Abstract
We isolated and characterized a novel light-regulated cDNA from the short-day plant Pharbitis nil that encodes a protein with a leucine (Leu) zipper motif, designated PNZIP (Pharbitis nil Leu zipper). The PNZIP cDNA is not similar to any other gene with a known function in the database, but it shares high sequence homology with an Arabidopsis expressed sequence tag and to two other sequences of unknown function from the cyanobacterium Synechocystis spp. and the red alga Porphyra purpurea, which together define a new family of evolutionarily conserved Leu zipper proteins. PNZIP is a single-copy gene that is expressed specifically in leaf photosynthetically active mesophyll cells but not in other nonphotosynthetic tissues such as the epidermis, trichomes, and vascular tissues. When plants were exposed to continuous darkness, PNZIP exhibited a rhythmic pattern of mRNA accumulation with a circadian periodicity of approximately 24 h, suggesting that its expression is under the control of an endogenous clock. However, the expression of PNZIP was unusual in that darkness rather than light promoted its mRNA accumulation. Accumulation of PNZIP mRNA during the dark is also regulated by phytochrome, since a brief exposure to red light in the middle of the night reduced its mRNA levels. Moreover, a far-red-light treatment at the end of day also reduced PNZIP mRNA accumulation during the dark, and that effect could be inhibited by a subsequent exposure to red light, showing the photoreversible response attributable to control through the phytochrome system.
Light is essential for normal plant growth and development not only as a source of energy but also as an environmental signal that regulates various developmental and metabolic processes. These light-regulated responses occur throughout the entire life cycle of the plant, including seed germination, seedling de-etiolation, leaf and chloroplast development, flowering, and eventually, senescence (Kendrick and Korenberg, 1994). The perception and transduction of the light signals are governed by at least three families of photoreceptors, including the phytochrome (red and far-red) receptors, blue-light receptors, and UV receptors (Deng, 1994; Quail et al., 1995; Chamovitz and Deng, 1996). In addition to light-regulated development and gene expression, it has been reported that certain light-inducible genes, especially nuclear-encoded photosynthetic genes, are controlled also by a circadian rhythm (Guiliano et al., 1988; Nagy et al., 1988; Taylor, 1989; Piechulla, 1993). Because all rhythmically regulated genes characterized to date are also light regulated, it has been proposed that there is an interaction between light and the endogenous circadian rhythm (Lumsden, 1991; Dunlap, 1996).
During the past few years many light-regulated genes from different species have been identified, and among the most extensively studied have been the genes that code for the chlorophyll a/b-binding protein of PSII (CAB), the small subunit of ribulose-1,5-bisphosphate carboxylase (rbcS), and chalcone synthase (CHS; for review, see Li et al., 1993; Terzaghi and Cashmore, 1995). Through the analysis of the promotors of these genes, multiple light-responsive elements were identified and used to isolate cDNA clones encoding proteins binding to these DNA sequences (Gilmartin et al., 1990; Li et al., 1993; Terzaghi and Cashmore, 1995). It is interesting that many of these DNA-binding proteins, including Arabidopsis GBF1–4, parsley CPRF1–3, and wheat HBP-1a, belong to the bZIP family of transcription factors (Weisshaar et al., 1991; Schindler et al., 1992; Feldbrugge et al., 1994; Menkens and Cashmore, 1994). However, although considerable progress has been made in identifying these promotor elements and DNA-binding proteins, it was found that at least two different promotor elements are required to allow light-regulated transcription and that most of the DNA-binding proteins isolated so far are themselves not light regulated, suggesting that other regulatory proteins, such as a possible repressor in the dark or an activator in the light, are required to regulate light-responsive transcription. Light-regulated gene expression may be accomplished through the coordination of multiple-component protein complexes and promotor elements, but only some of them have been isolated so far (Gilmartin et al., 1990; Li et al., 1993; Terzaghi and Cashmore, 1995).
To identify additional light-regulated genes that may be involved in the light signal transduction pathway, we isolated by differential hybridization several cDNAs corresponding to mRNAs in which abundance is altered after the transition of the short-day plant Pharbitis nil cotyledons to continuous darkness (Zheng et al., 1993; O'Neill et al., 1994). In this paper we describe the identification and characterization of a cDNA named PNZIP (for Pharbitis nil Leu zipper), which is a novel plant cDNA that is homologous to an Arabidopsis EST, which was fully sequenced by us, and to two more unknown ORFs from the cyanobacterium Synechocystis spp. and the red alga Porphyra purpurea. Together, these genes form a new family of evolutionarily highly conserved Leu zipper proteins. In P. nil, PNZIP mRNA accumulates during the dark specifically in the leaf mesophyll cells, and its expression is regulated by both phytochrome and a circadian clock. Given that PNZIP encodes a Leu zipper protein, it is possible that PNZIP may heterodimerize with other mesophyll bZIP DNA-binding proteins and thus act as a negative regulator of light-induced gene expression during darkness.
MATERIALS AND METHODS
Pharbitis nil Choisy strain Violet (Japanese morning glory) was used for all experiments. Seeds of this Pharbitis strain were originally obtained from Marutane Co., Ltd (Kyoto, Japan) and maintained as an inbred line for more than 12 generations at the University of California, Davis. Seeds were scarified in concentrated sulfuric acid for 45 min on a stirring plate at room temperature, rinsed well for 15 min under a continuous stream of deionized water, and then allowed to rehydrate in aerated, distilled water for 12 to 16 h. The germinated seeds were planted in trays containing a standard soil mixture and held in a growth chamber under continuous fluorescent light (250 μmol m−2 s−1; VHO/EW 185 W/1500 mA Philips, Mahwah, NJ) at 28°C until germination was complete. The 1st d of seedling emergence was designated d 1 in our experiments.
Photoperiodic and Light Treatments
Photoperiodic and light treatments were routinely initiated at 4:30 pm (0 h) on postgerminated 6-d-old seedlings grown under continuous light. These treatments included an extended dark treatment of up to 48 h; an NB with 10 min of red-light interruption given at 8 h into the dark period; and an end-of-day treatment with 10 min of red, far-red, red/far-red, or far-red/red light, prior to the transfer to darkness. For red-light treatments four fluorescent tubes (Philips F30T12/CW/RS) were filtered with a 3-mm-thick translucent red plexiglass sheet (Acrylite GP, color 210–0; CYRO Industries, Mt. Arlington, NJ) as described previously (Zheng et al., 1993; O'Neill et al., 1994). The far-red source consisted of two fluorescent tubes (F48T12/660 nm/VHO, Sylvania) filtered with a 3-mm-thick translucent far-red plexiglass sheet (FRS700, Rohm and Haas, Philadelphia, PA, dye no. 58015) as described by Li and Lagarias (1994). At the end of each treatment the seedling tissue was harvested in complete darkness directly into liquid nitrogen, transferred to light-proof plastic containers, and stored at −80°C for later use in RNA extraction.
For the end-of-day treatments, 10 plants were returned to the growth chamber for an additional 2 to 3 weeks of growth in continuous light to examine the effectiveness of the photoperiodic light treatment on floral induction. Flowering was assessed by determining the amount of flower buds per plant.
cDNA Library Construction
Total RNA was isolated as described previously (O'Neill, 1992). Poly(A+) RNA was isolated using paramagnetic oligo(dT) beads (Dynabeads, Dynal, Great Neck, NY) according to the manufacturer's suggestions. LiCl was removed from the poly(A+) RNA by two ethanol precipitations prior to first-strand cDNA synthesis. Libraries were constructed from 5 μg of poly(A+) RNA isolated from 6-d-old cotyledon tissue treated with 8 to 12 h of darkness. cDNA was constructed and cloned into the λZAPII phage vector (Stratagene) according to the manufacturer's procedures. The cotyledon-specific cDNA library contained approximately 3 × 106 clones, approximately 95% of which contained inserts.
cDNA Library Screening
Differential screening of the cotyledon-specific cDNA library was carried out as follows. cDNAs were labeled with [32P]dATP in 50-μL reverse-transcription reactions containing 5 μg of poly(A+) RNA, 1.5 μg of oligo(dT)12–18 mers (Pharmacia), 40 units of RNasin (Promega), 50 μm dATP, 500 μm dCTP, dGTP, and dTTP, 1× reverse transcriptase buffer (GIBCO-BRL), 10 mm DTT, 12.5 μL of [α-32P]dATP (6000 Ci/mmol), and 600 units of Superscript reverse transcriptase (GIBCO-BRL) at 37°C for 1 h. Approximately 0.5 × 106 clones from the cDNA library were plated, and replica filters (BA85 nitrocellulose, Schleicher & Schuell) were made from each plate and screened by differential hybridization using the 32P-labeled cDNA probes as previously described (O'Neill et al., 1994). Each filter set was hybridized with first-strand cDNA probes synthesized from either control or experimental poly(A+) RNA probes. The control probe was synthesized using poly(A+) RNA isolated from noninduced cotyledon tissues, whereas the experimental probe was synthesized using poly(A+) RNA isolated from cotyledon tissues that were photoperiodically induced by 14 h of dark treatment. Differentially hybridizing cDNA clones were identified by comparing the autoradiograms of replica filters hybridized with both the control and experimental probes. One cDNA clone corresponding to a differentially abundant mRNA is described in this paper.
Sequence Analysis
Nucleotide sequencing was carried out by constructing a nested set of deletion plasmids using the Erase-a-Base system (Promega) and sequencing the deletions by the dideoxynucleotide chain termination method (Sanger et al., 1977) using double-stranded DNA templates, 35S-dATP (Amersham), and Sequenase enzyme (United States Biochemical). The nucleotide sequence was determined from overlapping clones and from both strands. Sequence analysis and multiple sequence alignment (PileUp) were accomplished by using the Genetics Computer Group (University of Wisconsin, Madison) and BLAST (Altschul et al., 1990) computer programs. The GenBank accession numbers of PNZIP and ATZIP are U37437 and U38232, respectively.
DNA Gel-Blot Analysis
DNA was extracted from cotyledon tissue using the procedure described by Jofuku and Goldberg (1988). Ten micrograms of genomic DNA was digested with EcoRI, BamHI, HindIII, or SacI (Promega), separated on a 0.7% agarose gel, and blotted onto a Nytran membrane (Schleicher & Schuell). Blots were hybridized with a PNZIP cDNA probe labeled to high specific activity by random priming (Boehringer Mannheim) with [32P]dCTP at 37°C in 50% formamide, 5× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate), 0.05 m phosphate buffer, pH 7.0, 5× Denhardt's solution (1× Denhardt's is 0.02% Ficoll, 0.02% PVP, and 0.02% BSA), 0.2 mg/mL sheared denatured salmon testes DNA (type III, Sigma), and 0.2% SDS. Blots were washed three times for 20 min at 50, 55, and 60°C with 0.2× SSC and 0.1% SDS. Autoradiography was performed at −80°C using Kodak XAR-5 film and one intensifying screen (Cronex Lightning Plus, DuPont). Blots were exposed for up to 2 d.
RNA Gel-Blot Analysis
The methods for RNA extraction and RNA gel-blot hybridization have been described previously (O'Neill et al., 1994). Total RNA (30 μg/lane) was separated on a 0.8% formaldehyde agarose gel and blotted onto a Nytran membrane (Schleicher & Schuell). Blots were hybridized with a PNZIP cDNA probe labeled to high specific activity by random priming (Boehringer Mannheim) with [32P]dCTP at 42°C as described for DNA blot hybridization (see above). Blots were washed three times for 20 min at 55, 60, and 65°C with 0.2× SSC and 0.1% SDS solution and autoradiographed at −80°C using Kodak XAR-5 film and an intensifying screen (Cronex Lightning Plus). Blots were exposed for approximately 2 to 3 d.
In Situ Hybridization
Tissues from cotyledons, leaves, hypocotyls, and shoots were fixed for 4 to 6 h in 50 mm phosphate buffer, pH 7.0, 4% paraformaldehyde (Sigma), and 0.1% glutaraldehyde (Polysciences, Warrington, PA). Afterward, the tissues were rinsed in phosphate buffer alone and dehydrated through a graded series of ethanol (10–100%, v/v). The tissues were embedded in Paraplast Plus (Oxford Labware, St. Louis, MO), cut into 7-μm sections, and mounted on Superfrost Plus microscope slides (Fisher Scientific). For the synthesis of PNZIP antisense and sense transcripts, a 1008-bp XbaI-KpnI cDNA fragment within the ORF was ligated into pBluescript SK+ (Stratagene) and transcribed in vitro with digoxigenin-UTP using T3 or T7 polymerases (Boehringer Mannheim). Prehybridization, hybridization, washings, RNase treatment, and immunological detection of the incorporated digoxigenin-UTP were performed using a digoxigenin nucleic acid detection system (Boehringer Mannheim) according to the manufacturer's recommendations. Photographic images were recorded on Kodak Royal Gold 25 film, using a photomicroscope system (BX60, Olympus).
RESULTS
Isolation and Characterization of PNZIP cDNA
By differential hybridization screening of a P. nil cDNA library that was constructed from poly(A+) RNA isolated from dark-induced cotyledons, we identified and isolated a cDNA clone that corresponded to an mRNA in which abundance increased during the dark. After complete sequencing this cDNA was named PNZIP (see above). The full-length cDNA sequence of PNZIP consists of 1402 bp, with an ORF of 1110 bp. The PNZIP gene encodes a predicted polypeptide of 370 amino acids, with a predicted molecular mass of approximately 43 kD and a pI of 8.37.
The PNZIP protein contains a Leu zipper motif between amino acid residues 228 and 256 (Fig. 1). The Leu zipper begins with a Met residue that was reported to serve as the best substitute for Leu and is found in many known Leu zippers (Landschulz et al., 1988; Hu et al., 1990). This is followed by three periodic repetitions of Leu residues that appear every seventh position (Fig. 1). At the next seventh position after these Leu repeats, there is a Tyr residue that has a long aromatic side chain and was reported to functionally replace Leu residues in various Leu zippers (Hu et al., 1990; Kusano et al., 1995; Nantel and Quatrano, 1996). Altogether, the PNZIP Leu zipper contains five heptadic repeats of hydrophobic amino acids and contains the minimum of three Leu residues, which were reported to be required for dimerization (Landschulz et al., 1988; Hu et al., 1990).
Figure 1.
Alignment of the predicted polypeptides of P. nil PNZIP, A. thaliana ATZIP, Synechocystis spp. ORF, and P. purpurea ORF. Amino acids that are identical in at least three of the four different proteins are in closed boxes. The Leu zipper, hydrophilic domain, and hydrophobic domain are marked in black. The Leu residues in the Leu-rich region are in bold type, and the Arg and Lys residues in the possible nuclear localization domain are in bold and underlined. The GenBank accession numbers of PNZIP, ATZIP, Synechocystis spp. ORF, and P. purpurea ORF are U37437, U38232, D90899, and U38804, respectively.
Unlike bZIP transcription factors that have a basic domain adjacent to the Leu zipper (Vinson et al., 1989; Izawa et al., 1993), PNZIP does not contain a typical basic domain but, rather, has a hydrophilic domain (Fig. 1). It is possible that this hydrophilic region of PNZIP interacts with the basic domain of other bZIPs and thus supports possible heterodimerization between the Leu zipper motifs. The PNZIP protein also contains a hydrophobic domain rich in Ala, Ile, Leu, Met, and Val at the C terminus of the protein and a Leu-rich region between amino acid residues 143 and 159 (Fig. 1). These regions may be involved in selective protein-protein interactions (Kobe and Deisenhofer, 1994). PNZIP contains a possible nuclear localization signal that consists of the conserved basic amino acids Arg and Lys and appears between amino acid residues 113 and 121 (Raikhel, 1992; Fig. 1).
PNZIP Defines a New Family of Evolutionarily Conserved Proteins
Comparison of the PNZIP cDNA with the database of known sequences revealed no obvious similarity with any other genes of known function. However, by searching the Arabidopsis EST database with the PNZIP cDNA, we identified and fully sequenced an Arabidopsis cDNA that was homologous to PNZIP and was named ATZIP by us (for Arabidopsis thaliana Leu zipper). In addition, the PNZIP protein was also homologous to two other ORFs with an unknown function from Synechocystis spp. and P. purpurea that were just recently submitted to the GenBank database (Reith and Munholland, 1995; Kaneko et al., 1996).
Altogether, the proteins of the Pharbitis PNZIP, Arabidopsis ATZIP, and the ORFs from Synechocystis spp. (S.ORF) and P. purpurea (P.ORF) form a new family of evolutionarily highly conserved proteins (Fig. 1). All of the different proteins contain the Leu zipper motif, the hydrophilic domain adjacent to it, the C-terminal hydrophobic domain, the Leu-rich region, and the possible nuclear localization signal with only minor modifications, suggesting that these motifs serve a conserved evolutionary function (Fig. 1).
The pairwise identity between PNZIP and ATZIP is 86%, whereas that between PNZIP and the Synechocystis spp. ORF and P. purpurea ORF is only 60 and 59%, respectively. The pairwise similarity between PNZIP and ATZIP and the ORFs from Synechocystis spp. and P. purpurea are 94, 81, and 80%, respectively. The relationships between the different members of the PNZIP family as observed by phylogenetic comparison emphasizes that the higher plant proteins form a separate class from the cyanobacterial and red algal proteins (data not shown).
Genomic DNA Gel-Blot Hybridization Analysis of PNZIP
To determine whether PNZIP represents a single locus in the P. nil genome or whether it is a multicopy gene, genomic DNA gel-blot hybridization analysis was performed using the PNZIP cDNA sequence as a probe. The results showed that PNZIP hybridized to only one genomic restriction fragment, suggesting that it represents a single-copy gene (data not shown). A similar DNA gel-blot hybridization analysis of the Arabidopsis genome using the ATZIP cDNA as a probe revealed similar results (data not shown), suggesting that the PNZIP gene probably represents a single locus also in the genome of other plants.
Expression of the PNZIP Gene in Different Organs
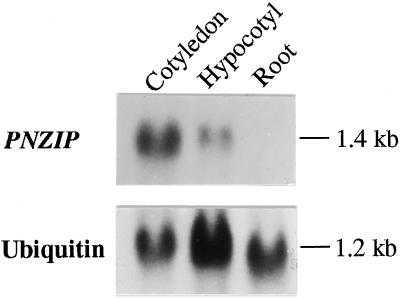
Since the PNZIP cDNA was isolated from a cDNA library that was constructed from poly(A+) RNA isolated from cotyledons of P. nil seedlings, we wanted to examine whether the expression of PNZIP was specific to the cotyledons or was also expressed in other organs. Figure 2 shows that a 1.4-kb transcript was strongly detected in the cotyledons, more weakly in the hypocotyl tissue, but could not be detected at all in the root. These results suggest that the PNZIP gene is expressed especially in cotyledons, less in other green vegetative tissues, but not at all in nonphotosynthetic tissues such as the roots.
Figure 2.
RNA gel-blot hybridization analysis of PNZIP mRNA accumulation in different P. nil organs. Each lane contained 30 μg of total RNA isolated from cotyledons, hypocotyls, or roots. Hybridization to a ubiquitin cDNA probe served as a control for equal loading of RNA in each lane. Numbers on the right indicate the approximate sizes of the mRNAs detected.
Regulation of PNZIP Gene Expression by a Circadian Rhythm and Phytochrome
PNZIP cDNA was initially isolated by a differential hybridization screening because of its increased mRNA levels during the dark. To further study the light and dark regulation of PNZIP gene expression and the possible role of the phytochrome in this process, we performed RNA gel-blot hybridizations using RNA isolated from plants grown under various light treatments.
Figure 3 shows the circadian regulation of PNZIP mRNA accumulation. P. nil seedlings were pretreated with continuous light, and on the evening of the 6th d they were transferred to complete darkness. The results show that levels of PNZIP mRNA increased markedly within 8 to 12 h of darkness and began to decline after 16 h, returning to its basal level after 20 to 24 h (Fig. 3). The pattern of PNZIP mRNA abundance appeared to be under the control of a circadian rhythm, since this pattern of accumulation and loss appeared to repeat at cycles of approximately 24 h. During a continuous dark treatment of 48 h, PNZIP mRNA levels peaked after 12 and 36 h and were at their lowest levels at 0, 24, and 48 h (Fig. 3).
Figure 3.
RNA gel-blot hybridization analysis of PNZIP mRNA levels during an extended dark treatment. Seedlings were grown in continuous light for 6 d and than transferred to darkness for 0 to 48 h. At the hours indicated, plants were harvested and their RNA was isolated. Each lane contained 30 μg of cotyledon total RNA. Hybridization to a ubiquitin cDNA probe served as a control for equal loading of RNA in each lane. Numbers on the right indicate the approximate sizes of the mRNAs detected.
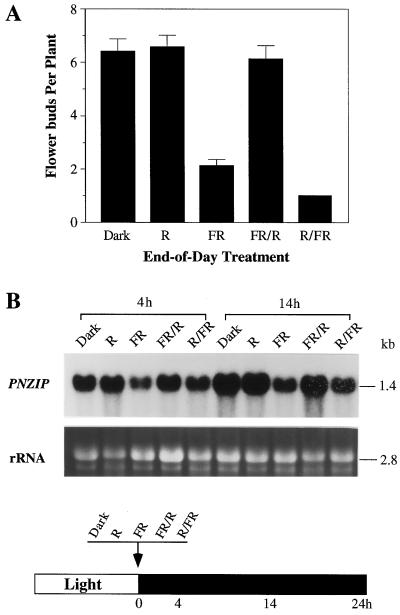
To study the possible role of phytochrome in the regulation of PNZIP gene expression, we examined the effects of an end-of-day treatment on PNZIP transcript accumulation. The end-of-day treatment consisted of a brief illumination with red or far-red light at the end of the main light period just before the transition to darkness. This end-of-day treatment is well known to affect flowering in different short-day plants including P. nil (Takimoto, 1967; Fig. 4A). When a red-light treatment was given at the end of the day, it had no effect on the normal accumulation of PNZIP mRNA during the dark as indicated after 4 or 14 h (Fig. 4B). However, when a far-red-light treatment was given, it reduced the accumulation of PNZIP mRNA as shown after both 4 and 14 h of darkness (Fig. 4B). The effects of both red- and far-red-light treatments were reversible when subsequent far-red- or red-light treatments were given shortly after them (Fig. 4B).
Figure 4.
Effects of end-of-day treatments of P. nil seedlings with red and far-red light on flowering and PNZIP mRNA accumulation during the dark. Seedlings were grown in continuous light for 6 d, and just before the transition to darkness at 0 h they were exposed for 10 min to red (R), far-red (FR), far-red and then red (FR/R), or red and afterward far-red (R/FR) light. A, Effects of end-of-day treatments on flower induction as measured after 3 weeks. Error bars indicate ses. B, RNA gel-blot hybridization analysis of PNZIP mRNA levels detected 4 and 14 h after the end-of-day treatments. Each lane contained 30 μg of cotyledon total RNA. A photograph of the amount of rRNA served as a control for equal loading of RNA in each lane. Numbers on the right indicate the approximate sizes of the mRNAs detected. The black and white bar at the bottom of the figure schematically presents the light/dark conditions and the timings of light treatments and RNA isolation.
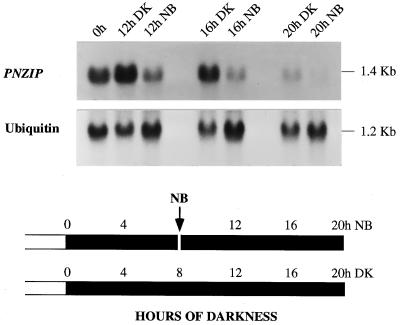
The role of phytochrome in the regulation of PNZIP gene expression was further investigated by examining the effect of a 10-min red-light NB treatment given precisely 8 h after the transition to darkness, a treatment that inhibits the photoperiodic induction of flowering. Figure 5 shows that the NB treatment decreased the accumulation of PNZIP mRNA during the darkness as detected after 12, 16, or 20 h.
Figure 5.
RNA gel-blot hybridization analysis of PNZIP mRNA accumulation during continuous darkness or following an NB treatment. Seedlings were grown in continuous light for 6 d and then treated with three durations of darkness (DK 12, 16, or 20 h) or were interrupted at the 8th h of dark treatment by a 10-min NB with red light. Each lane contained 30 μg of cotyledon total RNA. Hybridization to a ubiquitin cDNA probe served as a control for equal loading of RNA in each lane. Numbers on the right indicate the approximate sizes of the mRNAs detected. The black and white bars at the bottom of the figure schematically presents the light/dark conditions and the timings of the NB treatment and RNA isolation.
In Situ Localization of PNZIP mRNA Accumulation
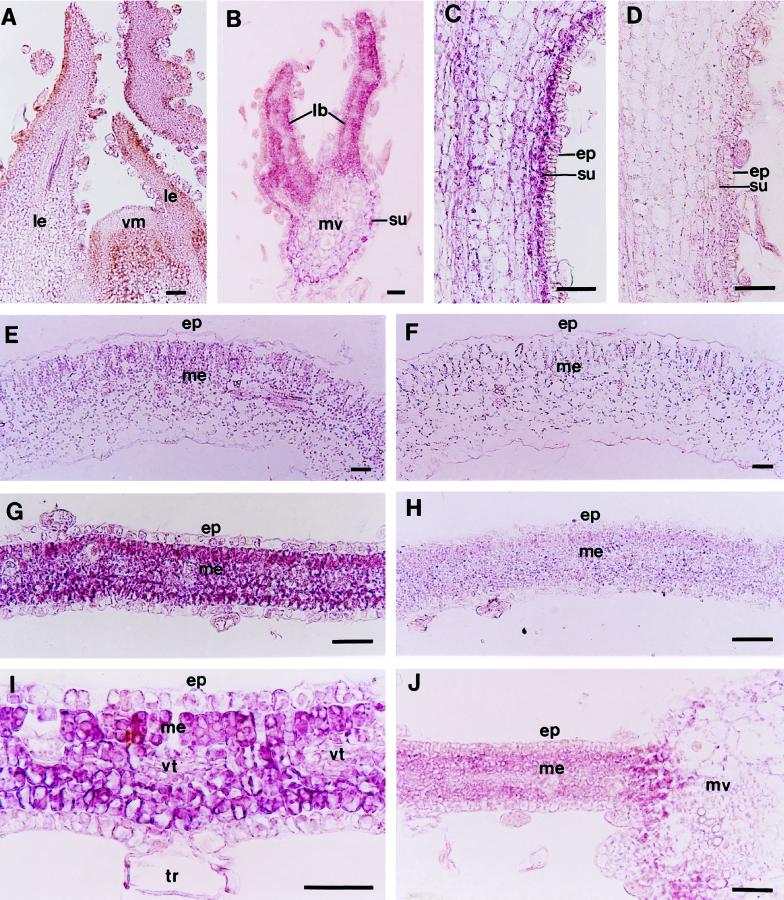
To define the spatial pattern of PNZIP gene expression in different tissues and following different light treatments, we conducted RNA in situ-hybridization experiments using a PNZIP digoxigenin-labeled antisense RNA as the probe. Figure 6A shows that PNZIP mRNA could not be detected in the shoot apical meristem or in the leaf primordia that covers it. However, in a transverse section through a young expanding leaf that was taken from the fourth or fifth position below the shoot apical meristem, PNZIP mRNA was found in the cells of the leaf blade that were beginning to differentiate into mesophyll cells and in the subepidermal cells of the main leaf vein (Fig. 6B). In the hypocotyl tissue PNZIP mRNA could be clearly detected in the subepidermal cells but not in the epidermis or other cortical cells (Fig. 6C). A control section of a hypocotyl hybridized with a PNZIP sense strand did not show any hybridization signal (Fig. 6D).
Figure 6.
In situ localization of PNZIP mRNA in different P. nil organs and following various light treatments. Seedlings were grown in continuous light for 6 d and then were exposed to 12 h of darkness or were interrupted at the 8th h by a 10-min NB with red light. Afterward, longitudinal and transverse sections (7 μm thick) through the shoot meristem, hypocotyl, cotyledon, and leaf tissues were collected and hybridized with a PNZIP antisense RNA probe (A–C, E, G, I, and J) or a PNZIP sense RNA probe as a control (D, F, and H). Probes were labeled with digoxigenin-UTP. The transcript-specific hybridization signal is shown in purple. A, Vegetative shoot meristem of a 7-d-old seedling. B, Young expanding leaf taken from the fourth or fifth position below the shoot meristem. C, Hypocotyl. D, Hypocotyl hybridized with a PNZIP sense RNA probe as a control. E, Cotyledon. F, Cotyledon hybridized with a PNZIP sense RNA probe as a control. G, Leaf. H, Leaf hybridized with a PNZIP sense RNA probe as a control. I, Higher magnification of a young leaf. J, Young leaf after an NB treatment. ep, Epidermis; lb, leaf blade; le, leaf; me, mesophyll; mv, main vein; su, subepidermis; tr, trichome; vm, vegetative meristem; and vt, vascular tissue. Bar = 25 μm.
In the cotyledons PNZIP mRNA accumulated specifically in the leaf mesophyll and could be detected especially in the palisade parenchyma and spongy mesophyll cells (Fig. 6E). A control section of a cotyledon hybridized with a PNZIP sense strand did not show any hybridization signal (Fig. 6F). The strongest accumulation of PNZIP mRNA was observed in the leaf tissue and was specific to the leaf mesophyll cells but not in the epidermis (Fig. 6G). Again, a control section of a leaf hybridized with a PNZIP sense strand did not show any detected hybridization signal (Fig. 6H). Figure 6I is an enlargement of the leaf tissue, showing more clearly the specific accumulation of PNZIP mRNA in the mesophyll cells but not in the epidermal cells, vascular tissue, or trichomes. After an NB treatment, the accumulation of PNZIP mRNA in the leaf mesophyll cells was strongly reduced (Fig. 6J).
DISCUSSION
PNZIP Defines a New Family of Evolutionary Conserved Leu Zipper Proteins
The Leu zipper motif serves as a dimerization domain by allowing the Leu side chains from one helix to interdigitate with those of a matching helix of a second polypeptide. The characteristic features of a Leu zipper are the periodic repetitions of at least four Leus or alternative substitutes that appear at every seventh position along the protein (Landschultz et al., 1988; Vinson et al., 1989; Hu et al., 1990). The PNZIP protein contains a Met residue at position 228, which, similar to Leu, has a long hydrophobic side chain that is bulky at its tip, and afterward contains three periodic repetitions of Leu residues followed by a Tyr residue. All together, the PNZIP Leu zipper contains five periodic repetitions of hydrophobic amino acids that are separated by charged or polar amino acids and is likely to promote a functional dimerization domain.
It is interesting that all of the domains detected in PNZIP, including the Leu zipper, the Leu-rich region, the hydrophilic and hydrophobic domains, and the possible nuclear localization signal, were found also to be highly conserved in an Arabidopsis EST cDNA that was fully sequenced by us and in two other unknown ORFs from Synechocystis spp. and P. purpurea. For example, the only difference in the Leu zipper motif is the replacement of the third Leu with a Met in the P. purpurea ORF (Fig. 1). The hydrophilic region, on the other hand, appeared to be completely conserved among all of the proteins (Fig. 1).
The highly conserved primary sequence between the higher plant PNZIP and ATZIP and the lower photosynthetic cyanobacterial and red algal ORFs suggests that these proteins may serve a conserved evolutionary function. Since all of these proteins contain a Leu zipper motif, it is possible that they can dimerize with other Leu zipper proteins However, since these members of the PNZIP protein family are distinct from other known Leu zipper proteins, it is also possible that they share some unique type of interactions and functions. Despite the overall similarities, the higher plant proteins PNZIP and ATZIP form a separate class that distinguishes them from that of the cyanobacterial and red algal proteins. It is possible that they have evolved an additional function that is specific to higher plants, such as the regulation of flower development (O'Neill, 1992).
PNZIP Is Expressed in the Mesophyll Cells and Is Regulated by Phytochrome and a Circadian Rhythm
RNA gel-blot hybridization analysis revealed that PNZIP mRNA accumulated mainly in the cotyledons and only slightly in the hypocotyl but not at all in the roots (Fig. 2). More detailed analysis of the pattern of PNZIP gene expression by RNA in situ hybridization confirmed these results and explored more clearly that PNZIP mRNA accumulated in the cotyledons and leaves (Fig. 6, E and G). Within the leaf tissue, PNZIP mRNA accumulated specifically in the photosynthetically active mesophyll cells but not in the nonphotosynthetic tissues such as the epidermis, trichomes, and vascular tissues (Fig. 6I).
When plants were held in continuous darkness, PNZIP exhibited a rhythmic pattern of mRNA accumulation with a circadian periodicity of approximately 24 h (Fig. 3). This pattern for PNZIP is unusual in that darkness rather than light promoted its mRNA accumulation, in a manner opposite to that reported for most other light-regulated genes and especially for the genes encoding proteins of the light-harvesting complex of PSI/PSII (Guiliano et al., 1988; Nagy et al., 1988; Millar and Kay, 1991; Piechulla, 1993). Therefore, although PNZIP is expressed in the photosynthetically active mesophyll cells, it is expressed in a manner opposite to that of most of the photosynthetic genes. Other genes that exhibit an unusual circadian rhythm pattern are the Arabidopsis Gly-rich proteins Ccr1 and Ccr2 and the Arabidopsis and maize catalase genes CAT3 and Cat3, respectively, which peak at the end of the light period and at the beginning of the dark phase (Redinbaugh et al., 1990; Carpenter et al., 1994; Zhong and McClung, 1996). Another gene that has been identified in which mRNA accumulation is suppressed by light is the Arabidopsis HMG1; however, it is not under the control of a circadian rhythm (Learned, 1996).
The accumulation of PNZIP mRNA during the dark is regulated by phytochrome, an observation that is supported by several different sets of results. For example, a far-red illumination at the end of the day reduced the levels of PNZIP mRNA accumulation, whereas a subsequent irradiation with red light reversed this response (Fig. 4). Moreover, a brief NB treatment of red light during the middle of the night markedly reduced the levels of PNZIP mRNA accumulation as indicated by both RNA gel-blot hybridization (Fig. 5) and RNA in situ hybridization (Fig. 6J).
It is interesting that a red-light treatment given at the end of the light period had no observable effect on PNZIP mRNA accumulation during the dark, whereas the same treatment given during the dark phase markedly reduced its transcript accumulation (Figs. 4 and 5). This suggests that a red-light treatment is not effective by itself but, rather, only when applied during a particular phase of the circadian rhythm. Since phytochrome is known to be involved in the oscillation of the circadian rhythm (Lumsden, 1991), the inhibitory effects of a far-red illumination at the end of the day and a red-light NB treatment during the middle of the night may be indirect and may have a role in shifting the phase of the circadian clock.
Possible Role of PNZIP in Light-Regulated Gene Expression
Leu zipper proteins without a basic domain are present in both animal and plant kingdoms (Kageyama and Pasten, 1989; Ron and Habener, 1992; Vatten et al., 1992; Bange et al., 1994; Sun et al., 1996). They have been demonstrated to act as negative transcriptional regulators by forming nonfunctional heterodimeric complexes with those transcription factors that contain the basic domain. For example, the rat CHOP Leu zipper protein negatively regulates transcription by heterodimerizing with the C/EBP and LAP bZIP transcription factors (Ron and Habener, 1992). A similar mechanism for the negative regulation of gene expression by forming competitive heterodimeric complexes was also reported for helix-loop-helix proteins (Van Doren et al., 1991).
According to this role for Leu zipper proteins and the specific accumulation of PNZIP mRNA in the mesophyll cells during the dark period, at the same time that the mRNA levels of many photosynthetic genes are declining suggests that PNZIP may be involved in the negative regulation of these genes during the dark. The transcription factors that bind to the G-box element of the Arabidopsis rbcS promotor belong to the bZIP class, and at least GBF1 and GBF2 were reported to be expressed ubiquitously in the light and in the dark (Schindler et al., 1992). Therefore, it is possible that PNZIP may negatively regulate rbcS gene expression during the dark by forming nonfunctional heterodimers with these transcription factors.
For the future, we suggest that further research be done with Arabidopsis using ATZIP, the Arabidopsis homolog of PNZIP. Examining its possible protein-protein interactions with GBF1 and GBF2 and directly assessing its function by altering its expression in transgenic plants or by obtaining a null mutant will provide further information about the direct role of this gene in the regulation of light-induced gene expression.
ACKNOWLEDGMENTS
We thank A. Bui, J. Nadeau, and D. Van Tassel for their assistance. We also thank Dr. C.J. Lagarias (University of California, Davis) for the use of the red- and far-red-light filters and for his help with the light treatments. The Arabidopsis Biological Resource Center (Ohio State University, Columbus) is acknowledged for the gift of the EST clone.
Abbreviations:
- bZIP
basic Leu zipper
- EST
expressed sequence tag
- NB
night break
- ORF
open reading frame
Footnotes
This research was supported by grant no. IBN-9317249 to S.D.O. from the National Science Foundation Developmental Mechanisms Program.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bange FC, Vogel U, Flohr T, Kiekenbeck M, Denecke B, Bottger EC. IFP 35 is an interferon-induced leucine zipper protein that undergoes interferon-regulated cellular redistribution. J Biol Chem. 1994;269:1091–1098. [PubMed] [Google Scholar]
- Carpenter CD, Kreps JA, Simon AE. Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz DA, Deng XW. Light signaling in plants. Crit Rev Plant Sci. 1996;15:455–478. [Google Scholar]
- Deng XW. Fresh view of light signal transduction in plants. Cell. 1994;76:423–426. doi: 10.1016/0092-8674(94)90107-4. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Genetics and molecular analysis of circadian rhythms. Annu Rev Genet. 1996;30:579–601. doi: 10.1146/annurev.genet.30.1.579. [DOI] [PubMed] [Google Scholar]
- Feldbrugge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B. Functional analysis of a light-responsive plant bZIP transcription factor. Plant Cell. 1994;6:1607–1621. doi: 10.1105/tpc.6.11.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelnik J, Chua NH. Molecular light switches for plant genes. Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilano G, Hoffman NE, Ko K, Scolnick PA, Cashmore AR. A light-entrained circadian clock controls transcription of several plant genes. EMBO J. 1988;7:3635–3642. doi: 10.1002/j.1460-2075.1988.tb03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, O'Shea EK, Kim PS, Sauer RT. Sequence requirements for coiled-coils: analysis with λ repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH. Plant bZIP protein DNA binding specificity. J Mol Biol. 1993;230:1131–1144. doi: 10.1006/jmbi.1993.1230. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, Goldberg RB. Analysis of plant gene structure. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford, UK: IRL Press; 1988. pp. 37–66. [Google Scholar]
- Kageyama R, Pastan I. Molecular cloning and characterization of a human DNA binding factor that represses transcription. Cell. 1989;59:815–825. doi: 10.1016/0092-8674(89)90605-3. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S and others. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants, Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- Kusano T, Berberich T, Harada M, Suzuki N, Sugawara K. A maize DNA-binding factor with a bZIP motif is induced by low temperature. Mol Gen Genet. 1995;248:507–517. doi: 10.1007/BF02423445. [DOI] [PubMed] [Google Scholar]
- Landschultz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Learned RM. Light suppresses 3-hydroxy-3-methylglutaryl coenzyme A reductase expression in Arabidopsis thaliana. Plant Physiol. 1996;110:645–655. doi: 10.1104/pp.110.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Washburn T, Chory J. Regulation of gene expression by light. Curr Opin Cell Biol. 1993;5:455–460. doi: 10.1016/0955-0674(93)90011-e. [DOI] [PubMed] [Google Scholar]
- Li L, Lagarias CJ. Phytochrome assembly in living cells of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:12533–12539. doi: 10.1073/pnas.91.26.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden PJ. Circadian rhythms and phytochrome. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:351–371. [Google Scholar]
- Menkens AE, Cashmore AR. Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA. 1994;91:2522–2526. doi: 10.1073/pnas.91.7.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA. Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell. 1991;3:541–550. doi: 10.1105/tpc.3.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua NH. A circadian clock regulates transcription of the wheat Cab-1 gene. Genes Dev. 1988;2:376–382. [Google Scholar]
- Nantel A, Quatrano RS. Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem. 1996;271:31296–31305. doi: 10.1074/jbc.271.49.31296. [DOI] [PubMed] [Google Scholar]
- O'Neill SD. The photoperiodic control of flowering: progress towards understanding the mechanism of induction. Photochem Photobiol. 1992;56:789–801. [Google Scholar]
- O'Neill SD, Zhang XS, Zheng CC. Dark and circadian regulation of mRNA accumulation in the short-day plant Pharbitis nil. Plant Physiol. 1994;104:569–580. doi: 10.1104/pp.104.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B. ‘Circadian clock’ directs the expression of plant genes. Plant Mol Biol. 1993;22:533–542. doi: 10.1007/BF00015982. [DOI] [PubMed] [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbaugh MG, Sabre M, Scandalios JG. Expression of the maize Cat3 catalase gene is under the influence of a circadian rhythm. Proc Natl Acad Sci USA. 1990;87:6853–6857. doi: 10.1073/pnas.87.17.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith ME, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun DS, Chang AC, Jenkins NA, Gilbert DJ, Copeland NG, Chang NA. Identification, molecular characterization, and chromosomal location of the cDNA encoding a novel leucine zipper motif-containing protein. Genomics. 1996;36:54–62. doi: 10.1006/geno.1996.0425. [DOI] [PubMed] [Google Scholar]
- Takimoto A (1967) The spectral dependence of photoperiodic responses. In S Imamura, ed, Physiology of Flowering in Pharbitis nil. Japanese Society of Plant Physiologists, Tokyo, Japan, pp 73–93
- Taylor WC. Transcriptional regulation by a circadian rhythm. Plant Cell. 1989;1:259–264. doi: 10.1105/tpc.1.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Vatten NC, Lu G, Feri RJ. A maize protein associated with the G-box binding complex has homology to brain regulatory proteins. Plant Cell. 1992;4:1295–1307. doi: 10.1105/tpc.4.10.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Armstrong GA, Block A, Silva OC, Hahlbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promotor element with functional relevance in light responsiveness. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng CC, Bui AQ, O'Neill SD. Abundance of an mRNA encoding a high mobility group DNA-binding protein is regulated by light and an endogenous rhythm. Plant Mol Biol. 1993;23:813–823. doi: 10.1007/BF00021536. [DOI] [PubMed] [Google Scholar]
- Zhong HH, McClung CR. The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet. 1996;251:196–203. doi: 10.1007/BF02172918. [DOI] [PubMed] [Google Scholar]