Abstract
Methicillin sensitive Staphylococcus aureus is an important bacterial pathogen associated with hospital- and community-acquired infections leading to endocarditis, skin tissue infection and pneumonia. The objective of this study was to determine both the genetic characteristics of methicillin-sensitive S. aureus (MSSA) strains, and the occurrence of virulence factors produced by S. aureus strains isolated from UMMC and healthy students in the University from year 2009. Out of 429 nasal swab samples, 67 were MSSA. The prevalence of 21 different virulence genes among 67 Malaysian clinical and community MSSA strains was determined by PCR, and their genetic features were assessed by PCR-RFLP of coa gene, agr types, spa typing and PFGE. The five predominant virulence genes were ica (79 %), efb and fnbA (61 % each), sdrE (57 %) and hlg (45 %). Toxin genes (enterotoxin, etd and pvl) were significantly more common (P < 0.05) in clinical strains compared to community strains. Three agr genotypes were observed: agr type I (45 %), agr type III (25 %) and agr type II (19 %). All 67 MSSA strains were distinguished into 26 profiles by PCR-RFLP of coa, 55 pulsotypes and 21 spa types. Four novel spa types (t7312, t7581, t7582 and t7583) were observed. In conclusion, different virulence profiles were observed in MSSA strains in Malaysia where toxin genes were more prevalent among clinical strains. No correlation between DNA profiles (coa-RFLP, PFGE and spa) and virulotypes was observed. The Malaysian MSSA strains from clinical and community sources were genetically diverse and heterogeneous.
Keywords: Staphylococcus aureus, Virulence genes, PFGE, spa and agr
Introduction
Staphylococcus aureus is one of the most important bacterial pathogens responsible for food-borne poisoning and toxin-mediated disease [1]. It produces different types of virulence factors, which are involved in attachment, persistence, tissue penetration and sepsis [2].
Enterotoxins are short secreted proteins that are usually heat-resistant, and usually resistant to most of the proteolytic enzymes produced by the human body [3]. There are more than 20 different types of enterotoxins (SEA to SEE, SEG to SEI, SEIJ, SEIK-SEIQ, SER to SET, SEW, SEIU) being reported [4]. An accessory gene regulator (agr) is known to be a global regulator of staphylococcal virulon which coordinates the expression of secretion and cell-associated virulence factors [5].
MRSA has evolved from methicillin-susceptible S. aureus (MSSA) via acquisition of mobile genetic elements called staphylococcal cassette chromosome mec (SCCmec) [6, 7]. Hence; MSSA is known to be a potential reservoir for these MRSA strains.
Several methods are available for subtyping S. aureus, include PCR-RFLP of coa gene [8], pulsed-field gel electrophoresis (PFGE) [9], and spa typing [10]. PCR-RFLP of coa gene is based on the restriction digest of the heterogeneity of the coagulase region that contains the 81 bp tandem repeats at 3′coding region [11] whereas PFGE is based on digestion of chromosomal DNA with the restriction enzyme followed by agarose gel electrophoresis [12]. On the other hand, spa typing is based on the sequence analysis of polymorphic region X of S. aureus protein A is reported to be a highly effective tool in subtyping S. aureus [12].
Detailed report on the genetic characterisation of MSSA strains in Malaysia is scanty. The only report in Malaysia was carried out by Ghasemzadeh-Moghaddam et al. [13] who reported diverse genotypes of MSSA strains isolated from six different populations of diverse geographical areas.
The objective of this study was to determine both the genetic characteristics of methicillin-sensitive 67 S. aureus (MSSA) strains, and the occurrence of virulence factors produced by MSSA strains isolated from UMMC and healthy students in the University.
Materials and Methods
Bacterial Strains
Sixty-seven non-repeat MSSA strains obtained from 29 in-patients admitted to University Malaya Medical Centre (UMMC) and 38 healthy undergraduate students from University Malaya (UM) in year 2009 were studied. The 38 MSSA strains from healthy students were collected from S. aureus carrier study involving 400 volunteer students.
All strains were isolated from nasal swabs. The strains were identified by standard biochemical methods, confirmed by BBL Staphyloside-Latex Test (BD, USA) and were checked for purity before analysis. All the strains were cultured in Tryptone Soy Broth and stored in cryo-vials containing 25 % v/v glycerol (Invitrogen, USA) at −85 °C.
PCR Detection of Virulence Genes
Detection of 21 different virulence genes including enterotoxins, sea to see, seg to sej and tst [14]; exfoliative-toxin, eta, etb [14] and etd [15]; cytotoxin, pvl [16]; adhesion genes, cna, hlg, ica and sdrE [17], efb [18], fnbA and fnbB [19] were performed by PCR using conditions as described earlier [14–19].
agr Typing by Multiplex PCR
Multiplex PCR for agr types was performed using specific primers and conditions described by Lina et al. [20].
Genotyping of MSSA Strains by PCR-RFLP of coa Gene, PFGE and spa Typing
PCR amplification of coa gene was performed using genomic DNA, primers and cycling parameters as described by Hookey et al. [21]. The amplicon of coa was digested with 10U of AluI enzyme (Promega, USA) for 1-h at 37 °C and the digested products were separated in 1.5 % agarose gel at 90 V for 3-h.
PFGE was performed according to Thong et al. [9]. The chromosomal DNA was digested with 10U SmaI for 2-h followed by separation on CHEF DRIII (Bio-Rad, USA) in 0.5× Tris–borate-EDTA at 14 °C for 22-h with pulsed-time of 5–60 s. Both PCR-RFLP of coa gene and PFGE gels were photographed under UV light after staining with ethidium-bromide (0.5 μg/ml).
Cluster analyses of PCR-RFLP for coa gene and PFGE profiles of MSSA strains were analyzed with BioNumerics 6.0 (Applied Maths, Belgium) based on unweighted-pair group method with arithmetic averages (UPGMA) with a position tolerance of 0.15. All DNA profiles were assigned arbitrary designation, and differences were defined by Dice coefficient of similarity, F.
spa typing was performed as described by Harmsen et al. [22]. The amplicons of spa were purified using Megaquick-DNA purification kit (Intron Biotechnology, Korea) and sequenced. Nucleotide sequences of spa type were analyzed using BioNumerics 6.0.
Cluster analysis setting for minimum-spanning tree was set to 25 % duplicate extension, 25 % duplicate creation, 50 % gap extension cost, 250 % gap creation cost, maximum duplication length of three-repeats and bin grouping distance of 0.5 %. Based on the interpretation scheme recommended by Shore et al. [23], two strains are considered closely related if two spa types are at a MST distance of ≤3 (corresponding to >98.5 % similarity).
Statistical Analysis
Statistica 8.0 software was used for data analysis. Comparison of certain variables was determined by Fisher’s exact test. Associations between different virulence factors were determined by Spearman’s rank order correlation coefficient test. P value <0.05 (two-tailed) was taken as the level of significance for Fisher’ exact test whereas R-value was taken as type of association between the variables. Breakpoints for the association of virulence factors are defined as follows: perfect association (R = 1), no association (R = 0) and invert correlation with (R = −1).
Results and Discussion
Prevalence of Virulence Genes Among MSSA Strains
Among the 21 virulence genes tested, ica gene was detected in a majority (79 %) of the strains. efb, fnbA, sdrE and hlg genes were detected in 41 (61 %), 41 (61 %), 38 (57 %) and 30 (45 %) strains, respectively. Based on Spearman’s rank correlation test, correlations between efb and fnbA (R = 1, P < 0.05), fnbA and sdrE (R = 0.478, P < 0.05), fnbA and hlg (R = 0.71, P < 0.05), fnbA and ica (R = 0.118, P < 0.05), fnbA and enterotoxin (R = 0.38, P < 0.05) were observed. This indicates that efb-positive strains are most likely to harbour fnbA, ica, hlg, sdrE and enterotoxin’s genes. Presence of efb, fnb, ica and hlg genes are needed for pathogenesis and adherence of S. aureus in host tissue [2].
A total of 39 strains was tested positive for at least one type of staphylococcal enterotoxins, etd or pvl gene. No sed, see, sej, eta, etb and etd gene was detected. The distribution of virulence genes in community and clinical MSSA strains is summarized in Table 1. There was a significant difference in the prevalence of seh gene between clinical and community strains (P = 0.0229), and this concurred with the result reported by Ghasemzadeh-Moghaddam et al. [13]. There was no significant difference in the prevalence of other virulence genes, efb, fnbA, hlg, ica, sdrE, sea, seb, sec, seg, sei,etd, tst and pvl, between clinical and community strains. However, Ghasemzadeh-Moghaddam et al. [13] reported that sea, seb, seg,sei and fnbA were significantly more prevalent among community than clinical strains. The discrepancy between the distribution of these six virulence genes might be related to the sources of the community strains as the previous study examined strains from six different populations, including aboriginals, university student and staff, kindergarten kids and village people whereas the present study only examined community strains from healthy undergraduate students from one institution.
Table 1.
Prevalence of virulence genes, agr and spa types in Malaysian MSSA strains
| Gene | Clinical (n = 29) | Community (n = 38) | P value |
|---|---|---|---|
| Enterotoxin | |||
| sea | 6 | 3 | 0.1604 |
| seb | 3 | 0 | 0.0763 |
| sec | 6 | 5 | 0.5116 |
| seg | 3 | 0 | 0.0763 |
| seh | 9 | 3 | 0.0229 |
| sei | 6 | 3 | 0.1604 |
| etd | 0 | 1 | 1.0000 |
| tst | 1 | 5 | 0.2235 |
| Adhesion | |||
| Efb | 18 | 23 | 1.000 |
| fnbA | 18 | 23 | 1.000 |
| hlg | 14 | 16 | 0.6300 |
| ica | 24 | 29 | 0.5613 |
| sdrE | 17 | 21 | 0.0806 |
| Cytotoxin | |||
| pvl | 2 | 7 | 0.2802 |
| agr types | |||
| agr type I | 9 | 21 | 0.0818 |
| agr type II | 4 | 9 | 0.3650 |
| agr type III | 11 | 6 | 0.0503 |
| Un-typeable | 5 | 2 | 0.2251 |
PVL toxin (encoded by pvl gene) that causes necrotic suppurative skin lesions [24] was common among community than clinical strains. This is not surprising as pvl gene is often associated with community strains (MRSA and MSSA) and is considered as a marker for community-acquired infections [6].
Toxic shock syndrome toxin-1 (encoded by tst) that causes neonatal toxic shock syndrome-like exanthematous disease and Staphylococcal purpura fulminans [25, 26] was detected in five community and one clinical strain.
The number of MSSA strains harbouring toxin genes were significantly higher (P < 0.05) in clinical strains when compared to community strains. This is a cause of concern as enterotoxins are often associated with food poisoning and several allergic and autoimmune diseases [1, 3].
Occurrence of agr Types
Three agr types were observed: agr type I (45 %), agr type II (25 %) and agr type III (19 %). No agr type IV was observed. Seven strains did not belong to any agr type. This concurred with the report by Perez-Vazquez et al. [27] that agr types in MSSA were more diverse as 35 (31 %), 40 (35.4 %), 35 (31 %) and 3 (2.7 %) of their MSSA strains were subtyped into agr types I, II, III and IV. Besides, no significant difference between the distribution of agr types in clinical and community MSSA strains were observed.
Genetic Relatedness of MSSA Strains based on PCR-RFLP of coa Gene, PFGE and spa Typing
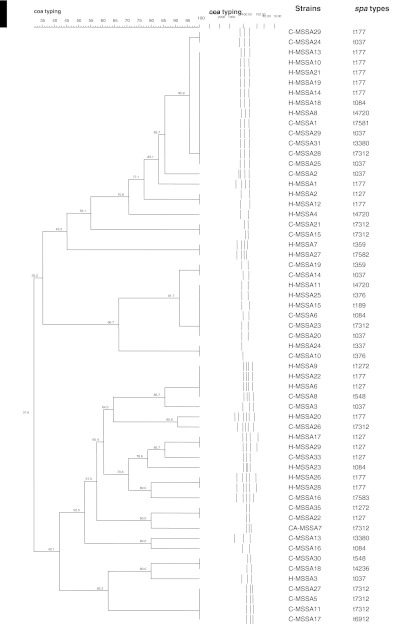
Three different typing approaches were used to subtype the 67 MSSA strains. Based on PCR-RFLP of coa, the 59 strains were subtyped into 27 different restriction profiles (F = 0.3–1.0) (Fig. 1), eight strains could not be typed due to the absence of coa gene. Sixteen strains were genetically related (shared 80 % similarity) and among them, 12 strains were indistinguishable, although they were from two different sources (clinical and community) (Fig. 1).
Fig. 1.
Dendrogram of PCR-RFLP of coa gene of clinical and community MSSA strains
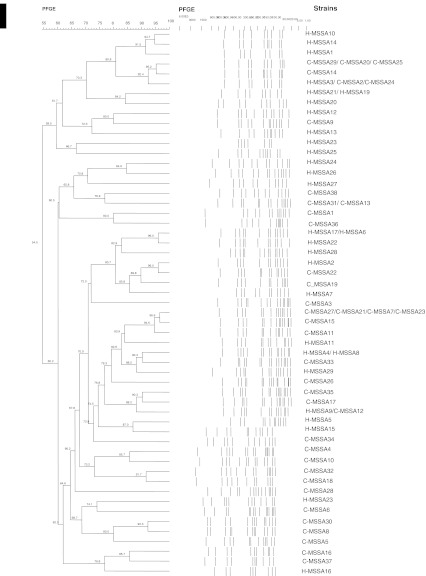
PFGE differentiated the 67 strains to 55 pulsed-field profiles (PFPs) (F = 0.54–1.0) (Fig. 2) with 11 strains (4 clinical, 7 community) being clonally related (shared ≥80 % similarity). Different virulence profiles were observed for the clonally related strains, suggesting that presence of enterotoxin and cytotoxin genes were not associated with any particular PFGE profiles.
Fig. 2.
Dendrogram of PFGE SmaI-digested chromosomal DNA profiles of clinical and community MSSA strains
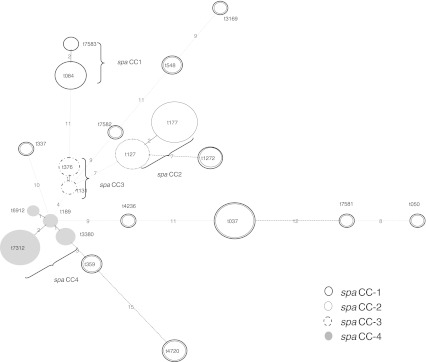
The 67 MSSA strains were subtyped into 21 different spa types, including four novel spa types (t7312, t7581, t7582 and t7583) (Fig. 3). Seven spa types (t084, t1272, t177, t127, t376, t359 and t037) were present in both clinical and community strains. Five other spa types (t7583, t337, t7582, t189 and t4720) were unique for clinical strains while nine spa types (t3169, t548, t131, t6912, t7312, t3380, t4236, t7581 and t050) were only present in community strains. spa types t037, t127, t131, t177, t050 and t189 have been reported elsewhere [13, 28] while spa types t3169, t376, t6912, t337, t4236, t3380, t7312, t7581, t7582 and t7583 are new in Malaysia (http://ridom.spa.server.com).
Fig. 3.
Minimal spanning tree analysis for the spa types of 67 MSSA strains
A minimum spanning tree based on the degree of similarity between spa-type repeats successions showed four spa clonal complexes (spaCC) arbitrarily named spaCC1 to spaCC4 (Fig. 3). Fourteen strains from four spa types (t7312, t189, t6912 and t3380) were closely related (shared ≥98.5 % similarity), and all four spa types shared three spa-type repeats succession (12-21-17) although they only shared 60.2 % similarity by PFGE. Different virulence profiles were observed for strains with indistinguishable profiles, suggesting that presence of enterotoxin and cytotoxin genes were not associated with any particular spa types. This is because virulence genes such as enterotoxin genes are often carried by mobile genetic elements such as plasmids, pathogenicity islands, SCCmec and prophages [29].
Both PFGE and spa typing were more discriminative than PCR-RFLP of coa gene. Twelve strains that exhibited identical PCR-RFLP of coa profiles were distinguishable by spa typing and PFGE. Discrepancies in the grouping of the strains by the three subtyping methods might be due to different target sites as PCR-RFLP target the coa gene, spa typing examines the variability in spa gene whereas PFGE targets large genomic fragment after digestion with restriction enzymes.
In conclusion, different virulence profiles were observed in MSSA strains in Malaysia where toxin genes were more prevalent among clinical strains. However, virulence genes studied were not source-specific and no correlation between DNA profiles (i.e. PCR-RFLP of coa gene, PFGE and spa types) and virulotypes was observed. The Malaysian MSSA strains from clinical and community sources were genetically diverse and heterogeneous.
Acknowledgments
This work was funded by PPP Grant (PV046/2011B) from University of Malaya and PulseNet Grant (57-02-03-1015) from JJID, Japan. LKT is supported by University of Malaya Fellowship.
References
- 1.Plata K, Rosato AE, Wegrzyn G. Staphylococcus aureus as an infectious agent: overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim Pol. 2009;56:597–612. [PubMed] [Google Scholar]
- 2.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortega E, Abriouel H, Lucas R, Galvez A. Multiple roles of Staphylococcus aureus enterotoxins: pathogenicity, superantigenic activity, and correlation to antibiotic resistance. Toxins. 2010;2:2117–2131. doi: 10.3390/toxins2082117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argudin MA, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2:1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. Agr function in clinical Staphylococcus aureus strains. Microbiol. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranovich T, Zarake H, Shabana II, Nevzorova V, Turcutyuicov V, Suzuki H. Molecular characterization and susceptibility of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from hospitals and the community in Vladivostok, Russia. Clin Microbiol Infect. 2010;16:575–582. doi: 10.1111/j.1469-0691.2009.02891.x. [DOI] [PubMed] [Google Scholar]
- 7.Chonngtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006;50:1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitani N, Koizumi A, Sano R, Masutani T, Murukawa K, Mikasa K, Okamoto Y. Molecular typing of methicillin-resistant Staphylococcus aureus by PCR-RFLP and its usefulness in an epidemiological study of an outbreak. Jpn J Infect Dis. 2005;58:250–252. [PubMed] [Google Scholar]
- 9.Thong KL, June J, Liew FY, Yusof MY, Hanifah YA. Antibiograms and molecular subtypes of methicillin-resistant Staphylococcus aureus in local teaching hospital, Malaysia. J Microbiol Biotechnol. 2009;19:1265–1270. [PubMed] [Google Scholar]
- 10.Ghaznavi-Rad E, Shamsudin MN, Sekawi Z, Khoon LY, Azoz MN, Hamat RA, Othman N, Chong PP, Belkum A, Ghasemzadeh-Moghaddam H, Neela V. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol. 2010;48:867–872. doi: 10.1128/JCM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himabindu M, Muthamilselvan DS, Bishi DK, Verma RS. Molecular analysis of coagulase gene polymorphism in clinical isolates of methicillin resistant Staphylococcus aureus by restriction fragment length polymorphism based genotyping. Am J Infect Dis. 2009;5:170–176. [Google Scholar]
- 12.Deurenburg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect Dis. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 13.Ghasemzadeh-Moghaddam H, Ghaznavi-Rad E, Sekawi Z, Liew YK, Aziz MN, Hamat RA, Melles DC, Belkum A, Shamsudin MN, Neela V. Methicillin-susceptible Staphylococcus aureus from clinical and community sources are genetically diverse. Intern J Med Microbiol. 2011;301:347–353. doi: 10.1016/j.ijmm.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesmw X, Etienne J, Vandenesh F. Relationship between Staphylococcus aureus genetic background, virulence factors, agr types (alleles), and human disease. Infect Immun. 2002;70:631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hisata K, Kuwahara-Arai K, Yamanoto M, Ito T, Nakatomi Y, Cui L, Baba T, Terasawa M, Sotozono C, Kimoshita S, Yamashiro Y, Hiratmasu K. Dissemination of methicillin-resistant Staphylococcus among healthy Japanese children. J Clin Microbiol. 2005;43:3364–3372. doi: 10.1128/JCM.43.7.3364-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. Involvement of panton-valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1129–1133. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 17.Kumar JD, Negi YK, Gaur A, Khanna D. Detection of virulence genes in Staphylococcus aureus strains from paper currency. Intern J Infect Dis. 2009;13:e450–e455. doi: 10.1016/j.ijid.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Moore PCL, Lindsay JA. Genetic variation among hospital strains of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J Clin Microbiol. 2001;39:2767–2860. doi: 10.1128/JCM.39.8.2760-2767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arciola CR, Campoccia D, Gemberini S, Baldassarri L, Montanaro L. Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopaedic infections associated to different types of implant. FEMS Microbiol Lett. 2005;246:81–86. doi: 10.1016/j.femsle.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 20.Lina G, Boutitie F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microbiol. 2003;69:18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hookey JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen D, Clause H, Witte W, Rothaganger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a University Hospital Setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shore AC, Rossney AS, Kinnevey PM, Brennam OM, Creamer E, Sherlock O, Dolan A, Cunney R, Sullivan DJ, Goering RV, Humphreys H, Coleman DC. Enhanced discrimination of highly clonal ST22-methicillin-resistant Staphylococcus aureus IV strains achieved by combining spa, dru, and pulsed-field gel electrophoresis typing data. J Clin Microbiol. 2010;48:1839–1852. doi: 10.1128/JCM.02155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain A, Robinson G, Malkin J, Duthie M, Kearns A, Perera N. Purpura fulminans in a child secondary to panton-valentine leukocidin-producing Staphylococcus aureus. J Med Microbiol. 2007;56:1407–1409. doi: 10.1099/jmm.0.47374-0. [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi K, Takahashi N, Piao C, Totsuka K, Nishida H, Uchiyama T. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains causing neonatal toxic shock syndrome-like exanthematous disease in neonatal and perinatal wards. J Clin Microbiol. 2003;41:3001–3006. doi: 10.1128/JCM.41.7.3001-3006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kravitz GR, Dries DJ, Peterson ML, Schlievert PM. Purpura fulminans due to Staphylococcus aureus. Clin Infect Dis. 2005;40:941–947. doi: 10.1086/428573. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Vazquez M, Vindel A, Marcos C, Oteo J, Cuevas O, Trincado P, Bautista V, Grundmann H, Campos J. Spread of invasive Spanish Staphylococcus aureus spa-type t067 associated with a high prevalence of the aminoglycoside-modifying enzyme gene ant(4′)-Ia and the efflux pump genes msrA/msrB. J Antimicrob Agents. 2008;63:21–31. doi: 10.1093/jac/dkn430. [DOI] [PubMed] [Google Scholar]
- 28.Vasanthakumari N, Subeh A, Alshrari D, Ghaznavi-Rad E, Moghaddam A, Belkum A, Alreshidi MA, Selamat N, Shamsudin MN. Highly dynamic transient colonization by Staphylococcus aureus in healthy Malaysian student. J Med Microbiol. 2009;58:1531–1532. doi: 10.1099/jmm.0.011692-0. [DOI] [PubMed] [Google Scholar]
- 29.Hu DL, Omoe K, Inoue F, Kasai T, Yasujima M, Shinagawa K, Nakane A. Comparative prevalence of superantigenic toxin genes in methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Med Microbiol. 2008;57:1106–1112. doi: 10.1099/jmm.0.2008/002790-0. [DOI] [PubMed] [Google Scholar]