Abstract
Commensal bacteria are necessary for the development and maintenance of a healthy immune system. Harnessing the ability of microbiota to affect host immunity is considered an important therapeutic strategy for many mucosal and non-mucosal immune-related conditions, such as inflammatory bowel diseases (IBD), celiac disease, metabolic syndrome, diabetes and microbial infections. In addition to well-established immunostimulatory effects of the microbiota, the presence of individual mutualistic commensal bacteria with immunomodulatory effects has been described. These organisms are permanent members of the commensal microbiota and affect host immune homeostasis in specific ways. Identification of individual examples of such immunomodulatory commensals and understanding their mechanisms of interaction with the host will be invaluable in designing therapeutic strategies to reverse intestinal dysbiosis and recover immunological homeostasis.
Introduction
Mucosal surfaces are colonized by a complex and dynamic microbial ecosystem termed “microbiota”. An astounding number and diversity of microbes, including fungi, bacteria, archaea, and viruses, are present in any given moment throughout our bodies. The vast majority of these organisms are not disease-causing invaders, but have made the host their one-and-only home. Through co-evolution, these “commensals” have established one of the most impressive examples of mutualistic relationship in the natural world, in which both microbes and their animal host depend on each other for optimal survival. Indeed, we rely on our microbiota for many basic physiologic and metabolic functions, as well as proper immune functions. Commensals provide immune protection in several ways. They can defend their mucosal home by directly combating invading pathogens or by mobilizing host anti-microbial immune defenses. They can also affect host immunity in a more inconspicuous, but equally important, way by directing the development of host immune cell subsets at steady state and therefore affecting mucosal and systemic innate or adaptive immune responses. Recent studies have identified examples of commensal bacterial species with such immunomodulatory roles. The existence of multiple members of the microbiota that affect host immune homeostasis in different ways means that differences in the composition of this community may contribute to individual differences in immune responses during infection, autoimmunity, cancer or other immunological conditions. Unveiling the underlying cellular and molecular mechanisms of each of these examples holds the promise to lead to exciting new ways for regulating mucosal immunity.
Microbial-host interactions in the intestine
Decades of studies in germ-free (GF) animals have established the importance of microbiota for proper host immune function (Macpherson and Harris, 2004). GF animals were first created more than a century ago (Nuttal and Thierfeledr, 1895–1896) and long-term husbandry of GF rats has been possible since the 1940’s (Reyniers et al., 1946). However, until recently, the composition of gut microbial communities remained largely unknown. Advances in high-throughput sequencing in the last few years have led to extensive cataloguing of the human microbiota (HMP, 2012). In addition to correlating changes in microbiota composition with disease, this has allowed for the identification of commensal species with specific immune effects. The vast majority of these studies have focused on the bacterial component of the microbiota, however characterization of the fungal and viral components and their function is under way (Iliev et al., 2012; Reyes et al., 2012).
The individual’s microbiota composition is dynamic. It changes with age and fluctuates with environmental changes, such as geographical location, diet, antibiotic use, or influx and efflux of external microbes (Clemente et al., 2012). In addition, vastly different microbial communities reside in different parts of the body (Costello et al., 2009). Based on their colonization ability, bacteria in the gut can be transient or permanent. Transient bacteria represent microbes that are introduced during adult life from the external environment and do not permanently colonize the intestinal tract for different reasons, such as lack of appropriate adaptations for colonization or inability to compete with the resident microbiota. Many food-associated microbes, including pathogens and conventional commercial probiotics, are part of this category. Transient organisms can affect the immune system in different ways and be innocuous, pathogenic, or even beneficial, e.g. ingestion of probiotic-containing foods. These organisms have not co-evolved and therefore do not establish a mutualistic relationship with the host, but rather try to survive in the gut environment despite established host defenses. Indeed, many acute intestinal pathogens have developed strategies to forcefully colonize the intestine, which in most cases induces a strong immune response aimed at clearing the pathogen. The pathologic immunological consequences of this inflammatory response may persist for years after the clearance of the transient organisms as in post-infectious irritable bowel syndrome (Spiller and Garsed, 2009).
In contrast to transient bacteria, permanent bacteria are long-term members of the microbial community. Their colonization occurs in successive waves during ontogeny and they have developed evolutionary adaptations to establish a permanent relationship with the host. In most cases they have co-evolved with the host and are not normally found as free-living organisms. These are the true commensal bacteria. Commensals have multiple effects on the immune system of the host. On one hand the presence of a large number of “innocuous” bacteria has immunostimulatory effects. Commensals stimulate general recruitment of immune cells to the mucosa, as well as generation and maturation of organized gut-associated lymphoid tissues (Macpherson and Harris, 2004). They also stimulate protective epithelial cell functions, such as mucus and anti-microbial peptide secretion (Hooper and Macpherson, 2010). On the other hand, recent studies have identified the presence of commensal species with immunomodulatory effects. These effects are specific for individual bacteria or groups of bacteria, i.e. for specific components of the microbiota. They involve reversible changes in differentiation or effector function of host immune cell subsets. In this way, microbiota composition can influence the type and robustness of host immune responses. Here, we refer to such permanent microbiota members with immunomodulatory effects as autobionts. In contrast to transient pathogens or pathobionts (see below), the immune effects of autobionts are more subtle because they do not cause any overt change in the health state of the host. Rather, they help maintain and regulate the host’s healthy immune steady state. Permanent microbiota members that can demonstrate detrimental effects under special conditions are called pathobionts. Pathobionts colonize the host, but do not cause disease with a full complement of normal microbiota. However, they can expand and cause disease if microbiota or host immune homeostasis are perturbed (for example after antibiotic treatment or under conditions of intestinal inflammation). The immune effects and characteristics of different intestinal bacteria are summarized in Table 1.
Table 1.
Non-pathogenic bacterial members of the intestinal microbiota with immune effects
| Concept | Examples* | Association with Host | Immune Effects | Mechanisms* | |
|---|---|---|---|---|---|
| Probiotics |
|
Bifidobacterium spp; Lactobacillus spp | Transient | Innocuous Immunostimulatory |
Cytokine induction, TLR activation, Pathobiont and pathogen suppression, Lactic acid Short-chain fatty acids |
| Autobionts |
|
Bacteroides fragilis Clostridia XIVa and IV SFB Faecalibacterium prauznitsii |
Permanent Host-dependent Symbiotic |
Immunomodulatory | Largely Unknown (TLR2, metabolites (?), antigens (?), effects on IEC function (?)) |
| Pathobionts |
|
Helicobacter hepaticus Clostridium difficile Prevotela spp. Klebsiella spp Bilophila wadsworthia |
Permanent Pa rasitic/I nfectious |
Innocuous Detrimenta |
Invasive mechanisms, Spore formation, Toxins |
Examples and mechanistic studies have been performed in mice (with the exception of F. prauznitsii and C. difficile). See text for details and references. (?) Speculative mechanisms
In contrast to pathogens, pathobionts, and even probiotics, very little is known about the mechanisms by which autobionts exert their immunomodulatory effects. Until recently, one reason for this was the lack of specific examples of immunomodulatory commensals. Another reason is the difficulty in culturing these organisms ex vivo and the relative lack of genetic tools to study their genome function. In this review we focus on recently described examples of immunomodulatory commensals and speculate on potential cellular and molecular mechanisms involved in their interaction with the host and the establishment of a healthy steady immune state. Knowledge of these mechanisms can benefit the development of future therapies for intestinal diseases (Clemente et al., 2012).
The intestinal microbial community influences immunity
During steady state conditions, the microbiota affects the development and function of various immune cell populations, including IgA-secreting plasma cells, Th17 cells, regulatory T (Treg) cells, invariant natural killer T (iNKT) cells, γδT cells, NK cells, macrophages, dendritic cells (DCs), and innate lymphoid cells (ILCs) (Honda and Littman, 2012). For example, IgA+ plasma cells in gut lymphoid tissues and lamina propria (LP) are greatly reduced in GF or antibiotics-treated conventional animals (Macpherson and Harris, 2004). In another example, the abundance and function of CD4+ T cells expressing the FoxP3 transcription factor (Tregs) or interleukin (IL)-17 (Th17 cells) in the intestinal mucosa at steady state are affected by the microbiota. In GF or antibiotic-treated mice, the percentages of both Tregs and Th17 cells are markedly reduced, and expression of the immune suppressive cytokine IL-10 in Treg cells is severely reduced (Atarashi et al., 2008; Atarashi et al., 2011; Ivanov et al., 2008). These reductions are quickly restored by transplantation of intestinal or fecal microbiota from conventionally raised/specific pathogen free (SPF) mice. As described below, the development of these CD4+ T cell subsets is differentially regulated by certain components of the intestinal microbiota including Bacteroides fragilis, Clostridia species, and segmented filamentous bacteria (SFB) (Atarashi et al., 2011; Ivanov et al., 2009; Round and Mazmanian, 2010). Th17 cells contribute to host defense against infection with pathogenic microbes but can also augment harmful autoinflammatory functions, whereas Tregs play critical roles in immune suppression. Therefore, under healthy conditions Th17 cells and Tregs should co-exist in a well-regulated balance. The particular combinations and relative abundances of the corresponding commensal species would generate distinct immune environments and immune responses in the host.
In addition to inducing development or recruitment of host immune cell subsets, microbiota may also affect the function of these subsets. For example, gut microbiota provides an environment not only for the accumulation of IgA+ cells but also for the functional maturation of IgA+ plasma cells by inducing the generation of iNOS+ IgA+ plasma cells (Fritz et al., 2012). iNOS+ IgA+ plasma cells are absent in GF mice, but present in conventionally-raised mice. This subset plays critical roles in the enhancement of IgA+ plasma cell development and the host defense against enteric pathogens, such as Citrobacter rodentium (Fritz et al., 2012). Commensal bacteria also affect NK cell function. Even though NK cell numbers are normal in GF mice, NK cell priming and anti-viral activity is deficient in the absence of microbiota (Ganal et al., 2012). This effect is due to microbiota-directed introduction of epigenetic changes and induction of type I Interferons from monocytic macrophages, which are required for proper NK cell priming (Ganal et al., 2012).
Microbiota-controlled immune effects may also play role in regulation of the microbiota itself. Commensal-induced IgA+ plasma cells contribute to controlling microbiota abundance and composition. For instance, mice carrying a knock-in mutation of activation-induced cytidine deaminase (AID) (AIDG23S), which can mediate normal IgA class switching but cannot induce somatic hypermutation and high-affinity IgA responses, exhibit excessive proliferation of anaerobic bacteria in the small intestine (Wei et al., 2011). Similar overgrowth of microbiota was also reported in programmed cell death-1 (PD-1) deficient mice, which have increased follicular helper T (TFH) cells development, and therefore low-affinity IgA-producing plasma cell are aberrantly selected in germinal centers (Kawamoto et al., 2012). Thus, microbiota is required for the development of fully functional IgA+ cells, which in turn function to maintain microbial homeostasis in the gut.
The microbiota also plays suppressive roles in immune cell function and accumulation in the gut. For example, it represses constitutive production of IL-22 in lymphoid tissue inducer (LTi) cells and NKp46+ cells (both of which are RORγt+ ILCs, see detailed review by Tait Wonjo and David Artis in this issue) through epithelial expression of IL-25 (Sawa et al., 2011). Another example is that early exposure to gut microbiota provides an epigenetic suppressive marking in the regulatory element of CXCL16 gene in the host, resulting in life long suppression of CXCL16 expression in gut and lung (Olszak et al., 2012). This suppression is accompanied by reduced abundance of iNKT cells in the colon and lung at steady state and affects host resistance to colitis and asthma (Olszak et al., 2012). Taken together, intestinal microbiota provides diverse signals for activation and suppression of the immune system, thereby having the ability to skew host immune status towards either effector or regulator dominance.
Commensals have immunomodultory functions not only in the intestine. For example, resident commensals in the skin induce local Th17 and Th1 responses that are crucial in protection from bacterial infections (Naik et al., 2012). More importantly, these responses were compartmentalized to the skin and were independent of gut commensals (Naik et al., 2012).
Commensals influence pathogenesis of many diseases. Because this community is evolutionarily established to sustain healthy immune steady state, any major perturbations in its composition may have negative effects and perpetuate the cycle of chronic inflammation, allergy, or metabolic syndrome. Such, “dysbiosis” can be induced by diet, pharmacological agents, infection, inflammation, and host genetics (Honda and Littman, 2012). Once established, the dysbiotic microbiota may become stable and transplantable to GF or even conventionally raised animals. Indeed, cohousing of wild-type mice with disease-prone mutant mice, such as Tbet−/−Rag−/−, Nrlp6−/−, or Asc−/− mice, results in transfer of dysbiotic microbiota and predisposition of the wild-type mice to disease, including colitis and metabolic syndrome (Elinav et al., 2011; Garrett et al., 2007; Henao-Mejia et al., 2012).
Dysbiosis may lead to elimination of beneficial bacteria or outgrowth of pathobionts. Pathobionts are permanent members of the microbiota, present at low levels and innocuous under normal conditions. They can become pathogenic if allowed sufficient expansion due to loss of microbiota or immune homeostasis. For instance, overgrowth of members of Prevotellaceae and TM7 has been implicated in host susceptibility to DSS colitis in Nrlp6−/− mice (Elinav et al., 2011). In Tbet−/−Rag−/− mice, Proteus mirabilis and Klebsiella pneumoniae were identified to, at least in part, be responsible for the phenotype of spontaneous colitis (Garrett et al., 2010). Most importantly, disease susceptibility was transferable to wild-type mice.
The composition of the gut microbiota is also altered by diet. Milk fat- and taurocholic acid-rich diets induced marked increase in Bilophila wadsworthia colonization, which is associated with enhancement of Th1 responses and acceleration of colitis development in Il10−/− mice (Devkota et al., 2012). It should be noted that depending on the host genotype, otherwise innocuous symbionts may become pathogenic. Indeed, in an inflammatory bowel disease (IBD) mouse model with deficiencies in IL-10 and TGF-β signaling, Bacteroides thetaiotaomicron, a well-characterized symbiotic species, potently induces colitis (Bloom et al., 2011).
Autobionts – mutualistic immunomodulatory microbes
As discussed above, there is abundant evidence that microbiota directs host immunity. Regulation of immune responses by influencing development, differentiation, or effector function of different cells of the immune system is especially interesting, because it results not simply from the presence of innocuous bacteria, but from the biological activity of commensals. In many cases these effects are functionally distinct, e.g. induction of Th17 cells vs Tregs, and depend on the activity of different members of the commensal community. The relative abundance of these “autobionts” can direct the general type of immunity in the host mucosa of an individual at a given time. Currently there are relatively few specific examples of commensals with immunomodulatory effects as discussed below. More are surely going to be identified in the future.
Despite considerable species diversity, the intestinal microbiota in most mammals consists of bacteria belonging to two major phyla – Gram-negative Bacteroidetes and Gram-positive Firmicutes. This probably reflects evolutionary adaptations of these phyla to survive in the gut environment and both of them contain important immunomodulatory commensals.
Bacteorides fragilis promotes Treg function
B. fragilis was the first commensal to be implicated in affecting T helper cell balance by promoting Th1 development systemically (Mazmanian et al., 2005). However, further studies showed that B. fragilis also affects mucosal T cell homeostasis by promoting regulatory T cell function (Round and Mazmanian, 2010). B. fragilis is a Gram-negative member of the phylum Bacteroidetes. It is not a very abundant member of the gut microbiota, however the genus Bacteroides is well represented in human gut and has a superior ability to utilize the nutrients in the gut microenvironment (Flint et al., 2008). For example, the genome of the prototypical dominant commensal member of the class, B. thetaiotaomicron, contains several hundred proteins involved in harvesting and metabolizing of dietary polysaccharides (Sonnenburg et al., 2005). These are probably adaptations that have allowed Bacteroides to establish mutualistic relationship with the host, by 1) being able to flourish in the plant polysaccharide-enriched gut environment and 2) being able to provide biological “by-products” necessary for the well-being of the host. Different Bacteroidetes species produce different beneficial biological “by-products”, which may have helped establish them as permanent symbionts. For example, B. thetaiotaomicron is one of the major producers of short-chain fatty acids (SCFAs), which are necessary for proper host metabolic and immune functions. B. fragilis seems to have pronounced immunomodulatory functions. B. fragilis is not normally present in conventionally raised specific pathogen-free (SPF) mice and colonization with B. fragilis protects mice from colitis in the T cell transfer and 2,4,6-trinitrobenzene sulfonic acid (TNBS) models (Mazmanian et al., 2008). This protection is due to the expansion of immune suppressive IL-10 producing Tregs by the bacteria (Round and Mazmanian, 2010). The introduction of B. fragilis as a permanent member of the microbiota also affects T cell homeostasis in the absence of inflammation. Colonization of SPF or GF mice with B. fragilis leads to an induction of IL-10 production by Foxp3+ Tregs even at steady state (Round and Mazmanian, 2010). Thus, B. fragilis colonization modulates intestinal T cell homeostasis by boosting Treg function. This anti-inflammatory effect of the bacteria likely represents an evolutionary adaptation for establishing mutualism. Indeed, when Tregs were depleted, B. fragilis could not efficiently colonize host tissues (Round et al., 2011). Most importantly, the identification of B. fragilis as a modulator of Treg function allowed for investigation of the bacterial and host mechanisms involved. It was found that the anti-inflammatory effects of B. fragilis require the expression of bacterial capsular polysaccharide A (PSA). PSA-deficient B. fragilis mutants were incapable of inducing IL-10 production by Tregs and did not provide protection from colitis (Mazmanian et al., 2008; Round and Mazmanian, 2010). Instead, lack of PSA expression led to expansion of Th17 cells and loss of the mutualistic ability of B. fragilis to colonize host tissues (Round et al., 2011). Moreover, treatment of mice with purified PSA is sufficient to replicate the effects of the bacteria, including induction of IL-10 production by Tregs, suppression of Th17 cell production, protection in colitis models, and colonization of the host (Mazmanian et al., 2008; Round et al., 2011; Round and Mazmanian, 2010). These studies represent an elegant example of how investigating an immunomodulatory commensal and its effect on the host immune system can lead to elucidation of underlying molecular mechanisms and identification of clinically relevant immunomodulatory molecules.
Cluster IV and XIVa Clostridia induce Treg differentiation
Intestinal Clostridia are a heterogeneous group that forms the core of Firmicutes of the normal commensal microbiota. Clostridia are Gram-positive, rod-shaped, endospore-forming bacteria. They are a highly heterogeneous class that is composed of at least 19 clusters based on genomic similarity (Collins et al., 1994). The prototypical Clostridia from cluster I contain frequent environmental toxin-producing members, such as Clostridium perfringens, C. difficile, and C. tetani. These Clostridia may be present in the intestine, but usually as transient pathogens or, at best, pathobionts (e.g., C. difficile). In contrast, most commensal intestinal Clostridia are non-toxinogenic members of clusters XIVa and IV. They are typically described as fusiform-shaped bacteria and constitute 10–40% of the total microbiota (Frank et al., 2007). Cluster XIVa includes the genera Clostridium, Eubacterium, Ruminococcus, Coprococcus, and Roseburia. The cluster IV group includes species belonging to the Clostridium, Faecalibacterium, and Ruminococcus genera. Clostridia colonize the mucus layers in vicinity of the epithelium, in contrast to Bacteroidaceae, Enterococcaceae, and Lactobacillaceae, which colonize in regions of the central lumen, suggesting unique influences of Clostridia on host physiology (Nava and Stappenbeck, 2011). Indeed, the cluster XIVa Lachnospiraceae family is significantly less abundant in IBD patients compared to healthy subjects (Frank et al., 2007). Loss of mucosa-associated Clostridia and cluster IV Clostridia, particularly Faecalibacterium prausnitzii, is observed in IBD patients (Sokol et al., 2008). Although it remains unclear whether the decrease in Clostridia is a cause or effect of chronic inflammation, it is likely that maintenance of the Clostridia community is necessary to prevent IBD. In addition to the role in the intestinal (local) immune homeostasis, Clostridia also affect systemic immunity. Indeed, it has been shown that reduction of Clostridia clusters XIVa and IV by neonatal vancomycin treatment, promotes airway hypersensitivity in a mouse model (Russell et al., 2012). Furthermore, decreased abundances of clusters IV and XIVa Clostridia have been associated with atopy during childhood (Candela et al., 2012).
The levels of Clostridia clusters IV and XIVa in adult mouse colon microbiota were found to correlate with the numbers of Tregs (Atarashi et al., 2011; Russell et al., 2012). Consistent with a role in colonic Treg induction, GF mice colonized with 46 strains of Clostridia clusters XIVa and IV showed accumulation of Treg cells in the colon (Atarashi et al., 2011). The 46 strains of Clostridia were originally isolated from the sporulating microbiota fraction of conventional mice based on their capacity to normalize the enlarged cecum in GF mice (Itoh and Mitsuoka, 1985). These Clostridia were found to induce near steady state numbers of colonic Tregs in GF mice, in contrast to other intestinal bacteria including Treg-associated autobionts such as B. fragilis (Atarashi et al., 2011). Whereas most Treg cells in the colon of GF mice were Helioshi, which has been proposed as a marker for thymically derived Treg cells (Thornton et al., 2010), Treg cells in mice colonized with 46 strains of Clostridia clusters XIVa and IV were mostly Helioslo (Atarashi et al., 2011), suggesting that Clostridia robustly trigger peripheral differentiation of induced Treg (iTreg) cells. Consistent with these findings, colonization of GF mice with altered Schaedler flora (ASF), a defined bacterial cocktail containing eight enteric species that includes Clostridium clostridioforme, induces the accumulation of Treg cells in the colon (Geuking et al., 2011). Treg cells in the intestine exhibit different characteristics from those in secondary lymphoid organs, and they express CD103, killer cell lectin-like receptor G1 (KLRG1), granzyme B (Gzmb), IL-10 and IL-35 (Feuerer et al., 2010). In particular, IL-10 plays an indispensable role in the suppression of aberrant activation of Th17 cells, myeloid cells, andγδT cells in the intestine. Indeed, Treg-specific disruption of IL-10 results in severe colitis (Rubtsov et al., 2008), as does a Treg-specific deficiency of STAT3, which regulates many of the above-mentioned genes (Chaudhry et al., 2009). STAT3-deficient Tregs lack IL-10 expression and the mice develop spontaneous, Th17-mediated, commensal microbiota-dependent fetal colitis (Chaudhry et al., 2009). Colonization of GF mice with the 46 strains of Clostridia clusters XIVa and IV, induced not only increase in Treg numbers, but also high levels of IL-10 production (Atarashi et al., 2011). In humans, F. prausnitzii, which belongs to Clostridium cluster IV, increases IL-10 expression in peripheral blood mononuclear cells in vitro (Sokol et al., 2008). Therefore, autochthonous Clostridia constitutively induce accumulation and functional activation of Tregs in the colon and the relative abundance of Clostridia in the microbiota may strongly affect the immune status of the host.
SFB induce Th17 cell differentiation
T cell homeostasis in the intestine can be defined as the balance between T cell subsets that promote immune responses and T cell subsets that subdue immune responses. IL-17-producing Th17 cells are pro-inflammatory CD4 T cells that contribute to disease pathogenesis in a number of chronic autoimmune inflammatory conditions, including IBD, multiple sclerosis, rheumatoid arthritis, psoriasis, and certain cancers. At the same time Th17 cells are crucial for efficient immune responses against mucosal pathogens, including viruses, bacteria, and fungi. Th17 cells differentiate from naïve T cells under the combined effects of TGF-β and pro-inflammatory cytokines, such as IL-6, IL-23, and IL-1β. The latter group of cytokines is upregulated in secondary lymphoid tissues during certain infections, which instructs Th17 cell differentiation in this context. However, at steady state, in the absence of infection or overt inflammation, Th17 cells are highly enriched in the intestinal LP and are not present in secondary lymphoid tissues (Ivanov et al., 2006). In the gut, Th17 cells co-exist with Foxp3+ Tregs. The two subsets share common developmental pathway and alternative differentiation fates. This includes shared dependence on TGF-β and direct interaction and mutual functional inhibition of the two master transcriptional regulators, RORγt (Th17 cells) and FoxP3 (Tregs) (Zhou et al., 2008). This overlap of cytokine and transcriptional networks results in an elegant balance between two functionally opposing T cell subsets. Importantly, this balance is flexible and can be quickly reversed depending on the required immune response. One of the most important factors controlling the homeostasis between these two T cell subsets in the gut is the composition of intestinal microbiota. Th17 cells are not present in GF mice, but are induced upon colonization with the full complement of gut bacteria from SPF mice (Atarashi et al., 2008; Ivanov et al., 2008). In contrast, colonization with culturable intestinal isolates, including Treg-inducing commensals, such as B. fragilis or the mix of 46 Clostridia described above, does not lead to Th17 cell induction (Ivanov et al., 2009; Round et al., 2011), arguing that commensal microbiota contains unknown Th17 cell-inducing bacteria. This was further supported by the discovery that C57BL/6 mice from a colony at the Jackson Laboratory did not contain gut Th17 cells due to the lack of Th17 cell-inducing bacteria and total microbiota from these mice could not induce Th17 cells in GF animals, in contrast to microbiota from C57BL/6 mice from a colony at Taconic Farms (Ivanov et al., 2008). Comparison of the microbiota between these two colonies revealed that Taconic B6 mice are highly enriched in segmented filamentous bacteria (SFB), which are absent from Jackson B6 mice (Ivanov et al., 2009). Interestingly, mono-colonization of GF animals with SFB or introduction of SFB into Jackson B6 mice induces Th17 cell differentiation in the LP (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009), identifying SFB as a Th17 cell-inducing autobiont.
SFB are Gram-positive anaerobic bacteria that are known to permanently colonize the intestinal tract of many animal species. SFB or SFB-like bacteria have been described in invertebrates, such as termites and cockroaches, and in vertebrate animals such as fish, chickens, rabbits, mice, rats, cats, dogs, sheep, cows, pigs, zebras and monkeys (Klaasen et al., 1992). However, SFB have not yet been detected in humans (Sczesnak et al., 2011). SFB are members of the Firmicutes phylum and based on 16S rRNA sequence, were assigned to Clostridia (Snel et al., 1995). The similarity to Clostridia was later confirmed by the full SFB genomic sequence (Sczesnak et al., 2011). In fact, more than 60% of SFB ORFs are derived from Clostridial ORFs (Sczesnak et al., 2011). Despite this close similarity, SFB do not cluster with any of the sequenced Clostridial genomes. Moreover, the SFB genome is almost three times smaller than the average Clostridial genome. Such genome reduction is typical for obligate symbionts and therefore may be a mutualistic evolutionary adaptation to establish SFB as a permanent member of the gut microbiota.
Even though SFB induce Th17 cells, they do not cause any overt pathology, such as intestinal inflammation. It is unclear why SFB-induced Th17 cells do not induce inflammation. It could be due to control by Tregs and the opposing effects of Treg-inducing autobionts. However, SFB-monocolonized mice do not develop spontaneous colitis, which argues against suppressive effects of other bacteria. Another possibility is that either the levels or the effector function of SFB-induced Th17 cells are not sufficient for induction of inflammation. Indeed, naturally occurring intestinal Th17 cells do not cause colitis in the T cell transfer model of colitis in contrast to transfer of Th17 cells isolated from colitic mice (Ono et al., 2012). Small intestinal Th17 cells, presumably induced by microbiota, even had some regulatory activity in this model (Ono et al., 2012). Thus, Th17 cells induced by SFB may be qualitatively different from pathogenic Th17 cells induced during colitis. The lack of pathological changes in the face of induction of a “pro-inflammatory” T cell subset may reflect an evolutionary adaptation that has helped preserve SFB as part of the microbiota. This also suggests that SFB, SFB-induced Th17 cells, and other SFB immunomodulatory effects are beneficial for the host. Indeed, SFB colonization leads to improved protection against infections with intestinal pathogens, as shown for Citrobacter rodentium in mice and enteropathogenic E. coli in rabbits (Heczko et al., 2000; Ivanov et al., 2009). Th17 cells are important mediators of protection against intestinal infections, which could be the underlying mechanism of SFB-mediated protection. However, SFB have other more general immunostimulatory effects, such as induction of IgA production, general CD4 T cell and IEL accumulation (Umesaki et al., 1995), and stimulation of anti-microbial peptide production from epithelial cells (Ivanov et al., 2009). Therefore, whether the protective effects of SFB are mediated by Th17 cells remains to be established. SFB are currently the only known commensal species, and hence the only autobiont, that can induce Th17 cells, even though indirect evidence suggests that other Th17 cell-inducing commensals may exist (Atarashi et al., 2008; Ivanov et al., 2009).
Mechanisms of commensal immunomodulatory effects
As defined and discussed here, autobionts are commensal bacteria that modulate immune homeostasis by affecting the development, differentiation or effector function of different immune cell subsets. Whether these effects represent a natural immune response to the presence of the bacteria or a mutualistic adaptation has not been clearly established in all cases. However, the immune changes induced by autobionts possess several characteristics that distinguish them from conventional immune responses against pathogens or other commensals. Firstly, the response is unique for the bacteria. For example SFB is the only known commensal to induce Th17 cells. Secondly, the response itself is specialized and usually restricted to a certain immune cell subset. For example, the presence of commensal bacteria induces a general recruitment of all lymphoid subsets to the gut and maturation of secondary lymphoid structures. A pathogen invasion will induce a full cascade of pro-inflammatory immune responses, such as release of a battery of pro-inflammatory cytokines and infiltration of multiple immune cell subsets. In contrast, B. fragilis predominantly induces IL-10 production from Tregs, Clostridia from clusters IV and XIVa induce Foxp3+ Tregs, and SFB induce specifically Th17 cells. Most importantly, the effects of autobionts are “subtle” and do not present as overt immune changes, such as immune deficiency or inflammation. Invasive intestinal pathogens also induce Th17 cells, but in contrast to SFB, this induction is part of the general tissue damaging inflammatory immune response with induction and infiltration of multiple inflammatory subsets. Thus, the effects of autobionts are fundamentally different from those of pathogens or innocuous commensals. Therefore, characterization of the molecular and cellular mechanisms underlying these effects is of particular interest and may be extremely relevant for the development of immunomodulatory therapies with low side effects.
Modification of Pattern-Recognition Receptor (PRR) signaling
Toll-like receptors (TLRs) and Nod-like receptors (NLRs) are crucial innate immune receptors for the general detection of bacteria. They are also crucial for the establishment of intestinal homeostasis by the microbiota. Loss of TLR signaling negatively affects intestinal epithelial cell (IEC) regeneration after injury and aggravates intestinal inflammation (Rakoff-Nahoum et al., 2004). Ligation of TLRs by commensals may also inhibit immune activation by IECs through various mechanisms (Cerf-Bensussan and Gaboriau-Routhiau, 2010). Deficiencies in PRR signaling also control homeostasis at the level of microbiota composition. Indeed, genetic deficiencies of Nod1, Nod2, Nlrp3, and Nlrp6 have all been shown to affect microbiota homeostasis, which leads to increased disease susceptibility (Bouskra et al., 2008; Elinav et al., 2011; Henao-Mejia et al., 2012; Petnicki-Ocwieja et al., 2009). Interestingly, all these studies involved perturbations of NLR signaling. In contrast, a recent comprehensive study demonstrated that loss of TLR signaling does not affect microbiota composition and that the distinct microbiota composition of TLR-deficient mice is due to housing isolation rather than defective innate immunity (Ubeda et al., 2012). Regardless of its effects on microbiota composition, TLR activation is likely to be involved in mediating immunomodulatory effects of commensals in general. The effects of B. fragilis and PSA on Th1 and Treg function require TLR2 signaling (Round et al., 2011). TLR2 expression seems to be required on Tregs themselves in vitro, although whether this is the case in vivo remains to be investigated (Round et al., 2011). In contrast, induction of Tregs by the 46 strains of Clostridia occurred normally after colonization of GF Myd88−/− mice, which lack the signaling adaptor for several TLRs (Atarashi et al., 2011), although whether it is controlled by other PRRs has not been investigated. Whether TLR-signaling is involved in the induction of Th17 cells by SFB, is not completely clear. Th17 cells are still present in normal numbers in the LP of SPF Myd88−/− mice, as well as in Myd88−/−Trif−/− mice, which lack both TLR signaling adaptors and therefore all TLR signaling. or Rip2−/− mice which lack downstream Nod1/2-signaling (Atarashi et al., 2008; Ivanov et al., 2009; Ivanov et al., 2008). However, full genome sequencing of SFB has revealed that SFB harbor flagellar assembly proteins with flagellins capable of binding to TLR5 (Kuwahara et al., 2011). Moreover, there are reports that TLR signaling is important for intestinal Th17 cell generation. The numbers of Th17 cell-bound RORγt+ CD4 T cells, as identified by GFP reporter expression, were decreased in the absence of MyD88 (Shaw et al., 2012). In support of this, Tlr9−/− mice have decreased numbers of LP Th17 cells (Hall et al., 2008). An additional complication is potential heterogeneity of responses through different TLRs mediated by different commensals. For example, it is possible that activation of some TLRs inhibits, while activation of other TLRs promotes Th17 cell differentiation. In this scenario, ablation of signaling through both activating and inhibitory TLRs in Myd88−/− mice will not lead to considerable changes in Th17 cell number. The precise role of PRRs in the Th17 cell inductive capacity of total microbiota and specific commensals, such as SFB, awaits controlled colonization experiments.
Modification of LP immune cell function and antigen specificity
Autobionts are non-invasive members of the luminal microbiota, however they affect immune cell function in the LP and even systemically. In most cases, the affected host cells are not in direct contact with the bacteria. Rather, they interact with bacterial products or are affected by the activity of bacteria-detecting cells. The nature of the immune cells involved in inducing Th17 or Treg cells by commensals is an area of active investigation. Naïve T cells differentiate into effector cells upon appropriate activation by a cognate antigen in the context of an appropriate cytokine environment. Therefore, T cell modulating commensals, such as SFB and Clostridia, must specifically affect one, or both, of these processes. The specificity can be provided by induction of a unique cytokine environment, appropriate level of TCR stimulation by commensal-deriver antigens, or both. These signals can be transmitted to T cells by the activation of unique subsets of immune cells in the LP. Dendritic cells (DCs) are major modulators of T cell responses, because they can serve as potent antigen-presenting cells (APCs) and at the same time contribute to the local cytokine environment through detection of microbial substances by their PRRs. The intestinal LP harbors distinct subsets of intestinal DCs (iDCs) and several of them have been implicated in regulating intestinal T cell homeostasis and in particular the Th17/Treg balance (Bogunovic et al., 2012; Varol et al., 2010). iDCs can produce the major cytokines involved in Th17 and Treg differentiation, including IL-6, IL-23, and TGF-β, and have been directly implicated in regulating T cell homeostasis (Swiatczak and Rescigno, 2012). Indeed iDCs have been shown to induce Treg and Th17 cell differentiation in vitro. CD103+ iDCs induce Treg differentiation in vitro through the production of the vitamin A-derived nuclear hormone receptor agonist retinoic acid (RA) (Coombes et al., 2007; Sun et al., 2007). A number of different iDC subsets have been shown to induce Th17 cell differentiation in vitro, including CD70hi, TLR5+, and CD11b+CD103+ DCs (Atarashi et al., 2008; Denning et al., 2011; Uematsu et al., 2008). However, the exact contribution of any of these iDC subsets, or even iDCs in general for Th17 or Treg induction in vivo is not completely clear. Probably the most direct data come from studies of DC-specific deletion of various molecules. DC-specific deletion of Notch2 leads to a specific loss of the CD103+CD11b+ iDC subset with corresponding decrease in Th17, but not Treg, cell differentiation, which seems to implicate this subset in Th17 rather than Treg induction (Lewis et al., 2011). iDCs express the integrin αvβ8, which is an important activator of intestinal TGF-β, a cytokine crucial for both Th17 and Treg differentiation. DC-specific deletion of integrin αv or β8 leads to significant disruption in Treg and Th17 cell differentiation and development of colitis (Acharya et al., 2010; Lacy-Hulbert et al., 2007; Paidassi et al., 2011; Travis et al., 2007). In addition, DC-specific loss of the transcription factors T-bet and Stat3, the cytokine IL-10, and the Wnt signaling mediator β-catenin, all lead to disrupted Th17/Treg balance and chronic colitis (Garrett et al., 2007; Manicassamy et al., 2010; Melillo et al., 2010). Therefore, although a role for iDCs in regulating gut T cell homeostasis seems obvious, the exact contribution of different DC subsets in vivo will need to be examined further. In contrast, the role of iDCs in microbiota-mediated immune effects is not known. Even though CD103+ iDCs express RA-producing enzymes and are capable of promoting Treg induction in vitro, whether the in vivo effects of autobionts, such as Clostridia or B. fragilis are mediated through iDCs remains to be investigated. In the case of Th17 cells, SFB colonization or colonization with other Th17 cell-inducing bacteria stimulates production of Th17 cell-regulating cytokines, e.g. IL-23, IL-6, and TGF-β from LP DCs (Atarashi et al., 2008; Ivanov et al., 2009). CD103+CD11b+ DCs isolated from mice with Th17 cell-inducing microbiota, were the only gut DC subset capable of inducing Th17 cells in vitro (Denning et al., 2011) and this was dependent on microbiota effects, because the same cells isolated from Jackson B6 mice (that lack SFB and Th17 cells) were deficient in Th17 cell induction. The exact role of DCs in SFB-mediated induction of Th17 cells in vivo remains to be established.
LP DCs may mediate T cell induction by commensals in several ways. They may sample commensal-derived antigens, either by directly contacting the bacteria through extension of dendrites into the gut lumen (Rescigno et al., 2001), or by detecting bacterial products that gain access to the LP. They may then present these antigens to intestinal T cells to induce commensal-specific T cells. Alternatively, LP DCs may be conditioned by commensals indirectly, e.g. by cytokines produced by other cells in response to the bacteria. For example, SFB attachment may induce cytokine production by IECs that may modulate DC function. Indeed, SFB induce serum amyloid A (SAA) production from IECs, which in turn may stimulate DC production of IL-23 (He et al., 2006; Ivanov et al., 2009), a cytokine required for Th17 cell maintenance.
Another important question that will help shed light into autobiont-specific mechanisms is: what are the antigen specificities of autobiont-induced Th17 and Treg cells? If autobionts provide antigens to induce autobiont-specific T cells, identification of the APCs involved can help identify cells that receive signals from the commensal as well as how commensals or their antigens are detected by the immune system. At the same time, characterization of Treg or Th17 cell-inducing autobiont-derived antigens will identify potential commensal genes involved in the process, which may help pave the path for design and development of microbial products as immunomodulatory therapeutics. On the other hand, if most autobiont-induced T cells are not specific for the bacteria, autobionts are mostly affecting cytokine environment in the gut. A recent study using high-throughput sequencing demonstrated that a large proportion of inducible colonic Tregs recognize commensal-derived antigens (Lathrop et al., 2011). It has also been shown that Foxp3+ Tregs with TCRs that are specific for flagellins related to those of Clostridia cluster XIVa contribute to the induction of IgA+ B cells in the intestine (Cong et al., 2009). Still, it is currently not known whether Tregs induced by commensal Clostridia are mostly specific for Clostridial antigens. In the case of Th17 cells, one study found that Th17 cells still develop in the gut of non-commensal TCR-transgenic (Tg) mice, which argues that presentation of commensal antigens is not required for Th17 cell induction (Lochner et al., 2011). Surprisingly, T cell activation and Th17 cell differentiation still occurred in the absence of cognate antigen (Lochner et al., 2011), which may reflect potential cross-reactivity of the Tg TCRs to unknown endogenous or commensal-derived antigens. In contrast, in another study, transferred activated TCR Tg T cells did not differentiate into Th17 cells in the LP (Hand et al., 2012). More importantly, these studies were performed in mice with undefined commensal microbiota, which may contain multiple Th17 cell-inducing species, including pathobionts such as Helicobacter spp (Muller and Solnick, 2011). Therefore, the role of individual Th17 cell-inducing commensals, such as SFB, has not been investigated and whether they preferentially induce commensal-specific Th17 cells is not known.
Modification of intestinal epithelial cell (IEC) function
IECs form a single cell barrier that separates the microbiota from host tissues and immune cells. IECs also represent a cell type to which both microbiota and immune cells have direct access and may, therefore, play a major role in transmitting microbiota’s immunomodulatory signals. There are multiple examples of how commensal bacteria affect IEC function. Signals from microbiota are constantly being processed by IECs to maintain proper barrier function (Hooper and Macpherson, 2010; Rakoff-Nahoum et al., 2004). At the same time IEC produced cytokines are crucial in modulating LP immune responses (He et al., 2007; Saenz et al., 2008). Exactly how these two types of events are connected is not completely clear. However, it is logical to imagine a scenario where immunomodulatory commensals modify IEC function either by directly binding to IECs or by secreting substances that ligate IEC receptors on the apical side. Following IEC activation, IEC-derived cytokines will be secreted on the basolateral side to influence the activity of LP immune cells, such as DCs, that in turn will affect T cell homeostasis. Such sequence of events is highly likely especially for mucosa-associated autobionts. SFB are a prototypical example. SFB colonize the intestine of many animal species in the post-weaning period. The majority of microbiota are kept away from direct contact with the epithelium by a layer of mucus enriched in anti-microbial substances (Johansson et al., 2008; Vaishnava et al., 2011). SFB differ from most commensal in that they can penetrate the mucosal layer and adhere tightly to IECs in the terminal ileum as well as to follicle-associated epithelium (FAE) of Peyer’s Patches (Klaasen et al., 1992). The host and bacterial molecules involved in this adherence are unknown. However the interaction resembles ligation of a surface receptor on the IEC apical site by a bacterial adhesin. Actin cytoskeletal reorganization is seen in the IEC at the point of contact (Jepson et al., 1993), which implies initiation of downstream signaling events and modification of IEC function by the adhering commensal. Indeed, SFB are known to affect gene-expression in IECs, including MHC class II expression, surface fucosylation, and anti-microbial peptide production (Ivanov et al., 2009; Umesaki et al., 1995). They also seem to induce cytokine-like molecules such as SAA, which can modify the function of LP cells (Ivanov et al., 2009). Whether adherence to IECs or any of these effects on IEC function are required for the Th17 cell-inductive capacity of SFB is not yet known. SFB interaction with IECs may also bring the bacteria in close proximity to sampling cells, such as M cells in PPs and intestinal villi or interdigitating DCs (Rescigno et al., 2001). In any case, the strong association of SFB with IECs is a unique feature of this commensal and is likely to be involved in its immunomodulatory effects. Whether other mucosa-associated commensals also have strong immunomodulatory effects remains to be investigated.
Modification of host metabolic functions
Autobionts may influence the balance of intestinal immune cells by generating immunomodulatory metabolites. Microbiota is integral for the generation of dietary metabolites (Holmes et al., 2012) and plays major role in metabolic conditions, such as obesity (Turnbaugh et al., 2006), metabolic syndrome (Vijay-Kumar et al., 2010), and nonalcoholic fatty liver disease (Henao-Mejia et al., 2012). In addition to energy harvest, microbial metabolites have profound effects on immune responses. A comparison of sera from GF and conventional mice by mass spectrometry revealed that the microbiota play critical roles in metabolizing tryptophan, phenylalanine and tyrosine, and producing tryptophan metabolites such as indoxyl sulfate and the antioxidant indole-3-propionic acid (IPA), and other organic acids containing phenyl groups, such as phenyl sulfate (Wikoff et al., 2009). Importantly, these aromatic hydrocarbons are known to influence the differentiation of T cells and ILCs (Qiu et al., 2012; Veldhoen et al., 2008). The aryl hydrocarbon receptor (AhR) is a receptor for tryptophan derivatives and has been implicated in immune regulation by tryptophan metabolites. AhR is expressed in differentiating Th17 cells, and AhR ligands upregulate IL-22 expression and boost Th17 cell differentiation (Veldhoen et al., 2008). Other reports have shown that AhR activation can result in control of pro-inflammatory responses and expansion of the Treg compartment (Gandhi et al., 2010; Quintana et al., 2008). Thus, the control of Th17 and Treg differentiation by AhR seems to be complex. AhR is also expressed by IL-22-producing ILCs and IELs, and AhR−/− mice have fewer IL-22-producing ILCs and IELs in the intestine (Li et al., 2011; Qiu et al., 2012). Tryptophan metabolism in general, may be important for T cell homeostasis. In this context, it is noteworthy that IECs from mice colonized with 46 strains of Clostridia express high levels of indoleamine 2,3-dioxygenase (IDO) (Atarashi et al., 2011), an enzyme that degrades tryptophan and has been implicated in Treg induction (Matteoli et al., 2010). Furthermore, it has been shown that uptake of dietary amino acids, including tryptophan, by IECs via the amino acid transporter B0AT1 complexed with angiotensin-converting enzyme 2 (ACE2) is essential for the production of antimicrobial peptides and control of gut microbial ecology (Hashimoto et al., 2012). Therefore, it will be interesting to address the involvement of amino acid metabolites in commensal-mediated immune modulation.
Bacteroidetes and Clostridia employ a wide variety of enzymes capable of degrading polysaccharides and oligosaccharides into short chain fatty acids (SCFAs), such as acetic acid, propionic acid, and butyric acid (Flint et al., 2008). SCFAs influence host physiology, including gut motility and colonocyte development. In addition, SCFAs contribute to maintenance of intestinal immune homeostasis. Indeed, butyrate metabolism is impaired in patients with ulcerative colitis, and topical sodium butyrate treatments or butyrate enemas are effective therapies (Scheppach et al., 1992; Vernia et al., 2003). GF mice are highly susceptible to dextran sulfate sodium (DSS)-mediated colitis, but treatment of GF mice with acetate in drinking water markedly improves colitis disease indices (Maslowski et al., 2009). SCFAs are known to act as extracellular signaling molecules that activate G-protein coupled receptors, such as GPR41 and GPR43. GPR41 is expressed by a subset of IECs and controls microbiota effects on energy harvest and obesity (Samuel et al., 2008). GPR43 is expressed by neutrophils and eosinophils, and transmits microbial-derived immunomodulatory signals. Gpr43−/− neutrophils have an intrinsic hyper-reactive phenotype, including hyper-production of reactive oxygen species (ROS), and high chemotactic activity (Maslowski et al., 2009). It is not known whether SCFA-signaling is involved in the induction of Th17 cells and Tregs by autobionts. However, considering that Bacteroidetes and Firmicutes are high producers of SCFAs (Flint et al., 2008), SCFA-GPR signaling may represent an important molecular pathway activated by commensals to regulate immune and inflammatory responses and maintain the mucosal barrier.
Therapeutical use of intestinal microbes or their products/effects
Alteration of the gut microbiota in humans via fecal transplants of stool from healthy donors has been effective in a large number of case studies of refractory C. difficile infection (Khoruts and Sadowsky, 2011). While fecal transplantation has established a proof of principle for the feasibility of manipulating human microbiota as a therapeutic strategy (Khoruts and Sadowsky, 2011), treatment with specific microorganisms is more desirable for medical purposes. Many of the conventional probiotic microorganisms were traditionally used to preserve food products by fermentation and are not necessarily part of the normal microbiota (O’Toole and Cooney, 2008). In other words, no probiotic has been rationally isolated from the microbiota with the purpose of boosting specific arms of the host immune system or to fix a particular commensal dysbiosis. As described earlier, several lines of evidence suggest that dysbiotic microbiota can be a highly stable complex and therefore refractory to treatment with individual transient probiotic strains. Indeed, despite promising data from animal models, most probiotics tested to date have demonstrated, at best, mediocre effects in the clinic, particularly for treatment of IBD. Thus, there is compelling need to identify more robust therapeutic organisms that are compatible to the host and can affect the host immune system in a well-controlled, physiological fashion. Ideally, these organisms will be able to induce broader changes to the microbial ecosystem and have the ability to ameliorate dysbiosis (Figure 1). Such therapeutic organisms may include some traditional probiotics, but immunomodulatory autobionts represent the most promising candidates. Autobionts are capable of becoming permanent members of the microbiota and therefore may be able to restore dysbiosis and inhibit the growth of pathogens or pathobionts. Indeed, SFB colonization can reduce replication of enteropathogenic E. coli in rabbits (Heczko et al., 2000), Salmonella enteritidis in rats (Garland et al., 1982), and Citrobacter rodentium in mice (Ivanov et al., 2009). Colonization with indigenous Clostridia and Lactobacilli isolated from conventional mice was shown to effectively control E. coli expansion in the large intestine of mice (Itoh and Freter, 1989). At the same time, autobionts have focused and independent effects on the immune system that are relatively more subtle and aimed at sustaining intestinal immune homeostasis. SFB and Clostridia induce controlled expansion of Th17 cells or Tregs independently of each other and even in GF mice, and the induced Th17 or Treg cells do not exceed the levels in SPF mice and do not cause overt inflammatory disease or immune deficiency. Therefore, the mechanisms controlling autobiont-induced immune effects are more “physiological”. Characterization of the cellular and molecular mechanisms responsible for the effects in each example of autobiont-host interaction is crucial in designing therapies that will not rely on colonization with live bacteria, but instead use specific small molecules which mimic their beneficial effects. For example, even though SFB have not yet been found in human metagenomic databases (Sczesnak et al., 2011) the molecular and cellular mechanisms of SFB immune modulation may still be preserved and taken on by other commensals in humans. Characterization of such molecular mechanisms will allow for development of rationally designed combinations of targeting agents to boost the immune system in a desired way or correct effects of intestinal dysbiosis to treat various immune diseases.
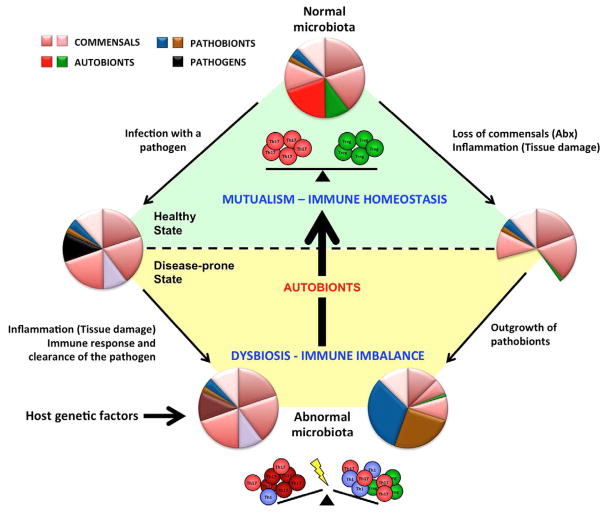
Figure 1. Mutualistic commensals with immunomodulatory effects sustain healthy immune homeostasis.
Autobionts are permanent members of the normal commensal microbiota. They control immune homeostasis in the lamina propria by, for example, inducing different subsets of effector T cells (control of T cell homeostasis). The relative proportions of Th17 and Treg cells depend on the relative presence of different autobionts, e.g. in different individuals, different intestinal locations, or at different stages in ontogeny. These mutualistic interactions sustain the healthy steady state. Loss of autobionts and general dysbiosis perturbs also the immune balance of the host. Dysbiosis may occur in multiple ways. Invasive intestinal pathogens cause transient infections, but may lead to longterm perturbations of the microbiota due to strong inflammatory responses against the pathogen. Antibiotic (Abx) treatments or inflammation caused by physical damage to the mucosa may also lead to dysbiosis and the outgrowth of pathobionts, which are permanent members of the microbiota, but do not cause disease in the presence of autobionts. Host genetic factors may also initiate or perpetuate dysbiosis. Dysbiosis leads to loss of the immunomodulatory effects of autobionts and results in a perturbed immune balance, which under appropriate conditions may manifest itself in disease. The disease state augments dysbiosis in a vicious circle. Autobionts and conventional probiotics are both mircoorganisms with beneficial effects. Probiotics have transient effects and can boost host immunity. Autobionts are part of the normal microbiota and have developed evolutionary adaptations to colonize the host, regulate host immunity and establish a healthy immune state. Autobionts can therefore reverse dysbiosis as well as immune homeostasis.
Acknowledgments
This work was supported by grants to K.H. from Japan Science and Technology Agency for CREST, the Japan Society for the Promotion of Science NEXT program, and the Waksman Foundation of Japan Inc., and by grants to I.I.I. from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the Crohn’s and Colitis Foundation of America (CCFA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharya M, Mukhopadhyay S, Paidassi H, Jamil T, Chow C, Kissler S, Stuart LM, Hynes RO, Lacy-Hulbert A. alphav Integrin expression by DCs is required for Th17 cell differentiation and development of experimental autoimmune encephalomyelitis in mice. The Journal of clinical investigation. 2010;120:4445–4452. doi: 10.1172/JCI43796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M, Mortha A, Muller PA, Merad M. Mononuclear phagocyte diversity in the intestine. Immunol Res. 2012 doi: 10.1007/s12026-012-8323-5. [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G, Masetti R, Ricci G, Pession A, Brigidi P. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. Journal of immunology. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature reviews Microbiology. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, Rubino S, Robertson SJ, Larijani M, Gosselin J, Ivanov II, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, Kozoriz D, Weiner HL, Quintana FJ. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland CD, Lee A, Dickson MR. Segmented filamentous bacteria in the rodent small intestine: their colonization of growing animals and possible role in host resistance to Salmonella. Microb Ecol. 1982;8:181–190. doi: 10.1007/BF02010451. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M, Maynard CL, Elson CO, 3rd, Belkaid Y. Acute Gastrointestinal Infection Induces Long-Lived Microbiota-Specific T Cell Responses. Science. 2012 doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- He R, Shepard LW, Chen J, Pan ZK, Ye RD. Serum amyloid A is an endogenous ligand that differentially induces IL-12 and IL-23. J Immunol. 2006;177:4072–4079. doi: 10.4049/jimmunol.177.6.4072. [DOI] [PubMed] [Google Scholar]
- Heczko U, Abe A, Finlay BB. Segmented filamentous bacteria prevent colonization of enteropathogenic Escherichia coli O103 in rabbits. J Infect Dis. 2000;181:1027–1033. doi: 10.1086/315348. [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, Nicholson JK. Therapeutic modulation of microbiota-host metabolic interactions. Sci Transl Med. 2012;4:137rv136. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Freter R. Control of Escherichia coli populations by a combination of indigenous clostridia and lactobacilli in gnotobiotic mice and continuous-flow cultures. Infect Immun. 1989;57:559–565. doi: 10.1128/iai.57.2.559-565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Mitsuoka T. Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Lab Anim. 1985;19:111–118. doi: 10.1258/002367785780942589. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jepson MA, Clark MA, Simmons NL, Hirst BH. Actin accumulation at sites of attachment of indigenous apathogenic segmented filamentous bacteria to mouse ileal epithelial cells. Infection and immunity. 1993;61:4001–4004. doi: 10.1128/iai.61.9.4001-4004.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–489. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- Khoruts A, Sadowsky MJ. Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol. 2011;4:4–7. doi: 10.1038/mi.2010.79. [DOI] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS microbiology reviews. 1992;8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, Itoh T, Nakayama-Imaohji H, Ichimura M, Itoh K, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy-Hulbert A, Smith AM, Tissire H, Barry M, Crowley D, Bronson RT, Roes JT, Savill JS, Hynes RO. Ulcerative colitis and autoimmunity induced by loss of myeloid alphav integrins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15823–15828. doi: 10.1073/pnas.0707421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, Ng D, Klinakis A, Charo IF, Jung S, Gommerman JL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Lochner M, Berard M, Sawa S, Hauer S, Gaboriau-Routhiau V, Fernandez TD, Snel J, Bousso P, Cerf-Bensussan N, Eberl G. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J Immunol. 2011;186:1531–1537. doi: 10.4049/jimmunol.1001723. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- Manicassamy S, Reizis B, Ravindran R, Nakaya H, Salazar-Gonzalez RM, Wang YC, Pulendran B. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science. 2010;329:849–853. doi: 10.1126/science.1188510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoli G, Mazzini E, Iliev ID, Mileti E, Fallarino F, Puccetti P, Chieppa M, Rescigno M. Gut CD103+ dendritic cells express indoleamine 2,3-dioxygenase which influences T regulatory/T effector cell balance and oral tolerance induction. Gut. 2010;59:595–604. doi: 10.1136/gut.2009.185108. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Melillo JA, Song L, Bhagat G, Blazquez AB, Plumlee CR, Lee C, Berin C, Reizis B, Schindler C. Dendritic cell (DC)-specific targeting reveals Stat3 as a negative regulator of DC function. Journal of immunology. 2010;184:2638–2645. doi: 10.4049/jimmunol.0902960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Solnick JV. Inflammation, immunity, and vaccine development for Helicobacter pylori. Helicobacter. 2011;16(Suppl 1):26–32. doi: 10.1111/j.1523-5378.2011.00877.x. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2:99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttal GHF, Thierfeledr F. Thierisches Leben ohne Bacterien im Verdauungskanal. Z Physiol Chem. 1895–1896;21:109–121. [Google Scholar]
- O’Toole PW, Cooney JC. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip Perspect Infect Dis. 2008;2008:175285. doi: 10.1155/2008/175285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Kanai T, Sujino T, Nemoto Y, Kanai Y, Mikami Y, Hayashi A, Matsumoto A, Takaishi H, Ogata H, et al. T-helper 17 and Interleukin-17-Producing Lymphoid Tissue Inducer-Like T Cells Make Different Contributions to Colitis in Mice. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.07.108. [DOI] [PubMed] [Google Scholar]
- Paidassi H, Acharya M, Zhang A, Mukhopadhyay S, Kwon M, Chow C, Stuart LM, Savill J, Lacy-Hulbert A. Preferential expression of integrin alphavbeta8 promotes generation of regulatory T cells by mouse CD103+ dendritic cells. Gastroenterology. 2011;141:1813–1820. doi: 10.1053/j.gastro.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nature reviews Microbiology. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers JA, Trexler PC, Ervin RF. Rearing germ-free albino rats. Lobund reports. 1946:1–84. [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR, Jr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, Cua D, Di Santo JP, Eberl G. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature immunology. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, Richter F, Dusel G, Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, Littman DR, Ivanov II. The genome of Th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host & Microbe. 2011;10:1–13. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC, Collins MD. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. International journal of systematic bacteriology. 1995;45:780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979–1988. doi: 10.1053/j.gastro.2009.02.074. [DOI] [PubMed] [Google Scholar]
- Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatczak B, Rescigno M. How the interplay between antigen presenting cells and microbiota tunes host immune responses in the gut. Seminars in immunology. 2012;24:43–49. doi: 10.1016/j.smim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, Reizis B, Melton AC, Masteller E, Tang Q, Proctor JM, Wang Y, Bernstein X, Huang X, Reichardt LF, et al. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. The Journal of experimental medicine. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, Nishiyama M, Sato S, Tsujimura T, Yamamoto M, Yokota Y, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nature reviews Immunology. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Vernia P, Annese V, Bresci G, d’Albasio G, D’Inca R, Giaccari S, Ingrosso M, Mansi C, Riegler G, Valpiani D, et al. Topical butyrate improves efficacy of 5-ASA in refractory distal ulcerative colitis: results of a multicentre trial. Eur J Clin Invest. 2003;33:244–248. doi: 10.1046/j.1365-2362.2003.01130.x. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]