Abstract
Paraoxonase (PON1) is an A-esterase capable of hydrolyzing the active metabolites (oxons) of a number of organophosphorus (OP) insecticides such as parathion, diazinon and chlorpyrifos. PON1 activity is highest in liver and in plasma. Human PON1 displays two polymorphisms in the coding region (Q192R and L55M) and several polymorphisms in the promoter and the 3’-UTR regions. The Q192R polymorphism imparts differential catalytic activity toward some OP substrates, while the polymorphism at position –108 (C/T) is the major contributor of differences in the levels of PON1 expression. Both contribute to determining an individual's PON1 “status”. Animal studies have shown that PON1 is an important determinant of OP toxicity. Administration of exogenous PON1 to rats or mice protects them from the toxicity of specific OPs. PON1 knockout mice display a high sensitivity to the toxicity of diazoxon and chlorpyrifos oxon, but not of paraoxon. In vitro catalytic efficiencies of purified PON192 alloforms for hydrolysis of specific oxon substrates accurately predict the degree of in vivo protection afforded by each isoform. Evidence is slowly emerging that a low PON1 status may increase susceptibility to OP toxicity in humans. Low PON1 activity may also contribute to the developmental toxicity and neurotoxicity of OPs, as shown by animal and human studies.
Keywords: Paraoxonase 1, genetic polymorphisms, organophosphates
Introduction
The important discoveries that certain organophosphorus (OP) insecticides could be enzymatically hydrolyzed by plasma (Mazur, 1946), and that this reaction is catalyzed by enzymes which were named “A-esterases”, were reported in the 1940s and 1950s (Mazur, 1946; Aldridge, 1953). Decades later it was shown that recombinant paraoxonase/arylesterase catalyzes the hydrolysis of paraoxon, the active metabolite of the OP insecticide parathion (Gan et al. 1991). Earlier studies indicated that the plasma hydrolytic activity toward paraoxon was distributed polymorphically in human populations (Playfer, 1976; Eckerson et al. 1983; Mueller et al. 1983), suggesting a possible genetically-based differential susceptibility to OP toxicity. The molecular basis of the polymorphisms of paraoxonase 1 (PON1), and their role in the toxicity of OP compounds have since been extensively investigated. Furthermore, novel important roles of PON1 in the metabolism of oxidized lipids, in the bioactivation or detoxication of certain drugs, and in the hydrolysis of quorum sensing factor, have also emerged, highlighting the importance of this enzyme in a number of biomedical fields. These latter aspects of PON1 are not discussed in this review, but are the topic of several recent publications (Costa and Furlong, 2002; Mackness et al. 2002; Costa et al. 2003a; Draganov and La Du, 2004; Ozer et al. 2005; Ng et al. 2005; Camps et al. 2009a).
PON1 and its human polymorphisms
PON1 is a member of a family of proteins that also includes PON2 and PON3, the genes of which are clustered in tandem on the long arms of human chromosome 7 (q21.22) or mouse chromosome 6 (Primo-Parmo et al. 1996). PON1 is synthesized in the liver and a portion is secreted into the plasma, where it associates with high density lipoproteins (HDL) (Sorenson et al. 1999; Deakin et al. 2002). Low levels of PON1 may also be expressed in a number of tissues, primarily in epithelia (Marsillach et al. 2008). PON1 received its name from its ability to hydrolyze paraoxon, its first and most studied substrate. However, PON1 also hydrolyzes the active metabolites of other OP insecticides (e.g. chlorpyrifos oxon, diazoxon), as well as nerve agents such as sarin and soman. However, several other OP and carbamate insecticides are not hydrolyzed by PON1. Furthermore, only PON1 has OP esterase activity, while all three PONs are lactonases displaying overlapping but distinct substrate specificities for lactone hydrolysis (Draganov et al. 2005). For example, all three PONs can hydrolyze a number of acyl-homoserine lactones (acyl-HCL), molecules which mediate bacterial quorum-sensing signals, important in regulating expression of virulence factors and in inducing a host inflammatory response (Draganov et al. 2005; Teiber et al. 2008).
The crystal structure for a recombinant PON1 (rPON1) has been elucidated, and has been shown to be a six-bladed β-propeller, which in the central tunnel contains two calcium ions, one of which is essential for enzyme activity and the other for stability (Harel et al. 2004). However, this rPON1 differs from the human enzyme by at least 51 aminoacids (Trovaslet-Leroy et al. 2011), and the OP esterase activity of human PON1 and rPON1 are substantially different (Otto et al. 2009). Thus, the 3D structure of human PON1 remains to be determined.
As mentioned, studies in the early 1980s indicated that the plasma paraoxonase activity in human populations exhibited a polymorphic distribution, and individuals with high, intermediate, or low paraoxonase activity could be identified (Eckerson et al. 1983; Mueller et al. 1983). Within a decade, human PON1 was purified, cloned and sequenced (Gan et al. 1991; Furlong et al. 1991; Hassett et al. 1991), and its major polymorphisms were identified (Humbert et al. 1993; Adkins et al. 1993). Two common polymorphisms are present in the PON1 coding sequence: a Gln(Q)/Arg(R) substitution at position 192, and a Leu(L)/Met(M) substitution at position 55 (Humbert et al. 1993; Adkins et al. 1993). The gene frequency of PON1Q192 ranges from 0.75 for Caucasians of Northern European origin to 0.31 for some Asian populations (Brophy et al. 2002). Additional polymorphisms have been found in the non-coding region of the PON1 gene; a most significant one is that at position –108, with the –108C allele providing levels of PON1 about twice as high as those seen with the –108T allele (Leviev and James, 2000; Suehiro et al. 2000; Brophy et al. 2001a; 2001b). More than 160 single nucleotide polymorphisms, some in the coding regions and others in introns and regulatory regions of the gene, have been identified by complete re-sequencing of PON1 from several individuals (Jarvik et al. 2003). These polymorphisms have for the most part not been yet characterized, but may affect splicing efficiency, message stability or efficiency of polyadenylation. A few of them, however, have explained discrepancies found when comparing PON1 status (see below) and PCR analysis of codon 192 (Jarvik et al. 2003).
The coding region polymorphisms of PON1 have been investigated for effects on the catalytic efficiencies of hydrolysis of specific substrates. The L/M polymorphism at position 55 does not affect catalytic activity, but has been associated with plasma PON1 protein levels, with PON1M55 being associated with low plasma PON1 (Blatter Garin et al. 1997; Mackness et al. 1998). However, this appears to result primarily from linkage disequilibrium with the low efficiency –108T allele of the –108 promoter region polymorphism (Brophy et al. 2002). In contrast, the Q192R polymorphism significantly affects the catalytic efficiency of PON1. This polymorphism is substrate-dependent, as for example, the PON1R192 alloform hydrolyzes chlorpyrifos oxon and paraoxon more rapidly than PON1Q192 in vitro, while both PON1 alloforms hydrolyze diazoxon with the same efficiency (Li et al. 2000).
Establishing the PON1 status of individuals
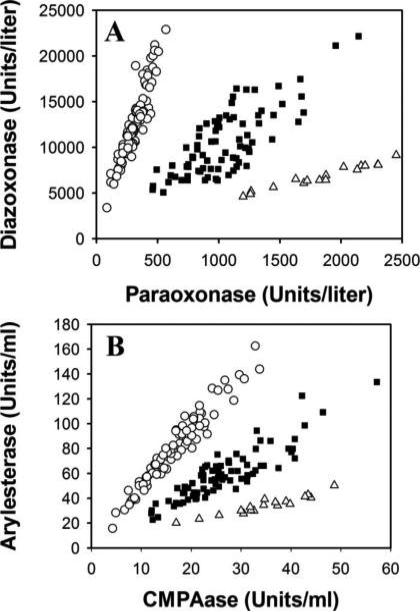
While most studies investigating the association of PON1 polymorphisms with diseases have examined the single nucleotide polymorphisms (Q192R, L55M, C-108T) with PCR-based assays, it has become apparent that measurements of an individual's PON1 function (plasma activity) takes into account all polymorphisms that might affect activity. This functional genomic analysis is accomplished through the use of a high-throughput enzyme assay involving two PON1 substrates (usually diazoxon and paraoxon) (Richter et al. 2004), or two non-toxic compounds (phenylacetate at high salt, and 4-(chloromethyl) phenylacetate at low salt) (Richter et al. 2008; 2009) (Fig. 1). This approach provides a functional assessment of the plasma PON1192 alloforms, including the plasma level of PON1 for each individual, thus encompassing the two factors that affect PON1 levels or activity (position 192 amino acid and plasma alloform levels). This approach has been referred to as the determination of PON1 “status” for an individual (Richter and Furlong, 1999). For adequate risk prediction, it is important to know two variables of PON1 (192 genotype and level), as high catalytic efficiency and high concentrations of PON1 are the two determinants of PON1 protection. In a given population, plasma PON1 activity can vary up to 40-fold (Eckerson et al. 1983; Mueller et al. 1983; Davies et al. 1996; Richter and Furlong, 1999), and differences in PON1 protein levels up to 13-fold are also present within a single PON1192 genotype in adults (Davies et al. 1996). Several studies investigating the role of PON1 in cardiovascular disease have provided evidence that PON1 status (encompassing genotype and activity levels) is a much better predictor of disease than PON1 genotype alone (Jarvik et al. 2000; Mackness et al. 2001; Zhao et al. 2012; Bayrak et al. 2012).
Fig. 1.
Comparison of two protocols for determining PON1 status. A. Assay utilizing paraoxon and diazoxon, two highly toxic substrates. B. Assay utilizing two non toxic substrate: phenylacetate and chloromethyl phenylacetate (CMPA). Genotypes (○, PON1QQ192; ■, PON1QR192; Δ, PON1RR192) are clearly identified in both assays, together with enzymatic activity for each genotype. Reproduced with permission from Richter et al. (2008).
PON1 modulates the toxicity of OPs: studies in animal models
While PON1 has been known for decades to hydrolyze a number of OP substrates in vitro, evidence of the role played by this enzyme in modulating the toxicity of OPs in vivo has emerged only in the past twenty years. Initially, exogenous PON1 was directly injected into rats or mice to determine whether increasing plasma PON1 levels would protect against OP toxicity. Administration (by the i.v. route) of the purified rabbit PON1 (as rabbits have high PON1 activity; Aldridge 1953; Costa et al. 1987) to rats, increased serum PON1 activity toward paraoxon by 9-fold, and that toward chlorpyrifos-oxon by 50-fold (Costa et al. 1990). Upon challenge with an acute dose of oxons given by various routes (including the dermal route which is relevant for occupational exposure), administration of PON1 afforded significant protection (as indicated by a lower degree of brain and diaphragm acetylcholinesterase (AChE) inhibition) particularly in case of chlorpyrifos oxon (Costa et al. 1990). Similarly in mice, administration of rabbit PON1 (i.v.) increased serum chlorpyrifos oxonase activity by 30-40 fold, and protected animals toward AChE inhibition by dermally applied chlorpyrifos oxon (Li et al. 1993). Administration of exogenous PON1 provided protection also against the toxicity of the parent compound, chlorpyrifos, given by the dermal route. Furthermore, PON1 exerted a protective effect even when given after (0.5-3.0 h) dermal administration of chlorpyrifos, suggesting a potential therapeutic use in OP poisoning, possibly in combination with other conventional treatments (Li et al. 1995).
PON1 knockout and PON1 transgenic animals have more recently provided new information on the role of PON1 in modulating OP toxicity. PON1 knockout (PON1-/-) mice, produced by targeted disruption of exon 1 of the PON1 gene, have no detectable hydrolytic activity toward paraoxon and diazoxon, and very limited chlorpyrifos-oxonase activity in plasma and liver (Shih et al. 1998; Li et al. 2000). PON1-/- mice do not differ from wild-type animals in their sensitivity to demeton-S-methyl, an OP insecticide which is not a substrate for PON1 (Li et al. 2000). In contrast, PON1-/- mice are dramatically more sensitive than wild-type animals to the acute toxicity of chlorpyrifos oxon and diazoxon, and also show a slight increase in sensitivity to the toxicity of chlorpyrifos and diazinon (Shih et al. 1988; Li et al. 2000). The most surprising finding of these studies was that PON1-/- mice did not show an increased sensitivity to paraoxon, the substrate after which the enzyme was named, in spite of having no paraoxonase activity in plasma and liver (Li et al. 2000).
Administration of exogenous PON1 to PON1-/- mice, to restore serum PON1, also restored resistance to OP toxicity. In these experiments, instead of rabbit PON1 as in the earlier experiments, purified human PON1Q192 or PON1R192 was injected, by the i.v. route, into PON1-/- mice to yield equal plasma PON1 levels (Li et al. 2000). PON1R192 provided significantly better protection than PON1Q192 toward chlorpyrifos oxon, as also confirmed in a subsequent study by Cowan et al. (2001), who administered recombinant adenoviruses containing PON1-LQ or PON1-LR genes to BALB/c mice before challenge with chlorpyrifos oxon. In contrast, both alloforms were equally effective in protecting against the toxicity of diazoxon (Li et al. 2000), while neither PON1R192 nor PON1Q192 afforded protection against paraoxon toxicity (Li et al. 2000). The results of these in vivo experiments have been explained by in vitro kinetic analyses of substrate hydrolysis by purified human PON1 alloforms. Indeed, in the case of chlorpyrifos oxon, the catalytic efficiency of both PON1 alloforms is very high, and is higher for the PON1R192 alloform than the PON1Q192 alloform. In the case of diazoxon, the catalytic efficiency is still high (albeit lower than with chlorpyrifos oxon), and no alloform-specific differences are evident. With paraoxon, the PON1R192 alloform is much more efficient than the PON1Q192 alloform; however, its overall catalytic efficiency is too low to protect against exposure, confirming the hypothesis (Chambers et al. 1994; Pond et al. 1995) that PON1 may not degrade paraoxon efficiently in vivo. Further support for these conclusions has derived from studies in PON1 transgenic mice. TgHuPON1R192 mice (i.e. mice expressing human PON1R192 on a knockout background) were significantly less sensitive to the toxicity of chlorpyrifos oxon than tgHuPON1Q192 mice, though they have the same level of PON1 protein in liver and plasma (Cole et al. 2005). Furthermore, tgHuPON1R192 mice are also less sensitive than tgHuPON1Q192 mice to the toxicity of the parent compound chlorpyrifos (Cole et al. 2005). Injection of PON1-/- mice with engineered recombinant human PON1 protected them against the toxicity of diazoxon administered either before or after the enzyme (Stevens et al. 2008).
These animal experiments indicate that PON1 exerts protection against OP toxicity, and that protection varies depending on the specific OP compound. In the case of chlorpyrifos oxon, both the level of expression and the Q192R genotype are important determinants of susceptibility, highlighting the importance of assessing PON1 status in potentially exposed individuals. With diazoxon, protection or susceptibility is dictated primarily by the level of expression of PON1, independently of the Q192R genotype, stressing the importance of knowing PON1 levels. In contrast, PON1 status does not seem to play an important role in modulating sensitivity to paraoxon toxicity.
A further facet of the role of PON1 in modulating OP toxicity is its potential influence on the outcome of combined OP exposures. Jansen et al. (2009) showed that three OPs (chlorpyrifos oxon, diazoxon, and paraoxon) potentiate the toxicity of malaoxon, and that the degree of potentiation is dependent upon PON1 status. Malaoxon is not a substrate of PON1, but is metabolized in vivo by carboxylesterase (CarE), and inhibition of CarE potentiates maloxon toxicity (Cohen and Murphy, 1971). Chlorpyrifos oxon, diazoxon, and paraoxon, at doses that caused only minimal inhibition of brain AChE, significantly inhibited CarE activity. All these three OPS potentiated the toxicity of malaoxon. However, the degree of potentiation differed depending on PON1 status. For chlorpyrifos oxon and diazoxon, a much greater potentiation was observed in PON1-/- mice, which are unable to detoxify these compounds. When comparing tgHuPON1Q192 and tgHuPON1R192 transgenic mice in the same experimental paradigm, the potentiation of maloxon toxicity by chlorpyrifos oxon was more pronounced in tgHuPON1Q192 animals, because of their lower ability to detoxify this compound. In contrast, no differences were observed in case of diazoxon. With paraoxon, potentiation of malaoxon toxicity was similar in all mouse genotypes. These results show that low doses of major OP insecticides can potentiate the toxicity of malaoxon, and that PON1 status can greatly influence the outcome of the interaction between the OPs and malaoxon, in a PON1 substrate-dependent manner (Jansen et al. 2009).
PON1 status and susceptibility to OP toxicity: human studies
As discussed in the previous section, animal studies have clearly indicated that PON1 modulates the toxicity of some OPs. Given that the Q192R polymorphism and the level of PON1 expression determine PON1 status, one would expect the latter to modulate susceptibility of humans to OPs. Evidence for a role of PON1 in OP toxicity in humans is still limited, though important findings have been emerging in the past few years, related to exposures to OP nerve agents or to OP insecticides.
Studies in Japan, in individuals exposed to the nerve agent sarin in two terrorist attacks which caused several deaths and thousands of injuries (Suzuki et al. 1995), failed to observe protection from acute sarin poisoning by the PON1Q192 genotype which confers high hydrolyzing activity toward sarin (Davies et al. 1996; Yamada et al. 2001). However, the range of sarinase activity among individuals with the QQ or QR genotype ranges from 0 to 758 U/L (Davies et al. 1996), and no determinations of PON1 status were done in the Yamada et al. (2001) study. Furthermore, the catalytic efficiency of sarin hydrolysis by PON1 is low, a situation thus similar (though reversed) to that of paraoxon. Finally, these individuals were exposed to very high doses of sarin, as all died within 48 h, and this would overcome any potential protection afforded by the PON1Q192 genotype (Yamada et al. 2001).
Various studies investigated PON1 polymorphisms in military personnel deployed in the Persian Gulf area in 1990-91, who were exposed to low levels of sarin and of OP insecticides, in addition, however, to several other biological and chemical agents (IOM, 2000; 2003). Haley et al. (2000) reported that PON1R192 homozygotes or PON1Q/R192 heterozygotes were more likely to have neurologic symptoms than individuals homozygous for PONQ192. Low activity of the plasma PON1Q192 isoform also appeared to better correlate with illness than the PON1 genotype or the activity levels of the PON1R192 genotype (Haley et al. 2000). This small study may suggest low PON1 as a risk factor for illness in Gulf War veterans, though further confirmation in a larger population is needed (Furlong, 2000). In another study on Gulf War veterans, plasma paraoxonase activity and levels of PON1 protein were lower in veterans than in a control group, and these decreases were independent of the PON1 genotype (Mackness et al. 2000). A third study found that individuals who were deployed to the Gulf had 25-35% lower median PON1 values than two other groups, and these differences were not explained by differences in PON1192 genotypes. However, PON1192 genotype and activity were not associated with specific symptoms; hence the findings remain somewhat inconclusive (Hotopf et al. 2003).
A series of studies investigated the potential role of PON1 in modulating chronic central and/or peripheral nervous system abnormalities, at times referred to as “dipper's flu”, associated with exposure of sheep dippers to diazinon. In an initial study, diazoxonase activity was lower in cases than referents (Cherry et al. 2002); however, in a follow-up study in the same populations, serum activity toward diazoxon did not differ between cases and controls (Mackness et al. 2003). Nevertheless, when the two groups were divided into quintiles according to the capacity of their serum to hydrolyze diazoxon, sheep dippers in the lowest quintile had a greater risk of reporting ill health than those in the other quintiles (Mackness et al. 2003). A further study also suggested that the risk associated with PON1 polymorphisms may vary with genotypes of CYP2D6 (which bio-activates diazinon) (Povey et al. 2007). On the basis of these and of additional similar findings (O'Leary et al. 2005; Mackenzie Ross et al. 2010; Cherry et al. 2011) it is suggested that diazinon has contributed to the ill health of those sheep dippers who had a lower ability to detoxify diazoxon.
In one study of South African workers, symptoms consistent with chronic OP toxicity were significantly more likely among subjects with the QQ or QR genotypes than the RR genotype (Lee et al. 2003). However, no indication of which OP was involved in exposure was provided. Another occupational study reported that Brazilian agricultural workers homozygotes for PON1Q192 presented higher genotoxic effects (as determined by a higher lymphocyte micronucleus frequency) than other workers (da Silva et al. 2008). However, this population of workers was exposed to a large number of pesticides; among the few OPs, none was a known PON1 substrate. Thus, the significance of this finding is obscure, at best. In a similar study in India, agricultural workers exposed to unknown OP insecticides were reported to have higher DNA damage in lymphocytes if they had low PON1 status (only identified as QQ/MM genotypes) (Singh et al. 2011). An additional study investigated the association between PON1 status, OP exposure and thyroid function in Spain (Lacasana et al. 2010). Results indicate that impaired thyroid function in OP-exposed agricultural workers was greater in individuals with lower PON1 activity.
Residential exposure to OPs in a rural community was found to be associated with an increased risk of Parkinson's disease in carriers of the PON155M variant (low PON activity). Of interest is that the odds ratios for diazinon (2.2) and for chlorpyrifos (2.6) were statistically significant, while no increased risk was observed for parathion (Manthripragada et al. 2010). In a similar study, residential exposure to diazinon and chlorpyrifos was associated with an increased risk of brain tumors in children with low PON1 activity (Searles Nielsen et al. 2005).
In another recent study in pesticide handlers in Washington State, exposed to various OPs (most notably chlorpyrifos) and carbamates, low PON1 catalytic efficiency (Q192) and low plasma PON1 activity were associated with increased degrees of plasma butyrylcholinesterase (BuChE) inhibition from baseline levels (Hoffman et al. 2009) (Table 1). In contrast to these findings, however, in workers involved in the manufacturing of chlorpyrifos and exposed to this OP, the decreased plasma BuChE activity was not associated with low PON1 status (Albers et al. 2010).
Table 1.
Changes in plasma BuChE activity from baseline in pesticide handlers after stratification by PON1Q192R genotype and plasma PON1 activity
| Q192R Genotype | PON1 activity |
||
|---|---|---|---|
| High | Moderate | Low | |
| RR | +0.53% | -0.11% | -8.22%* |
| QR | -2.06% | -6.17%* | -7.58%* |
| -9.47%* | -7.23%* | -12.15%* | |
PON1 activity was determined as arylesterase activity: high (>145 U/mL); moderate (124-145 U/mL); low (<124 U/mL).
Significantly different from reference group (RR high), p<0.05 or more. Total n was 163; n in individual groups ranged from 7 to 29. Adapted from Hoffman et al. (2009).
Overall, the human studies available so far provide initial evidence that low PON1 status may increase susceptibility to adverse effects of certain OP insecticides. Nevertheless, while animal studies indicate that PON1 status is an important determinant of sensitivity to acute toxicity of certain OPs, it is still uncertain whether PON1 status may play a significant role at lower dose levels of exposure. For example, using a physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model, Timchalk et al. (2002) carried out a Monte Carlo analysis of the human PON1Q192R polymorphism, and calculated theoretical brain concentrations of chlorpyrifos oxon for each PON1192 genotype, following different single doses of chlorpyrifos. The results suggested that PON1 status may influence chlorpyrifos oxon brain dosimetry, and hence toxicity, at high chlorpyrifos doses, but not at lower doses, closer to the Reference dose (RfD) (Timchalk et al. 2002). Such conclusion was also reached by Cole et al. (2005). Further studies in which exposure to OPs and PON1 status are carefully characterized would be needed to ascertain individual susceptibility to OPs’ adverse effects upon chronic low level exposure. Without such confirmatory studies, genetic testing in the work place to screen for individual at high risk for OP-induced adverse health effects appears to be premature (Battuello et al. 2004).
PON1 and the developmental toxicity and neurotoxicity of OPs
In addition to genetic polymorphisms, age is also an important determinant of PON1 activity. Studies in rodents have shown that activity of PON1 in serum and liver is very low at birth, and increases up to postnatal day 21, with a parallel increase in liver mRNA (Mortensen et al. 1996; Li et al. 1997; Moser et al. 1998; Karanth and Pope, 2000). A similar increase was also seen in transgenic mice expressing either the human PON1R192 or the PON1Q192 alloforms under the control of the human PON1 regulatory sequences, indicating conservation of the developmental regulatory elements between human and mouse PON1 (Cole et al. 2003). Low PON1 activity during development may represent a relevant risk factor for increased susceptibility to the toxicity of certain OP insecticides. The toxicity of OPs is influenced by age, as young animals are more sensitive than adults to their acute cholinergic effects (Harbison, 1975; Pope and Liu, 1997; Moser et al. 1998). Such increased susceptibility is likely due to a lower metabolic detoxication in young animals (Benke and Murphy, 1975; Murphy, 1982). In case of chlorpyrifos, it has been shown that a lower hydrolytic detoxication by PON1 accounts for the differential age-related sensitivity in acute toxicity (Mortensen et al. 1996; Moser et al. 1998; Padilla et al. 2000).
Recent studies in mice have shown that administration of chlorpyrifos oxon (various dose levels on postnatal days 4-21) causes no inhibition of brain AChE in wild-type and tgHuPON1R192 mice, but significant inhibition in tgHuPON1Q192 and in PON1-/- mice (Cole et al. 2011). Thus, low PON1 levels during early development contribute to the greater sensitivity of young animals to the acute toxicity of certain OPs, and a low PON1 status (exemplified here by the PON1-/- and the tgHuPON1Q192 mice) may further exacerbate the neurotoxic effects of OPs. Postnatal repeated exposure to chlorpyrifos oxon also caused significant alterations in gene expression in the cerebellum, particularly in the domains of mitochondrial-related genes, oxidative stress, and synaptic transmission, with more pronounced effects in PON1-/- and tgHuPON1Q192 mice (Cole et al. 2011).
Studies in humans have also shown that serum PON1 activity is very low at birth and increases over time, reaching a plateau between 6 months and a few years of age, depending on the study (Augustinsson and Barr, 1963; Ecobichon and Stephens, 1973; Mueller et al. 1983; Cole et al. 2003; Chen et al. 2003; Holland et al. 2006; Huen et al. 2010; Smith et al. 2011). PON1 activity may be even lower before birth, as indeed indicated by data showing a 24% lower activity in premature babies (33-36 weeks of gestation) compared to term babies (Ecobichon and Stephens, 1973), and by very low levels of PON1 expression in human fetal liver (Parker-Katiraee et al. 2008). Furthermore, an expectant mother with low PON1 status would not be able to provide protection for her fetus against exposure to some OPs (Cole et al. 2003).
In a study in New York City, offspring of mothers with low PON1 activity exposed in utero to chlorpyrifos were found to have smaller head circumference compared to those born to mothers with high PON1 activity or those not exposed to chlorpyrifos (Berkowitz et al. 2004). This finding would suggest that prenatal exposure to chlorpyrifos may be detrimental in offspring of mothers with low PON1 activity. Another study examined PON1 status in 130 women and their newborns (Furlong et al. 2006). Among newborns, levels of PON1 (measured as arylesterase activity) varied by 26-fold, and among mothers, by 14-fold. On average, newborns’ PON1 levels were four-fold lower than the mothers’ PON1 levels. Average PON1 levels in newborns were comparable with hPON1 levels in transgenic mice expressing PON1Q192 or PON1R192, allowing for prediction of relative sensitivity to diazoxon and chlorpyrifos oxon. For diazoxon, since it is hydrolyzed by both hPON1 alloforms with the same catalytic efficiency (Li et al. 2000), the range of sensitivity would be 26-fold in newborns and 14-fold in mothers, with an average fourfold difference between mothers and newborns, and a range of 65-fold from the most sensitive newborn to the most resistant mother (Furlong et al. 2006). For chlorpyrifos oxon, since tgHuPON1Q192 mice were found to be 2.0-2.5-fold more sensitive than tgHuPON1R192 mice (Cole et al. 2005), the overall predicted variability in sensitivity between newborns and mothers would be estimated to range between 131 and 165-fold (Furlong et al. 2006). These observations suggest that most newborns, as well as many mothers, would be expected to be more susceptible to the acute effects of certain OPs, due to their PON1 status. These extrapolations would be relevant at high oxon exposure levels; whether they may apply to lower level of exposure to the parent insecticides needs to be established (Timchalk et al. 2002; Cole et al. 2005).
Additional studies provide evidence that a low PON1 status may be associated with adverse effects upon developmental exposure to OP insecticides. In infants born to OP-exposed Mexican-American women in California, shorter gestational age and smaller head circumference was associated with low PON1 levels (Harley et al. 2011). In the same population, low PON1 status in children at age two was associated with poorer scores in various tests for mental and psychomotor development (Eskenazi et al. 2010). Similar results were reported for a population of New York children (the Mount Sinai Children's Environmental Health Study); prenatal OP exposure was associated with decrements in mental development at 12 and 24 months and at 6-9 years, particularly when mothers had a low PON1 status (Engel et al. 2011).
Dietary and pharmacological approaches for a positive modulation of PON1
Given the role of PON1 in protecting against toxic OP exposures and various diseases, particularly cardiovascular disease (Costa et al. 2003; Androutsopoulus et al. 2011), and its decrease in a number of pathological conditions (Camps et al. 2009a), it is not surprising that continuous efforts are devoted at identifying factors that may increase PON1 activity and/or expression (Costa et al. 2005; 2011; Camps et al. 2009b; Schrader and Rimbach, 2011). As said, a major determinant of PON1 activity is represented by genetic polymorphisms; in addition, age and gender may also play a role, with lower activity in early development and in aging, and a higher activity in females (Costa et al. 2005).
A number of drugs have been shown to increase PON1 levels and/or activity. These include several cardiovascular drugs, in particular the hypolipidemic statins, and some anti-diabetic drugs (e.g. rosiglitazone) (Costa et al. 2005; 2011). However, since such pharmaceutical drugs also have potential adverse health effects, chronic administration to healthy individuals with the purpose of increasing PON1 would not be recommended. Most attention has thus been devoted to identifying dietary intervention strategies that may increase PON1. Since PON1 is inactivated by oxidative stress (Nguyen and Sok, 2003) several studies have examined the effects of antioxidants. Two vitamins, ascorbic acid (Vit. C) and alpha-tocopherol (Vit. E), appear to increase PON1 activity in animals but results in humans are inconclusive (see Costa et al. 2011 for references). Of particular interest are the results of in vitro, and of animal and human studies, with various polyphenols. These studies indicate that quercetin, green tea catechins, resveratrol, and pomegranate juice polyphenols are capable of increasing PON1 activity and/or levels in both animals and humans (see Costa et al. 2011 for references). Low alcohol consumption also has a positive effect on PON1 levels (Rao et al. 2003).
In all cases, increases of PON1 are relatively small, less than 2-fold, most often of about 50% or less. However, as knowledge of the mechanisms regulating PON1 expression increases, it may be possible to target specific signal transduction pathways or transcription factors to provide a higher stimulatory effect. An alternative or complimentary approach would be that of administering exogenous PON1. As discussed earlier, PON1 confers protection toward the toxicity of OP insecticides by acting as a catalytic scavenger. Important issues to be addressed in this regard are: 1) engineering PON1 with higher catalytic efficiency (Stevens et al. 2008); 2) solving the problem of immunogenicity (Trovaslet-Lery et al. 2011); 3) devising novel modes of PON1 delivery (Zhang et al. 2010); and 4) addressing the issue of the relatively low half-life of exogenous PON1 (Li et al. 2003; 2005).
Conclusions
Polymorphisms in the PON1 gene influence both the quality and the quantity of PON1 (i.e. PON1 status). Evidence provided by in vitro and by animal studies indicates that PON1 plays a relevant role in the metabolism of certain OPs and modulates their acute toxicity. Though these studies provide strong evidence that PON1 levels and, in some cases, the Q192R polymorphism, determine the efficiency with which an individual will detoxify a specific OP, further proof in human populations is still needed. In particular, additional studies are needed where PON1 status is correlated with the degree of exposure (e.g. AChE/BuChE inhibition, OP-protein adducts) and/or with signs and symptoms of toxicity.
Acknowledgements
Research by the authors was supported by grants from the National Institutes of Health (ES04696, ES07033, ES09883, ES09601/EPA-R826886).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins S, Gan KN, Mody M, LaDu BN. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am. J. Hum. Genet. 1993;53:598–608. [PMC free article] [PubMed] [Google Scholar]
- Albers JW, Garabrant DH, Berent S, Richardson RJ. Paraoxonase status and plasma butyrylcholinesterase activity in chlorpyrifos manufacturing workers. J. Expos. Sci. Environ. Epidemiol. 2010;20:79–89. doi: 10.1038/jes.2009.9. [DOI] [PubMed] [Google Scholar]
- Aldridge WN. Serum esterases I. Two types of esterase (A and B) hydrolyzing p-nitrophenyl acetate, proprionate and butyrate and a method for their determination. Biochem. J. 1953;53:110–117. doi: 10.1042/bj0530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androutsopoulos VP, Kanavouras K, Tsatsakis AM. Role of paraoxonase 1 (PON1) in organophosphate metabolism: implications in neurodegenerative diseases. Toxicol. Appl. Pharmacol. 2011;256:418–424. doi: 10.1016/j.taap.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Augustinsson KB, Barr M. Age variation in plasma arylesterase activity in children. Clin. Chim. Acta. 1963;8:568–573. doi: 10.1016/0009-8981(63)90106-2. [DOI] [PubMed] [Google Scholar]
- Battuello K, Furlong CE, Fenske RA, Austin MA, Burke W. Paraoxonase polymorphisms and susceptibility to organophosphate pesticides. In: Khoury MJ, Little J, Burke W, editors. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. Oxford University Press; New York: 2004. pp. 305–321. [Google Scholar]
- Bayrak A, Bayrak T, Tokgozoglu SL, Vlokan-Salanci B, Deniz A, Yavuz B, Alikasifoglu M, Demirpence E. Serum PON1 activity but not Q192R polymorphism is related to the extent of atheriosclerosis. J. Atheroscl. Thromb. 2012;2012 doi: 10.5551/jat.11320. in press. [DOI] [PubMed] [Google Scholar]
- Benke GM, Murphy SD. The influence of age in the toxicity and metabolism of methylparathion and parathion in male and female rats. Toxicol. Appl. Pharmacol. 1975;31:254–269. doi: 10.1016/0041-008x(75)90161-1. [DOI] [PubMed] [Google Scholar]
- Berkowitz GS, Wetmur JG, Birman-Deych E, Obel J, Lapinski RH, Godbold JH, Holzman IR, Wolff MS. In utero pesticide exposure, maternal paraoxonase activity, and head circumference. Environ. Health Perspect. 2004;112:388–391. doi: 10.1289/ehp.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter Garin MC, James RW, Dussoix P, Blanche M, Passa P, Froguel P, Ruiz J. Paraoxonase polymorphism Met-Leu 54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J. Clin. Invest. 1997;99:62–66. doi: 10.1172/JCI119134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy VH, Jarvik GP, Furlong CE. PON1 polymorphisms. In: Costa LG, Furlong CE, editors. Paraoxonase (PON1) in Health and Disease: Basic and Clinical Aspects. Kluwer Academic Publishers; Norwell, MA: 2002. pp. 53–77. [Google Scholar]
- Brophy VH, Hastings MD, Clendenning JB, Richter RJ, Jarvik JP, Furlong CE. Polymorphisms in the human paraoxonase (PON1) promoter. Pharmacogenetics. 2001a;11:77–84. doi: 10.1097/00008571-200102000-00009. [DOI] [PubMed] [Google Scholar]
- Brophy VH, Jampsa RL, Clendenning JB, McKinstry LA, Furlong CE. Effects of 5' regulatory – region polymorphisms on paraoxonase gene (PON1) expression. Am. J. Hum. Genet. 2001b;68:1428–1436. doi: 10.1086/320600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Marsillach J, Joven J. The paraoxonases: role in human diseases and methodological difficulties in measurement. Crit. Rev. Clin. Lab. Sci. 2009a;46:83–106. doi: 10.1080/10408360802610878. [DOI] [PubMed] [Google Scholar]
- Camps J, Marsillach J, Joven J. Pharmacological and lifestyle factors modulating serum paraoxonase-1 activity. Mini Rev. Med. Chem. 2009b;9:911–920. doi: 10.2174/138955709788681591. [DOI] [PubMed] [Google Scholar]
- Chambers JE, Ma R, Boone JS, Chambers HW. Role of detoxication pathways in acute toxicity of phosphorothioate insecticides in the rat. Life Sci. 1994;54:1357–1364. doi: 10.1016/0024-3205(94)00515-x. [DOI] [PubMed] [Google Scholar]
- Chen J, Kumar M, Chan W, Berkowitz GS, Wetmur JG. Increased influence of genetic variation on PON1 activity in neonates. Env. Health Perspect. 2003;111:1403–1410. doi: 10.1289/ehp.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry N, Mackness MI, Durrington P, Povey A, Dippnall M, Smith T, Mackness B. Paraoxonase (PON1) polymorphisms in farmers attributing ill health to sheep dip. Lancet. 2002;359:763–764. doi: 10.1016/s0140-6736(02)07847-9. [DOI] [PubMed] [Google Scholar]
- Cherry N, Mackness M, Mackness B, Dippnall M, Povey A. “Dippers’ flu” and its relationship to PON1 polymorphisms. Occup. Environ. Med. 2011;68:211–217. doi: 10.1136/oem.2009.052126. [DOI] [PubMed] [Google Scholar]
- Cohen SD, Murphy SD. Carboxylesterase inhibition as an indicator of malathion potentiation in mice. J. Pharmacol. Exp. Ther. 1971;176:733–742. [PubMed] [Google Scholar]
- Cole TB, Jampsa RL, Walter BJ, Arndt TL, Richter RJ, Shih DM, Tward A, Lusis AJ, Jack RM, Costa LG, Furlong CE. Expression of human paraoxonase during development. Pharmacogenetics. 2003;13:1–8. doi: 10.1097/00008571-200306000-00007. [DOI] [PubMed] [Google Scholar]
- Cole TB, Walter J, Shih DM, Tward AD, Lusis AJ, Timchalk C, Richter RJ, Costa LG, Furlong CE. Toxicity of chlorpyrifos oxon in a transgenic mouse model of the human paraoxonase (PON1) Q192R polymorphism. Pharmacogenet. Genom. 2005;15:589–598. doi: 10.1097/01.fpc.0000167327.08034.d2. [DOI] [PubMed] [Google Scholar]
- Cole TB, Beyer RP, Bammler TK, Park SS, Farin FM, Costa LG, Furlong CE. Repeated developmental exposure of mice to chlorpyrifos oxon is associated with paraoxonase 1 (PON1) - modulated effects on cerebellar gene expression. Toxicol. Sci. 2011;123:155–169. doi: 10.1093/toxsci/kfr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG, Furlong CE, editors. Paraoxonase (PON1) in Health and Disease: Basic and Clinical Aspects. Kluwer Academic Publishers; Norwell, MA: 2002. p. 210. [Google Scholar]
- Costa LG, Richter RJ, Murphy SD, Omenn GS, Motulsky AG, Furlong CE. Species differences in serum paraoxonase correlate with sensitivity to paraoxon toxicity. In: Costa LG, Galli CL, Murphy SD, editors. Toxicology of Pesticides: Experimental, Clinical and Regulatory Perspectives. Springer-Verlag; Heidelberg: 1987. pp. 263–266. [Google Scholar]
- Costa LG, McDonald BE, Murphy SD, Omenn GS, Richter RJ, Motulsky AG, Furlong CE. Serum paraoxonase and its influence on paraoxon and chlorpyrifos-oxon toxicity in rats. Toxicol. Appl. Pharmacol. 1990;103:66–76. doi: 10.1016/0041-008x(90)90263-t. [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Jarvik GP, Furlong CE. Functional genomics of the paraoxonase (PON1) polymorphisms: effect on pesticide sensitivity, cardiovascular disease and drug metabolism. Annu. Rev. Med. 2003;54:371–392. doi: 10.1146/annurev.med.54.101601.152421. [DOI] [PubMed] [Google Scholar]
- Costa LG, Cole TB, Furlong CE. Paraoxonase (PON1): from toxicology to cardiovascular medicine. Acta Biomedica Suppl. 2005a;2:50–57. [PubMed] [Google Scholar]
- Costa LG, Vitalone A, Cole TB, Furlong CE. Modulation of paraoxonase activity. Biochem Pharmacol. 2005b;69:541–550. doi: 10.1016/j.bcp.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Costa LG, Giordano G, Furlong CE. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: the hunt goes on. Biochem. Pharmacol. 2011;81:337–344. doi: 10.1016/j.bcp.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan J, Sinton CM, Varley AW, Wians FH, Haley RW, Munford RS. Gene therapy to prevent organophosphate intoxication. Toxicol. Appl. Pharmacol. 2001;173:1–6. doi: 10.1006/taap.2001.9169. [DOI] [PubMed] [Google Scholar]
- da Silva J, Moraes CR, Heuser VD, Andrade VM, Silva FR, Kvitko K, Emmel V, Rohr P, Bordin DL, Andreazza AC, Salvador M, Henriques JAP, Erdtmann B. Evaluation of genetic damage in a Brazilian population occupationally exposed to pesticides and its correlation with polymorphisms in metabolizing genes. Mutagenesis. 2008;23:415–422. doi: 10.1093/mutage/gen031. [DOI] [PubMed] [Google Scholar]
- Davies H, Richter RJ, Kiefer M, Broomfield C, Sowalla J, Furlong CE. The human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nature Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- Deakin S, Leviev I, Gomeraschi M, Calabresi L, Franceschini G, James RW. Enzymatically active paraoxonase-1 is located at the external membrane of producing cells and released by a high-affinity, saturable, desorption mechanism. J. Biol. Chem. 2002;277:4301–4308. doi: 10.1074/jbc.M107440200. [DOI] [PubMed] [Google Scholar]
- Draganov DI, La Du BN. Pharmacogenetics of paraoxonases: a brief review. Naunyn-Schmiedeberg's Arch. Pharmacol. 2004;369:78–88. doi: 10.1007/s00210-003-0833-1. [DOI] [PubMed] [Google Scholar]
- Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, La Du BN. Human paraoxonases (PON1, PON2 and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005;46:1239–1247. doi: 10.1194/jlr.M400511-JLR200. [DOI] [PubMed] [Google Scholar]
- Ecobichon DJ, Stephens DS. Perinatal development of human blood esterases. Clin. Pharmacol. Ther. 1973;14:41–47. doi: 10.1002/cpt197314141. [DOI] [PubMed] [Google Scholar]
- Eckerson HW, Wyte CM, LaDu BN. The human serum paraoxonase/arylesterase polymorphism. Am. J. Hum. Genet. 1983;35:1126–1138. [PMC free article] [PubMed] [Google Scholar]
- Engel SM, Wetmur J, Chen J, Zhu C, Barr DB, Canfield RL, Wolff MS. Prenatal exposure to organophosphates, paraoxonase 1, and cognitive development in childhood. Environ Health Perspect. 2011;119:1182–1188. doi: 10.1289/ehp.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskenazi B, Huen K, Marks A, Harley KG, Bradman A, Barr DB, Holland N. PON1 and neurodevelopment in children from the CHAMACOS study exposed to organophosphate pesticides in utero. Environ. Health Perspect. 2010;118:1775–1781. doi: 10.1289/ehp.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong CE. PON1 status and neurologic symptom complexes in Gulf War Veterans. Genome Res. 2000;10:153–155. doi: 10.1101/gr.10.2.153. [DOI] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Chapline C, Crabb JW. Purification of rabbit and human serum paraoxonase. Biochemistry. 1991;30:10133–10140. doi: 10.1021/bi00106a009. [DOI] [PubMed] [Google Scholar]
- Furlong CE, Holland N, Richter RJ, Bradman A, Ho A, Eskenazi B. PON1 status of farmworkers mothers and children as a predictor of organophosphate sensitivity. Pharmacogenet. Genom. 2006;16:183–190. doi: 10.1097/01.fpc.0000189796.21770.d3. [DOI] [PubMed] [Google Scholar]
- Gan KN, Smolen AL, Eckerson HW, LaDu BN. Purification of human serum paraoxonase/arylesterase. Evidence for one esterase catalyzing both activities. Drug Metab. Dispos. 1991;19:100–106. [PubMed] [Google Scholar]
- Haley RW, Billecke S, LaDu BN. Association of low PON1 type Q (type A) arylesterase activity with neurologic symptom complexes in Gulf War veterans. Toxicol. Appl. Pharmacol. 2000;157:227–233. doi: 10.1006/taap.1999.8703. [DOI] [PubMed] [Google Scholar]
- Harbison RD. Perinatal development of human blood esterases. Clin. Pharmacol. Ther. 1975;14:41–47. doi: 10.1002/cpt197314141. [DOI] [PubMed] [Google Scholar]
- Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Revelli RBG, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nature Struct. Mol. Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- Harley KG, Huen K, Schall RA, Holland NT, Bradman A, Barr DB, Eskenazi B. Association of organophosphate pesticide exposure and paraoxonase with birth outcome in Mexican American weomen. PLoS One. 2011;6:e23923. doi: 10.1371/journal.pone.0023923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett C, Richter RJ, Humbert R, Chapline C, Crabb JW, Omiecinski CJ, Furlong CE. Characterization of DNA clones encoding rabbit and human serum paraoxonase: the mature protein retains its signal sequence. Biochemistry. 1991;30:10141–10149. doi: 10.1021/bi00106a010. [DOI] [PubMed] [Google Scholar]
- Hoffman JN, Keifer MC, Furlong CE, de Roos AJ, Farin FM, Fenske RA, Van Belle G, Checkoway H. Serum cholinesterase inhibition in relation to parapxonase-1 (PON1) status among organophosphate-exposed agricultural pesticide handlers. Environ. Health Perspect. 2009;117:1402–1408. doi: 10.1289/ehp.0900682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland N, Furlong CE, Bastaki M, Richter RJ, Bradman A, Huen K, Beckma K, Eskenazi B. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ. Health Perspect. 2006;114:985–991. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopf M, Mackness MI, Nikolau V, Collier DA, Curtis C, David A, Durrington P, Hull L, Ismail K, Peekman M, Unwin C, Wessely S, Mackness B. Paraoxonase in Persian Gulf veterans. J. Occup. Environ. Med. 2003;45:668–675. doi: 10.1097/01.jom.0000071506.96740.39. [DOI] [PubMed] [Google Scholar]
- Huen K, Harley K, Bradman A, Eskenazi B, Holland N. Longitudinal changes in PON1 enzymatic activities in Mexican-American mothers and children with different genotypes and haplotypes. Toxicol. Appl. Pharmacol. 2010;244:181–189. doi: 10.1016/j.taap.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase polymorphisms. Nature Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- IOM (Institute of Medicine) Gulf War and Health. Vol. 1. Depleted Uranium, Pyridostigmine Bromide, Sarin, Vaccines. National Academy Press; Washington, DC: 2000. [Google Scholar]
- IOM (Institute of Medicine) Gulf War and Health. Vol. 2. Insecticides and Solvents. National Academy Press; Washington, DC: 2003. [Google Scholar]
- Jansen KL, Cole TB, Park SS, Furlong CE, Costa LG. Paraoxonase 1 (PON1) modulates the toxicity of mixed organophosphorus compounds. Toxicol. Appl. Pharmacol. 2009;236:142–153. doi: 10.1016/j.taap.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik GP, Rozek LS, Brophy VH, Hatsukami TS, Richter RJ, Schellenberg GD, Furlong CE. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1192 or PON155 genotype. Arterioscler. Thromb. Vasc. Biol. 2000;20:2441–2447. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- Jarvik GP, Jampsa R, Richter RJ, Carlson CS, Rieder MG, Nickerson DA, Furlong CE. Novel paraoxonase (PON1) nonsense and missense mutations predicted by functional genomic assay of PON1 status. Pharmacogenetics. 2003;13:291–295. doi: 10.1097/00008571-200305000-00009. [DOI] [PubMed] [Google Scholar]
- Karanth S, Pope C. Carboxylesterase and A-esterase activities during maturation and aging: relationship to the toxicity of chlorpyrifos and parathion in rats. Toxicol. Sci. 2000;58:282–289. doi: 10.1093/toxsci/58.2.282. [DOI] [PubMed] [Google Scholar]
- Lacasana M, Lopez-Flores I, Rodriguez-Barranco M, Aguilar-Garduno C, Blanco-Munoz J, Perez-Mendez O, Gamboa R, Gonzales-Alzaga B, Bassol S, Cebrian ME. Intercation between organophosphate pesticide exposure and PON1 activity on thyroid function. Toxicol. Appl. Pharmacol. 2010;249:16–24. doi: 10.1016/j.taap.2010.07.024. [DOI] [PubMed] [Google Scholar]
- Lee BW, London L, Poulauskis J, Myers J, Christiani DC. Association between human paraoxonase gene polymorphism and chronic symptoms in pesticide-exposed workers. J. Occup. Environ. Med. 2003;45:118–122. doi: 10.1097/01.jom.0000052953.59271.e1. [DOI] [PubMed] [Google Scholar]
- Leviev I, James RW. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler. Thromb. Vasc. Biol. 2000;20:516–521. doi: 10.1161/01.atv.20.2.516. [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Furlong CE. Serum paraoxonase status: a major factor in determining resistance to organophosphates. J. Toxicol. Environ. Health. 1993;40:337–346. doi: 10.1080/15287399309531798. [DOI] [PubMed] [Google Scholar]
- Li WF, Furlong CE, Costa LG. Paraoxonase protects against chlorpyrifos toxicity in mice. Toxicol. Lett. 1995;76:219–226. doi: 10.1016/0378-4274(95)80006-y. [DOI] [PubMed] [Google Scholar]
- Li WF, Matthews C, Disteche CM, Costa LG, Furlong CE. Paraoxonase (PON1) gene in mice: sequencing, chromosomal localization and developmental expression. Pharmacogenetics. 1997;7:137–144. doi: 10.1097/00008571-199704000-00007. [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Richter RJ, Hagen T, Shih DM, Tward A, Lusis AJ, Furlong CE. Catalytic efficiency determines the in vivo efficacy of PON1 for detoxifying organophosphates. Pharmacogenetics. 2000;10:767–779. doi: 10.1097/00008571-200012000-00002. [DOI] [PubMed] [Google Scholar]
- Mackenzie Ross SJ, Brewin CR, Curran HV, Furlong CE, Abraham-Smith KM, Harrison V. Neuropsychological and psychiatric functioning in sheep farmers exposed to low levels of organophosphate pesticides. Neurotoxicol. Teratol. 2010;32:452–459. doi: 10.1016/j.ntt.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackness B, Mackness MI, Arrol T, Turkie W, Durrington PN. Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modifications. FEBS Lett. 1998;423:57–60. doi: 10.1016/s0014-5793(98)00064-7. [DOI] [PubMed] [Google Scholar]
- Mackness B, Durrington PN, Mackness MI. Low paraoxonase in Persian Gulf War veterans self-reporting Gulf War Syndrome. Biochem. Biophys. Res. Commun. 2000;276:729–733. doi: 10.1006/bbrc.2000.3526. [DOI] [PubMed] [Google Scholar]
- Mackness B, Davies GK, Turkie W, Lee E, Roberts DM, Hill E, Roberts C, Durrington PN, Mackness MI. Paraoxonase status in coronary heart disease: are activity and concentration more important than genotype? Arterioscler. Thromb. Vasc. Biol. 2001;21:1451–1457. doi: 10.1161/hq0901.094247. [DOI] [PubMed] [Google Scholar]
- Mackness MS, Mackness B, Durrington PN. Paraoxonase and coronary heart disease. Artheroscler. Suppl. 2002;3:49–55. doi: 10.1016/s1567-5688(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Mackness B, Durrington PN, Povey A, Thomson S, Dippnall M, Mackness M, Smith T, Cherry N. Paraoxonase and susceptibility to organophosphorus poisoning in farmers dipping sheep. Pharmacogenetics. 2003;13:81–88. doi: 10.1097/00008571-200302000-00004. [DOI] [PubMed] [Google Scholar]
- Manthripragada AD, Costello S, Cokburn MG, Bronstein JM, Ritz B. Paraoxonase 1, agricultural organophosphate exposure, and Parkinson disease. Epidemiology. 2010;21:87–94. doi: 10.1097/EDE.0b013e3181c15ec6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsillach J, Mackness B, Mackness M, Riu F, Beltran R, Joven J, Camps J. Immunohistochemical analysis of paraoxonases-1, 2 and 3 expression in normal mouse tissues. Free Rad. Biol. Med. 2008;45:146–157. doi: 10.1016/j.freeradbiomed.2008.03.023. [DOI] [PubMed] [Google Scholar]
- Mazur A. An enzyme in animal tissue capable of hydrolyzing the phosphorus-fluorine bond of alkyl fluorophosphates. J. Biol. Chem. 1946;164:271–289. [PubMed] [Google Scholar]
- Mortensen SR, Chanda SM, Hooper MJ, Padilla S. Maturational differences in chlorpyrifos-oxonase activity may contribute to age-related sensitivity to chlorpyrifos. J. Biochem. Toxicol. 1996;11:279–287. doi: 10.1002/(SICI)1522-7146(1996)11:6<279::AID-JBT3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Moser VC, Chanda SM, Mortensen SR, Padilla S. Age- and gender-related differences in sensitivity to chlorpyrifos in the rat reflect developmental profiles of esterase activities. Toxicol. Sci. 1998;46:211–222. doi: 10.1006/toxs.1998.2526. [DOI] [PubMed] [Google Scholar]
- Mueller RF, Hornung S, Furlong CE, Anderson J, Giblett ER, Motulsky AG. Plasma paraoxonase polymorphism: a new enzyme assay, population, family biochemical and linkage studies. Am. J. Hum. Genet. 1983;35:393–408. [PMC free article] [PubMed] [Google Scholar]
- Murphy SD. Toxicity and hepatic metabolism of organophosphate insecticides in developing rats. Banbury Report. 1982;11:125–136. [Google Scholar]
- Ng CJ, Shih DM, Hama SY, Villa N, Navab M, Reddy ST. The paraoxonase gene family and atherosclerosis. Free Rad. Biol. Med. 2005;38:153–163. doi: 10.1016/j.freeradbiomed.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Nguyen SD, Sok DE. Oxidative inactivation of paraoxonase 1 an antioxidant protein and its effect on antioxidant action. Free Rad. Res. 2003;37:77–83. [PubMed] [Google Scholar]
- O'Leary KA, Edwards RJ, Town MM, Boobis AR. Genetic and other sources of variation in the activity of serum paraoxonase/diazoxonase in humans: consequences for risk from exposure to diazinon. Pharmacogenet. Genom. 2005;15:51–60. doi: 10.1097/01213011-200501000-00008. [DOI] [PubMed] [Google Scholar]
- Otto TC, Harsch CK, Yeung DT, Magliery TJ, Cerasoli DM, Lenz DE. Dramatic differences in organophosphorus hydrolase activity between human and chimeric recombinant mammalian paraoxonase-1 enzymes. Biochemistry. 2009;48:10416–10422. doi: 10.1021/bi901161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer EA, Pezzulo A, Shih DM, Chun C, Furlong CE, Lusis AJ, Greenberg EP, Zabner J. Human and murine paraoxonase 1 are host modulators of pseudomonas aeruginosa quorum-sensing. FEMS Microbiol. Lett. 2005;253:29–37. doi: 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Padilla S, Buzzard J, Moser VC. Comparison of the role of esterases in the differential age-related sensibility to chlorpyrifos and metamidophos. Neurotoxicology. 2000;21:49–56. [PubMed] [Google Scholar]
- Parker-Katiraee L, Bousiaki E, Monk D, Moore GE, Nakabayashi K, Scherer SW. Dynamic variation in allele-specific gene expression of paraoxonase -1 in murine and human tissues. Hum. Mol. Genet. 2008;17:3263–3270. doi: 10.1093/hmg/ddn222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Playfer JR, Eze LC, Bullen MF, Evans DA. Genetic polymorphism and interethnic variability of plasma paraoxonase activity. J. Med. Genet. 1976;13:337–342. doi: 10.1136/jmg.13.5.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond AL, Chambers HW, Chambers JE. Organophosphate detoxication potential of various rat tissues via A-esterase and aliesterase activities. Toxicol. Lett. 1995;70:245–252. doi: 10.1016/0378-4274(95)03327-h. [DOI] [PubMed] [Google Scholar]
- Pope CN, Liu J. Age-related differences in sensitivity to organophosphorus pesticides. Environ. Toxicol. Pharmacol. 1997;4:309–314. doi: 10.1016/s1382-6689(97)10029-1. [DOI] [PubMed] [Google Scholar]
- Povey AC, Jury F, Dippnall WM, Smith AE, Thomson S, Mackness B, Mackness M, Durrington P, Cherry NM. GST, CYP and PON1 polymorphisms in farmers attributing ill health to organophosphate-containing sheep dip. Biomarkers. 2007;12:188–202. doi: 10.1080/13547500601043500. [DOI] [PubMed] [Google Scholar]
- Primo-Parmo SL, Sorenson RC, Teiber J, La Du BN. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–507. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- Rao MN, Marmillot P, Gong M, Palmer DA, Seef LB, Strader DB, Lakshman MR. Light, but not heavy alcohol drinking, stimulates paraoxonase by upregulating liver mRNA in rats and humans. Metabolism. 2003;10:1287–1294. doi: 10.1016/s0026-0495(03)00191-4. [DOI] [PubMed] [Google Scholar]
- Richter RJ, Furlong CE. Determination of paraoxonase (PON1) status requires more than genotyping. Pharmacogenetics. 1999;9:745–753. [PubMed] [Google Scholar]
- Richter RJ, Jampsa RL, Jarvik GP, Costa LG, Furlong CE. Determination of paraoxonase 1 status and genotypes at specific polymorphic sites. In: Maines M, Costa LG, Reed DJ, Hodgson E, editors. Current Protocols in Toxicology. John Wiley & Sons; New York: 2004. pp. 4.12.1–4.12.19. [DOI] [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, Furlong CE. Determination of paraoxonase 1 (PON1) status without the use of toxic organophosphate substrates. Circ. Cardiovasc. Genet. 2008;1:147–152. doi: 10.1161/CIRCGENETICS.108.811638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter RJ, Jarvik GP, Furlong CE. Paraoxonase 1 (PON1) status and substrate hydrolysis. Toxicol. Appl. Pharmacol. 2009;235:1–9. doi: 10.1016/j.taap.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C, Rimbach G. Determinants of paraoxonase 1 status: genes, drugs, nutrition. Curr. Med. Chem. 2011;18:5624–5643. doi: 10.2174/092986711798347216. [DOI] [PubMed] [Google Scholar]
- Searles Nielsen S, Mueller BA, DeRoos AJ, Viernes HM, Farin FM, Checkoway H. Risk of brain tumors in children and susceptibility to organophosphorus insecticide; the potential role of paraoxonase (PON1). Environ. Health Perspect. 2005;113:909–913. doi: 10.1289/ehp.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- Singh S, Kumar V, Thakur S, Banerjee BD, Rautela RS, Grover SS, Rawat DS, Pasha ST, Jain SK, Ichhpujani RL, Rai A. Paraoxonase-1 genetic polymorphisms and susceptibility to DNA damage in workers occupationally exposed to organophosphate pesticides. Toxicol. Appl. Toxicol. 2011;252:130–137. doi: 10.1016/j.taap.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Smith JN, Timchalk C, Bartels MJ, Poet TS. In vitro age-dependent enzymatic metabolism of chlorpyrifos and chlorpyrifos oxon in human hepatic microsomes and chlorpyrifos-oxon in plasma. Drug. Metab. Disp. 2011;39:1353–1362. doi: 10.1124/dmd.111.038745. [DOI] [PubMed] [Google Scholar]
- Sorenson RC, Bisgaier CL, Aviram M, Hsu C, Billecke S, LaDu BN. Human serum paraoxonase/arylesterase's retained hydrophobic N-terminal leader sequence associates with HDLs by binding phospholipids: apoprotein A-1 stabilizes activity. Arterioscler. Thromb. Vasc. Biol. 1999;19:2214–2225. doi: 10.1161/01.atv.19.9.2214. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Suzuki SM, Cole TB, Park SS, Richter RJ, Furlong CE. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc. Natl. Acad. Sci. USA. 2008;105:12780–12784. doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suehiro T, Nakamura T, Inoue M, Shiinoki T, Ikeda Y, Kumoin Y, Shindo M, Tanaka H, Hashimoto K. A polymorphism upstream from the human paraoxonase (PON1) gene and its association with PON1 expression. Atherosclerosis. 2000;150:295–298. doi: 10.1016/s0021-9150(99)00379-2. [DOI] [PubMed] [Google Scholar]
- Teiber JF, Horke S, Haines DC, Chowdhary PK, Xiao J, Kramer GL, Haley RW, Draganov DI. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-L-homoserine lactone. Infect. Immun. 2008;76:2512–2519. doi: 10.1128/IAI.01606-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchalk C, Kousba A, Poet TS. Monte Carlo analysis of the human chlorpyrifos oxonase (PON1) polymorphism using a physiologically based pharmacokinetic and pharmacodynamic (PBPK/PD) model. Toxicol. Lett. 2002;135:51–59. doi: 10.1016/s0378-4274(02)00233-3. [DOI] [PubMed] [Google Scholar]
- Trovaslet-Leroy M, Musilova L, Renault F, Brazzolotto X, Misik J, Novotny L, Froment MT, Gillon E, Loiodice M, Verdier L, Masson P, Rochu D, Jun D, Nachon F. Organophosphate hydrolases as catalytic bioscavengers of organophosphorus nerve agents. Toxicol. Lett. 2011;206:14–23. doi: 10.1016/j.toxlet.2011.05.1041. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Takatori T, Nagao M, Iwase H, Kurada N, Yanagida J, Shinozuka T. Expression of paraoxonase isoform did not confer protection from acute sarin poisoning in the Tokyo subway terrorist attack. Int. J. Leg. Med. 2001;115:82–84. doi: 10.1007/s004140100226. [DOI] [PubMed] [Google Scholar]
- Zhang C, Peng W, Wang M, Zhu J, Zang Y, Shi W, Zhang J, Qin J. Studies on protective effects of human paraoxonases 1 and 3 on atherosclerosis in apolipoprotein E knockout mice. Gene Ther. 2010;17:626–633. doi: 10.1038/gt.2010.11. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ma Y, Fang Y, Liu L, Wu S, Fu D, Wang X. Association between PON1 activity and coronary heart disease risk: a meta-analysis besed on 43 studies. Mol. Genet. Metab. 2012;105:141–148. doi: 10.1016/j.ymgme.2011.09.018. [DOI] [PubMed] [Google Scholar]