 |
| 16p13.3 | 60973L | M | 18,045 | Loss | Unknownd | MMP25,IL32 | 7p21.3 (42.5 kb) Gain THSD7A;9q22.31 (7.5 kb) loss MIR3910-1,MIR3910-2;10p12.33 (23.4 kb) loss not in any gene;20p13 (4.3 kb) loss PANK2 | |
| 20 | 17p13.3 | 68672 | M | 24,812 | Gain | Maternal | YWHAE exonic | 4q28.3 (79.3 kb) Gain LOC641365;6p21.2 (24.3 kb) Gain ZFAND3;21q21.1 (11.7 kb) loss C21orf34 |
| 17p13.3 | 50800L | M | 24,812 | Gain | Maternal | YWHAE exonic | 7q31.1-q31.31 (11 Mb) loss encompasses 32 genes including FOXP2 | |
| 21 | 17p13.1 | 59902L | M | 9003 | Loss | Maternal | MYH4 exonic | 2p21 (25.6 kb) loss EML4 |
| 17p13.1 | 114094L | F | 9003 | Loss | Maternal | MYH4 exonic | − | |
| 22 | 19p13.11 | 154267L | M | 17,268 | Gain | Paternal | RAB3A,MPV17L2 | 1p36.23 (11 kb) loss GPR157;4q21.1 (11.4 kb) gain not in any gene;7q31.33 (481.3 kb) loss not in any gene;8q21.3 (70 kb) loss LRRC69;13q34 (118.3 kb) gain F10,MCF2L,F7;14q31.3,14q31.2 (299.5 kb) loss not in any gene;Xp22.33 (262.1 kb) gain DHRSX |
| 19p13.11 | 66673 | F | 19,829 | Gain | Maternal | RAB3A,MPV17L2 | 1p36.22 (9.5 kb) loss PLOD1;5q11.2 (32.2 kb) gain not in any gene;5q21.3 (8.8 kb) loss FBXL17;7q11.22 (27 kb) loss not in any gene;7q31.1 (19.2 kb) loss not in any gene;8p22 (24.1 kb) loss not in any gene;14q31.3 (30.2 kb) gain not in any gene;16q22.3 (33.2 kb) gain not in any gene | |
| 23 | 19q13.32 | 44644 | M | 24,729 | Gain | Paternal | SAE1 exonic | 7q31.33 (11.1 kb) gain POT1;8p11.21 (15.9 kb) gain CHRNB3;12p13.33 (35.2 kb) loss CACNA2D4;16q22.3 (9.7kb) loss not in any gene;17p13.2 (12.4 kb) loss TM4SF5 |
| 19q13.32 | 124475 | M | 24,729 | Gain | Unknownd | SAE1 exonic | 17p13.1 (428.4 kb) gain MYH1,MYH8,MYH13,GAS7,MYH4 | |
| 19q13.32 | 45554 | M | 24,729 | Gain | Paternal | SAE1 exonic | 1q43,1q42.3 (96.9 kb) gain EDARADD;8q21.3 (82.1 kb) loss not in any gene;12q21.1 (20.5 kb) loss not in any gene;Xq27.3 (82.2 kb) gain not in any gene | |
| 24 | Xp22.2 | 55310 | M | 19,171 | Gain | Maternal | SYAP1 exonic | 3q13.33 (29.1 kb) gain ARHGAP31;7q21.12 (12.3 kb) loss not in any gene;10q23.1 (23.3 kb) loss NRG3 |
| Xp22.2 | 58294L | M | 19,171 | Gain | Maternal | SYAP1 exonic | 7p21.3 (57.3 kb) loss not in any gene;8p22 (89.9 kb) gain MTUS1;11q25 (41.3 kb) gain not in any gene;Xp11.3 (6.3 kb) loss not in any gene | |
| 25 | Xp21.1 | 100570L | M | 6261 | Loss | Maternal | DMD intronic | − |
| Xp21.1 | 91548L | M | 6917 | Loss | Maternal | DMD intronic | 2q31.2 (15.8 kb) loss PDE11A;Xp22.11 (124.1 kb) loss DDX53 | |
| 26 | 14q23.3 | 103018L | M | 36,180 | Loss | de novo | GPHN intronic | − |
ASD, autism spectrum disorder; CNV, copy number variations; F, female; M, male; qPCR, quantitative polymerase chain reaction; UTR, untranslated region.
The CNVs were validated by qPCR. However, accurate breakpoints have not been identified. The size of the CNVs shown is as detected by microarrays.
A CNV encompassing coding regions of the gene are defined as exonic, whereas those encompassing introns were defined as intronic. One of the CNVs intersected untranslated region of the gene (UTRs).
Other rare variants present in the individual.
Inheritance is unknown if the parents DNA were unavailable.
Figure 3 .
Pedigrees (A-Q) represent ASD families with overlapping/recurrent CNVs in novel loci and a de novo CNV event (from Table 4). The open symbols represent unaffected individuals, filled symbols represent individuals with ASD diagnosis and arrows indicate the probands. Individuals from which DNA was not available (N/A) for testing are denoted inside the symbols.
Figure 4 .
Genome browser views of a subset of novel rare CNVs occurring in two or more ASD cases or are de novo (from Table 4). The genome coordinates are from genome build 36 (hg18) and, in each panel, ASD case ID numbers are listed next to blue bars (denoting a duplication/gain) or red bar (denoting a deletion/loss). Each panel also shows, in separate tracks, the RefSeq genes, UCSC segmental duplications, and probe distributions for the different microarray platforms used for CNV detection (Affy500K, Affy6.0, Illumina 1M, and Agilent 1M). (A) Maternally inherited duplications at the 15q25.1 locus encompassing an exon of the CIB2 gene in three unrelated ASD cases. (B) Paternally inherited duplications at the 4q23 locus encompassing several exons of the DAPP1 gene in two unrelated ASD cases. (C) Maternally inherited duplications at the 17p13.3 locus encompassing exons of the YWHAE gene in two unrelated ASD cases. (D) Duplications at the 19q13.32 locus encompassing exons of SAE1 gene in four unrelated ASD cases, three of which are novel and the fourth (ASD case 90278) was previously detected using an Affymetrix 6.0 array. (E) A paternally inherited deletion and a maternally inherited duplication at the 7q32.3 locus encompassing exons of the PLXNA4 gene (F). A de novo deletion at the 14q23.3 locus encompassing an intronic region of the GPHN gene in one ASD case.
One of the ASD-specific CNVs was a maternally inherited duplication at 15q25.1 in three unrelated ASD cases (117395L, 94478, 132199L; Figure 4A) disrupting the exon of CIB2 (Calcium and integrin binding family member 2; 3/696 cases vs. 0/5139 controls; FET two-tailed p = 0.001691). The transcript and protein of CIB2 gene is found to be present mainly in the hippocampus and cortex of the brain (Blazejczyk et al. 2009). The encoded protein of this gene is shown to be involved in Ca2+ signaling, which controls a variety of processes in many cell types. In neurons, Ca2+ signaling maintains synaptic transmission, neuronal development and plasticity (Blazejczyk et al. 2009).
In another two unrelated ASD cases (115813L, 117463L), we identified a recurrent CNV of size 45.7 kb (2/696 cases vs. 0/5,139 controls; FET two-tailed p = 0.01421). The duplication disrupts four exons of the DAPP1 (dual adaptor of phosphotyrosine and 3-phosphoinositides) gene at the 4q23 (Figure 4B) and the gene is suggested to be involved in signal transduction processes.
Another interesting recurrent CNV of size 24.8 kb was detected in two unrelated ASD cases (68672, 50800L), a duplication at 17p13.3 disrupting two exons of YWHAE (tyrosine 3/tryptophan 5-monooxygenase gene) gene, which presumably act via haploinsufficiency (Figure 4C). It was maternally inherited in both ASD cases, and an equivalent event based on our definition of overlap was not present in controls (2/696 cases vs. 0/5139 controls; FET two-tailed p = 0.01421). YWHAE belongs to the 14-3-3 family of proteins, which mediate signal transduction, and is highly conserved in both plants and mammals. Only microduplications in YWHAE gene have been reported in ASD. It has been shown that the phenotype of patients with a 17p13.3 microduplication involving YWHAE gene show autistic manifestation, behavioral symptoms, speech and motor delay, subtle dysmorphic facial features, and subtle hand-foot malformations (Bruno et al. 2010). It is also noteworthy that there were larger CNVs found in two PDx controls to intersect YWHAE. The two ASD cases were both of Asian descent, and we also have found other cases and controls of Asian descent bearing YWHAE-CNVs (M. Gazzellone, unpublished data), suggesting that the role of this gene in ASD need to be further explored.
In three unrelated male ASD probands (44644, 124475, 45554), we observed a recurrent novel CNV, a 24.7-kb duplication encompassing two exons of the SAE1 (SUMO1 activating enzyme subunit 1) gene at the 19q13.32 locus (Figure 4D). The same CNV also was found in one control (3/696 cases vs. 1/5139 controls; FET two-tailed p = 0.00616). Interestingly, another duplication of size 50.8 kb disrupting six exons of SAE1 was observed in a fourth unrelated ASD case (90278) in the present study and was also detected by previous SNP microarray study (Lionel et al. 2011). The SAE1 gene is involved in protein sumoylation process and is shown to interact with the ARX gene, which is involved in Autistic disorder (Sherr 2003; Rual et al. 2005; Ewing et al. 2007; Gareau and Lima 2010; Wilkinson et al. 2010; Szklarczyk et al. 2011).
In two unrelated cases (69180, 59144), we identified overlapping CNVs impacting the PLXNA4 (plexin A4) gene at the 7q32.3 locus (Figure 4E). One CNV is a 14.2-kb loss encompassing an exon of the gene and based on our overlap criteria is not observed in 5,139 controls, while the second CNV is a 15.5-kb gain encompassing untranslated regions of the gene and is observed in 2 of 5139 controls. PLXNA4 is involved in axon guidance as well as nervous system development (Suto et al. 2003; Miyashita et al. 2004).
In the present study, only one de novo CNV was found (all other qPCR-validated CNVs, 116 of 117, were inherited from either parent) a 36-kb loss encompassing the intron of the GPHN (Gephyrin) gene at the 14q23.3 locus. This de novo CNV (Figure 4F) was found in a male ASD proband (103018L) and was not picked up on the previous SNP array (Pinto et al. 2010), and it was not found in any of the 5139 controls. Gephyrin is suggested to play a central organizer role in assembling and stabilizing inhibitory postsynaptic membranes in human brain (Waldvogel et al. 2003). In our other unpublished data, we have also identified another deletion encompassing several exons of GPHN in an unrelated ASD case and a de novo deletion encompassing exons of the gene in a schizophrenia case suggesting that GPHN gene could be a novel susceptibility gene playing a more general role in neurodevelopmental disorders. We believe the lack of novel, rare de novo CNVs captured in the present study is simply due to our study design because nearly all the de novo CNVs in this ASD cohort are relatively larger in size and therefore were already detected using SNP microarrays (Marshall et al. 2008; Pinto et al. 2010, Lionel et al. 2011, A. C. Lionel, unpublished data, i.e., not reported in the present study).
We also detected other novel, rare CNVs present in only one unrelated ASD case (Table 5) in previously identified genes associated with ASD such as CTNND2, CDH18, PARK2, NXPH1, MTHFD1, and NF1 [Table 5 (Williams and Hersh 1998; Marui et al. 2004; Glessner et al. 2009; Pinto et al. 2010; Griswold et al. 2011; Salyakina et al. 2011)]. Novel, rare CNVs occurring in genes known to be associated with other neurodevelopmental disorders (e.g., KIRREL3) or potentially playing a role in neurodevelopment also were found (e.g., CTNNA2, NDST1, SLC24A2, NFIB, APLP2, ATP2C2, CECR2, DAGLA, and UPB1).
Table 5. Other singleton novel rare CNVs.
| Chromosome | Sample | Gender | Size, bpa | CNV | Origin | Genesb | Other Rare Variantsc |
|---|---|---|---|---|---|---|---|
| 1p36.11 | 88253 | M | 16,863 | Loss | Maternal | WASF2 intronic | 9p21.3 (13.1 kb) loss KIAA1797;9q22.31 (10.7 kb) loss not in any gene |
| 1p34.1 | 94073 | F | 9205 | Loss | Paternal | PRDX1 exonic | 2q35 (6.8 kb) loss 40972;8p23.1 (74.5 kb) loss PINX1,MIR1322 |
| 2p21 | 115818L | M | 9068 | Loss | Paternal | PPM1B exonic | 16q22.3 (90.8 kb) gain not in any gene |
| 2p16.3 | 75420 | F | 10,491 | Gain | Maternal | STON1-GTF2A1L,STON1 | 2p16.3 (10.5 kb) gain STON1-GTF2A1L,STON1;2q14.1 (118.8 kb) gain DPP10;17q12 (84.3 kb) gain GPR179, MRPL45, LOC440434 |
| 2p12 | 100678L | F | 52,069 | Gain | Paternal | CTNNA2 exonic | 1p31.1 (53.5 kb) loss not in any gene;1q42.2 (5 kb) loss FAM89A;2p23.1 (7.4 kb) loss GALNT14;4p15.31 (19 kb) loss SLIT2;9p21.3 (65 kb) loss not in any gene;16q24.1 (12.4 kb) loss ATP2C2 |
| 2q11.2 | 47173L | F | 780,104 | Gain | Unknownd | 11 genes | 19q13.43 (843.3 kb) loss TRAPPC2P1, ZNF835,USP29,ZNF17, ZNF71, ZNF749, ZNF264, LOC147670, VN1R1, AURKC, PEG3-AS1, ZIM2, ZIM3, ZNF304, ZNF805, ZNF547, ZNF543, MIMT1, ZNF460, DUXA, ZNF548, PEG3;20q13.12 (15.1 kb) gain CD40 |
| 3p22.2 | 58016 | M | 38,181 | Gain | Paternal | LRRFIP2 exonic | 6p22.1 (9.5 kb) loss HIST1H2AG,HIST1H2BJ;19q13.31 (78.2 kb) gain TEX101;21q21.1 (70.9 kb) loss C21orf34 |
| 3p14.2 | L656 | M | 332,421 | Loss | Maternal | FHIT exonic | 1q32.1 (37 kb) loss MDM4;2p14 (26 kb) loss FBXO48,APLF;6q12 (16.3 kb) loss EYS;8q22.2 (38.3 kb) loss VPS13B;17q25.3 (46.2 kb) loss CYTH1;17q25.3 (32.7 kb) loss LOC100294362, ENDOV, RNF213;Xp11.3 (32.8 kb) loss CXorf36 |
| 3q13.31 | 52026 | M | 10,004 | Loss | Maternal | ZBTB20 intronic | 7p11.2 (81.7 kb) gain LOC650226, DKFZp434L192;7q36.2 (70.7 kb) loss DPP6;13q31.2,13q31.3 (74.9 kb) gain not in any gene |
| 3q13.33 | 55310 | M | 29,147 | Gain | Paternal | ARHGAP31 exonic | 7q21.12 (12.3 kb) loss not in any gene;10q23.1 (23.3 kb) loss NRG3;Xp22.2 (19.2 kb) gain SYAP1 |
| 3q25.1 | 50002 | F | 7969 | Gain | Unknownd | WWTR1 intronic | 1p36.22 (6.4 kb) loss UBE4B; 5p15.1,5p15.2 (300.2 kb) gain not in any gene;7p21.1 (20.7 kb) loss not in any gene;9p24.1 (10.2 kb) loss KDM4C;14q23.2 (8.4 kb) gain MTHFD1 |
| 3q26.2 | 46685 | M | 10,633 | Loss | Paternal | PRKCI exonic | 4q13.1 (32.6 kb) gain not in any gene |
| 3q29 | 76066 | M | 27,516 | Loss | Unknownd | KIAA0226 exonic | 17q25.3 (13.4 kb) gain NARF;22q11.21 (2762.8 kb) gain encompassing 65 genes |
| 4p15.31 | 100678L | F | 19,015 | Loss | Maternal | SLIT2 intronic | 1p31.1 (53.5 kb) loss not in any gene;1q42.2 (5 kb) loss FAM89A;2p23.1 (7.4 kb) loss GALNT14;2p12 (52.1 kb) gain CTNNA2;9p21.3 (65 kb) loss not in any gene;16q24.1 (12.4 kb) loss ATP2C2 |
| 4q21.1 | 62345L | M | 8527 | Loss | Paternal | SHROOM3 intronic | 5q21.1 (20.8 kb) loss not in any gene;14q21.2 (59.5 kb) loss not in any gene |
| 4q32.1 | 55360 | M | 16,194 | Gain | Maternal | FGA exonic | 9p21.3 (18 kb) gain IFNA22P;12p12.1 (18.9 kb) loss not in any gene;15q13.3 (36.3 kb) loss MIR211,TRPM1 |
| 5p15.2 | 51119 | M | 12,982 | Loss | Unknownd | CTNND2 intronic | — |
| 5p14.3 | 167532 | F | 16,492 | Gain | Maternal | CDH18 intronic | 11p15.4 (98.4 kb) loss LOC283299, OR5E1P, OR5P3, OR5P2;12q24.12 (12.7 kb) loss ATXN2 |
| 5q14.3 | 55262-L | M | 665,114 | Gain | Maternal | LYSMD3,POLR3G,CETN3,MBLAC2, GPR98 | 2q37.1 (119.8 kb) gain NMUR1,C2orf57;4q21.21 (22.7 kb) gain ANTXR2;14q22.2 (26.4 kb) gain CDKN3;22q13.31 (54.2 kb) loss not in any gene |
| 5q22.2 | 59269L | F | 25,834 | Loss | Maternal | MCC exonic | 22q12.1 (225.1 kb) loss TTC28;Xq11.1 (308.9 kb) gain MTMR8 |
| 5q31.3 | 66559 | M | 19,794 | Gain | Unknownd | TAF7,SLC25A2 | 6q27 (307.6 kb) gain C6orf118, PDE10A;11q12.1 (24.3 kb) loss OR5B2, OR5B12;16q21 (81.9 kb) gain not in any gene |
| 5q33.1 | 60560L | M | 54,785 | Gain | Paternal | NDST1 exonic | 20p13 (22.4 kb) gain SNPH;22q12.3 (77.4 kb) gain SLC5A4 |
| 6p22.1 | 58016 | M | 9549 | Loss | Paternal | HIST1H2AG,HIST1H2BJ | 3p22.2 (38.2 kb) gain LRRFIP2;19q13.31 (78.2 kb) gain TEX101;21q21.1 (70.9 kb) loss C21orf34 |
| 6q26 | 75744 | M | 17,712 | Loss | Paternal | PARK2 intronic | 21q21.3 (8.1 kb) loss NCRNA00189 |
| 7p21.3 | 68687 | M | 258,183 | Loss | Maternal | NXPH1 exonic | 1q32.1 (145.7 kb) gain CAMSAP1L1, C1orf106, GPR25;13q33.2 (63.9 kb) loss not in any gene;13q33.2 (69.6 kb) gain not in any gene |
| 8p21.2 | 88810 | M | 28,281 | Gain | Paternal | ADAM7 exonic | 6p22.1 (17.3 kb) gain not in any gene;19q13.43 (24.1 kb) gain SLC27A5,ZBTB45 |
| 9p22.3 | 131240 | M | 8341 | Loss | Paternal | NFIB intronic | 5p13.2 (51.9 kb) gain PRLR,AGXT2;6q25.1 (20.8 kb) loss RAET1L;8p12 (66.6 kb) loss NRG1;16p13.3 (16.5 kb) loss HBM, HBA1, HBA2, HBQ1 |
| 9p22.1 | 61180-L | M | 7974 | Loss | Paternal | SLC24A2 exonic | 3p21.31 (20.4 kb) loss MIR425, DALRD3, MIR191, QRICH1, IMPDH2, NDUFAF3;6q22.31 (296.4 kb) gain C6orf204, PLN, BRD7P3 |
| 9p13.3 | 59641L | F | 190,658 | Gain | Maternal | PTENP1,PRSS3,UBE2R2 | 6q24.1 (13.2 kb) loss TXLNB;17q24.1 (5.9 kb) loss CEP112;Xp21.1 (15.4 kb) gain DMD |
| 10q25.1 | 115816L | M | 9442 | Loss | Maternal | SORCS3 intronic | 11q23.3 (9.2 kb) loss CBL;15q21.3 (58.8 kb) loss not in any gene |
| 11q12.2 | 72816L | M | 15,159 | Loss | Paternal | DAGLA exonic | 7q31.31 (19.4 kb) gain not in any gene;8q22.1 (19.8 kb) loss not in any gene;12q12 (25 kb) gain KIF21A;14q24.2 (8.5 kb) gain RGS6;22q12.1 (25.6 kb) loss MIR1302-1,MYO18B |
| 11q23.3 | 96280L | M | 19,424 | Gain | Paternal | VPS11 exonic | 3p12.3 (123 kb) loss not in any gene;13q33.1 (74.2 kb) loss FGF14;15q11.2 (25.7 kb) loss not in any gene;Xp11.4 (39 kb) gain GPR82,GPR34,CASK |
| 11q24.2 | 138952L | F | 11,398 | Loss | Maternal | KIRREL3 intronic | 4p13 (71.3 kb) gain not in any gene;11q14.1 (28.8 kb) gain not in any gene;14q31.2 (27.9 kb) loss not in any gene |
| 11q24.3 | 98320L | M | 8428 | Loss | Maternal | APLP2 exonic | 1q31.1 (321.4 kb) loss not in any gene;4q13.1 (1326.4 kb) gain not in any gene;5q23.1 (42 kb) loss not in any gene;7q31.1 (98.6 kb) loss IMMP2L |
| 12q15 | 59794L | M | 9279 | Loss | Maternal | RAP1B intronic | 5q35.2 (8 kb) loss ATP6V0E1;14q32.2 (8.9 kb) loss WDR25;20p11.22 (148.7 kb) gain not in any gene |
| 12q24.33 | 50280 | M | 202,302 | Gain | Maternal | LOC100130238, GALNT9 | 3q21.3 (6.1 kb) loss CCDC48;12q24.33 (77.8 kb) gain SFSWAP |
| 14q23.2 | 50002 | F | 8427 | Gain | Unknownd | MTHFD1 exonic | 1p36.22 (6.4 kb) loss UBE4B;3q25.1 (8 kb) gain WWTR1;5p15.1,5p15.2 (300.2 kb) gain not in any gene;7p21.1 (20.7 kb) loss not in any gene;9p24.1 (10.2 kb) loss KDM4C |
| 15q24.2 | 62261L | M | 12,936 | Loss | Paternal | PTPN9 intronic | 5q14.1 (15.3 kb) gain BHMT2;8q24.21 (45.3 kb) gain GSDMC |
| 15q25.1 | 68388 | F | 7142 | Loss | Paternal | IL16 exonic | 1p32.2 (567.4 kb) gain DAB1;10q22.3 (12.9 kb) gain C10orf11;22q12.1 (8.4 kb) loss not in any gene |
| 16p13.3 | 47378 | M | 13,736 | Loss | Maternal | TRAP1 intronic | 2p23.1 (42.7 kb) loss CAPN14,EHD3;3p25.1 (16.1 kb) loss CAND2;21q21.2,21q21.1 (75 kb) loss not in any gene |
| 16p11.2 | 100564 | F | 546,709 | Gain | Maternal | 27 genes | 8q12.3 (17.9 kb) loss ASPH;10q21.3 (47.3 kb) loss not in any gene;17p11.2 (186.2 kb) loss LOC162632, CCDC144A, FAM106CP, KRT16P2;Xq21.1 (130.3 kb) gain not in any gene |
| 16q24.1 | 100678L | F | 12,352 | Loss | Maternal | ATP2C2 exonic | 1p31.1 (53.5 kb) loss not in any gene;1q42.2 (5 kb) loss FAM89A;2p23.1 (7.4 kb) loss GALNT14;2p12 (52.1 kb) gain CTNNA2;4p15.31 (19 kb) loss SLIT2;9p21.3 (65 kb) loss not in any gene |
| 16q24.2 | 68711 | M | 17,969 | Loss | Maternal | KLHDC4 exonic | 2p23.1 (61.4 kb) loss LCLAT1;4p15.1 (48 kb) loss not in any gene;6q21 (14.6 kb) gain not in any gene;7p21.3 (87.4 kb) loss VWDE;12p13.2 (8 kb) gain KLRK1, KLRC4-KLRK1 |
| 17p13.1 | 47387 | M | 7758 | Gain | Maternal | NDEL1, MYH10 | 2p23.1 (7.4 kb) loss GALNT14;2q22.1 (299.4 kb) loss THSD7B;5q31.3 (29.9 kb) loss PCDHB9, PCDHB8, PCDHB7, PCDHB16 |
| 17q11.2 | 87042 | F | 14,177 | Loss | Maternal | NF1 exonic | − |
| 17q25.3 | 103818L | M | 48,984 | Gain | Maternal | TIMP2 exonic | 8q24.13 (54.1 kb) loss not in any gene |
| 19q13.32 | 85287L | M | 17,455 | Loss | Maternal | BBC3,MIR3190, MIR3191 | 16q22.1 (36.5 kb) gain DDX19A, DDX19B;16q23.1 (16.2 kb) loss WWOX |
| 19q13.33 | 168753 | M | 61,936 | Loss | Maternal | SHANK1,CLEC11A | 4p15.33 (5.4 kb) loss not in any gene |
| 20p13 | 92540L | M | 15,645 | Loss | Paternal | SLC23A2 exonic | 2q34 (23 kb) loss SPAG16 |
| 22q11.21 | 118909L | F | 7277 | Loss | Unknownd | CECR2 exonic | 7q22.2 (12.4 kb) gain not in any gene;10q21.3 (8.1 kb) loss CTNNA3 |
| 22q11.22-q11.23 | MM0177-3 | M | 656,280 | Gain | Maternal | RAB36,FBXW4P1,RTDR1,GNAZ,MIR650,IGLL5,BCR | − |
| 22q11.23 | 154266L | M | 6784 | Loss | Maternal | UPB1 | 11p14.3 (138.6 kb) loss not in any gene |
| Xq28 | 100676L | M | 123,871 | Gain | Maternal | ZNF185,CETN2, NSDHL | 3q22.3 (11.5 kb) loss not in any gene;5p14.2 (12.2 kb) loss not in any gene;22q12.3 (35.6 kb) gain not in any gene |
ASD, autism spectrum disorder; CNV, copy number variations; F, female; M, male; qPCR, quantitative polymerase chain reaction; UTR, untranslated region.
These CNVs were validated by qPCR. However accurate breakpoints have not been identified. The size of the CNVs shown is as detected by microarrays.
A CNV encompassing coding regions of the gene are defined as exonic, whereas those encompassing introns were defined as intronic.
Other rare variants present in the individual.
Inheritance is unknown if the parents DNA were unavailable.
A paternally inherited 7.9-kb deletion disrupting an exon of the SLC24A2 [solute carrier family 24 (sodium/potassium/calcium exchanger), member 2] gene was observed at 9p22.1 in a male ASD case (61180-L; Figure 5A) but in none of the controls. The SLC24A2 gene may play a role in neuronal plasticity (Li et al. 2006).
Figure 5 .
Genome browser view of a subset of the novel rare CNVs found in one ASD case that impact one or more exons (Table 5). The genome coordinates are from genome build 36 (hg18) and, in each panel, the ASD case ID number is listed next to a red bar (denoting a deletion/loss). Each panel also shows, in separate tracks, the RefSeq genes and probe distributions for the different microarray platforms used for CNV detection (Affy500K, Affy6.0, Illumina 1M and Agilent 1M). (A) Paternally inherited deletion at the 9p22.1 locus impacting the 5′-end of the SLC24A2 gene. (B) Deletion at the 22q11.21 locus resulting in loss of an exon of the CECR2 gene. (C) Paternally inherited deletion at the 11q12.2 impacting the 3′-end of the DAGLA gene. (D) Maternally inherited deletion at the 22q11.23 impacting exons near the 5′-end of the UPB1 gene. (E) Pedigrees of ASD families for the CNVs in novel loci occurring in one ASD case that correspond to panels A-D. The open symbols represent unaffected individuals, filled symbols represent individuals with ASD diagnosis and arrows indicate the probands. Individuals from which DNA was not available (N/A) for testing are denoted inside the symbols.
In a female ASD case (118909L), we observed a 7.2-kb loss disrupting an exon of the CECR2 (cat eye syndrome chromosome region, candidate 2) gene at the 22q11.21 locus (Figure 5B), which was not observed in controls. We also detected a CNV in the same gene in ASD case 124632L reported in another study (Pinto et al. 2010). CECR2 is a chromatin remodeling factor that has been proposed to play a role in embryonic nervous system development (Banting et al. 2005).
In another unrelated male ASD proband (72816L), we have identified a 15-kb paternally inherited deletion (Figure 5C) disrupting seven exons of the DAGLA (diacylglycerol lipase, alpha) gene at the 11q12.2 locus that is not found in controls. DAGLA is known to synthesize an endocannabinoid that has been associated with retrograde synaptic signaling and plasticity (Gao et al. 2010).
Another interesting gene is UPB1 (ureidopropionase, beta) located at the 22q11.23 locus, in which a 6.7-kb deletion disrupting two exons has been found in a male ASD proband (154266L; Figure 5D). The deletion was not observed in any of 5139 controls. This gene is involved in the last step of the pyrimidine degradation pathway and deficiencies in UPB1 have been associated with developmental delay (van Kuilenburg et al. 2004).
Finally, our study includes analysis of two ASD cases not previously run on SNP microarrays. In both cases, large-sized CNVs previously associated with ASD were found, a 546-kb maternally inherited 16p11.2 duplication (ASD case 100564) and a 656-kb maternally inherited 22q11.22-q11.23 duplication (ASD case MM0177-3) that overlaps with the 22q distal deletion region (Figure S2).
Selection of ASD cases of European ancestry
To perform more robust downstream analyses (global rare CNV burden and gene-set association) on the ASD CNV findings in the present study, we used the SNP genotype data available on the 615 ASD cases to identify those of European ancestry based on MDS. We visualized the HapMap and ASD samples in a two-dimensional space using the first and second MDS components. The plot displayed three major clusters (Europeans, Eastern Asians, and Africans), with Mexicans and Indians distributed between the European and Eastern Asian cluster (Figure S3A). According to MDS components 1 and 2, ASD samples mostly clustered with Europeans, although some of them clearly clustered with other ethnicities. We defined a set of boundaries on the first and second MDS component, which identified all European samples, and only two Mexican samples (Figure S3B). Any ASD sample falling within these boundaries was considered to be European (505 of 615 total genotyped unrelated samples). We did not use the third MDS component, which is better at resolving ethnic subgroups but does not account faithfully for differences among major groups (e.g., Europeans and Eastern Asians were very close to each other; Figure S3C). The rare variants from 505 European ASD cases along with 1000 European PDx controls were used further for rare CNV burden and gene-set association analyses to avoid any confounding issues due to different ancestries of samples.
Global rare CNV burden analyses
To investigate global differences in CNV burden, we assessed the distribution of three CNV statistics in ancestry-matched European ASD case subjects compared with control subjects: (1) subject CNV number, (2) subject total CNV length, and (3) subject total number of CNV genes.
We observed significant differences in CNV number and total gene number for deletions but not for duplications (significance threshold: Wilcoxon test p value < 0.01; Table 6). In terms of magnitude, the total number of deletion genes was the largest difference found between the ASD and control subjects (ratio of means: 1.77). Considering that subjects were matched by platform type and other essential parameters, and also considering that previous authors found a stronger difference in deletion burden rather than duplication, the differences observed very likely translate to real biological differences. In addition, the relative difference in total gene number was larger than the relative difference in CNV number, an effect that is harder to explain by technical or experimental factors; because of the relatively large sample size, it is important to consider both the significance and magnitude of burden differences.
Table 6. Global burden of rare CNVs in European ASD cases vs. PDx controls.
| Type | Measure | Mean ASD | Mean Controls | Mean ratio ASD/controls | P-value |
|---|---|---|---|---|---|
| All | No. CNV | 2.680942 | 2.565848 | 1.05 | 0.0575976 |
| All | Total CNV size, kb | 124.50 | 112.92 | 1.10 | 0.1804119 |
| All | Total no. genes | 3.850107 | 2.560268 | 1.50 | 0.2345081 |
| Deletions only | No. CNV | 1.958225 | 1.773793 | 1.10 | 0.0046012a |
| Deletions only | Total CNV size, kb | 54.29 | 48.81 | 1.11 | 0.1043462 |
| Deletions only | Total number of genes | 1.741514 | 0.9834483 | 1.77 | 0.0087554a |
| Duplications only | No. CNV | 1.549383 | 1.601266 | 0.97 | 0.7716051 |
| Duplications only | Total CNV size, kb | 101.66 | 96.02 | 1.11 | 0.3778319 |
| Duplications only | Total no. genes | 3.490741 | 2.495253 | 1.40 | 0.8564186 |
Significant differences (P ≤ 0.01) are indicated.
We also assessed whether rare CNVs in genes that are causally implicated in ASD were enriched in cases over controls. The ASD gene list used comprised 110 genes compiled from the peer-reviewed literature (Betancur 2011). We observed significant enrichment of deletions impacting genes implicated in ASD [p = 8.896e-05, odds ratio = 20.59 with 95% confidence interval = 2.95-888.96] in ASD cases than controls (Table 7). There were 11 ASD cases with deletions in ASD candidate genes (Table S3) and all were experimentally validated by qPCR except three false-positive CNVs overlapping the SYNGAP1 gene. The one SYNGAP1 CNV that did validate is a de novo 112 kb deletion, which was described previously in the Pinto et al. 2010 study that disrupts SYNGAP1 and encompasses four other genes. After removing the three false-positive CNVs in the SYNGAP1 gene and testing for enrichment again, we still observed significant enrichment of deletions in genes implicated in ASD (p = 0.001585, odds ratio = 14.74 with 95% CI = 1.95−656.52) in ASD cases as compared with controls. The other ASD cases where we observed rare exonic loss in ASD candidate genes were PTCHD1 (Marshall et al. 2008; Pinto et al. 2010), VPS13B (Pinto et al. 2010), DMD (Pinto et al. 2010), DPYD (novel to this study and as described in Carter et al. 2011), SHANK2 (Pinto et al. 2010), NF1 (novel to this study) and NRXN1 (ILMN Omni 2.5 M array; A. C. Lionel, unpublished data).
Table 7. CNV burden in known ASD genes in cases vs. PDx controls.
| Type | Sizea | Case counts | Control counts | ASD % | CT % | p value |
|---|---|---|---|---|---|---|
| All | 110 | 14 | 15 | 4.307692 | 2.340094 | 0.0700680 |
| Deletions | 110 | 11 | 1 | 5.641026 | 0.288184 | 8.90E-05b |
| Duplications | 110 | 3 | 14 | 1.369863 | 3.030303 | 0.9480197 |
ASD, autism spectrum disorder; CNV, copy number variations; PDx, control cohort DNA samples.
Number of genes causally implicated in ASD (Betancur 2011).
Significant differences (p ≤ 0.01) are indicated; ASD and CT (controls) % are based on the total number of subjects with at least one rare exonic CNV of the respective type.
Gene-set association tests
We tested functional gene-sets for enrichment in ASD cases over controls to identify biological processes potentially involved in ASD. We found significant results only for deletions, which is in agreement with global burden results (Table 6). There were 23 gene-sets that had a permutation-based FDR < 25% in deletions and these were used to construct a functional map of ASD (Figure 6, Table S4). We identified several gene-set clusters, some of which were associated with ASD by earlier studies. For example, gene-sets involved in GTPase/Ras signaling pathways were previously reported in ASD (Meechan et al. 2009; Pinto et al. 2010). The novel pathway that has been discovered from this study is the gene-set enriched for nucleotide metabolism. The list of genes in the nucleotide metabolism pathway is shown in Table S5. To strengthen the conclusion that ASD variants impact genes involved in the nucleotide metabolism pathway, we performed experimental qPCR validation on this set of CNVs and found all were valid except one deletion arising due to cell line artifact in the FHIT gene. The confirmation of 14 of 15 genes (Table S5) based on qPCR validation of the microarray findings suggests that this pathway is not the result of false positives in the dataset.
Figure 6 .
A gene-set functional enrichment map of ASD. Each node represents gene-sets with its size proportional to the total number of genes in each set. The map shows gene sets enriched for deletions with enrichment significance (permutation-based FDR) represented as a node color gradient.
Discussion
Our high-resolution CGH data have identified multiple novel, rare CNVs in ASD cases that went undetected by SNP microarrays run on the same individuals; thus providing an additional valuable resource for ASD risk gene discovery and validation. Although hundreds of ASD susceptibility genes/loci (Betancur 2011) are now known, no single gene or locus accounts for more than 0.8% frequency of cases in a given cohort, with most contributing to less than 0.1% of ASD cases (Devlin and Scherer 2012). Moreover, other CNV studies (Pinto et al. 2010; Sanders et al. 2011) and recent exome sequencing studies (Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012) suggest that perhaps several hundred autism risk genes may exist. These data indicate that to delineate all ASD risk genes and alleles, different experimental strategies will likely be required when assessing even larger sets of ASD patient collections.
We find that the Agilent CGH array is sensitive for detection of many smaller (<30 kb) CNVs, which are often missed by SNP microarrays even when probe coverage is sufficient at these loci. The results of this study suggest that currently available microarray platforms are complementary (i.e., not all CNVs are captured by one platform/array design) and that the number and type of CNVs detected varies depending on microarray probe distribution, sample labeling and hybridization chemistries, and CNV detection algorithms used. The Agilent 1M array is designed exclusively for CNV detection and utilizes a more uniform probe distribution across the genome as compared with SNP arrays such as the Illumina 1M and Affymetrix 6.0 arrays. The median probe spacing of the Agilent 1M CGH array is 2.1 kb and a small proportion of probes correspond to regions that are non-unique in nature; that is, they map to multiple locations in the genome, with most targeted to segmentally duplicated regions. Therefore, the Agilent 1M CGH array is better at detecting CNVs in segmental duplicated regions compared to SNP arrays. Also, a better signal-to-noise ratio has been observed in CGH arrays compared to SNP arrays and is attributed to the use of longer probes [60-mers (Pinto et al. 2011)]. The SNP arrays are also biased toward known CNVs. However, in addition to copy number analysis, one of the advantages of using SNP arrays is the ability to genotype SNPs which is not possible using the CGH array from this study. Until a single technology (e.g., whole-genome sequencing) is sufficiently robust to capture most genetic variants, including structural variants such as CNVs, use of multiple platforms will be advantageous. In some instances, the identification of previously undetected CNV variants will reveal pathogenic events relevant for clinical diagnosis of ASD (Miller et al. 2010; Scherer and Dawson 2011).
The discovery of additional rare variants in this present study led to the identification of a novel disease pathway of triphosphate nucleotide metabolism; specifically, purine and pyrimidine metabolism as a potential biological process involved in ASD. Several genes found to harbor a rare exonic loss in cases but not in controls have been previously associated to a neuropsychiatric or neurodevelopmental phenotype in human [DPYD, UPB1 (van Kuilenburg et al. 2004, 2009)] and mouse [UPP1, TYMP (Haraguchi et al. 2002; López et al. 2009)] studies. Another interesting functional category revealed by this study is small GTPase signaling pathways, with genes such as ARHGAP15, CDH13, NF1, RALGDS, SYNGAP1, and VAV3 harboring rare losses in cases but not in controls (Table S2). Rare mutations in the ARHGAP15 gene have been reported to be associated with ASD (O’Roak et al. 2011); NF1 and SYNGAP1 genes are also implicated in ASD (Williams and Hersh 1998; Marui et al. 2004; Pinto et al. 2010). The protein encoded by the CDH13 gene is one of numerous cadherins expressed in the brain, which have been shown to regulate many neural processes (Redies et al. 2012) and CDH13 has been implicated in ADHD through genome-wide association and extended pedigree linkage studies (Lesch et al. 2008; Rivero et al. 2012). The RALGDS and VAV3 genes are involved in multiple signaling pathways, including nerve growth factor receptor signaling pathway (Ferro and Trabalzini 2010; Keilhoff et al. 2012). Therefore, the genes CDH13, RALGDS, and VAV3 could represent interesting examples of ASD candidate genes for further follow-up. One of the genes, DLC1, contributed to both the “cytoskeleton and contractile fiber” and “small GTPase signaling” pathways. Rare exonic loss in this gene was observed in an ASD case but not in any of the controls. The DLC1 gene is involved in multiple phenotypes in mouse, including nervous system development (Sabbir et al. 2010) and thus is an interesting ASD candidate gene for further study. Examples of some of the genes contributing to the “cytoskeleton and contractile fiber” pathway are DMD, EPB41L2, MYH7, PGM5, and TRIM32, where we observed rare exonic losses in cases but not in controls. The gene DMD has been shown to be associated with ASD in several studies (Wu et al. 2005; Hinton et al. 2009; Erturk et al. 2010). The TRIM32 gene is suggested to be involved in muscle and nervous system development mouse phenotypes (Kudryashova et al. 2009) and deletions in this gene are also reported in cases with attention deficit hyperactivity disorder (Lionel et al. 2011). The gene EPB41L2 binds glutamate (Shen et al. 2000) and dopamine receptors (Binda et al. 2002) and is also suggested to be involved in mouse reproductive and nervous system development phenotypes (Ivanovic et al. 2012). The PGM5 gene interacts with the DMD gene (Wakayama et al. 2000; Szklarczyk et al. 2011). Thus, we believe this set of ‘Cytoskeleton and Contractile fiber’ pathway genes, which are impacted by ASD-specific variants in the present study, warrant further follow-up.
Whereas much of the focus in identifying ASD genes has been in studying de novo events, assessing the role of both autosomal and X-linked rare inherited CNVs also has been fruitful, yielding new susceptibility loci such as NRXN3 (Vaags et al. 2012), SHANK1 (Sato et al. 2012), and the Xp11.2 PTCHD1-PTCHD1AS region (Noor et al. 2010). However, in taking de novo and rare inherited variants together, no one gene accounts for more than 1% of the etiology in ASD highlighting the complex genetic heterogeneity of the disorder. It is, therefore, essential to capture the entire spectrum of genetic variation contributing to ASD risk to account for unexplained heritability. Moreover, given the rarity of some of the putative risk variants identified in this study, it is likely that high resolution genome-wide scans of tens of thousands of ASD cases will be needed to validate and contextualize these findings. The results from the present study will complement ASD genetic risk factors being identified through current whole exome and genome sequencing efforts. The public availability of the new rich resource of rare CNVs uncovered in this study will serve as an important resource for further prioritization of putative ASD risk genes for subsequent genetic and functional characterization.
Supplementary Material
Acknowledgments
We thank Sylvia Lamoureux, Quen Tran, and The Centre for Applied Genomics (TCAG) for technical assistance. This work was supported by grants from NeuroDevNet, the University of Toronto McLaughlin Centre, Genome Canada, and the Ontario Genomics Institute, the Canadian Institutes for Health Research (CIHR), the Canadian Institute for Advanced Research, the Canada Foundation for Innovation, the government of Ontario, and The Hospital for Sick Children Foundation. A.P. is supported by a fellowship from the CIHR Autism Training Program. S.W.S. holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children. Two provisional patent applications originating from this data have been filed with the U.S. Patent and Trademark Office.
Footnotes
Communicating editor: R. Cantor
Literature Cited
- Ardlie K. G., Lunetta K. L., Seielstad M., 2002. Testing for population subdivision and association in four case-control studies. Am. J. Hum. Genet. 71: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Genome Project Consortium Szatmari P., Paterson A. D., Zwaigenbaum L., Roberts W., et al. , 2007. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39: 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banting G. S., Barak O., Ames T. M., Burnham A. C., Kardel M. D., et al. , 2005. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum. Mol. Genet. 14: 513–524 [DOI] [PubMed] [Google Scholar]
- Betancur C., 2011. Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res. 1380: 42–77 [DOI] [PubMed] [Google Scholar]
- Bierut L. J., Agrawal A., Bucholz K. K., Doheny K. F., Laurie C., et al. , 2010. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA 107: 5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda A. V., Kabbani N., Lin R., Levenson R., 2002. D2 and D3 dopamine receptor cell surface localization mediated by interaction with protein 4.1N. Mol. Pharmacol. 62: 507–513 [DOI] [PubMed] [Google Scholar]
- Blazejczyk M., Sobczak A., Debowska K., Wisniewska M. B., Kirilenko A., et al. , 2009. Biochemical characterization and expression analysis of a novel EF-hand Ca2+ binding protein calmyrin2 (Cib2) in brain indicates its function in NMDA receptor mediated Ca2+ signaling. Arch. Biochem. Biophys. 487: 66–78 [DOI] [PubMed] [Google Scholar]
- Bruno D. L., Anderlid B. M., Lindstrand A., van Ravenswaaij-Arts C., Ganesamoorthy D., et al. , 2010. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J. Med. Genet. 47: 299–311 [DOI] [PubMed] [Google Scholar]
- Carter M. T., Nikkel S. M., Fernandez B. A., Marshall C. R., Noor A., et al. , 2011. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin. Genet. 80: 435–443 [DOI] [PubMed] [Google Scholar]
- Christian S. L., Brune C. W., Sudi J., Kumar R. A., Liu S., et al. , 2008. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol. Psychiatry 63: 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S., Yau C., Taylor J. M., Mirza G., Butler H., et al. , 2007. QuantiSNP: an objective bayes hidden-markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 35: 2013–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook E. H., Jr, Scherer S. W., 2008. Copy-number variations associated with neuropsychiatric conditions. Nature 455: 919–923 [DOI] [PubMed] [Google Scholar]
- Curtis C., Lynch A. G., Dunning M. J., Spiteri I., Marioni J. C., et al. , 2009. The pitfalls of platform comparison: DNA copy number array technologies assessed. BMC Genomics 10: 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Scherer S. W., 2012. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 22: 229–237 [DOI] [PubMed] [Google Scholar]
- Erturk O., Bilguvar K., Korkmaz B., Bayri Y., Bayrakli F., et al. , 2010. A patient with duchenne muscular dystrophy and autism demonstrates a hemizygous deletion affecting dystrophin. Am. J. Med. Genet. A. 152A: 1039–1042 [DOI] [PubMed] [Google Scholar]
- Ewing R. M., Chu P., Elisma F., Li H., Taylor P., et al. , 2007. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro E., Trabalzini L., 2010. RalGDS family members couple ras to ral signalling and that’s not all. Cell. Signal. 22: 1804–1810 [DOI] [PubMed] [Google Scholar]
- Gao Y., Vasilyev D. V., Goncalves M. B., Howell F. V., Hobbs C., et al. , 2010. loss of retrograde endocannabinoid signaling and reduced adult neurogenesis in diacylglycerol lipase knock-out mice. J. Neurosci. 30: 2017–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau J. R., Lima C. D., 2010. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J. T., Wang K., Cai G., Korvatska O., Kim C. E., et al. , 2009. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert T. A., Cahan P., Edwin D., Selzer R. R., Richmond T. A., et al. , 2007. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 3: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold A. J., Ma D., Sacharow S. J., Robinson J. L., Jaworski J. M., et al. , 2011. A de novo 1.5 mb microdeletion on chromosome 14q23.2–23.3 in a patient with autism and spherocytosis. Autism Res. 4: 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi M., Tsujimoto H., Fukushima M., Higuchi I., Kuribayashi H., et al. , 2002. Targeted deletion of both thymidine phosphorylase and uridine phosphorylase and consequent disorders in mice. Mol. Cell. Biol. 22: 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester S. D., Reid L., Nowak N., Jones W. D., Parker J. S., et al. , 2009. Comparison of comparative genomic hybridization technologies across microarray platforms. J. Biomol. Tech. 20: 135–151 [PMC free article] [PubMed] [Google Scholar]
- Hinton V. J., Cyrulnik S. E., Fee R. J., Batchelder A., Kiefel J. M., et al. , 2009. Association of autistic spectrum disorders with dystrophinopathies. Pediatr. Neurol. 41: 339–346 [DOI] [PubMed] [Google Scholar]
- International HapMap 3 Consortium Altshuler D. M., Gibbs R. A., Peltonen L., Altshuler D. M., et al. , 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovic A., Horresh I., Golan N., Spiegel I., Sabanay H., et al. , 2012. The cytoskeletal adapter protein 4.1G organizes the internodes in peripheral myelinated nerves. J. Cell Biol. 196: 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont M. L., Sanlaville D., Redon R., Raoul O., Cormier-Daire V., et al. , 2006. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J. Med. Genet. 43: 843–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilhoff G., Wiegand S., Fansa H., 2012. Vav deficiency impedes peripheral nerve regeneration in mice. Restor. Neurol. Neurosci. 30: 463–479 [DOI] [PubMed] [Google Scholar]
- Korn J. M., Kuruvilla F. G., McCarroll S. A., Wysoker A., Nemesh J., et al. , 2008. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 40: 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Nikolaus S., von Eberstein H., Croucher P. J., El Mokhtari N. E., et al. , 2006. PopGen: Population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 9: 55–61 [DOI] [PubMed] [Google Scholar]
- Kudryashova E., Wu J., Havton L. A., Spencer M. J., 2009. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 18: 1353–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W. R., Johnson M. D., Kucherlapati R., Park P. J., 2005. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics 21: 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Scherer S. W., 2010. The clinical context of copy number variation in the human genome. Expert Rev. Mol. Med. 12: e8. [DOI] [PubMed] [Google Scholar]
- Lesch K. P., Timmesfeld N., Renner T. J., Halperin R., Roser C., et al. , 2008. Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J. Neural Transm. 115: 1573–1585 [DOI] [PubMed] [Google Scholar]
- Li X. F., Kiedrowski L., Tremblay F., Fernandez F. R., Perizzolo M., et al. , 2006. Importance of K+-dependent Na+/Ca2+-exchanger 2, NCKX2, in motor learning and memory. J. Biol. Chem. 281: 6273–6282 [DOI] [PubMed] [Google Scholar]
- Lionel A. C., Crosbie J., Barbosa N., Goodale T., Thiruvahindrapuram B., et al. , 2011. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci. Transl. Med. 3: 95ra75. [DOI] [PubMed] [Google Scholar]
- López L. C., Akman H. O., Garcia-Cazorla A., Dorado B., Marti R., et al. , 2009. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum. Mol. Genet. 18: 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C. R., Noor A., Vincent J. B., Lionel A. C., Feuk L., et al. , 2008. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marui T., Hashimoto O., Nanba E., Kato C., Tochigi M., et al. , 2004. Association between the neurofibromatosis-1 (NF1) locus and autism in the japanese population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 131B: 43–47 [DOI] [PubMed] [Google Scholar]
- McCarroll S. A., Kuruvilla F. G., Korn J. M., Cawley S., Nemesh J., et al. , 2008. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 40: 1166–1174 [DOI] [PubMed] [Google Scholar]
- Meechan D. W., Tucker E. S., Maynard T. M., LaMantia A. S., 2009. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc. Natl. Acad. Sci. USA 106: 16434–16445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G. D., 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. T., Adam M. P., Aradhya S., Biesecker L. G., Brothman A. R., et al. , 2010. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 86: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T., Yeo S. Y., Hirate Y., Segawa H., Wada H., et al. , 2004. PlexinA4 is necessary as a downstream target of Islet2 to mediate slit signaling for promotion of sensory axon branching. Development 131: 3705–3715 [DOI] [PubMed] [Google Scholar]
- Neale B. M., Kou Y., Liu L., Ma’ayan A., Samocha K. E., et al. , 2012. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A., Whibley A., Marshall C. R., Gianakopoulos P. J., Piton A., et al. , 2010. Disruption at the PTCHD1 locus on Xp22.11 in autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2: 49ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Deriziotis P., Lee C., Vives L., Schwartz J. J., et al. , 2011. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43: 585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Vives L., Girirajan S., Karakoc E., Krumm N., et al. , 2012. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnamenta A. T., Bacchelli E., de Jonge M. V., Mirza G., Scerri T. S., et al. , 2010. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol. Psychiatry 68: 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Pagnamenta A. T., Klei L., Anney R., Merico D., et al. , 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Darvishi K., Shi X., Rajan D., Rigler D., et al. , 2011. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat. Biotechnol. 29: 512–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redies C., Hertel N., Hubner C. A., 2012. Cadherins and neuropsychiatric disorders. Brain Res. 1470: 130–144 [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., et al. , 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero O., Sich S., Popp S., Schmitt A., Franke B., et al. , 2012. Impact of the ADHD-susceptibility gene CDH13 on development and function of brain networks. Eur. Neuropsychopharmacol. (in press) [DOI] [PubMed] [Google Scholar]
- Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., et al. , 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sabbir M. G., Wigle N., Loewen S., Gu Y., Buse C., et al. , 2010. Identification and characterization of Dlc1 isoforms in the mouse and study of the biological function of a single gene trapped isoform. BMC Biol. 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyakina D., Cukier H. N., Lee J. M., Sacharow S., Nations L. D., et al. , 2011. Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS ONE 6: e26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., Ercan-Sencicek A. G., Hus V., Luo R., Murtha M. T., et al. , 2011. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70: 863–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., Murtha M. T., Gupta A. R., Murdoch J. D., Raubeson M. J., et al. , 2012. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485: 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D., Lionel A. C., Leblond C. S., Prasad A., Pinto D., et al. , 2012. SHANK1 deletions in males with autism spectrum disorder. Am. J. Hum. Genet. 90: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf C. P., Sabo A., Sakai Y., Crosby J., Muzny D., et al. , 2011. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum. Mol. Genet. 20: 3366–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S. W., Dawson G., 2011. Risk factors for autism: translating genomic discoveries into diagnostics. Hum. Genet. 130: 123–148 [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., et al. , 2007. Strong association of de novo copy number mutations with autism. Science 316: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajee F. J., Zhong H., Nabi R., Huq A. H., 2003. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J. Med. Genet. 40: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Liang F., Walensky L. D., Huganir R. L., 2000. Regulation of AMPA receptor GluR1 subunit surface expression by a 4.1N-linked actin cytoskeletal association. J. Neurosci. 20: 7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Dies K. A., Holm I. A., Bridgemohan C., Sobeih M. M., et al. , 2010. Clinical genetic testing for patients with autism spectrum disorders. Pediatrics 125: e727–e735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr E. H., 2003. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): One gene leads to many phenotypes. Curr. Opin. Pediatr. 15: 567–571 [DOI] [PubMed] [Google Scholar]
- Shinawi M., Cheung S. W., 2008. The array CGH and its clinical applications. Drug Discov. Today 13: 760–770 [DOI] [PubMed] [Google Scholar]
- Stewart A. F., Dandona S., Chen L., Assogba O., Belanger M., et al. , 2009. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the ottawa heart genomics study. J. Am. Coll. Cardiol. 53: 1471–1472 [DOI] [PubMed] [Google Scholar]
- Suto F., Murakami Y., Nakamura F., Goshima Y., Fujisawa H., 2003. Identification and characterization of a novel mouse plexin, plexin-A4. Mech. Dev. 120: 385–396 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., et al. , 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39: D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaags A. K., Lionel A. C., Sato D., Goodenberger M., Stein Q. P., et al. , 2012. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet. 90: 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg A. B., Meinsma R., Beke E., Assmann B., Ribes A., et al. , 2004. Beta-ureidopropionase deficiency: an inborn error of pyrimidine degradation associated with neurological abnormalities. Hum. Mol. Genet. 13: 2793–2801 [DOI] [PubMed] [Google Scholar]
- van Kuilenburg A. B., Meijer J., Mul A. N., Hennekam R. C., Hoovers J. M., et al. , 2009. Analysis of severely affected patients with dihydropyrimidine dehydrogenase deficiency reveals large intragenic rearrangements of DPYD and a de novo interstitial deletion del(1)(p13.3p21.3). Hum. Genet. 125: 581–590 [DOI] [PubMed] [Google Scholar]
- Venkatraman E. S., Olshen A. B., 2007. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23: 657–663 [DOI] [PubMed] [Google Scholar]
- Wakayama Y., Inoue M., Kojima H., Murahashi M., Shibuya S., et al. , 2000. Aciculin and its relation to dystrophin: Immunocytochemical studies in human normal and duchenne dystrophy quadriceps muscles. Acta Neuropathol. 99: 654–662 [DOI] [PubMed] [Google Scholar]
- Waldvogel H. J., Baer K., Snell R. G., During M. J., Faull R. L., et al. , 2003. Distribution of gephyrin in the human brain: an immunohistochemical analysis. Neuroscience 116: 145–156 [DOI] [PubMed] [Google Scholar]
- Wilkinson K. A., Nakamura Y., Henley J. M., 2010. Targets and consequences of protein SUMOylation in neurons. Brain Res. Brain Res. Rev. 64: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. G., Hersh J. H., 1998. Brief report: the association of neurofibromatosis type 1 and autism. J. Autism Dev. Disord. 28: 567–571 [DOI] [PubMed] [Google Scholar]
- Winchester L., Yau C., Ragoussis J., 2009. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomics Proteomics 8: 353–366 [DOI] [PubMed] [Google Scholar]
- Wu J. Y., Kuban K. C., Allred E., Shapiro F., Darras B. T., 2005. Association of duchenne muscular dystrophy with autism spectrum disorder. J. Child Neurol. 20: 790–795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.