Abstract
Pathogenic Klebsiella pneumoniae, resistant to beta-lactam and quinolone drugs, is widely recognized as important bacteria causing array of diseases. The resistance property is obtained by acquisition of plasmid encoded blaTEM, blaSHV, blaCTX-M, QNRA, QNRB and QNRS genes. The aim of this study was to document the prevalence and association of these resistant genes in K. pneumoniae infecting patients in India. Approximately 97 and 76.7 % of the 73 K. pneumoniae isolates showed resistance towards beta-lactam and quinolone drugs respectively. Bla genes were detected in 74 % of K. pneumoniae isolates; with prevalence in the following order: blaTEM > blaSHV > blaCTXM. QNR genes were detected in 67 % samples. Chi-square analysis revealed significant association between presence of bla and qnr genes in our study (P value = 0.000125). Sequence analysis of some blaTEM, blaSHV, blaCTX-M and QNRB PCR products revealed presence of blaTEM1 (GenBank accession: JN193522), blaTEM116 (JN193523 and JN193524), blaSHV11, blaCTXM72 variants (JF523199) and QNRB1 (JN193526 and JN193527) in our samples.
Keywords: Klebsiella pneumoniae, beta-Lactamase, Quinolone, Resistance, India
Introduction
Multi-drug resistant Klebsiella pneumoniae is one of the important nosocomial and community acquired opportunistic bacteria causing urinary tract infection, pneumonia, septicemia, soft tissue infection etc. [1]. Resistance against beta-lactam and quinolone group of drugs, widely used for their treatment is mainly acquired by intra or inter species exchange of transferable plasmid encoded antibiotic resistant genes (viz blaTEM, blaSHV, blaCTX-M, QNR A, QNR B, and QNR S) [2]. Currently ≥150 TEM, ≥88 SHV, ≥69 CTX-M types variants has been described [3]. Six QNR A (QNR A1–QNR A6), 20 QNR B (QNR B1–QNR B20) and 3 QNR S variants (QNR S1–QNR S3) have been identified worldwide. Antibiotic sensitivity assays have revealed high frequencies of quinolone drug resistance in extended spectrum beta lactamase (ESBL) producing clinical isolates [4]. However, genetic evidence of this association is limiting and prevalence of these resistance genes in K. pneumoniae infecting Indian patients is not known [5–7].
This study demonstrated prevalence of both bla and qnr genes circulating in pathogenic K. pneumoniae of Indian origin. The occurrence of blaTEM and QNR B genes was observed in 52 %, blaCTX-M and QNR A genes in 37 % and blaSHV in 42 % samples individually. We hereby report significant genotypic association between these two groups of resistance genes (bla and QNR) in our samples as evidenced by Chi-square analysis. Sequence analysis of some of the genes indicated presence of blaTEM1, blaTEM116, blaSHV11, blaCTXM72 variant and QNRB1 circulating in pathogenic Klebsiella. Identification of these variants circulating in pathogenic bacteria might be helpful in proper therapeutic management of infected patients.
Materials and Methods
Bacterial Isolates
Klebsiella pneumoniae specimens (n = 73) were collected from clinical isolates of patients visiting Calcutta School of Tropical Medicine, Kolkata for 1 year (Table 1). Among the 73 isolates, 53 were isolated from urine, 10 from blood, 8 from throat swab and 2 from wound pus (data not shown). Forty-one of the patients with Klebsiella infection were female and the rest were male. Samples were processed for gram-stain and culture. Inoculation was done on nutrient agar, blood agar and Mac Conkey’s agar media and incubated overnight at 37 °C. Identification of K. pneumoniae isolates was done by standard biochemical methods [8].
Table 1.
Antibiotic sensitivity and resistant gene status of K. pneumoniae isolates
| Patient’s ID | blaTEM | blaSHIV | blaCTX-M | QNR A | QNR B | QNR S | ESBL/non ESBL | Quinolone resistance | Cephalosporin resistance |
|---|---|---|---|---|---|---|---|---|---|
| PP13 | − | − | − | + | − | − | + | + | + |
| U195/Dy8 | − | − | − | − | − | − | + | + | + |
| B39 | − | − | − | + | − | − | + | + | + |
| B40 | + | − | + | + | + | − | + | + | + |
| JG | + | − | − | − | − | − | + | + | + |
| SZ | + | + | − | + | + | − | − | + | + |
| IM | + | + | − | + | + | − | − | + | |
| U492(AP-1) | + | − | − | − | + | − | − | + | + |
| PP12 | + | + | − | + | + | − | + | + | + |
| Dy5 | − | − | − | − | + | − | + | + | |
| UCS(PP15) | + | + | + | + | + | − | + | + | + |
| Dy2 | − | − | − | − | − | − | + | + | |
| TS(JBC-6) | + | + | − | − | − | − | + | + | |
| NBC-4 | + | + | − | + | + | − | + | + | + |
| Hch-8 | − | + | − | − | + | − | + | − | + |
| SZ2 | − | − | − | − | + | − | − | − | + |
| AA | + | − | − | + | + | − | − | + | + |
| Dy-7 | + | + | − | + | + | − | − | + | − |
| Dp-1 | − | − | − | − | − | − | + | + | + |
| Dy-3 | + | + | − | − | + | − | + | + | + |
| PP16 | + | − | + | − | + | − | − | − | − |
| SP-O | + | + | − | + | − | − | + | − | |
| Lu-5K | − | − | − | − | − | − | + | + | + |
| T-216 | + | − | − | − | + | − | + | + | |
| B271 | + | − | + | + | − | + | + | + | |
| Lc-8 | − | − | − | − | − | − | + | + | + |
| AP-10 | + | + | + | + | + | − | − | + | + |
| SD | + | + | − | + | + | − | + | + | + |
| JBC-12 | + | + | + | + | + | − | + | + | + |
| B06 | + | − | + | + | + | − | − | + | + |
| HAD-1 | + | + | + | + | − | − | − | + | + |
| S.PATRA(NEW) | + | + | + | + | + | − | − | + | + |
| JBC-6 | + | + | + | + | − | − | + | + | |
| B-88 | + | − | − | + | + | − | + | + | + |
| SC1 | + | + | − | + | + | − | + | − | + |
| B86A | + | − | − | − | + | − | + | − | + |
| JNC-14 | − | − | − | − | − | − | + | + | |
| SP1 | − | − | − | − | − | − | + | − | + |
| B06 (2) | − | − | + | − | + | − | + | + | + |
| LU-6 | − | + | − | − | + | − | − | − | + |
| AP-9 | − | − | − | − | − | − | + | + | + |
| SC2 | + | + | − | − | + | − | + | − | + |
| SC3 | + | − | − | − | + | − | + | + | + |
| PM | − | − | − | − | − | − | + | + | + |
| DY-5 | − | − | + | − | − | − | − | + | + |
| JBC-6(NEW) | − | + | − | + | − | − | + | − | + |
| HAD-2 | − | − | − | − | − | − | − | + | + |
| SM | − | − | − | + | + | − | + | + | + |
| LU-10 | − | − | − | − | − | − | − | + | + |
| K17 | − | + | − | + | − | − | + | + | + |
| DY-4 | − | + | + | − | + | − | − | + | + |
| JNC-7 | − | + | + | + | + | − | − | + | + |
| Lu-5 | − | − | − | − | + | − | + | + | + |
| Lcab-5 | − | + | + | − | − | − | + | − | + |
| TS | + | + | + | − | − | − | + | − | + |
| DY-10 | − | + | + | − | − | − | + | + | |
| JNC-7 | + | + | + | − | − | − | + | − | + |
| HAD-2 | + | + | + | − | − | + | + | + | |
| K9 | − | − | + | − | − | + | + | + | |
| B01 | − | − | + | − | − | + | + | + | |
| BA | + | − | + | − | − | − | + | + | + |
| AP-5 | + | − | + | − | + | − | + | + | + |
| HAD-1 | − | − | − | − | − | − | − | − | + |
| BO5 | − | − | + | − | − | − | + | + | + |
| K13 | + | + | + | + | + | − | + | + | + |
| HB | − | + | − | − | + | − | + | − | + |
| PP4 | + | − | + | − | − | − | − | + | + |
| TM | − | − | − | − | − | − | − | + | + |
| LD | − | − | − | − | − | − | + | + | |
| HAD-1 | − | − | − | − | − | − | − | + | |
| BH | + | + | + | − | + | − | − | + | + |
| DY-2 | + | + | − | − | + | − | − | + | + |
| K-CABIN | + | − | − | − | + | − | − | + | + |
+: Presence of the specific parameters, −: absence of the specific parameters
Susceptibility Assay for beta-Lactum and Quinolone Antibiotics
Antibiotic susceptibility of these Klebsiella isolates was determined according to Kirby–Bauer disc diffusion method and results were interpreted according to guidelines of Clinical and Laboratory Standard Institute (CLSI: former NCCLS, 1993) [9]. The following beta-lactam and quinolone antibiotics were used (disc: 6 mm: μg): ceftazidim (30), ceftazidim and clavulenic acid (30/10), cefotaxim (30), cefotaxim and clavulenic acid (30/10), cefpodoxim (10), nalidixic acid (30), ciprofloxacin (5), prulifloxacine (10) and levofloxacine (5) (HiMedia Lab Ltd., India). K. pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as positive and negative controls respectively in each phenotypic ESBL test [10]. K. pneumoniae B1 was used as quinolone drug resistant positive control [11].
Preparation of Plasmid DNA
Plasmid DNA was extracted from the K. pneumoniae isolates by alkaline lysis method [12]. Briefly, 3 ml overnight culture of patient-sample isolated K. pneumoniae was lysozyme and RNase treated followed by alkaline lysis, phenol/chloroform extraction and isopropanol precipitation of the plasmid DNA. Integrity of the plasmids was checked by agarose gel electrophoresis.
PCR Amplification of blaTEM, blaSHV, blaCTX-M, QnrA, QnrB and QnrS Genes
Amplification of blaTEM, blaSHV, blaCTX-M, QNR A, QNR B and QNR S genes were performed in thermal cycler (Applied Biosystems, USA) using primers previously mentioned [13, 14]. Briefly each reaction was carried out in 20 μl reaction volume using 1x PCR buffer (Fermentus, USA), 20 pmol of primers (Integrated DNA Technologies, USA), 1 mM of each dNTPs, 1 unit of Taq polymerase (Fermentus, USA) and 100 ng plasmid DNA. The MgCl2 concentration varied between 1 and 2 mM. Thermocycling parameters were as follows: an initial denaturation of 94 °C for 60 s, 30 cycles of denaturation at 94 °C for 30 s, primer annealing between 50 and 60 °C for 30 s, and extension at 72 °C for 60 s. PCR products were separated on 1.2 % agarose gels and visualized under UV transilluminator (UVP, USA). Some randomly chosen PCR products of blaTEM (TS, SP old, JBC6), blaSHV (HB), blaCTX-M (B06 and B271) and QNR B (K13 and HB) genes were purified with a QIAquick PCR purification kit (Qiagen, USA) and sequenced using automated DNA sequencer (ABI Prism 377; Applied Biosystems, USA). The nucleotide sequences and deduced protein sequences were analyzed by the programs BLAST (http://www.ncbi.nlm.nih.gov/) and Clustal W of the European Bioinformatics Institute (http://www.cbi.ac.uk/). The sequences were deposited in GenBank-Database. Chromatograms were visually inspected for double peaks as signs of presence of different variants of bla/QNR genes in the same PCR product.
Statistical Analysis
Chi-square analysis was performed to determine association between presence of bla/QNR genes with antibiotic resistance-pattern and also between presence of bla genes with that of QNR genes. Probability values of P < 0.05 was considered statistically significant.
Results
In order to get overviews of antibiotic resistance pattern of pathogenic Klebsiella infecting Indian patients and their genetic background, 73 K. pneumoniae isolates were obtained from Calcutta School of Tropical Medicine (CSTM), Kolkata, India (Table 1). Male: female ratio among the patients was 1:1.28. Fifty eight samples were collected from patients undergoing treatment in inpatient department, whereas 15 isolates were collected from patients visiting outpatient department of CSTM.
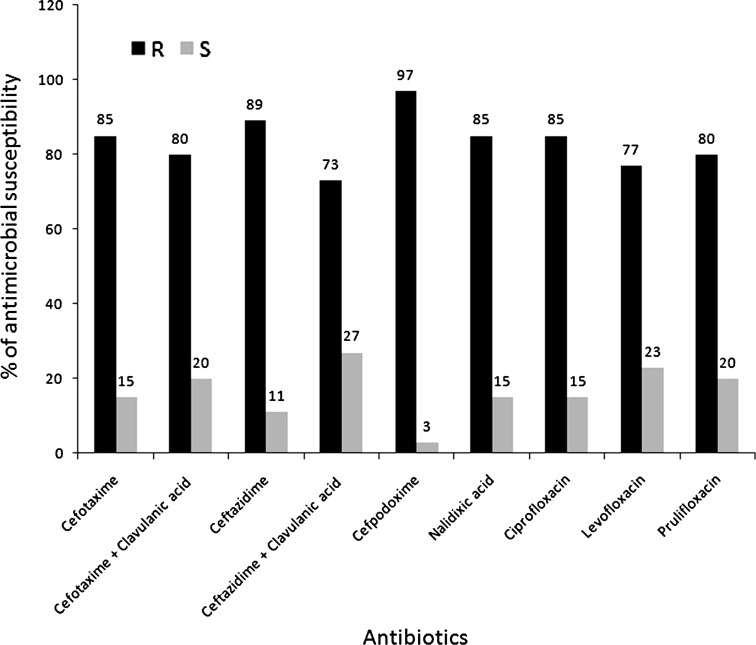
Antibiotic sensitivity of the samples was mentioned in Fig. 1. Approximately 97 % of them (71/73) showed resistance to at least one cephalosporin drugs. Frequency of resistance towards 3rd generation cephalosporin drugs were as follows: cefpodoxim (97 %), ceftazidime (89 %), ceftazidime and clavulenic acid (73 %), cefotaxime (85 %), cefotaxime and clavulenic acid (80 %). Out of the 73 isolates, 43 (58.9 %) were ESBL as evidenced by their antibiotic sensitivity assay against 3rd generation cephalosporin alone and in combination with clavulenic acid. Resistance to at least one quinolone group of drugs was found in 76.71 % isolates (56/73). Resistance rate to quinolone drugs were as follows: nalidixic acid (85 %), ciprofloxacin (85 %), prulifloxacine (80 %) and levofloxacine (77.6 %). 75.34 % (55/73) of K. pneumoniae showed resistance to both beta-lactam and quinolone group of antibiotics (at least one in each group) whereas two samples (# Hch8 and PP16) showed susceptibility towards all drugs studied.
Fig. 1.
Percentage of β-lactam and quinolone drug sensitivity of K. pneumoniae isolates. Antibiotics in mg; cefotaxime (30), cefotaxime/clavulanic acid (30/10), ceftazidime (30), ceftazidime/clavulanic acid (30/10), and cefpodoxime (10), nalidixic acid (30), ciprofloxacin (5), prulifloxacin (10) and levofloxacin (5); black bars indicate % of K. pneumoniae resistant to the antibiotic, gray bars indicate % of K. pneumoniae sensitive to the antibiotic. R resistance to that antibiotic, S sensitive to that antibiotic
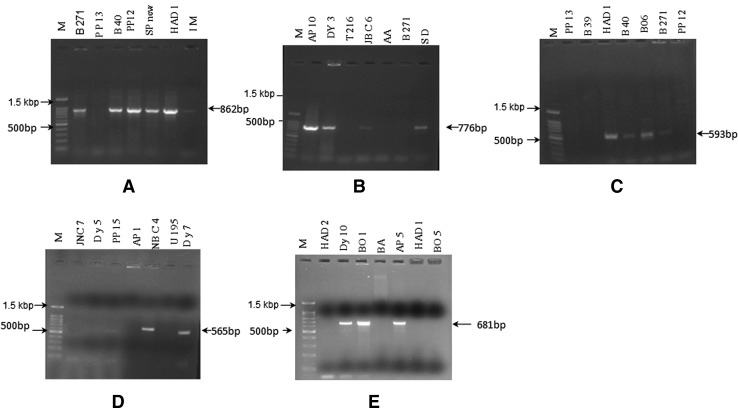
Genes blaTEM were detected in 52 % (38/73), blaSHV in 45 % (33/73) and blaCTX-M in 37 % (27/73) of the Klebsiella isolates (Fig. 2, Table 1). Overall 74 % (54/73) samples harbored at least one bla genes studied. Ten samples, mostly of UTI origin (9/10), showed presence of all the three bla genes (# PP15, AP10, JBC12, HAD1, SP/new, TS, JNC7, HAD2, K13 and BH). Genes QNRA were detected in 37 % (27/73), QNRB in 56 % (41/73) of the Klebsiella isolates. QNRS was not present in any of the samples. Both QNRA and QNRB genes were detected in 19 of them. All the three bla genes and two QNR genes were detected in five samples (viz. # PP15, AP10, JBC12, SP/new and K13).
Fig. 2.
Representative photograph of blaTEM (a), blaSHV (b), blaCTX-M (c), QNRA (d) and QNRB (e) genes in pathogenic K. pneumoniae isolates
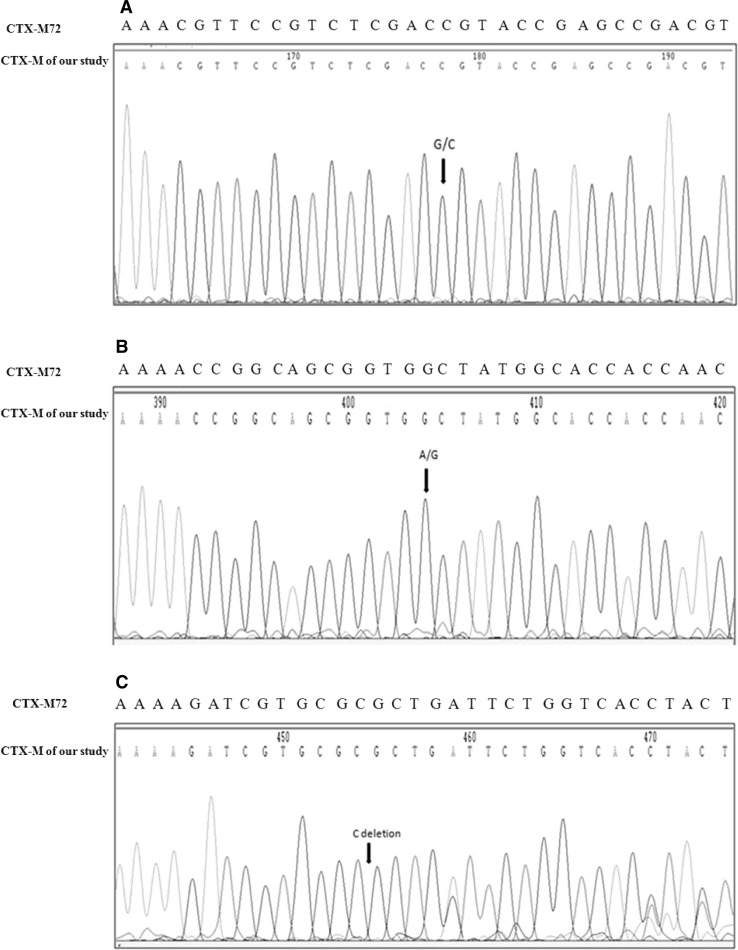
The analysis of the nucleotide sequences of three blaTEM PCR products revealed presence of two types of blaTEM genes–blaTEM-1 and blaTEM-116, in both the samples TS (GenBank accession: JN193523) and SP/old (GenBank accession: JN193524)—as evidenced by presence of double peaks in their discriminatory regions; whereas JBC6 (GenBank accession: JN193522) harbored only blaTEM1. The sequence of blaCTX-M PCR products of B06 and B271 (both these K. pneumoniae were isolated from blood of two unrelated patients suffering from septicemia—GenBank accession: JF523199) were similar to blaCTX-M72 (GenBank accession: AY847148.1)—except in three positions—at codon 167 (GGT → CGT), at codon 242 (GAC → GGC) and at codon 259 (one C deletion resulting in single frame shift and premature termination at codon 260) (Figs. 3 and 4). The sequences of QNRB PCR products [K13 (GenBank accession: JN193526) and HB (GenBank accession: JN193527)] revealed presence of QNRB1 among them.
Fig. 3.
Partial chromatogram of blaCTX-M in B271 sample
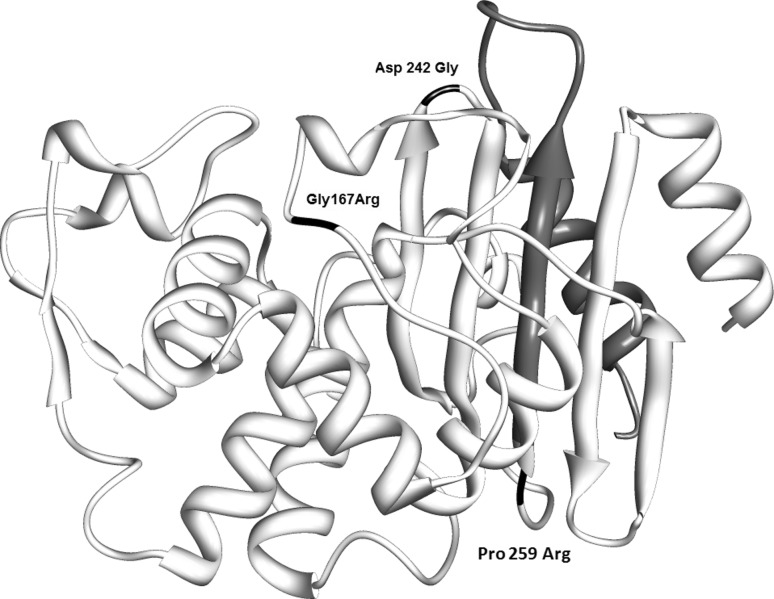
Fig. 4.
Location of mutations in CTX-M72 predicted protein structure. Amino acid substitutions are indicated in black. The region marked grey after codon 259 was absent. The diagram was drawn using UCSF CHIMERA (43)
Chi-square analyses among different genotypic and phenotypic characteristics revealed significant association between presence of bla genes to that of QNR genes in the Klebsiella samples studied (P value = 0.000125) (Table 2). No significant association was found among other characteristics of the Klebsiella isolates.
Table 2.
Chi-square analysis of presence of bla and qnr genes among K. pneumoniae iolates
| K. pneumoniae | Bla gene+ | Bla gene− |
|---|---|---|
| QNR gene+ | 43 | 6 |
| QNR gene− | 11 | 13 |
Discussion
Multidrug-resistant K. pneumoniae is the most common species of pathogenic Enterobacteriaceae in many parts of the world [17, 18]. During our one year study, most of the K. pneumoniae was isolated from patients with urinary tract infection at inpatient department. The prevalence of Klebsiella among hospitalized patients has been reported by other investigators from different countries including France, Turkey, Brazil and India [2, 3, 5, 19]. Similar to our study (58.9 % ESBL), reports from other parts of India and other Asian countries have demonstrated comparable prevalence of ESBL K. pneumoniae (40–83.3 and 60 % respectively) whereas much lower prevalence is noted in western countries [5–7, 20]. Unlike our report of 76.71 % quinolone resistance, it has been reported in lower frequency (4.9–11.9 %) among Klebsiella isolates of other countries [16, 19, 21]. Comparatively higher percentage of antibiotic resistance in Indian patients indicated the importance of antibiotic resistance in Indian health care system. Hence correct identification of resistant genes circulating among pathogenic bacteria is required for proper surveillance of their transmission.
Few studies have been done in India regarding the occurrence of bla and QNR genes in pathogenic K. pneumoniae [5, 6]. In our samples, occurrence of bla genes was observed in the following order: blaTEM > blaSHV > blaCTX-M. Similar to our report QNR genes were less prevalent than bla genes among pathogenic bacteria in other countries [4, 22, 23]. Significant association between the presence of bla and QNR genes (as observed in the study) is of serious concern since quinolone resistant ESBL bacteria has limited treatment options. Higher resistance against Ciprofloxacin in ESBL Klebsiella has been previously reported by Paterson et al. and Lautenbach et al. [17, 24].
The sequences of our blaTEM1 PCR products were identical to that of blaTEM1 (FJ668715.1) previously reported by Leinberger et al. in K. pneumoniae from Germany [15]. Our blaTEM116 sequences were identical to that isolated from K. pneumoniae and K. oxytoca in Australia (AY265882.1). Our blaSHV11 was identical to that isolated from K. pneumoniae in China by Zhou et al. (AF187732.1). The blaCTX-M PCR products had novel sequence that closely matched with blaCTXM-72 reported from China by Cheng et al. (AY847148.1), except at three positions—codons 167, 242 and 259 of blaCTXM-72 [25]. Mutation at codon 167 resulted in Glycine → Arginine substitution in the constituent omega loop (161 to 179 codon), situated at the floor of catalytic active site of CTX-M enzyme [26]. Both codons 242 and 259 encode amino acids constituent of α/β domain of CTX-M protein. Mutation at codon 242 resulted in Aspartic acid → Glycine substitution at the wall of its catalytic active site. The C deletion at codon 259 resulted in single frame shift mutation (CCG → CGC) of Proline → Arginine at this codon, followed by a premature termination (TGA) at codon 260. Such premature truncation of antibiotic resistant genes was previously reported in case of AmpD beta lactamase [27]. However, more resistant genes need to be genotyped in order to identify the strain variants circulating in our patient population. This study highlights the potential risk of spread of multidrug resistant bacteria among Indian patients—thus demanding their pre-screening before treatment and development of new drugs for effective treatment of these resistant pathogenic bacteria.
Acknowledgments
The authors are grateful to Dr. Chinmay Kumar Panda, Chittaranjan National Cancer Institute and Dr. Sila Chakraborty, Calcutta Medical College for providing technical assistance.
Footnotes
Anusri Tripathi and Sudip Kumar Dutta contributed equally to this study.
References
- 1.Taneja N, Chatterjee SS, Singh M, et al. Pediatric urinary tract infections in a tertiary care center from north India. Ind J Med Res. 2010;131:101–105. [PubMed] [Google Scholar]
- 2.Oktem IMA, Gulay Z, Bicmen M, et al. Qnr prevalence in extended spectrum beta lactamase-positive enterobacteriaceae isolates from Turkey. Jpn J Infect Dis. 2008;61:13–17. [PubMed] [Google Scholar]
- 3.Dropa M, Balsalobre LC, Lincopan N, et al. Extended-spectrum β-lactamase among enterobacteriaceae isolated in a public hospital in Brazil. Revista do Instituto de Medicina Tropical de São. 2009;51:203–209. doi: 10.1590/s0036-46652009000400005. [DOI] [PubMed] [Google Scholar]
- 4.Lal P, Kapil A, Das BK, et al. Occurrence of TEM & SHV gene in extended spectrum b-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res. 2007;125:173–178. [PubMed] [Google Scholar]
- 5.Goyal A, Prasad KN, Prasad A, et al. Extended spectrum β-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian J Med Res. 2009;129:695–700. [PubMed] [Google Scholar]
- 6.Lavilla S, González-López JJ, Sabaté M, et al. Prevalence of qnr genes among extended-spectrum b-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2007;61:291–295. doi: 10.1093/jac/dkm448. [DOI] [PubMed] [Google Scholar]
- 7.Jemima SA, Verghese S. Multiplex PCR for blaCTX-M & blaSHV in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian J Med Res. 2008;128:313–317. [PubMed] [Google Scholar]
- 8.Farmer JJ. Enterobacteriaceae: introduction and identification. In: Murray PR, Baron EJ, Pfaller MA, Jorgensen JH, Yolken RH, editors. Manual of Clinical Microbiology. 8. Washington, DC: ASM Press; 2003. pp. 636–653. [Google Scholar]
- 9.Clinical and Laboratory Standards Institute (CLSI) (2006) Performance standards for antimicrobial susceptibility testing. CLSI approved standard M100-S16 16th informational supplement. Clinical and Laboratory Standards Institute, Wayne
- 10.Jean SS, Teng LJ, Hsueh PR, et al. Antimicrobial susceptibilities among clinical isolates of extended-spectrum cephalosporin-resistant gram-negative bacteria in a Taiwanese University Hospital. J Antimicrob Chemother. 2002;49:69–76. doi: 10.1093/jac/49.1.69. [DOI] [PubMed] [Google Scholar]
- 11.Mugnaioli C, Luzzaro F, Luca FD, et al. CTX-M-Type extended-spectrum lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob Agents Chemother. 2006;50:2700–2706. doi: 10.1128/AAC.00068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Martínez JM, Briales A, Velasco C, et al. Mutational analysis of quinolone resistance in the plasmid-encoded pentapeptide repeat proteins QnrA, QnrB and QnrS. J Antimicrob Chemother. 2009;63:1128–1134. doi: 10.1093/jac/dkp111. [DOI] [PubMed] [Google Scholar]
- 13.Mena A, Plasencia V, Garcı′a L, et al. Characterization of a large outbreak by CTX-M-1 producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J Clin Microbiol. 2006;44:2831–2837. doi: 10.1128/JCM.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamang MD, Yong Seol S, Oh JY, et al. Plasmid-mediated quinolone resistance determinants qnrA, qnrB, and qnrS among clinical isolates of enterobacteriaceae in a Korean hospital. Antimicrob Agents Chemother. 2008;52:4159–4162. doi: 10.1128/AAC.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leinberger DM, Grimm V, Rubtsova M, et al. Integrated detection of extended-spectrum-beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV, and CTX-M genes. J Clin Microbiol. 2010;48:460–471. doi: 10.1128/JCM.00765-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacoby GA, Walsh KE, Mills DM, et al. QnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lautenbach E, Strom BL, Bilker WB, et al. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum b-lactamase–producing Escherichia coli and Klebsiella pneumonia. Clin Infect Diseases. 2001;33:1288–1294. doi: 10.1086/322667. [DOI] [PubMed] [Google Scholar]
- 18.Muzaheed DY, Adams-Haduch JM, Shivannavar CT, et al. Faecal carriage of CTX-M-15-producing Klebsiella pneumoniae in patients with acute gastroenteritis. Indian J Med Res. 2009;129:599–602. [PubMed] [Google Scholar]
- 19.Minarini LAR, Laurent P, Cattoir V, et al. Plasmid-mediated quinolone resistance determinants among enterobacterial isolates from outpatients in Brazil. J Antimicrob Chemother. 2008;62:474–478. doi: 10.1093/jac/dkn237. [DOI] [PubMed] [Google Scholar]
- 20.Potron A, Poirel L, Bernabeu S, et al. Nosocomial spread of ESBL-positive enterobacter cloacae co-expressing plasmid-mediated quinolone resistance Qnr determinants in one hospital in France. J Antimicrob Chemother. 2009;64:653–654. doi: 10.1093/jac/dkp222. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Tamang MD, Yong SS, Oh JY, et al. Plasmid-mediated quinolone resistance determinants qnrA, qnrB and qnrS among clinical isolates of enterobacteriaceae in a Korean hospital. Antimicrob Agents Chemother. 2008;52:4159–4162. doi: 10.1128/AAC.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang A, Yang Y, Lu Q, et al. Presence of qnr gene in Escherichia coli and Klebsiella pneumonia resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect Dis. 2008;8:68. doi: 10.1186/1471-2334-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng J, Wang Q, Chen Y, et al. Phenotypic and molecular characterization of a novel beta-lactamase carried by Klebsiella pneumoniae, CTX-M-72, derived from CTX-M-3. J Gen App Microbiol. 2009;55:207–216. doi: 10.2323/jgam.55.207. [DOI] [PubMed] [Google Scholar]
- 26.Delmas J, Robin F, Carvalho F, et al. Prediction of the evolution of ceftazidime resistance in extended-spectrum β-lactamase CTX-M-9. Antimicrob Agents Chemother. 2006;50:731–738. doi: 10.1128/AAC.50.2.731-738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett PM, Chopra I. Molecular basis of lactamase induction in bacteria. Antimicrob Agents Chemother. 1993;37:153–158. doi: 10.1128/AAC.37.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]