Abstract
Bacillus circulans MTCC 7906, an extracellular alkaline protease producer was genetically characterized. B. circulans genomic DNA was isolated, oligonucleotide primers specific to alkaline protease gene of B. circulans were designed and its PCR amplification was done. The purified PCR product and pTrcHisA vector were subjected to restriction digestion with NcoI and HindIII and transformed into Escherichia coli DH5-α competent cells. The recombinant expression of alkaline protease gene studied by inducible expression and analysis by SDS-PAGE, established that the alkaline protease protein had an estimated molecular size of 46 kDa. Gene sequencing of the insert from selected recombinant clone showed it to be a 1329 bp gene encoding a protein of 442 amino acids. The sequence was blasted and aligned with known alkaline protease genes for comparison with their nucleotide and amino acid sequences. This identified major matches with three closely related subsp. of B. subtilis (B. subtilis subsp. subtilis strain 168, B. subtilis BSn5 and B. subtilis subsp. spizizenii strain W23). The insert also showed a number of substitutions (mutations) with other sp. of Bacillus which established that alkaline protease of B. circulans MTCC 7906 is a novel gene. The phylogenetic analysis of alkaline protease gene and its predicted amino acid sequences also validated that alkaline protease gene is a novel gene and the same has been accessioned in GenBank with accession number JN645176.1.
Keywords: Cloning, Bacillus circulans, Alkaline protease, Sequencing, Phylogenetic analysis
Introduction
Proteases represent one of the three largest groups of industrial enzymes and account for about 60 % of the total global enzyme market [1]. They have wide range of commercial usage in detergent, leather, food, feed, pharmaceutical based industries [2–4]. Alkaline proteases that are active in alkaline pH and high temperatures are industrially more important, especially in detergent industry [5, 6]. Although these enzymes have been found in many viruses, fungi and animals, bacterial proteases (Bacillus) have more importance [1, 7] because of their wide temperature, pH tolerance and thermal stability [8].
Bacillus circulans MTCC 7906, an isolate from vegetable waste produced an extracellular alkaline protease, which had an optimal activity at 60 °C temperature and 9.0 pH, the suitable conditions for commercial laundry detergents. In order to make B. circulans MTCC 7906 strain commercially viable, this strain needs to be characterized at molecular level so that it may be genetically manipulated for higher alkaline protease production. A number of alkaline protease encoding bacterial genes have been cloned and expressed in heterologous hosts so as to increase the yield and selectivity in the bioprocess for serine alkaline protease (SAP) production [1]. To maximize industrial enzyme production, the host organism is generally manipulated to carry multiple copies of the gene. This can be accomplished by cloning the gene on a replicating plasmid, but preferentially this is achieved by amplification of the gene on the chromosome, as such amplification offers more stable clones [9].
In this paper we report the cloning of alkaline protease gene and its expression in Escherichia coli DH5-α host cells. The complete nucleotide sequence of alkaline protease gene was determined, which provides clues for novel structural features of alkaline protease gene of B. circulans MTCC 7906.
Materials and Methods
Bacterial Strain, Plasmids and Media
Bacillus circulans MTCC 7906 was aerobically cultured in modified Reese medium [10]. 100 ml of Reese medium inoculated with 2 % broth culture of B. circulans was incubated at 28 °C on an orbital shaker at 120 rpm for 24 h. The exponentially growing bacterial cells were centrifuged aseptically at 10,000 rpm for 10 min to collect the bacterial pellet. The latter was used for isolation of total DNA. E. coli DH5-α used as host strain was cultured in Luria–Bertani (LB) and the plasmid pTrcHisA (4.4 kb, invitrogen) was used as the cloning vector.
DNA Isolation and Designing of Primers
The total DNA was isolated from bacterial cell mass as per Cubero et al. [11]. To determine the complete nucleotide sequence of the alkaline protease gene (BcAP), a pair of primers BcAP-F and BcAP-R was designed (Table 1). For this purpose, information was derived on conserved sequences around coding nucleotide sequences (upstream and downstream) of the complete alkaline protease genes of three Bacillus sp. (GenBank accession numbers CP002468.1, CP002183.1 and AL009126.3). The individual nucleotide sequences were aligned by ‘CLC Sequence Viewer 6.5.2’ (CLC Bio A/S) software to identify the conserved sequences. Thereafter the sequences were processed by a primer designing software ‘FastPCR’ (Primer Digital Ltd., Finland) and appropriate primer pair selected in the identified conserved sequence region for amplification of complete structural gene region of alkaline protease (BcAP-F & BcAP-R).The sequences in these primers were also manipulated to include suitable restriction sites NcoI site-CCATGG in BcAP-F primer, HindIII site-AAGCTT in BcAP-R. The designed primers were custom synthesized through facility of ‘Integrated DNA Technologies, Inc’, Coralville, IA, USA.
Table 1.
Nucleotide sequence of primers designed for amplification of complete alkaline protease gene from B. circulans
| Primer | Nucleotide sequence, 5′→3′ | Size (bp) | Tm (°C) | Anealing temp (°C) | Expected amplicon (bp) | Restriction sites included |
|---|---|---|---|---|---|---|
| BcAP-F | GAGGAAAAACCATGGTCGGATACTCT | 26 | 58.2 | 55.5–61.5 | ~1,360 bp | NcoI |
| BcAP-R | AGAATTGCAAGCTTTTGATGATGTTTA | 27 | 54.0 | 54.2–60.2 | HindIII |
28 PCR cycles
PCR Amplification of Alkaline Protease Gene
Polymerase chain reactions were carried out in a programmable DNA thermal cycler (Mastercycler Gradient-eppendorf™). The 20 μl of PCR reaction mixture contained 100 ng of template (bacterial) DNA solution, 1 mM dNTPs mix, 10 μM of each primer, 5.0 U Taq Polymerase (MBI, Fermentas) and 1.5 mM MgCl2 in 1X Taq reaction buffer. The amplified DNA product along with a DNA marker (100 bp ladder plus, MBI Fermentas) was separated by electrophoresis using 1.0 % agarose gel in TAE. The gel was stained with ethidium bromide and the banding profile recorded using UV-gel Documentation system (Ultra Cam). The amplified DNA product (~1,360 bp) was purified from the gel band (using ‘QIA Quick Gel Extraction Kit’ of Qiagen).
Construction of the Cloning Vector Containing Alkaline Protease Gene
The purified PCR product and plasmid pTrcHisA were both double-restricted with restriction enzymes NcoI and HindIII (Fermentas Life Sciences). The restriction reaction mixture (30 μl) was incubated at 37 °C for overnight. The processed DNA fragment (alkaline protease gene) was ligated into linearised pTrcHisA vector and the ligation reaction product transformed into competent E. coli DH5-α host cells using ‘PCR Product Cloning Kit’ (Fermentas Life Sciences). For this purpose the insert and vector (molar ratio of 5/1, respectively), T4 DNA ligase (5 units/μl) and 3 μl of buffer (5X) were added at final volume of 15 μl and incubated at 16 °C overnight.
After the screening of transformed colonies on LB agar ampicillin culture medium, plasmid preparation was accomplished by alkaline lysis method [12]. The recombinant plasmids were confirmed with appropriate restriction enzymes (NcoI and HindIII).
Nucleotide Sequence of Cloned Alkaline Protease Gene
The information on nucleotide sequence of the targeted insert DNA (alkaline protease) from B. circulans now cloned in recombinant plasmid was obtained through Custom Sequencing Services of ‘M/S Xcelris, Ahmedabad’.
Analysis of Alkaline Protease Gene/Protein Sequences for Phylogenetic Relationships
Nucleotide Sequences
The nucleotide sequence of alkaline protease gene was aligned with the available sequences of the previously reported Bacillus sp. from different countries (available in GenBank database) using ‘CLC Free workbench software, ver 6.5.2’ of CLC Bio A/S. Based upon the ‘Multiple Alignment’, genetic relatedness dendrograms (phylogenetic tree) were developed for the B. circulans under study with already reported Bacillus sp./strains using ‘Tree Function’ of the above software. Additional information on level of phylogenetic relatedness of the alkaline protease sequence of B. circulans strain under study was obtained using ‘Nucleotide blast tool’ of ‘National Center for Biotechnology Information’.
Predicted Amino Acid Sequences
For determining the predicted amino acid sequences of alkaline protease gene, the corresponding nucleotide sequences were translated into ‘+1 open reading frame’ using ‘Translate into proteins’ function of the ‘CLC’ bio workbench, ver. 6.5.2.
Gene Expression in E. coli DH5-α Induced with IPTG
100 μl of overnight grown culture of E. coli DH5-α host cells, one of the five cloned alkaline protease plasmids that is PR1, was used to inoculate 10 ml of fresh LB or LB-Amp broth, respectively and allowed to grow in the orbital shaker (200 rpm) set at 37 °C for 2 h. After 2 h of growth, culture was divided into two 5 ml lots. One set of 5 ml of the culture was induced with 5 mM of IPTG and incubation was continued. Additional samples of induced alkaline protease clone were also obtained after 3, 6, 9, 12 and 15 h of growth to detect the expressed product with SDS-PAGE.
Results and Discussion
Amplification and Cloning of the Alkaline Protease Gene
Using total DNA of B. circulans as template, the PCR amplification with BcAP-F and BcAP-R primers resulted in the amplification of 1,360 bp DNA fragment (Fig. 1). The ligation of processed alkaline protease DNA fragment into linearlised pTrcHisA vector and subsequent transformation of the ligation product into competent E. coli DH5-α cells resulted in selective growth of a number of clones on LB-Amp agar. Plasmid preparation was performed for a number of clones. The agarose gel electrophoresis profile of miniprep plasmid from seven individual clones (PR1–PR7) is shown in Fig. 2. Since the adopted procedure allowed only unidirectional ligation, every recombinant clone was expected to be appropriate clone carrying alkaline protease gene from B. circulans. However, it was observed that whereas plasmids from five clones were of increased size (5.730 kb), plasmids from two of the other clones (PR2 and PR7) were of the size equal to that of pTrcHisA vector (4.4 kb). Most likely the latter clones included insert DNA of very low size(s) that were present in the processed vector and/or insert DNA preparations as unknown contaminants. Therefore, only five clones (PR1, PR3–PR6) were considered to be desired clones containing cloned alkaline protease gene DNA for further studies.
Fig. 1.

PCR amplification of alkaline protease gene from B. circulans genomic DNA. M is 100 bp DNA ladder plus. Three lanes (1, 2, 3) showing amplified alkaline protease gene (~1,360 bp)
Fig. 2.
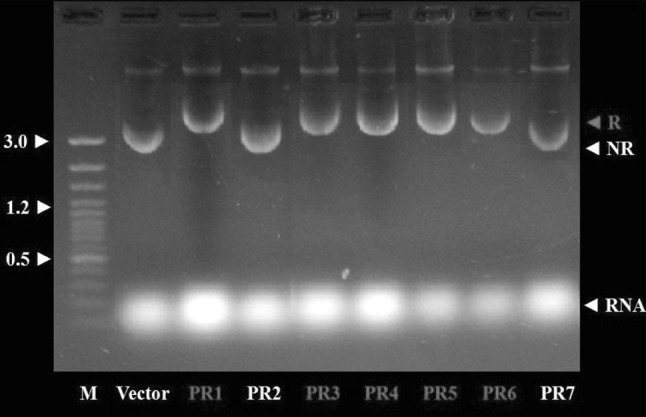
Plasmid profile of seven clones from recombinant progeny. M is 100 bp ladder plus, R is recombinant (PR1, PR3–PR6), NR is non recombinant (PR2 and PR7)
Analysis of Recombinant Clones for Cloned Alkaline Protease Fragment Insert
Restriction Analysis of Recombinant Clones
The combined restriction of the seven recombinant plasmids with NcoI and HindIII (unique bordering sites) resulted in release of single fragment of ~1.30 kb (similar to the size of alkaline protease amplicon) from the vector (~4.40 kb) in case of only those five individual recombinant clones which had earlier shown to be of increased size plasmids (Fig. 3). This established the presence of possible alkaline protease gene fragments based upon their size in all the five clones. These clones were suitable for custom sequencing. The determined sequence of this alkaline protease fragment was submitted to GenBank and has been assigned a GenBank accession #JN645176.1.
Fig. 3.
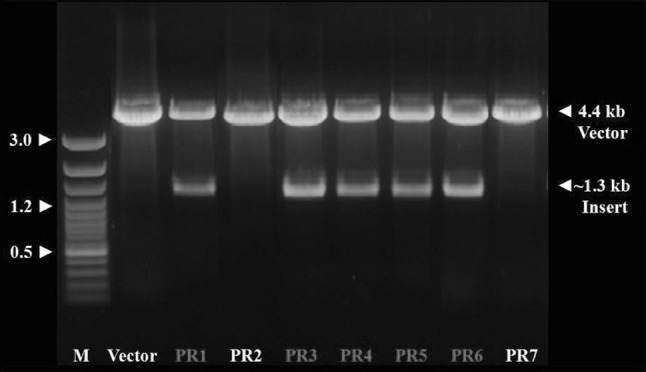
Restriction release of cloned fragments from alkaline protease clones (five) by double digestion with NcoI and HindIII restriction enzymes. M is 100 bp ladder plus, PR2 and PR7 are non recombinant clones
Genetic Analysis of Cloned Alkaline Protease Gene with Genbank Database
The nucleotide sequence of alkaline protease gene was processed by NCBI Blast function (available at www.ncbi.nlm.nih.gov/BLAST/) for ‘multiple alignments’ with GenBank database (Table 2). In this table, ‘score values’ for an alignment represent a sum of ‘substitutions’ and ‘gap scores’. Higher values for ‘Max score’ and ‘Total score’ signify higher identities amongst aligned sequences including the query sequence under study. This ‘BLAST’ identified homologous alkaline protease genes from three different subsp. of B.subtilis, with maximum identity of 99, 98 and 93 % from B. subtilis subsp. subtilis strain 168, B. subtilis BSn5 and B. subtilis subsp. spizizenii strain W23, respectively.
Table 2.
Alkaline protease gene sequences producing significant alignments with BcAP
| GenBank accession number | Description | Max score | Total score | Query coverage (%) | Max identity (%) |
|---|---|---|---|---|---|
| AL009126.3 | B. subtilis subsp. subtilis strain 168 complete genome | 2,449 | 2,449 | 100 | 99 |
| CP002468.1 | B. subtilis BSn5 complete genome | 2,366 | 2,366 | 100 | 98 |
| CP002183.1 | B. subtilis subsp. spizizenii strain W23 complete genome | 1,977 | 1,977 | 99 | 93 |
Information derived from www.ncbi.nlm.nih.gov
From these results, it became evident that alkaline protease gene under study (B. circulans) is a novel gene that is closely related to alkaline protease genes from B. subtilis subsp. subtilis strain 168 and B. subtilis BSn5. The GenBank database lacks in data on alkaline protease gene sequences from B. circulans and this is the first report to the best of our knowledge on nucleotide sequencing on the same. However, the data suggested that the alkaline protease gene from B. circulans MTCC 7906 though novel is conserved amongst B. circulans and some specific strains of B. subtilis.
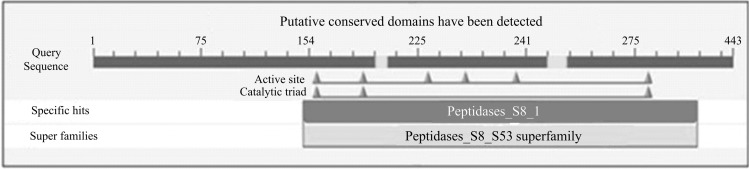
The corresponding predicted protein (amino acid) sequence of BcAP gene was also blasted into NCBI Blast (Protein) function (available at www.ncbi.nlm.nih.gov/BLAST/) for identifying homologous proteins (Table 3). The blast search identified specific conserved domains (active site and catalytic triads) in the protein sequence under study with specific hit with Peptidase_S8-1 type proteases that are subtilases, serine endo- and exo-peptidases categorized under Peptidase-S8–S53 superfamily (Fig. 4). Peptidase_S8-1 type proteases to which BcAP protein was found to belong are known to have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin [13].
Table 3.
Alkaline protease gene proteins producing significant alignments with BcAP
| Accession number | Description | Max score | Total score | Query coverage (%) | Max identity (%) |
|---|---|---|---|---|---|
| NP_389608.1 | B. subtilis subsp. subtilis strain 168 | 918 | 918 | 100 | 99.77 |
| YP_004207781.1 | B. subtilis BSn5 | 905 | 905 | 100 | 98.19 |
| ZP_06873415.1 | B. subtilis subsp. spizizenii strain W23 | 877 | 877 | 100 | 96.60 |
| YP_003973171.1 | B. atrophaeus 1942 | 806 | 806 | 100 | 88.00 |
| YP_003920391.1 | B. amyloliuefaciens DSM7, aprX | 751 | 751 | 100 | 80.99 |
| YP_079346.1 | B. licheniformis ATCC 14580 | 690 | 690 | 99 | 71.36 |
Information derived from www.ncbi.nlm.nih.gov
Fig. 4.
Specific conserved domains in the protein sequence of alkaline protease gene
Phylogenetic Analysis of Alkaline Protease Gene with Genbank Database
Multiple nucleotide sequence alignment of seven alkaline protease genes showed that nucleotide sequence of B. circulans alkaline protease gene have major matches with gene sequences available for the three closely protease subsp. of B. subtilis (B. subtilis subsp. subtilis strain 168, B. subtilis BSn5 and B. subtilis subsp. spizizenii strain W23) except for only minor mismatches, which all mere represented by single base substitutions (Fig. 5). The low level of polymorphism amongst alkaline protease from B. circulans and above subsp. of B. subtilis suggests that most of the alkaline proteases in this sp./strain have coevolved due to substitution mutations. In some other strains of Bacillus (B. atrophaeous 1942, B. amyloliquefaciens DSM7 and B. licheniformis ATCC14580), higher levels of similar substitutions have led to high degree of polymorphism (sequence divergence). However, all the seven alkaline protease genes from different Bacillus sp./strains are highly conserved with a size of 1,329 bp having multiple alignments showing no addition or deletion gaps. Multiple amino acid sequence alignment of seven alkaline protease genes showed many single amino acid substitutions that were randomly distributed all over the sequence (Fig. 6). All the seven alkaline protease gene proteins are highly conserved with a size of 442 amino acids. The gene encoding subtilisin Carlsberg from B. licheniformis has been cloned in pBR322 vector by Jacob et al. [14] that has a 1,137 bp open reading frame encoding 379 amino acids. Also B. lentus and B. alcalophilus alkaline protease genes have been cloned and sequenced, which both encode 380 amino acid proteins [9, 15].
Fig. 5.
Multiple alignment of alkaline protease gene sequences of different sp./strains showing substitution of single base(s)
Fig. 6.
Multiple alignment of amino acid sequences of different sp./strains showing presence of substitutive amino acid
Construction of the Phylogenetic Tree
In order to determine the effect of overall differences on genetic relatedness, quantitative relationship was derived in terms of ‘phylogenetic tree’. The phylogenetic relationship based upon alkaline protease nucleotide sequences (Fig. 7) resulted in distribution of all the reported and B. circulans into three separate ‘Clades’, indicating a divergent and independent evolution of alkaline protease genes in different Bacillus sp./strains. These clades which diverged from each other were:
Clade 1: All with a genetic divergence of up to 41.35 pns (percent nucleotide substitutions).
Clade 2: Due to very low genetic divergence of only 3.38 pns, the alkaline proteases in this clade appeared to have co-evolved.
Clade 3: This clade with a maximum genetic divergence of 21.69 pns also included the alkaline protease gene under study i.e. from B. circulans MTCC 7906.
Fig. 7.
Bootstrap dendrograms of the 22 different alkaline protease genes from different Bacillus sp./strains, generated using distance based ‘UPGMA algorithm’. Bar indicates distance expressed as nucleotide substitutions per 100 nucleotide length (indicator of percentage substitutions). The frequency of each group in dendrograms is presented behind the respective node
In clade 3, Bacillus sp. appeared to have evolved in sequential evolution in the order of B. licheniformis ATCC 14580, B. amyloliquefaciens DSM7 aprX, B.atrophaeus 1942 with preceding sp. serving as evolutionary parent for the succeeding species. The results also suggested that B. atrophaeus 1942 subsequently served as evolutionary parent of B. subtilis subsp. spizizenii strain W23, B. subtilis subsp. subtilis strain 168, B.subtilis BSn5 and B. circulans MTCC 7906.
Recombinant Expression of Cloned Alkaline Protease Gene in E. coli
The expression of the cloned BcAP gene designed to provide the expression of only full length gene protein in recombinant pTrcHisA vector is presented in Fig. 8. The SDS-PAGE resolution pattern of different proteins revealed that the uninduced cultures of E. coli DH5-α (lane 2) and BcAP clone (lane 3) produced a similar pattern of proteins. However, a single additional protein of ~46 kDa was produced in case of BcAP clones that were induced with IPTG for 3, 6, 9, 12 and 15 h (lanes 5–9, respectively). The size of the expressed protein was in agreement to the predicted size of 442 amino acids encoded by an open reading frame of 1,329 nucleotides to give an estimated size of ~46 kDa for alkaline protease of B. circulans MTCC 7906. These bands occupied the position that was similar in size to that observed in case of partially purified alkaline protease (lane 4, obtained from the DEAE cellulose column) of B. circulans. This established that the cloned BcAP gene was successfully cloned in E. coli DH5-α.
Fig. 8.
Lane 1: PageRuler Prestained protein ladder, 10–170 kDa. Lane 2: Uninduced cells of E. coli DH5-α. Lane 3: Uninduced BcAP clones. Lane 4: Partial purified alkaline protease (active fractions eluted and pooled from the DEAE cellulose column) of B. circulans. Lanes 5–9: BcAP clone induced for 3, 6, 9, 12 and 15 h, respectively
References
- 1.Gupta R, Beg QK, Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 2002;59:15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- 2.Bhaskar N, Sudeepa ES, Rashmi HN, Selvi AT. Partial purification and characterization of protease of Bacillus proteolyticus-CFR3001 isolated from fish processing waste and its antibacterial activities. Bioresour Technol. 2007;98:2758–2764. doi: 10.1016/j.biortech.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 3.Jellouli K, Bellaaj OG, Ayed HB, Manni L, Agrebi R, Nasri M. Alkaline-protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergent additive and for shrimp waste deproteinization. Process Biochem. 2011;46:1248–1256. doi: 10.1016/j.procbio.2011.02.012. [DOI] [Google Scholar]
- 4.Deng A, Wu J, Zhang Y, Zhang G, Wen T. Purification and characterization of a surfactant-stable high-alkaline protease from Bacillus sp. B001. Bioresour Technol. 2010;101:7100–7106. doi: 10.1016/j.biortech.2010.03.130. [DOI] [PubMed] [Google Scholar]
- 5.Maurer KH. Detergent proteases. Curr Opin Biotechnol. 2004;15:330–334. doi: 10.1016/j.copbio.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Kumar CG, Takagi H. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol Adv. 1999;17:561–594. doi: 10.1016/S0734-9750(99)00027-0. [DOI] [PubMed] [Google Scholar]
- 7.Rao MB, Tanksale AM, Ghatge MS, Deshpande VV. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genckal H, Tari C. Alkaline protease production from alkalophilic Bacillus sp. Isolated from natural habitats. Enzyme Microb Technol. 2006;39:703–710. doi: 10.1016/j.enzmictec.2005.12.004. [DOI] [Google Scholar]
- 9.Jorgensen PL, Tangney M, Pedersen PE, Hastrup S, Diderichsen B, Jorgensen ST. Cloning and sequencing of an alkaline protease gene from Bacillus lentus and amplification of the gene on the B. lentus chromosome by an improved technique. Appl Environ Microbiol. 2000;66:825–827. doi: 10.1128/AEM.66.2.825-827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaur S, Vohra RM, Kapoor M, Beg QK, Hoondal GS. Enhanced production and characterization of a highly thermostable alkaline protease from Bacillus sp. P-2. World J Microbiol Biotechnol. 2001;17:125–129. doi: 10.1023/A:1016637528648. [DOI] [Google Scholar]
- 11.Cubero OF, Crespo A, Fatehi J, Bridge PD. DNA extraction and PCR amplification method suitable for fresh, herbarium-store lichenized and other fungi. Plant Syst Evol. 1999;216:243–249. doi: 10.1007/BF01084401. [DOI] [Google Scholar]
- 12.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler BA, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Deweese SC, Fong JH, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs M, Eliasson M, Uhlen M, Flock JI. Cloning, sequencing and expression of subtilisin Carlsberg from Bacillus licheniformis. Nucleic Acids Res. 1985;13:8913–8926. doi: 10.1093/nar/13.24.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laan JC, Gerritse G, Mulleners LJ, Hoek RA, Quax WJ. Cloning, characterization and multiple chromosomal integration of a Bacillus alkaline protease gene. Appl Environ Microbiol. 1991;57:901–909. doi: 10.1128/aem.57.4.901-909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]