The immune receptor FLAGELLIN SENSING2 (FLS2) plays important roles in plant resistance to bacterial pathogens and is internalized from the plasma membrane via the endocytic pathway. Ligand-activated FLS2 traffics into late endosomal compartments, and this process requires the function of components distinct from nonactivated FLS2 that recycles between early endosomes and the plasma membrane.
Abstract
The activity of surface receptors is location specific, dependent upon the dynamic membrane trafficking network and receptor-mediated endocytosis (RME). Therefore, the spatio-temporal dynamics of RME are critical to receptor function. The plasma membrane receptor FLAGELLIN SENSING2 (FLS2) confers immunity against bacterial infection through perception of flagellin (flg22). Following elicitation, FLS2 is internalized into vesicles. To resolve FLS2 trafficking, we exploited quantitative confocal imaging for colocalization studies and chemical interference. FLS2 localizes to bona fide endosomes via two distinct endocytic trafficking routes depending on its activation status. FLS2 receptors constitutively recycle in a Brefeldin A (BFA)–sensitive manner, while flg22-activated receptors traffic via ARA7/Rab F2b– and ARA6/Rab F1–positive endosomes insensitive to BFA. FLS2 endocytosis required a functional Rab5 GTPase pathway as revealed by dominant-negative ARA7/Rab F2b. Flg22-induced FLS2 endosomal numbers were increased by Concanamycin A treatment but reduced by Wortmannin, indicating that activated FLS2 receptors are targeted to late endosomes. RME inhibitors Tyrphostin A23 and Endosidin 1 altered but did not block induced FLS2 endocytosis. Additional inhibitor studies imply the involvement of the actin-myosin system in FLS2 internalization and trafficking. Altogether, we report a dynamic pattern of subcellular trafficking for FLS2 and reveal a defined framework for ligand-dependent endocytosis of this receptor.
INTRODUCTION
Receptor-mediated endocytosis (RME) plays important roles in development, growth, and pathogen defense, regulating the levels and distribution of cell surface receptors (Geldner and Robatzek, 2008; Kleine-Vehn and Friml, 2008; Frei dit Frey and Robatzek, 2009; Sorkin and von Zastrow, 2009). In plants, these include members of the Leu-rich repeat receptor-like kinase (RLK) family, such as BRASSINOSTEROID INSENSITIVE1 (BRI1), which is responsible for brassinosteroid hormone signaling (Li and Chory, 1997); FLAGELLIN SENSING2 (FLS2), which mediates the perception of a conserved domain of bacterial flagellin (Gómez-Gómez and Boller, 2000; Chinchilla et al., 2006); and BRI1-ASSOCIATED KINASE1 (BAK1; also called SERK3 for SOMATIC EMBRYO RECEPTOR KINASE3), which forms a complex with BRI1 and FLS2 and is required for downstream signaling (Li et al., 2002; Chinchilla et al., 2007b). Localization studies using BRI1 fused to the green fluorescent protein (GFP) showed that BRI1-GFP is found both at the plasma membrane and in endosomal compartments. These endosomal compartments can be labeled with the endocytic tracer FM4-64 and the trans-Golgi network (TGN) and early endosome (EE) marker VHAa1 (Geldner et al., 2007). Subcellular localization of BRI1 is sensitive to Brefeldin A (BFA). BFA is an inhibitor of endosomal recycling to the plasma membrane, acting by targeting the ADP-ribosylation factor–GTP exchange factor GNOM and leading to the accumulation of so-called BFA bodies (Geldner et al., 2003; Robinson et al., 2008). Plasma membrane and BFA-sensitive endosomal localization patterns were also described for BAK1/SERK3 (Russinova et al., 2004), SERK1 (Kwaaitaal et al., 2005), and the non-Leu-rich repeat RLK CRINKLY4 (ACR4) (Gifford et al., 2005; Karlova et al., 2006), demonstrating that many described receptors undergo constitutive recycling. Coexpression of BRI1 and BAK1/SERK3 in protoplasts revealed distinct endosomal populations either containing both receptors together or each individually (Russinova et al., 2004). This highlights that RME is directed through different endosomal compartments, and intracellular receptor localization is transient in nature and depends on the maturation of these highly dynamic membrane compartments along the endocytic pathways (Robatzek, 2007; Geldner and Robatzek, 2008; Irani and Russinova, 2009). The nature of these different endosomes and the extent to which they contribute to the receptor’s function remain mostly unknown.
FLS2 functions as an important cell surface receptor in plant immunity against bacterial infection and triggers plant defenses following elicitation by bacterial flagellin or the flagellin-derived peptide flg22 (Zipfel et al., 2004). Impairment of FLS2 internalization correlates with altered flg22 responses (Robatzek et al., 2006; Salomon and Robatzek, 2006; Chinchilla et al., 2007a). Thus, it is essential to identify the key steps of the endocytic trafficking pathway of FLS2 that can regulate aspects of plant immunity. Endosomal markers for FLS2 compartments and a dissection of the FLS2 endosomal trafficking pathway have not been described so far. FLS2-GFP is translocated from the plasma membrane into small mobile vesicles only when flg22 is applied, unlike BRI1 endocytic trafficking (Geldner et al., 2007), which is constitutive and independent of its ligand brassinosteroid (Robatzek et al., 2006). The internalization of FLS2 requires BAK1/SERK3 and can be abolished by single amino acid substitutions in the FLS2 kinase domain that may be subject to posttranslational modifications, such as phosphorylation and ubiquitination (Robatzek et al., 2006; Salomon and Robatzek, 2006; Chinchilla et al., 2007b). Flg22-induced subcellular trafficking is sensitive to Wortmannin (Wm), a phosphatidylphosphate-3-kinase inhibitor that interferes with vesicle formation from the plasma membrane and the maturation of late endosomes (LEs) and multivesicular bodies (MVBs), resulting in their enlargement (Tse et al., 2004; Wang et al., 2009). Considering that flg22-activated FLS2-GFP signals decrease over time and that FLS2 internalization is insensitive to BFA, ligand-induced FLS2 endosomal trafficking must occur in a different pathway to that of BRI1. Notably, most RLKs, including BRI1 and BAK1, carry the conserved tetrapeptide YxxΦ motif in their cytoplasmic domains, which are known to mediate the endocytosis of cell surface proteins (Geldner and Robatzek, 2008; Reyes et al., 2011). This motif is not present in FLS2, providing additional evidence for differences in endosomal trafficking between receptors (Geldner and Robatzek, 2008; Reyes et al., 2011).
In plants, the occurrence of endocytic trafficking is well established. A key example is root cell polarity, which is based on the asymmetric localization of the PIN FORMED (PIN) proteins (Kleine-Vehn and Friml, 2008). Plasma membrane–resident PIN proteins enter an endocytic trafficking pathway in a clathrin-dependent manner, localize to TGN/EE compartments, and recycle through a BFA-sensitive pathway, resulting in their polar localization at one side of the cell (Geldner et al., 2001, 2003; Dhonukshe et al., 2007). PIN proteins can also enter endocytosis for vacuolar degradation (Petrásek et al., 2006; Kleine-Vehn et al., 2008; Laxmi et al., 2008); thus, endosomes resemble a population of varying distinct compartments transiently present in specific trafficking pathways. Endocytosis of PIN proteins depends on ARA7/Rab F2b (Dhonukshe et al., 2008), and members of this Rab5 GTPase family are known to label distinct endosomal compartments (Ueda et al., 2001; Ueda and Nakano, 2002; Pfeffer, 2005). In Arabidopsis thaliana, the Rab5 homologs ARA6/Rab F1 and ARA7/Rab F2b localize to different endosomal populations with a small degree of overlap (Ueda et al., 2004; Ebine et al., 2011). ARA6/Rab F1 localizes mainly to LE, while ARA7/Rab F2b is found at both EE and LE compartments, thus representing suitable markers for these endosomal populations (Ueda et al., 2004; Ebine et al., 2011). Defining the endosomal pathways of cell surface proteins is important for understanding their function, in particular in ligand-induced RLK signaling.
In this study, we defined the endocytic trafficking pathway of the A. thaliana FLS2 receptor in the absence of flg22 ligand and upon flg22-induced activation. The transient behavior of FLS2 endocytosis raises technical challenges for studying this process in detail. To address this issue, we employed cutting-edge quantitative, high-throughput, live-cell imaging techniques and developed EndomembraneQuantifier and EndomembraneCoLocQuantifier algorithms and software implementations for quantifying and identifying colocalized small membrane objects, respectively. With EndomembraneCoLocQuantifier, we were able to follow the spatial and temporal dynamics of FLS2 subcellular trafficking in detached Arabidopsis cotyledons by evaluating endosomal numbers over time compared with those of ARA6/Rab F1 and ARA7/Rab F2b endosomal markers. In addition, experiments involving FM4-64 colocalization and membrane trafficking inhibitors showed that activated FLS2 receptors traffic along the endosomal pathway and mature into LE/MVB compartments in a BFA-insensitive manner. By contrast, FLS2 recycles via a BFA-sensitive endocytic route in the absence of flg22. Overall, our study demonstrates that trafficking of FLS2 is subject to targeted endosomal sorting that is dependent on the activation status of the receptor.
RESULTS
Nonactivated FLS2 Constitutively Recycles via a BFA-Sensitive Endocytic Pathway in a BAK1-Independent Manner
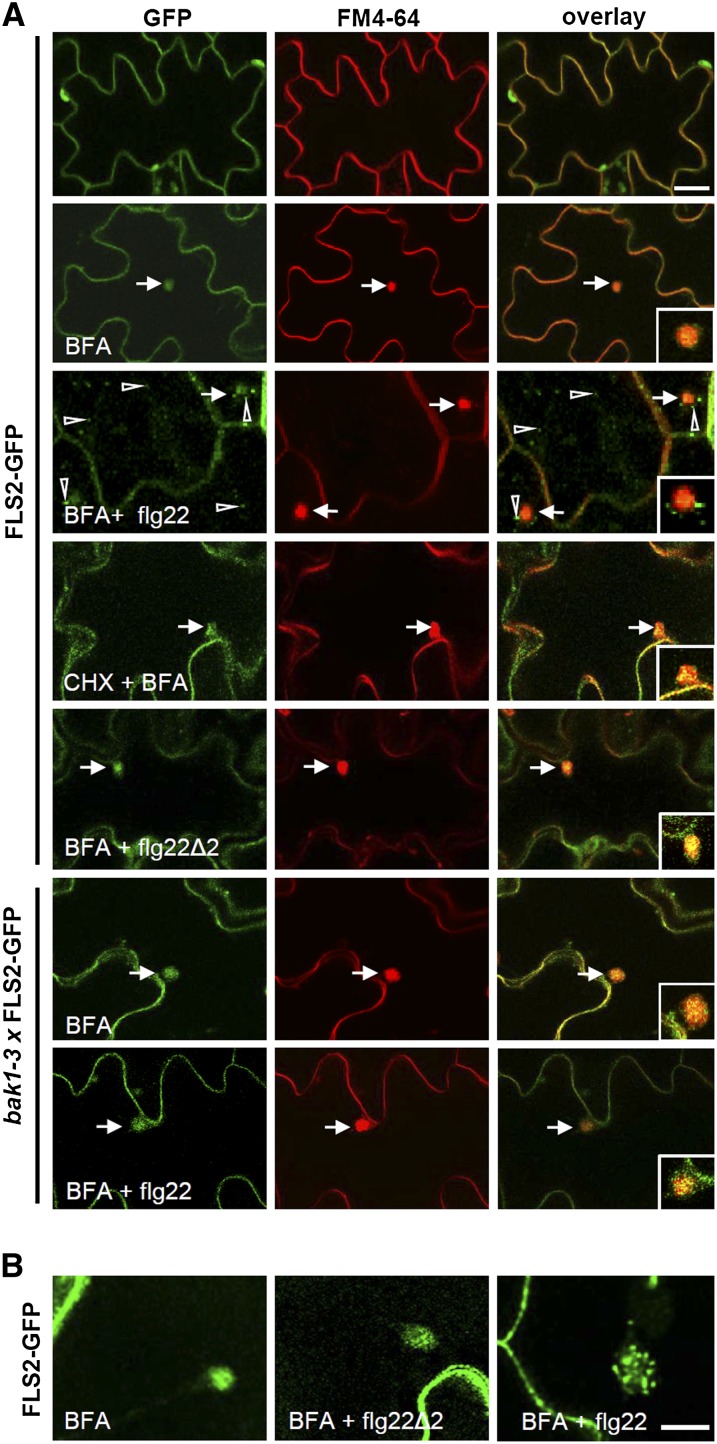
Plasma membrane–resident FLS2 accumulates into mobile vesicles upon elicitation with its ligand flg22 (Robatzek et al., 2006), but in its nonactivated state, FLS2-GFP is mainly present at the cell surface where it colocalized with the lipophilic dye FM4-64, which stains the plasma membrane (Figure 1A) (Bolte et al., 2004). Because the constitutive recycling of plasma membrane proteins is a well-known process in plant cells and an essential component of their function and regulation, we examined whether FLS2-GFP recycles between the plasma membrane and TGN/EE compartments. BFA inhibitor treatments affect secretory pathways, and in both root and cotyledon cells cause the accumulation of recycled proteins and TGN/EE but not LE or MVBs in BFA bodies (see Supplemental Figure 1 online; Langhans, et al., 2011). Despite the absence of flg22, FLS2-GFP accumulated in BFA bodies as revealed by FM4-64 costaining as a positive marker for plasma membrane–derived EE in Arabidopsis cotyledons (Figure 1A). Inhibition of protein synthesis by cyclohexamide treatments (see Supplemental Figure 2 online) did not interfere with BFA body accumulation of FLS2-GFP (Figure 1A). As previously described, BFA treatment did not inhibit flg22-induced FLS2 endocytosis (Robatzek et al., 2006), and some endosomes showing FLS2-GFP signals upon flg22 treatment did not colocalize with BFA bodies (Figures 1A and 1B). Instead, we observed that flg22-induced FLS2-GFP vesicles localized around the BFA body, similar to patterns of known markers labeling LE compartments in the presence of BFA (Figures 1A and 1B; see Supplemental Figure 1 online). When treating with BFA and the antagonistic peptide flg22Δ2, which binds but does not activate FLS2 (Bauer et al., 2001), FLS2-GFP accumulated solely in BFA bodies and no additional FLS2-GFP vesicles were present (Figures 1A and 1B). Furthermore, FLS2-GFP recycling is independent of its coreceptor BAK1. BFA body accumulation occurred in the bak1-3 mutant background (Chinchilla et al., 2007b), whether samples were treated with flg22 or not (Figure 1A). These data show that nonactivated FLS2 receptors are subject to constitutive endocytic cycling between the plasma membrane and EE compartments regardless of BAK1 and that flg22-activated FLS2 receptors are likely transported from BFA-insensitive EE compartments to LE/MVBs.
Figure 1.
Nonactivated FL2-GFP Recycles between the Plasma Membrane and Endosomal Compartments.
Standard confocal micrographs of the indicated Arabidopsis transgenic lines expressing FLS2-GFP show optical sections of cotyledon epidermis.
(A) FLS2-GFP subcellular localization in the presence of the indicated chemicals, ligands, and genotypes. Costaining with FM4-64 highlights the plasma membrane and BFA bodies. Arrows indicate BFA bodies, arrowheads FLS2 endosomes; inset images show details of BFA bodies. Bar = 20 µm; bar in inset = 5 µm.
(B) Spinning disc confocal images of FLS2-GFP subcellular localization upon BFA treatment show clear differences in BFA formation in the absence and presence of flg22Δ2 or flg22. Bar = 10 µm.
Flg22-Induced FLS2 Vesicles Are Bona Fide Endosomes and Colocalize with LE Compartments
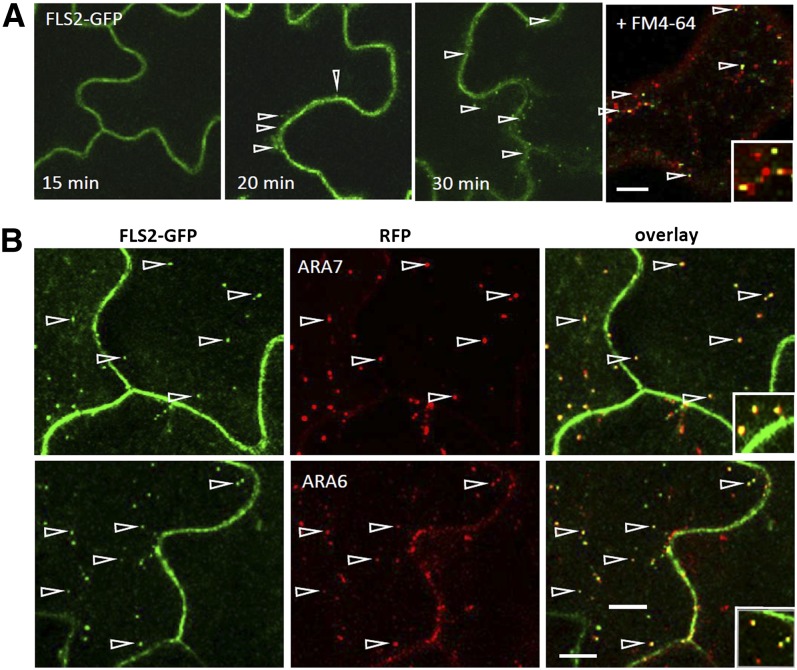
To determine the identity of the flg22-induced FLS2 vesicles, we investigated colocalization of FLS2 compartments with known endosomal markers. To further examine the utility of the endocytic tracer FM4-64 in this study, we established that time-dependent staining of different endocytic compartments by this dye is comparable in Arabidopsis cotyledons (see Supplemental Figure 3 online) to that observed in protoplasts and roots (Bolte et al., 2004; Jelínková et al., 2010). To trace FLS2-GFP compartments, we treated cotyledons of Arabidopsis seedlings expressing a functional FLS2-GFP with flg22 for 15 min to induce RME (Göhre et al., 2008) and stained the cotyledons with FM4-64. FLS2-GFP vesicles colocalized with FM4-64–labeled compartments after 30 min of flg22 treatment (Figure 2A). As FM4-64 can only label vesicles that originate from the plasma membrane, this demonstrates that FLS2-GFP is internalized via endocytic uptake and localized to bona fide endosomes. Colocalization experiments using FLS2-GFP lines crossed into red fluorescent protein (RFP)-ARA7/Rab F2b and ARA6/Rab F1-RFP–expressing plants revealed considerable overlap of these LE markers with FLS2-GFP endosomes after 30 to 50 min of flg22 elicitation (Figure 2B), confirming that activated FLS2 receptors are targeted to LE compartments.
Figure 2.
FLS2-GFP Differentially Colocalizes to Known Endosomal Markers.
Standard confocal micrographs of Arabidopsis transgenic lines expressing FLS2-GFP show optical sections of cotyledon epidermis untreated or treated with flg22.
(A) Flg22-induced FLS2-GFP internalization in a time-dependent manner and costaining with the endocytic tracer FM4-64. Arrowheads point at FLS2 endosomes. Bar = 10 µm.
(B) FLS2-GFP colocalization with ARA6/Rab F1-RFP and RFP-ARA7/Rab F2b endosomal markers in crossed transgenic lines. Arrowheads point at colocalizing endosomes. FLS2-GFP is shown in green, FM4-64, ARA6/Rab F1-RFP, and RFP-ARA7/Rab F2b in red, and the overlay indicating colocalization in yellow. Inset pictures show details (10 × 10 µm). Bar = 10 µm.
Activated FLS2 Traffics via ARA6/Rab F1– and ARA7/Rab F2b–Positive Endosomal Compartments in a Time-Dependent Manner
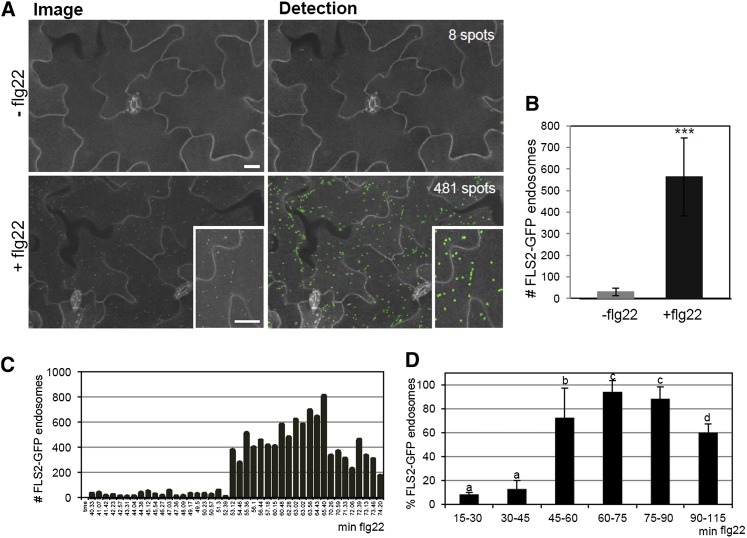
Flg22-induced FLS2 endocytosis is a transient phenomenon. To determine the precise dynamics of its subcellular trafficking pathway, we performed detailed time-course analysis. We applied our recently established high-throughput confocal imaging techniques, which successfully allow the quantification of spot-like structures in images of Arabidopsis leaves (Salomon et al., 2010). We developed a new image analysis algorithm called EndomembraneQuantifier, which subtracts background signals (e.g., those derived from chloroplasts, deeper tissues, and guard cells), and detects genuine endomembrane compartment signals with high accuracy (Figures 3A and 3B; see Supplemental Figure 4 online and Methods). This improves extensively upon our previously described endomembrane script (Salomon et al., 2010), which suffered from discovering redundant background signals when monitoring FLS2-GFP endocytosis (Chinchilla et al., 2007b; Salomon et al., 2010). Among the new features in EndomembraneQuantifier, we included codes to use image files acquired with two channels (GFP and chlorophyll autofluorescence) and to record the time of measurement, which enables the temporal analysis of image data sets (see Supplemental Data Set 1 and Supplemental Table 1 online). We applied quantitative imaging to monitor flg22-induced FLS2 endocytosis over time. FLS2-GFP endosomes appeared 30 to 45 min after flg22 application and their numbers increased significantly during time, reaching a maximum at 60 to 75 min (Figures 3C and 3D). FLS2-GFP endosomal numbers then decreased over prolonged incubation times of 90 to 115 min (Figures 3C and 3D). This demonstrates that these endosomes are dynamic and transient.
Figure 3.
Time Dependency of flg22-Induced FLS2-GFP Endocytosis Measured by Quantitative Imaging.
(A) High-throughput confocal micrographs of Arabidopsis transgenic lines expressing FLS2-GFP show cross sections of cotyledon epidermis untreated or treated with flg22 (image) and the respective computational spot detection. Inset pictures show details of FLS2-GFP endosome detection. Bars = 30 µm.
(B) Quantification of FLS2-GFP endosomal numbers per image area in the absence and presence of flg22. Error bars represent sd; control n = 78, flg22 n = 140 images. Asterisks indicate statistical significance of P value ≤ 0.001 based on Student’s t test analysis.
(C) FLS2-GFP endosomal numbers per image area in response to flg22 treatment over time. Example of one experiment; each time point depicts one well (n = 2 to 4 per time point, total analyzed images per plate, n = 155).
(D) Percentage of flg22-induced FLS2-GFP endosomes over time. Spot maximum per experiment was set to 100%, and the percentage of spot number per time interval compared with the maximum was calculated; 15 to 30 min, n = 157; 30 to 45 min, n = 250; 45 to 60 min, n = 369; 60 to 75 min, n = 237; 75 to 90 min, n = 202; 90 to 115 min, n = 86 images per different time interval. In total, 1301 images were analyzed; error bars = sd (four biological replicates); a, b, and c represent statistical significance (P value ≤ 0.05) between pairwise tested groups based on Student’s t test.
[See online article for color version of this figure.]
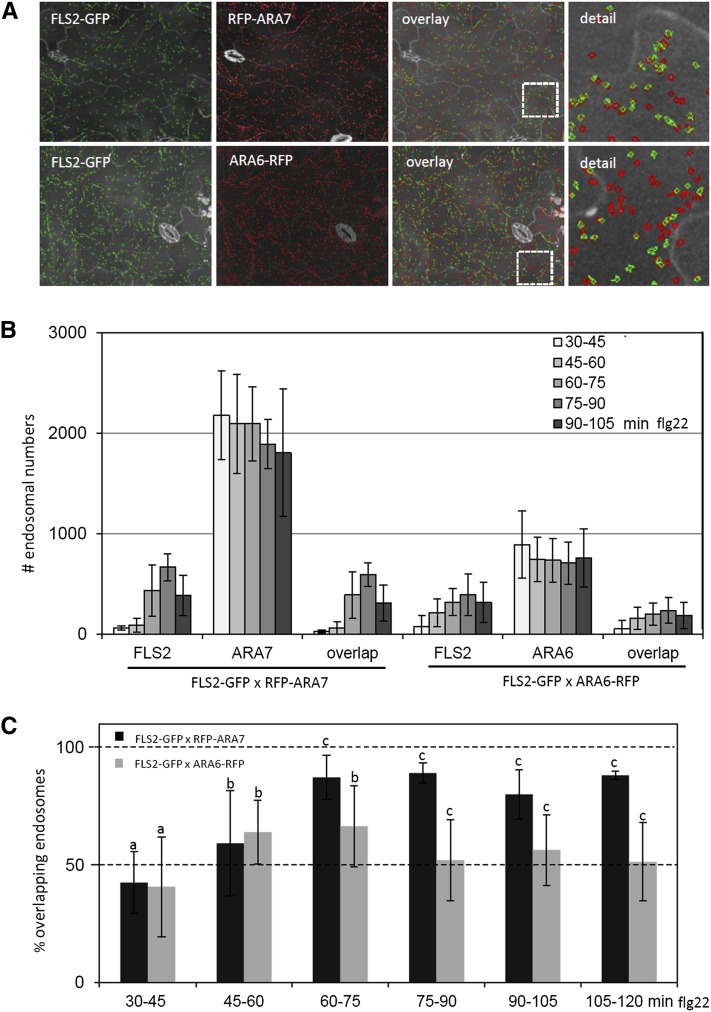
To compare FLS2 trafficking to that of known endosomal markers, we crossed plants expressing FLS2-GFP to those expressing RFP-ARA7/Rab F2b or ARA6/Rab F1-RFP and developed the EndomembraneCoLocQuantifier script to analyze the individual and overlapping endosomal numbers of GFP- and RFP-labeled endosomes (Figure 4). The EndomembraneCoLocQuantifier algorithm includes a code to use image files acquired with three channels (GFP, RFP, and autofluorescence signals) (see Supplemental Data Set 2 and Supplemental Table 2 online). These features allow the detection of spot-like objects labeled with two different fluorophores using the coordinates of the detected objects in the two separated channels to make counts of colocalized objects. In this algorithm, the objects are superimposed and all objects in one channel that converge with objects in the second channel are considered colocalized (see Supplemental Figure 5 online). Manual reinspection of images using GFP markers in channel one and RFP markers in channel two confirmed that overlapping GFP- and RFP-labeled compartments were precisely and robustly identified by the procedure. EndomembraneCoLocQuantifier was applied to maximum projections of 21 planes taken 1 µm apart and to consecutive projections of three planes each 1 µm apart. There was no significant difference between the percentage of overlapped spots identified in a maximum projection compared with those identified in the three plane analyses (see Supplemental Figures 6A and 6B online). However, object detection was more precise and efficient based on maximum projections, which was therefore chosen for our high-throughput colocalization quantification. Since imaging 21 planes for both GFP and RFP signals occurred consecutively and was associated with a time lapse of 120 ms between each plane, the position of endosomes differed slightly between planes; thus, we considered endosomes to be colocalized when their outlines either merged completely or simply intersected (see Supplemental Figure 6C online).
Figure 4.
Flg22-Induced FLS2-GFP Endosomes Colocalize to ARA6/Rab F1– and ARA7/Rab F2b–Positive Compartments in a Time-Dependent Manner.
(A) High-throughput confocal micrographs of Arabidopsis FLS2-GFP transgenic lines expressing RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP show computational spot detection of cross sections of the cotyledon epidermis treated with flg22. FLS2-GFP endosomes are shown in green and ARA6/Rab F1-RFP and RFP-ARA7/Rab F2b endosomes are shown in red, in single and overlay images.
(B) Quantification of endosomal numbers in percentage of total image area of the FLS2-GFP × RFP-ARA7/Rab F2b and FLS2-GFP × ARA6/Rab F1-RFP lines upon flg22 elicitation over time (FLS2-GFP × RFP-ARA7 in total n = 72, n = 7 to 10 per time point), FLS2-GFP × ARA6-RFP in total n = 125, n = 15 to 22 per time point; error bar represents sd.
(C) Percentage of FLS2-GFP, ARA6/Rab F1-RFP, and RFP-ARA7/Rab F2b endosomes over time flg22 treatment (FLS2-GFP × RFP-ARA7 in total n = 72, n = 7 to 10 per time point), FLS2-GFP × ARA6-RFP in total n = 125, n = 15 to 22 per time point; error bar represents sd; a, b, and c represent statistical significance (P value ≤ 0.05) between pairwise tested groups based on Student’s t test.
Quantification of the endosomal markers revealed significantly more RFP-ARA7/Rab F2b- than ARA6/Rab F1-RFP–labeled compartments (Figure 4B). This is in agreement with previously observed quantitative differences between distinct endomembrane compartments and the localization patterns described for ARA7/Rab F2b and ARA6/Rab F1 (Salomon et al., 2010; Ebine et al., 2011). Importantly, transcript levels of both UBQ10 promoter-driven RFP-tagged ARA7/Rab F2b and ARA6/Rab F1 are comparable between the transgenic lines, and differences in RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP protein abundances are likely to reflect differences in the respectively labeled endosomal compartments (see Supplemental Figure 7 and Supplemental Methods online). Furthermore, FLS2-GFP is expressed at similar levels in these crossed RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP lines.
We triggered FLS2-GFP endocytosis with flg22 and determined time-dependent colocalization with RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP using quantitative imaging. No significant differences in RFP-ARA7/Rab F2b endosomal numbers were detected during flg22 elicitation (Figure 4B). At 60 to 75 min, when FLS2 endocytosis was at its maximum, ∼90% of the FLS2-GFP endosomes were classified as colocalized with RFP-ARA7/Rab F2b compartments (Figure 4C). This level of colocalization remained relatively stable over time and showed no significant decrease up to 120 min upon flg22 elicitation. By contrast, a significantly lower proportion of FLS2-GFP endosomes was classified as colocalized with ARA6/Rab F1-RFP compartments. About 60% of the FLS2-GFP and ARA6/Rab F1-RFP signal colocalized in the same endosomes from 30 to 45 min to 60 to 75 min after flg22 elicitation (Figure 4C). This proportion then dropped to 50% and remained at this level until 120 min. These data demonstrate that during endocytic trafficking, ∼50% of activated FLS2 receptors are constantly in close association with LE compartments.
Flg22-Induced FLS2 Endosomal Trafficking Is Differentially Affected by the Inhibitors Tyrphostin A23, Endosidin 1, Wortmannin, and Concanamycin A
Perturbation by known chemical inhibitors is a well-established approach to probe the endomembrane trafficking in a controlled manner and can overcome limitations frequently encountered by genetic analysis of membrane trafficking mutants (Drakakaki et al., 2009; Hicks and Raikhel, 2010; Drakakaki et al., 2011). We used a series of inhibitors combined with quantitative imaging to further dissect the flg22-induced FLS2 endocytic pathway. As our colocalization studies revealed FLS2 at LE compartments, we considered inhibitors that can interfere with LE endocytic trafficking. Prolonged incubation with the phosphatidylphosphate-3-kinase inhibitor Wm caused an enlargement of FLS2-GFP endosomes (see Supplemental Figure 8A online). Since Wm additionally affects LE/MVBs by inducing homotypic fusions and thereby enlarging these compartments (see Supplemental Figure 8A online; Wang et al., 2009), the observed increase in FLS2-GFP endosomal size further confirms FLS2 trafficking via LE compartments. Another effect of Wm is the inhibition of endocytic uptake at the plasma membrane (Emans et al., 2002; Ito et al., 2012). Treatment with Wm also resulted in about a 70% decrease in flg22-induced FLS2-GFP endosomal numbers (Figure 5; see Supplemental Figure 9 online), which shows that FLS2 internalizes from the plasma membrane and is in agreement with previous findings (Robatzek et al., 2006). Another well-established membrane trafficking inhibitor is Concanamycin A (ConcA), which is known to block vacuolar transport (Dettmer et al., 2006). ConcA targets the vacuolar ATPase activity at the TGN/EE, which is required for MVB/LE biogenesis (Scheuring et al., 2011). When treated with ConcA, the number of flg22-induced FLS2-GFP endosomes increased more than twofold on average (Figure 5; see Supplemental Figure 9 online). Colocalization studies with FM4-64 and FLS2-GFP in the presence of ConcA revealed clustering of these endosomal compartments around small roundish structures (see Supplemental Figure 8B online). Taken together, these data suggest that the flg22-induced FLS2 endosomes mature into LE/MVBs, which likely target the activated FLS2 receptor for vacuolar degradation.
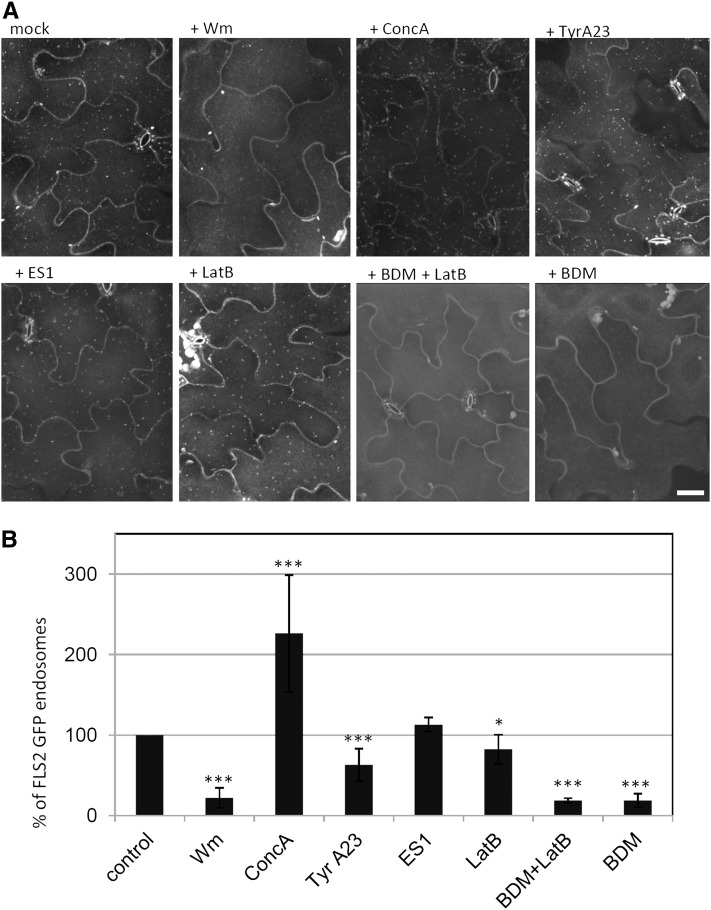
Figure 5.
Chemical Interference of flg22-Induced FLS2 Endosomes.
(A) High-throughput confocal micrographs of Arabidopsis FLS2-GFP transgenic lines show maximum projections of cotyledon epidermis treated with flg22 for 50 to 60 min and the indicated chemical inhibitors. Bar = 30 µm.
(B) Quantification of endosomal numbers in percentage of total image area of flg22-induced FLS2-GFP endosomes in the presence of the indicated chemical inhibitors. In total, 1306 images were analyzed (control n = 616, Wm n = 85, ConcA n = 71, TyrA23 n = 100, ES1 n = 38, LatB n = 154, BDM + LatB n = 105, BDM n = 137); the mean of three biological replicates is depicted; error bars represent sd; asterisks indicate statistical significance of *P value ≤ 0.05, **P value ≤ 0.01, and ***P value ≤ 0.001 based on Student’s t test.
Next, we focused on inhibitors known to interfere with RME. Tyrphostin A23 (TyrA23) is a well-described inhibitor of clathrin-mediated endocytosis in protoplasts, roots, and leaves affecting the endocytosis of PIN proteins and cell surface receptors (Ortiz-Zapater et al., 2006; Dhonukshe et al., 2007; Konopka et al., 2008; Kaiserli et al., 2009). Surprisingly, TyrA23 did not fully block flg22-induced FLS2-GFP endocytosis, but instead reduced FLS2-GFP endosomal numbers to 60% (Figures 5A and 5B). Endosidin 1 (ES1) was also reported to interfere with RME and to specifically inhibit endocytic trafficking of BRI1, PIN2, and AUX1 but not PIN1 and PIN7 (Robert et al., 2008). Similar to TyrA23, ES1 treatments did not block internalization of FLS2-GFP, but in contrast with TyrA23 did not affect endosomal numbers (Figures 5A and 5B). However, we observed that ES1 application reduced the mobility of the FLS2-GFP endosomes (see Supplemental Figure 10A online). This is reminiscent of the reduced vesicle trafficking by alterations of the cytoskeleton (see Supplemental Figure 10A online) (Voigt et al., 2005b). We therefore examined transgenic lines expressing GFP-ACTIN BINDING DOMAIN, which labels the actin cytoskeleton in the presence of ES1. We detected strong bundling and evidence of depolymerization of the actin cytoskeleton (see Supplemental Figure 10A online). Consistent with our data, ES1 was recently identified as an actin stabilizing molecule (Tóth et al., 2012).
We further addressed the role of the actin cytoskeleton on FLS2 endocytosis using the known actin cytoskeleton inhibitor Latrunculin B (LatB) (Voigt et al., 2005a), which exhibited only a minor impact on FLS2-GFP internalization (Figures 5A and 5B). This result is in contrast with previous findings (Robatzek et al., 2006), and we therefore tested whether this effect could be due to the different tissues (true leaves versus cotyledons) and experimental conditions used in these studies. Nonetheless, no actin filaments were visible after a 1-h incubation with LatB in both cotyledons and primary leaves, revealing no significant differences between these tissues (see Supplemental Figure 10B online). Although FLS2 was internalized in the presence of LatB, the FLS2-GFP endosomes remained close to the plasma membrane and immobile as detected by high-resolution imaging (see Supplemental Figure 10 online). A number of recent studies showed that the activity of myosins is involved in the dynamics of plasma membrane processes (Konopka et al., 2008; Sattarzadeh et al., 2008), which led us to investigate their role during internalization. Treatment with the general myosin inhibitor 2,3-butanedione monoxime (BDM) (Radford and White, 2011) reduced the number of flg22-induced FLS2-GFP endosomes by ∼80% compared with the control. Thus, the BDM-mediated inhibition of the activity of myosins was sufficient to block FLS2 internalization from the plasma membrane and provides evidence for myosin-dependent endocytic events (Figure 5).
Trafficking Inhibitors Differentially Alter FLS2, ARA7/Rab F2b, and ARA6/Rab F1 Endosomes and Affect Their Overlap
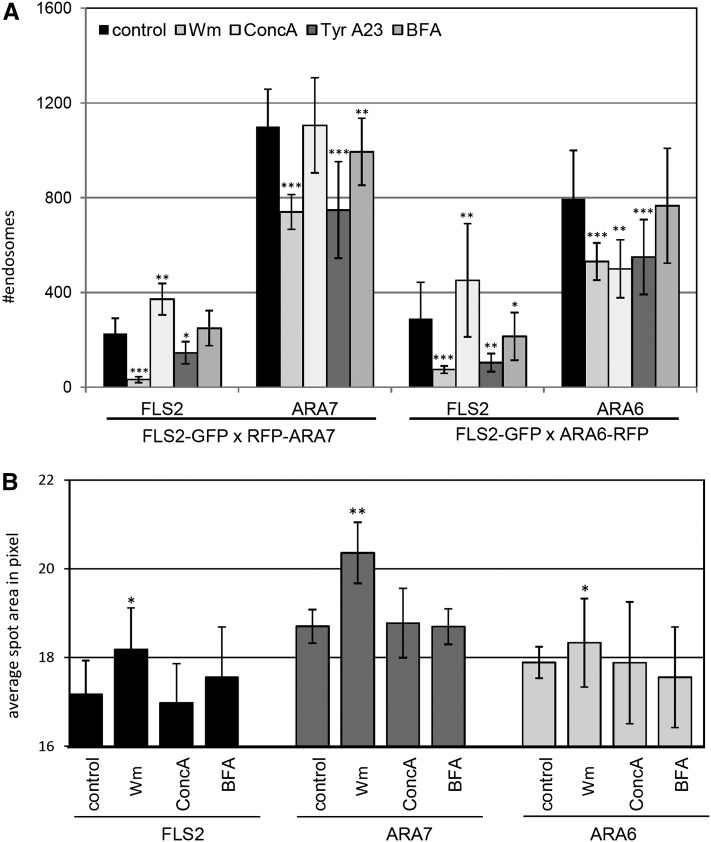
To further dissect the effects of chemical interference of membrane trafficking and FLS2 endocytosis, we combined inhibitor treatments with colocalization of the ARA7/Rab F2b and ARA6/Rab F1 endosomal markers by quantitative imaging (Figure 6), which was supported by qualitative assessment of the images for possible alterations in endosome morphology (see Supplemental Figure 11 online). This approach provides a useful tool to differentiate between the effects of chemical inhibition on ligand-induced RME and steady state endosomal populations within the same cell. As for FLS2-GFP transgenic plants, overall similar patterns of FLS2-GFP endosomal numbers in response to inhibitor treatments were obtained in the crossed dual marker lines that express FLS2-GFP and RFP-ARA7/Rab F2b or FLS2-GFP and ARA6/Rab F1-RFP (Figure 6A; see Supplemental Figure 11 online). In the presence of Wm, a decrease of ∼30% in RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP endosomal numbers was observed (Figure 6A). This coincided with enlarged endosomal areas for FLS2-GFP endosomes as well as RFP-ARA7– and ARA6-RFP–labeled endosomes (Figure 6B; see Supplemental Figure 11 online), which is in agreement with the described effects of Wm on the maturation of LE, and confirms that both ARA7/Rab F2b and ARA6/Rab F1 can be found at LE compartments. This enlargement was neither found in BFA- nor ConcA-treated cells and shows the specific effect of Wm on the morphology of LE (Figure 6B). For ConcA treatment, the distinct endosomal populations revealed differential effects. Although FLS2-GFP numbers were increased in both dual-color lines, ConcA did not affect the number of RFP-ARA7/Rab F2b endosomes, but it did cause a decrease in ARA6/Rab F1-RFP endosomal numbers (Figure 6A). This could be attributed to a block of endosomal trafficking from the TGN/EE to LE/MVB compartments (Scheuring et al., 2011).
Figure 6.
Quantification of flg22-Induced FLS2, ARA7/Rab F2b, and ARA6/Rab F1 Endosomes upon Chemical Interference.
(A) Percentage of FLS2-GFP, RFP-ARA7/Rab F2b, and ARA6/Rab F1-RFP endosomal numbers per 100% image area upon 30 to 75 min flg22 elicitation and in the presence of the indicated chemical inhibitors. Error bars represent sd; n = 20 to 44 images.
(B) Average spot area of FLS2-GFP, ARA6/Rab F1-RFP, and RFP-ARA7/Rab F2b endosomal numbers per image upon 40 to 90 min flg22 elicitation and in the presence of the indicated chemical inhibitors. Error bars represent sd; n = 10 to 20 images; asterisks indicate statistical significance of *P value ≤ 0.05, **P value ≤ 0.01, and ***P value ≤ 0.001 based on Student’s t test.
While flg22-induced FLS2-GFP endosomes remained mostly unaffected by BFA treatment, we detected a decrease of ∼10% of RFP-ARA7/Rab F2b endosomes following BFA treatment. By contrast, the number of ARA6/Rab F1-RFP endosomes was not affected in the presence of BFA. This suggests that a small but significant proportion of the ARA7/Rab F2b compartments comprise EE subject to endocytic recycling (Figure 6A). Interestingly, TyrA23 treatment caused a 30% decrease in FLS2-GFP endosomes, similar to that observed in RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP endosomal numbers (Figure 6A). This indicates that a significant proportion of these compartments are generated via AP2-dependent clathrin-mediated endocytic trafficking. When comparing the effect of TyrA23 on FLS2-GFP internalization between cotyledons and primary leaves, we did not detect any significant difference (see Supplemental Figure 12 online). Although we cannot generally exclude tissue-specific differences, these data suggest similar modes of action of chemical inhibition in cotyledons used in our study compared with published data mostly obtained from root meristem and protoplast cells and to some extent from leaves (Ortiz-Zapater et al., 2006; Dhonukshe et al., 2007; Robert et al., 2008; Drakakaki et al., 2011).
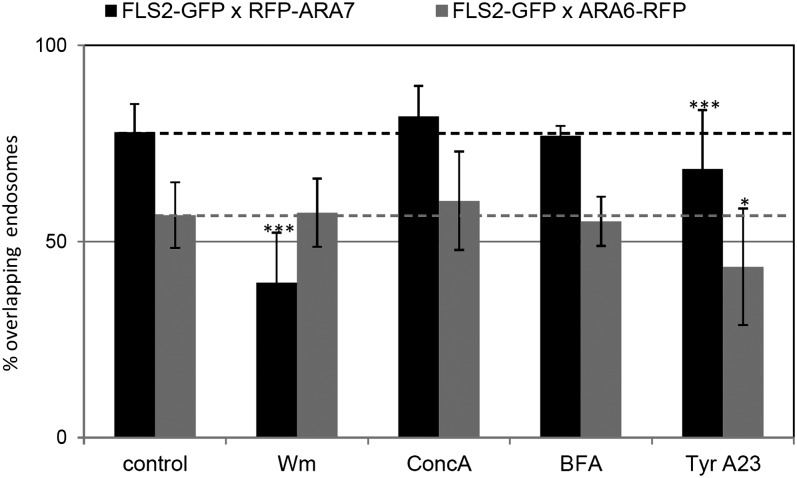
Based on these quantitative data, we determined the degree of overlap between FLS2-GFP and both RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP endosomes. Wm treatment affected the overlap of FLS2-GFP and RFP-ARA7/Rab F2b endosomal numbers, which was significantly reduced to ∼55% of colocalization, while the overlap between FLS2-GFP and ARA6/Rab F1-RFP endosomal numbers remained unaltered (Figure 7). Although Wm treatment decreased the numbers of both ARA7/Rab F2b and ARA6/Rab F1 endosomal compartments to a similar extent, the reduction was significantly higher for FLS2-GFP endosomes, possibly reflecting that FLS2 is internalized from the plasma membrane alongside a small proportion of RFP-ARA7/Rab F2b that localizes to EE. BFA and ConcA treatment did not alter the proportion of endosomal colocalization between FLS2-GFP and both markers (Figure 7). As BFA treatment did not alter FLS2-GFP endosomal numbers, the decrease in RFP-ARA7/Rab F2b compartments (Figure 6A) likely reflects a different endosome population. In the case of ConcA treatment, despite contrasting findings of increased FLS2-GFP endosomal numbers, unaltered RFP-ARA7/Rab F2b endosomal numbers, and decreased ARA6/Rab F1 endosomal numbers, the overlap between these compartments was not significantly affected (Figure 7). Treatment with TyrA23 leads to a small but significantly reduced overlap between FLS2-GFP and RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP endosomes, respectively (Figure 7). This is in line with the detected reduction in endosomal numbers of all three markers in the presence of TyrA23.
Figure 7.
Quantification of flg22-Induced FLS2 Endosomes Colocalizing to ARA7/Rab F2b– and ARA6/Rab F1–Positive Compartments upon Chemical Interference.
Percentage of overlap between FLS2-GFP, ARA6/Rab F1-RFP, and RFP-ARA7/Rab F2b endosomal numbers upon 30 to 75 min flg22 and chemical inhibitor treatments. Dashed lines relate to control values. Error bars represent sd; n = 10 to 20 images per time point. In total, between 90 and 120 images were analyzed. Asterisks indicate statistical significance of *P value ≤ 0.05, **P value ≤ 0.01, and ***P value ≤ 0.001 based on Student’s t test.
The Function of ARA7/Rab F2b GTPase Is Required for flg22-Induced FLS2 Endocytosis
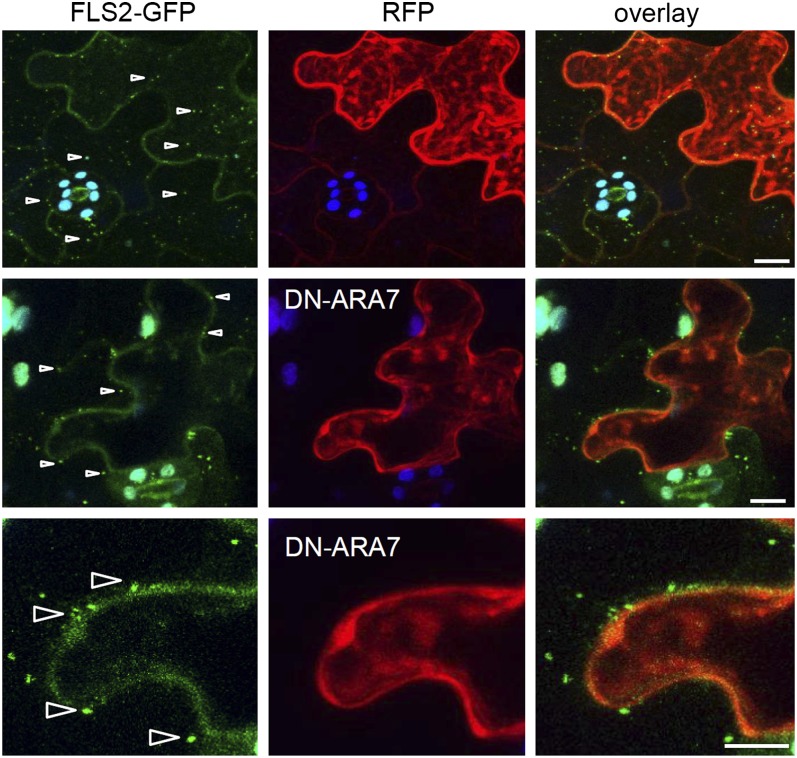
To investigate the significance of activated FLS2 trafficking via ARA7/Rab F2b–positive compartments, we studied flg22-induced endocytosis of FLS2-GFP in the presence of dominant-negative ARA7/Rab F2b (RFP-DN-ARA7/Rab F2b). Transient expression of RFP-DN-ARA7/Rab F2b was previously shown to reduce endocytosis in root meristem cells and impairs PIN1 and BRI1 endocytic trafficking (Dhonukshe et al., 2006, 2008; Irani et al., 2012). For transient expression, we used particle bombardment of Arabidopsis leaves and first confirmed that epidermal cells bombarded with soluble and endoplasmic reticulum–targeted monomeric RFP (mRFP) show normal plasma membrane localization of FLS2-GFP in the absence of flg22 and endocytosis of FLS2-GFP in the presence of flg22 (see Supplemental Figure 13 online). When transiently expressing RFP-ARA7/Rab F2b in FLS2-GFP plants, we detected colocalization of flg22-induced FLS2-GFP endosomes with RFP-ARA7/Rab F2b–positive compartments (see Supplemental Figure 13 online), as observed in our stable crossed lines (Figure 2). This demonstrates that endocytic trafficking of activated FLS2 is not generally influenced by bombardment-mediated transient expression, thus making this approach suitable to test for effects by DN-ARA7/Rab F2b. Epidermal cells transiently expressing RFP-DN-ARA7/Rab F2b were almost completely abolished in flg22-induced FLS2-GFP endocytosis (Figure 8). By contrast, neighboring nontransformed cells showed normal endocytosis of FLS2-GFP upon flg22 trigger. These findings revealed that activated FLS2 requires a functional Rab5 GTPase pathway for its endosomal trafficking.
Figure 8.
A Functional Rab5 GTPase Pathway Is Required for FLS2-GFP Endocytosis.
Standard confocal micrographs of Arabidopsis transgenic lines expressing FLS2-GFP show optical sections of flg22-treated leaf epidermal cells transiently expressing cytosolic RFP or dominant-negative RFP-ARA7/Rab F2b (DN-ARA7) after particle bombardment. RFP signals indicate successful transiently transformed cells. Flg22-induced FLS2-GFP endosomes are detected in RFP-expressing cells but not when DN-ARA7 is present. However, neighboring cells show FLS2-GFP endosomes. Arrowheads point at FLS2-GFP endosomes. FLS2-GFP is shown in green, RFP and RFP-DN-ARA7/Rab F2b in red, and autofluorescence from chloroplasts in blue. Similar results were obtained from eight transformed cells of at least two independent experiments. Bars = 10 µm.
DISCUSSION
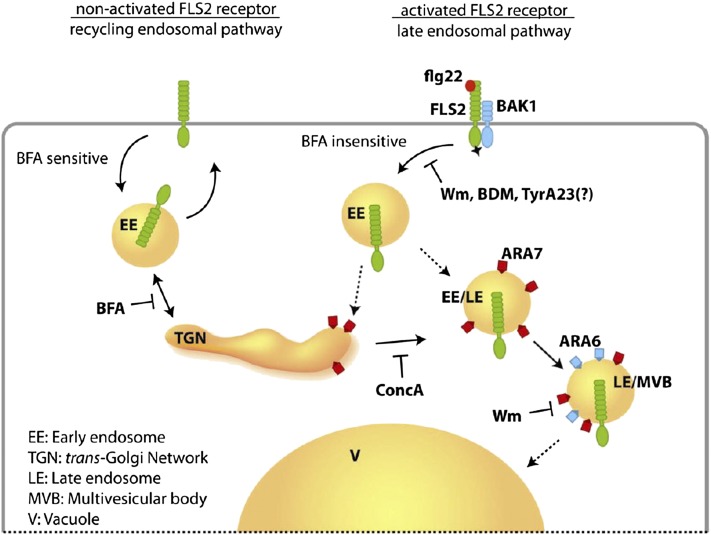
In this study, we resolved the endocytic pathway of FLS2 in an activation-state and time-dependent manner. In its nonactivated state, FLS2 enters the endocytic route and constitutively cycles between the plasma membrane and EE compartments via the common endocytic pathway under the control of GNOM, being the ADP-ribosylation factor-guanine-nucleotide exchange factor target of BFA (Geldner et al., 2003). Cycling via this common endocytic route is known for a number of plasma membrane proteins, including the PIN proteins, to mediate patterns of subcellular localization (Dhonukshe et al., 2007), and for cell surface receptors, such as BRI1, BAK1/SERK3, SERK1, and ACR4, to regulate the abundance of receptors at the plasma membrane and maintain a constant pool of signaling receptors (Russinova et al., 2004; Reyes et al., 2011). The latter may be particularly relevant for BRI1 recognizing an endogenous ligand (Geldner et al., 2007), while BAK1/SERK3 endocytic recycling may allow the retrieval of this protein from the activated receptor complex to allow de novo interactions with other ligand binding receptors (Russinova et al., 2004; Geldner and Robatzek, 2008). In the case of FLS2, endocytic recycling occurs in the absence of both its ligand and its coreceptor BAK1 and is likely linked to the maintenance of steady state levels of FLS2 at the plasma membrane. When activated by flg22, FLS2 enters the endocytic pathway and accumulates in bona fide endosomes, as revealed by FM4-64 tracing. BFA treatment does not alter FLS2 endosome numbers, and flg22-induced FLS2 endosomes appear not to be part of the BFA body, demonstrating that activated FLS2 is endocytosed via a GNOM-insensitive pathway. Therefore, our results indicate that endocytosis of the same cargo, FLS2, can occur in both a GNOM-sensitive and -insensitive manner, depending on its activation status (Figure 9), which suggests differential regulation at the transition from a plasma membrane–resident receptor complex to endosomes.
Figure 9.
Schematic Model of the FLS2 Endocytic Routes.
Depending on its activation status, FLS2 enters two distinct endosomal pathways. The nonactivated receptor follows a recycling and BFA-sensitive endosomal pathway. FLS2 receptors activated by its ligand flg22 traffic via a BFA-insensitive pathway and are sequentially transported via ARA7/Rab F2b–and ARA6/Rab F1–positive and ConcA- and Wm-sensitive endosomes to the vacuole.
[See online article for color version of this figure.]
A similar observation of a regulatory switch between the recycling and degradation pathways of endocytosis was reported for the boron exporter BOR1 (Takano et al., 2005, 2010). Under steady state conditions and low boron concentrations, BOR1 localizes primarily to the plasma membrane with a small proportion undergoing constitutive endocytic recycling. In response to high boron, BOR1 is rapidly internalized and targeted for vacuolar degradation and thereby prevents boron toxicity (Takano et al., 2005). Also, although BRI1 is mainly subject to constitutive endocytic recycling, a proportion of endocytosed BRI1 is targeted for vacuolar degradation (Geldner et al., 2007). Thus, BOR1 and BRI1 subcellular trafficking provided evidence that distinct modes of RME coexist in plants but the respective pathways remain to be defined (Jürgens and Geldner, 2007).
To date, our understanding of RLK localization is based largely on qualitative information gained from interpretation and analysis of confocal image series. To measure quantitative differences in endosomal localization patterns over time, we developed a robust and sensitive method for dissecting endocytic trafficking by defined parameters. Determining the numbers and area of endomembrane compartments over time takes into consideration the transient nature of endosomes, exemplified by induced FLS2 endocytosis (Robatzek et al., 2006). In combination with chemical interference of membrane trafficking and comparative analysis of known endosomal markers, this enables defined dissection of endocytic trafficking. Previously, ARA7/Rab F2b has been described to localize to EE/TGN as well as to LE/MVBs compartments (Ebine et al., 2011), and while ARA7/Rab F2b– and ARA6/Rab F1–positive endosomes mostly overlapped (Ueda et al., 2004; Ebine et al., 2011), ARA6/Rab F1 was more specifically associated with LE/MVBs (Ueda et al., 2004; Ebine et al., 2011). Both TyrA23 and Wm treatments reduced ARA6/Rab F1 and ARA7/Rab F2b endosomal numbers, while BFA treatment specifically reduced ARA7/Rab F2b endosomal numbers. This observation is consistent with the notion that a proportion of ARA7/Rab F2b–positive compartments are EEs that incorporate into BFA bodies, and another proportion mature into LE/MVBs, while ARA6/Rab F1 more specifically localizes to LE/MVBs (Ebine et al., 2011). These localization patterns suggest that these two Rab5 GTPases have partially redundant but distinct roles in endosomal trafficking.
Our quantitative imaging approach allows the dissection of such partially overlapping but otherwise distinct pathways in time and thus provides comprehensive insights into the dynamics of the endosomal network. Applying this methodology to dissect the FLS2 trafficking pathway, we determined that induced FLS2 endocytosis peaks at ∼60 to 75 min after induction, coincident with the maximal overlap of ARA7/Rab F2b–positive endosomes. Maximal overlap with ARA6/Rab F1–positive endosomes occurred at ∼45 to 60 min and, in contrast with the sustained overlap between ARA7/Rab F2b and FLS2, decreased by 75 to 90 min of flg22 elicitation. The large overlap between ARA6/Rab F1 and ARA7/Rab F2b compartments and their transient behavior indicates that activated FLS2 is sequentially trafficked from ARA7/Rab F2b–positive compartments to endosomes labeled by both Rab5 GTPases. Using transient expression of DN-ARA7/Rab F2b to genetically interfere with endocytic trafficking confirmed that this Rab5 GTPase pathway is required for endocytosis of activated FLS2 receptors, similar to findings with PIN1 and BRI1 (Dhonukshe et al., 2008; Irani et al., 2012). The significant reduction in number and enlargement in area of flg22-induced FLS2 endosomes upon Wm treatment shows that activated FLS2 is sorted and accumulates at LE/MVBs, in agreement with previous data (Robatzek et al., 2006). It is currently thought that activated FLS2 is degraded in a ubiquitin-dependant manner by the PUB12 and PUB13 E3 ligases (Göhre et al., 2008; Lu et al., 2011), possibly in association with vacuolar degradation (Robatzek et al., 2006; Geldner and Robatzek, 2008). Our data show that ConcA treatments increase flg22-induced FLS2 endosomes, forming clusters around small round structures, putatively of vacuolar nature. Genetic interference of PUB12/13 will be required to determine the mechanics of FLS2 degradation with respect to FLS2 endocytic trafficking.
Chemical inhibition is often employed to dissect membrane trafficking and is widely used in plant cell biology (Geldner et al., 2003; Robert et al., 2008; Drakakaki et al., 2009; von Kleist and Haucke, 2011). ES1 was found to specifically trigger the aggregation of PIN2, AUX1, and BRI1 into SYP61- and VHAa1-positive endosomes (Robert et al., 2008), but it did not alter FLS2 subcellular localization, as it does for PIN1 and PIN7. We identified that ES1 disrupts the actin cytoskeleton, which is a significant observation and a likely explanation for the reduced motility of FLS2 endosomes observed following ES1 treatment. Likewise, our refined analysis now shows that the actin depolymerizing drug LatB does not inhibit internalization and instead impairs the motility of FLS2-GFP endosomes. As a consequence, these immobile FLS2-GFP endosomes remain in close proximity to the plasma membrane, which may have been overlooked in previous studies (Robatzek et al., 2006). However, the internalization of FLS2 endosomes was arrested upon interference with the general activity of myosins by BDM treatment. This provides evidence that the actin-myosin system is critical for FLS2 endocytosis, whereby myosin functions in internalizing endosomes and actin is required for intracellular trafficking.
The differential effect of BFA and ES1 on BRI1 and flg22-induced FLS2 trafficking, respectively, shows that endocytosis of both receptors is routed via distinct endosomal pathways. This raises an interesting issue in light of the observation that BAK1/SERK3 forms a complex with BRI1 and FLS2 in a ligand-dependent manner (Russinova et al., 2004; Chinchilla et al., 2007b). Differential regulation of RME is likely associated with sequence motifs present in the cytosolic domains of the cell surface receptors (Geldner and Robatzek, 2008). FLS2 lacks the conserved endocytic motif YxxΦ, while this is present in most plant RLKs and receptor-like proteins. The role of the YxxΦ motif in regulating RME in plants was revealed by mutational analysis inhibiting the function of the tomato (Solanum lycopersicum) xylanase receptor LeEix2 (Ron and Avni, 2004) and by TyrA23-mediated inhibition of endocytic trafficking of in planta expressed human transferrin receptor, hTfR (Ortiz-Zapater et al., 2006). Although described as an inhibitor of clathrin-mediated endocytosis, and thus expected to fully block RME processes in plants (Chen et al., 2011), TyrA23 treatment only partially impaired flg22-induced FLS2 endocytosis. In plants, TyrA23 interferes with the recognition of the YxxΦ residues, a motif involved in specific cargo recruitment to clathrin-coated vesicles and known to interact with μ2 adaptin in the AP2 complex (Banbury et al., 2003; Happel et al., 2004; Chen et al., 2011). This specific YxxΦ motif is absent from FLS2 (Geldner and Robatzek, 2008). Our data suggest that in general the uptake of 30 to 40% of endocytic vesicles in the Arabidopsis cotyledon seem to depend on the μ2 adaptin-AP2 complex, and additional adaptins-AP2 complexes could be involved in mediating FLS2 endosome biogenesis. Alternatively, we cannot exclude the possibility of endocytic pathways mediated by components other than clathrin (Bandmann and Homann, 2012; Li et al., 2012). In summary, using quantitative imaging combined with subcellular markers and chemical inhibition, we established seedling cotyledons as a suitable model for studying cell biological processes in plants. We identified dynamic and transient FLS2 endosomal pathways that revealed distinct endocytic trafficking routes of nonactivated and ligand-activated FLS2 receptors. These data provide a well-defined framework for future research to investigate the role of FLS2 endocytosis in flg22-triggered responses.
METHODS
Plant Materials and Growth Conditions
The following Arabidopsis thaliana transgenic plants were used in this study (accession Columbia-0, if not otherwise indicated): pFLS2:FLS2-GFP (Göhre et al., 2008), p35S:GFP-2xFYVE (Voigt et al., 2005b), p35S:PIN1-GFP (provided by J. Friml, Ghent), p35S:CLC-GFP (provided by S. Bednarek, Madison, WI), pUBQ10:mRFP-ARA7/Rab F2b (provided by K. Schumacher, Heidelberg, Germany), pUBQ10:ARA6/Rab F1-mRFP (provided by K. Schumacher, Heidelberg), and VTI12-YFP, SYP32-YFP (Geldner et al., 2009). Dual color lines expressing FLS2-GFP and RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP, respectively, were generated by crossings. For standard confocal microscopy, seedlings were grown for 6 to 8 d on sterile half-strength Murashige and Skoog plates supplemented with 1% Suc and 0.9% phytagel under 16 h light at 22°C. For high-throughput confocal imaging, seedlings were grown for 2 weeks on soil under controlled environments (12 h light and 60% humidity). Imaging was done using cotyledons.
Chemicals and Treatments
All chemicals were purchased from Sigma-Aldrich, if not otherwise indicated, and used at the following concentrations: Wm (10 mM in DMSO, working solution 30 µM), ES1 (provided by N. Raikhel and G. Hicks, Riverside, CA; 1.13 mM in DMSO, working solution 10 µM), BFA (10 mM in ethanol, working solution 30 µM), LatB (10 mM in ethanol, working solution 30 µM), ConcA (10 mM in DMSO, working solution 10 µM), TyrA23 (30 mM in DMSO, working solution 100 µM), BDM (500 mM in water, 50 mM working solution), FM4-64 (Synapto RED, 5 mg/mL in water, working solution 1:2000), and cycloheximide (50 mM, working solution 50 µM). Detached cotyledons were vacuum infiltrated for 2 min in inhibitor solutions, followed by 60 min incubation at room temperature. Flg22 or flg22Δ2 (EZBiolab; in water, working solution 10 µM) was added to the inhibitor solutions, and imaging was done at different time points after the addition of flg22.
Particle Bombardment
For microprojectile bombardment assays, 5 μg pB7WG2.0.mRFP, pB7WG2.0.mRFPKDEL(RFP-ER; Thomas et al., 2008), mRFP-DN-ARA7 (Dhonukshe, et al., 2010), or pGreenRFP-ARA7 DNA was coated onto 1-µm gold particles and bombarded into 4- to 5-week-old leaves of pFLS2:FLS2-GFP-expressing plants using a Bio-Rad Biolistic PDS-1000/He particle delivery system. Bombardment sites were imaged 16 h after bombardment by confocal microscopy with or without flg22 treatment. For flg22 treatment, bombarded leaves were incubated for 40 to 50 min in 10 μM flg22. For each treatment, data were collected from at least two independent bombardment events and several independent plants.
Confocal Microscopy
Standard confocal laser microscopy was performed using the laser point-scanning microscope Leica SP5. GFP/FM4-64 was excited using the 488-nm argon laser, and fluorescence emissions were captured between 500 and 550 nm for GFP and between 580 and 640 nm for FM4-64. RFP was excited at 561 nm, and emission was taken between 580 and 620 nm. The sequential scan mode was used for simultaneously imaging of GFP/RFP. Images were processed using the LeicaLite and Adobe Photoshop CS4 software packages.
High-throughput confocal imaging was performed using the spinning disc automated Opera microscope (Perkin-Elmer Cellular Technologies) as described (Salomon et al., 2010). Excitation of the samples was performed at 488 nm for GFP and 561 nm for RFP; the emission spectrum for GFP was captured using the 540/570 band-pass filter, and RFP emission was detected using the 600/40 band-pass filter. The exposure time was set to 120 ms. Leaves were prepared in 96-well plates with optical glass bottoms (Greiner). Images of a consecutive series of 21 planes with a distance of 1 μm were taken and displayed as a maximum projection using Acapella software (Perkin-Elmer Cellular Technologies).
Image Processing and Data Analysis
Images taken by high content and high-throughput confocal microscopy were analyzed with the image processing software Acapella (version 2.0; Perkin-Elmer). Based on an earlier developed algorithm referred to as the Endomembrane script (Salomon et al., 2010), we designed an algorithm to robustly detect and quantify spots in one channel (GFP), which can subtract background autofluorescence signals in the same channel (EndomembraneQuantifier; see Supplemental Table 1 and Supplemental Data Set 1 online). In particular, spots were initially detected as described previously and then filtered based on their roundness, intensity, area, length, and width. The remaining spots are categorized into two groups (dark region and bright region) using a dynamic mask function (see explanation in Supplemental Figure 4 online). After that, the script calculates a contrast value between the average intensity of a spot and its surrounding area, based on which genuine spots from two regional groups are selected. Generally speaking, real spots in the bright-region group have considerably high contrast values, whereas real spots in the dark-region group have relatively low contrast. By combining the final results from two groups, we can separate genuine spots from noise and image artifacts effectively. Only spots passing our selection are retained (see Supplemental Figure 8 online). The procedure produces 25 output fields, including spot number per image, valid image area on pseudo-projection images, Opera imaging time, average intensity of the recognized spots, and total number of spots in a well (see Supplemental Table 1 online for details).
For the detection of endomembrane vesicles labeled with two different colors and their colocalization, we created a two-channel spots-overlapping detection algorithm (EndomembraneCoLocQuantifier; see Supplemental Table 2 and Supplemental Data Set 2 online). This algorithm consists of three parts: filtering background autofluorescence signals, selecting genuine spots from images in two channels (GFP and RFP), and overlapping these spots and output. The overlap detection places spots detected in each channel on a maximum projection image. If in this image a channel-one spot boundary touches or fully contains one or more channel-two spot boundaries (at least share two pixels ≥10% of spot size), the spot is considered to be overlapping (see details in Supplemental Figure 5 online). The procedure produces 40 output fields, which include valid image area, Opera imaging time, number of spots in a stack, overlapping spots per stack, and total number of overlapping spots per well. Our software implementation of our algorithms can be obtained from http://sourceforge.net/projects/robatzekimages/files/ and are provided as supplemental materials.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g46330 (FLS2), At4g19640 (ARA7/Rab F2b), and At3g54840 (ARA6/Rab F1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. BFA Body Formation in Arabidopsis Root and Cotyledon Cells.
Supplemental Figure 2. Cycloheximide Effects in Cotyledon Cells of FLS2-GFP Plants.
Supplemental Figure 3. FM4-64 Endocytic Tracing in Arabidopsis Cotyledons.
Supplemental Figure 4. Features of the EndomembraneQuantifier Script.
Supplemental Figure 5. Features of the EndomembraneCoLocQuantifier Script.
Supplemental Figure 6. Evaluation of the EndomembraneCoLocQuantifier.
Supplemental Figure 7. Expression Analysis of RFP-ARA7/Rab F2b and ARA6/Rab F1-RFP Transgenic Lines Expressing FLS2-GFP.
Supplemental Figure 8. Effects of Membrane Trafficking Inhibitors on Known Subcellular Markers and FLS2-GFP.
Supplemental Figure 9. Quantitative Imaging of FLS2 Endosomes upon Chemical Inhibition.
Supplemental Figure 10. Effects of Actin Inhibitors on FLS2-GFP Vesicle Mobility and Different Leaf Tissues.
Supplemental Figure 11. Effects of Chemical Interference on flg22-Induced FLS2 Endosomes Colocalizing to ARA7/Rab F2b and ARA6/Rab F1 and Positive Compartments.
Supplemental Figure 12. Effect of Tyrphostin A23 on flg22-Induced FLS2-GFP Endosomes Colocalizing to RFP-ARA7/Rab F2b-Positive Compartments in Cotyledon and Primary Leaves.
Supplemental Figure 13. Flg22-Induced Endocytosis of FLS2-GFP and Colocalization with RFP-ARA7/Rab F2b-Positive Compartments in Transient Transformed Cells by Particle Bombardment.
Supplemental Table 1. Output Parameter Produced by the EndomembraneQuantifier Script.
Supplemental Table 2. Output Parameter Produced by the EndomembraneCoLocQuantifier Script.
Supplemental Data Set 1. EndomembraneQuantifier Algorithm.
Supplemental Data Set 2. EndomembraneCoLocQuantifier Algorithm.
Supplementary Material
Acknowledgments
We thank Heidrun Häweker and Gildas Bourdais for technical assistance, Karin Schumacher, Natasha Raikhel, Glenn Hicks, Jiri Friml, Pankaj Dhonukshe, and Sebastian Bednarek for providing materials, and Malick Mbengue, Jens Müller, Sebastian Schornack, and Sophien Kamoun for reading the article and providing helpful and critical comments. M.B. was supported by a grant from the Deutsche Forschungsgemeinschaft, and the S.R. laboratory was supported by the Gatsby Charitable Foundation.
AUTHOR CONTRIBUTIONS:
S.R. designed the research and wrote the article. M.B. designed the research. M.B. and C.F. performed research and edited the article. M.B. analyzed data. J.Z. and D.M. contributed new analytic/computational tools.
Glossary
- RME
receptor-mediated endocytosis
- RLK
receptor-like kinase
- GFP
green fluorescent protein
- TGN
trans-Golgi network
- EE
early endosome
- BFA
Brefeldin A
- Wm
Wortmannin
- LE
late endosome
- MVB
multivesicular body
- RFP
red fluorescent protein
- ConcA
Concanamycin A
- TyrA23
Tyrphostin A23
- ES1
Endosidin 1
- LatB
Latrunculin B
- BDM
2,3-butanedione monoxime
References
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 [DOI] [PubMed] [Google Scholar]
- Bandmann V., Homann U. (2012). Clathrin-independent endocytosis contributes to uptake of glucose into BY-2 protoplasts. Plant J. 70: 578–584 [DOI] [PubMed] [Google Scholar]
- Bauer Z., Gómez-Gómez L., Boller T., Felix G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276: 45669–45676 [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Chen X., Irani N.G., Friml J. (2011). Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14: 674–682 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Boller T., Robatzek S. (2007a). Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 598: 358–371 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D., Felix G., Boller T. (2007b). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Baluska F., Schlicht M., Hlavacka A., Samaj J., Friml J., Gadella T.W., Jr (2006). Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev. Cell 10: 137–150 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dhonukshe P., Huang F., Galvan-Ampudia C.S., Mähönen A.P., Kleine-Vehn J., Xu J., Quint A., Prasad K., Friml J., Scheres B., Offringa R. (2010). Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137: 3245–3255 [DOI] [PubMed] [Google Scholar]
- Drakakaki G., Robert S., Raikhel N.V., Hicks G.R. (2009). Chemical dissection of endosomal pathways. Plant Signal. Behav. 4: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. USA 108: 17850–17855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Emans N., Zimmermann S., Fischer R. (2002). Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14: 71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidit Frey N., Robatzek S. (2009). Trafficking vesicles: pro or contra pathogens? Curr. Opin. Plant Biol. 12: 437–443 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Robatzek S. (2008). Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 147: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M.L., Robertson F.C., Soares D.C., Ingram G.C. (2005). ARABIDOPSIS CRINKLY4 function, internalization, and turnover are dependent on the extracellular crinkly repeat domain. Plant Cell 17: 1154–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J.W., Robatzek S. (2008). Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Happel N., Höning S., Neuhaus J.M., Paris N., Robinson D.G., Holstein S.E. (2004). Arabidopsis mu A-adaptin interacts with the tyrosine motif of the vacuolar sorting receptor VSR-PS1. Plant J. 37: 678–693 [DOI] [PubMed] [Google Scholar]
- Hicks G.R., Raikhel N.V. (2010). Advances in dissecting endomembrane trafficking with small molecules. Curr. Opin. Plant Biol. 13: 706–713 [DOI] [PubMed] [Google Scholar]
- Irani N.G., et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 8: 583–589 [DOI] [PubMed] [Google Scholar]
- Irani N.G., Russinova E. (2009). Receptor endocytosis and signaling in plants. Curr. Opin. Plant Biol. 12: 653–659 [DOI] [PubMed] [Google Scholar]
- Ito E., Fujimoto M., Ebine K., Uemura T., Ueda T., Nakano A. (2012). Dynamic behavior of clathrin in Arabidopsis thaliana unveiled by live imaging. Plant J. 69: 204–216 [DOI] [PubMed] [Google Scholar]
- Jelínková A., Malínská K., Simon S., Kleine-Vehn J., Parezová M., Pejchar P., Kubes M., Martinec J., Friml J., Zazímalová E., Petrásek J. (2010). Probing plant membranes with FM dyes: Tracking, dragging or blocking? Plant J. 61: 883–892 [DOI] [PubMed] [Google Scholar]
- Jürgens G., Geldner N. (2007). The high road and the low road: Trafficking choices in plants. Cell 130: 977–979 [DOI] [PubMed] [Google Scholar]
- Kaiserli E., Sullivan S., Jones M.A., Feeney K.A., Christie J.M. (2009). Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell 21: 3226–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Langowski L., Wisniewska J., Dhonukshe P., Brewer P.B., Friml J. (2008). Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol. Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M.A., de Vries S.C., Russinova E. (2005). Arabidopsis thaliana Somatic Embryogenesis Receptor Kinase 1 protein is present in sporophytic and gametophytic cells and undergoes endocytosis. Protoplasma 226: 55–65 [DOI] [PubMed] [Google Scholar]
- Langhans M., Förster S., Helmchen G., Robinson D.G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62: 2949–2957 [DOI] [PubMed] [Google Scholar]
- Laxmi A., Pan J., Morsy M., Chen R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li R., Liu P., Wan Y., Chen T., Wang Q., Mettbach U., Baluska F., Samaj J., Fang X., Lucas W.J., Lin J. (2012). A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 24: 2105–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Zapater E., Soriano-Ortega E., Marcote M.J., Ortiz-Masiá D., Aniento F. (2006). Trafficking of the human transferrin receptor in plant cells: Effects of tyrphostin A23 and brefeldin A. Plant J. 48: 757–770 [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pfeffer S. (2005). A model for Rab GTPase localization. Biochem. Soc. Trans. 33: 627–630 [DOI] [PubMed] [Google Scholar]
- Radford J.E., White R.G. (2011). Inhibitors of myosin, but not actin, alter transport through Tradescantia plasmodesmata. Protoplasma 248: 205–216 [DOI] [PubMed] [Google Scholar]
- Reyes F.C., Buono R., Otegui M.S. (2011). Plant endosomal trafficking pathways. Curr. Opin. Plant Biol. 14: 666–673 [DOI] [PubMed] [Google Scholar]
- Robatzek S. (2007). Vesicle trafficking in plant immune responses. Cell. Microbiol. 9: 1–8 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Jiang L., Schumacher K. (2008). The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S., Grunewald D., Stüber K., Schaaf S., MacLean D., Schulze-Lefert P., Robatzek S. (2010). High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol. 154: 1096–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S., Robatzek S. (2006). Induced endocytosis of the receptor kinase FLS2. Plant Signal. Behav. 1: 293–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarzadeh A., Franzen R., Schmelzer E. (2008). The Arabidopsis class VIII myosin ATM2 is involved in endocytosis. Cell Motil. Cytoskeleton 65: 457–468 [DOI] [PubMed] [Google Scholar]
- Scheuring D., Viotti C., Krüger F., Künzl F., Sturm S., Bubeck J., Hillmer S., Frigerio L., Robinson D.G., Pimpl P., Schumacher K. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23: 3463–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A., von Zastrow M. (2009). Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Miwa K., Yuan L., von Wirén N., Fujiwara T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J., Tanaka M., Toyoda A., Miwa K., Kasai K., Fuji K., Onouchi H., Naito S., Fujiwara T. (2010). Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc. Natl. Acad. Sci. USA 107: 5220–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R., Gerding-Reimers C., Deeks M.J., Menninger S., Gallegos R.M., Tonaco I.A., Hübel K., Hussey P.J., Waldmann H., Coupland G. (2012). Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana. Plant J. 71: 338–352 [DOI] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Nakano A. (2002). Vesicular traffic: An integral part of plant life. Curr. Opin. Plant Biol. 5: 513–517 [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B., et al. (2005b). Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur. J. Cell Biol. 84: 609–621 [DOI] [PubMed] [Google Scholar]
- Voigt B., Timmers A.C., Samaj J., Müller J., Baluska F., Menzel D. (2005a). GFP-FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur. J. Cell Biol. 84: 595–608 [DOI] [PubMed] [Google Scholar]
- von Kleist L., Haucke V. At the crossroads of chemistry and cell biology: Inhibiting membrane traffic by small molecules. Traffic http://dx.doi.org/10.1111/j.1600-0854.2011.01292.x [DOI] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S.K., Jiang L. (2009). Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.