Abstract
Microbial diversity of 1,000 m deep pelagic sediment from off Coast of Andaman Sea was analyzed by a culture independent technique, bacterial tag encoded FLX titanium amplicon pyrosequencing. The hypervariable region of small subunit ribosomal rRNA gene covering V6–V9, was amplified from the metagenomic DNA and sequenced. We obtained 19,271 reads, of which 18,206 high quality sequences were subjected to diversity analysis. A total of 305 operational taxonomic units (OTUs) were obtained corresponding to the members of firmicutes, proteobacteria, plantomycetes, actinobacteria, chloroflexi, bacteroidetes, and verucomicrobium. Firmicutes was the predominant phylum, which was largely represented with the family bacillaceae. More than 44 % of sequence reads could not be classified up to the species level and more than 14 % of the reads could not be assigned to any genus. Thus, the data indicates the possibility for the presence of uncultivable or unidentified novel bacterial species. In addition, the community structure identified in this study significantly differs with other reports from marine sediments.
Keywords: Metagenomics, 16S rRNA, Pyrosequencing, Microbial richness, Andaman Sea
Introduction
Ocean being the largest part of the biosphere, which covers approximately 71 % of the earth’s surface with an average depth of 3,800 m, harbors great numbers and diversity of microorganisms [1]. In the marine environment, microorganisms are not only present in the surface waters, but also in the lower and abyssal depths of the ocean [2]. The marine sediments occupy approximately 3.5 × 108 km2, which is 48.6 % of the earth’s crust, estimated to be the habitat of about 106 microbes/ml [3]. The deep ocean sediments, being the source for a myriad of microbial populations, play an important role in the ecological balance and marine biogeochemical cycles. These microorganisms have special nutritional requirements and physiological characteristics that enable them to thrive in complex conditions such as high salinity, high pressure, low temperature, poor-nutrient environment. For the past two decades, microbial populations of the deep-sea sediments were studied by culture independent 16S rRNA based microbial analysis [4]. Many projects have begun recently to delineate the abundance and diversity of microorganisms present in subsea floor biosphere and their biogeochemical importance [5]. Composition and diversity of the microbial populations in marine environments showed that the environmental microbial community structure is more diverse and complex than originally thought [6]. The increase in the speed and efficiency of the genomic data generation and the sequencing coast plummeted as the next-generation of highly efficient sequencing technologies are available [7]. The microbial diversity of various ecosystems such as deep mines [8], soil [9], deep marine biospheres [10], tidal flats [11], and human microflora [12] were analyzed by employing pyrosequencing technique. On the other hand, potential automated bioinformatics pipelines have been developed to analyze, achieve consistent, rapid and accurate taxonomic assignments of the 16S rRNA sequence reads [13].
In this study, we explored the microbial diversity of the 1,000 m deep-sea sediment from Andaman Sea, through the bacterial tag encoded FLX titanium amplicon pyrosequencing (bTEFAP) approach. The bTEFAP technique has been selected to determine the microbial community in the pelagic sediment avoiding the culture and cloning bias.
Materials and Methods
Sample Collection and Bacterial DNA Extraction
The sediment sample was collected at a depth of 1,000 m from a region 1.2 km away from the Ross Island of Andaman Sea (11º67′24.11″E, 92º77′61.8″N), by the National Institute of Ocean Technology (NIOT) vessels, Government of India. The sample was collected with the grab and stored immediately at −20 °C in a sterile air tight container, until further analysis. DNA was extracted by following a modified method of Zhou et al. [14]. A 5 g of sludge sample was mixed with DNA extraction buffer and proteinase K by horizontal shaking at 225 rpm for 30 min at 37 °C. Cells were lysed with 20 % SDS and the DNA was extracted with chloroform/isoamyl alcohol and precipitated with 100 % ethanol. The extracted DNA was purified using Qiagen DNeasy kit (Hilden, Germany), according to the manufacturer’s instructions and the DNA was stored at −20 °C until further analysis.
Pyrosequencing and Data Analysis
Polymerase chain reaction (PCR) was performed using the universal bacterial primers 926F 5′ AAACTYAAAKGAATTGRCGG 3′ and 1392R 5′ ACGGGCGGTGTGTRC 3′ covering the 460 bp (V6–V9 region) of the 16S rRNA gene [15]. The amplicon library was generated through one-step PCR consisting of hot start mixture and high fidelity Taq polymerase for 30 cycles and pyrosequencing was performed using 454 Roche FLX instrument following the titanium protocols at the Research and Testing Laboratory (Lubbock, TX, USA). The sequence reads were screened and filtered for quality and length using the programme, MOTHUR [16]. The chimeric sequences were removed by using chimera_check and chimera slayer [17]. High quality sequence reads were aligned against the rdp-trainset bacterial database and cluster linkage analysis and other ecological metrics were calculated using the ribosomal database project (RDP). OTUs and rarefaction curves were created from the aligned sequence reads by complete cluster linkage tool and also used to determine richness and diversity indexes Shannon–Weaver, Chao1 and evenness at each dissimilarity level by Shannon index calculator tools of RDP. The taxonomic assignments were given using the RDP classifier program [18] with a bootstrap score of 80 %. The sequences were analyzed by BLAST analysis against SILVA database and the results were represented using the MEGAN4 analysis tool [19].
Bacterial Classification
The bacterial populations were classified at their appropriate taxonomic level by BLASTN analysis performed through the web tool VITCOMIC [20]. Sequences with identity scores, to known or well characterized 16S rRNA gene sequence database and the taxonomic assignments were given on the basis of the BLAST average at different similarity level i.e., 80, 85, 90, 95 and 100 %. The overall taxonomic composition of the sample was represented by the vitcomic plot of dots in circle.
Results
Pyrosequencing Data
A total of 19,271 sequence reads were obtained from the Genome sequencer FLX system. The sequence has been deposited in the GenBank Sequence Read Archive with the accession number SRA048238.1. We have screened for the quality reads, ambiguous base, and homopolymers and also trimmed the primer sequence by using MOTHUR (http://www.mothur.org), which gave 18,206 high quality sequence reads with the average read length of 390 bp.
Microbial Diversity of Pelagic Sediment
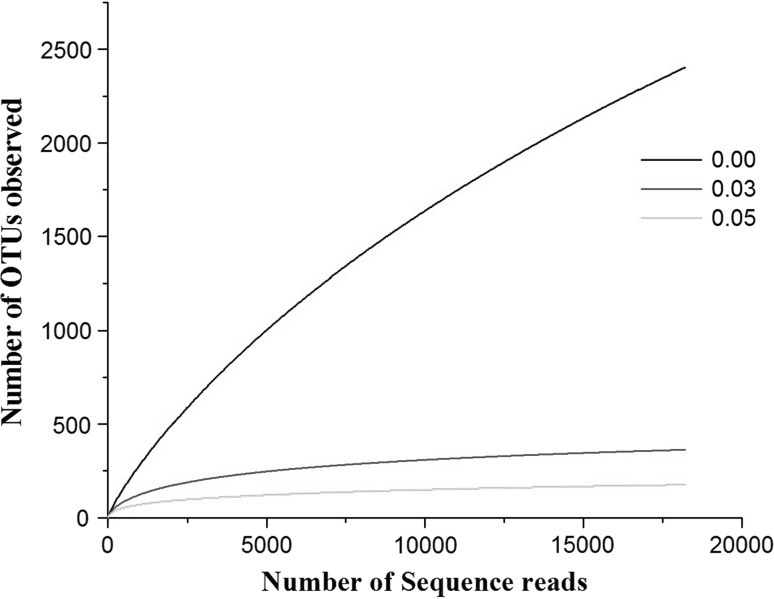
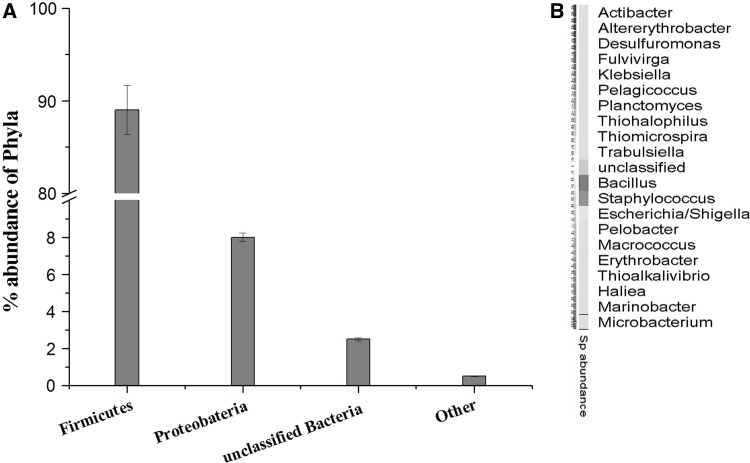
The richness of the microbial community present in the sample was estimated by rarefaction curves. The members of the sample or number of organisms assigned at each phylogenetic level depend on the number of sequences analyzed. The rarefaction curve is used to analyze whether complete diversity of the sample was obtained despite the number of sequences. The rarefaction curves based on the OTU values are good indications of the diversity within the sample as various percentage of dissimilarity level are known to differentiate at different taxonomic levels. At a distance level of 3 % is able to differentiate at species level, whereas at a distance level 5 % is able to differentiate at genus level and a distance level 10 % is able to differentiate at family/class level [21]. The rarefaction curves (Fig. 1) indicates that the bacterial richness of the sediment sample was almost completely revealed i.e., very close to the true microbial diversity of the sample. A total of 305 OTUs by clustering at 3 % dissimilarity level (0.03 distance units) and 143 OTUs by clustering a 5 % dissimilarity level (0.05 distance units) were observed. For the total microbial communities clustered at 3 % dissimilarity level, the number of OTUs obtained was very close to the number of OTUs estimated and the statistical estimates of the species richness like Chao1 and ACE diversity indices were obtained (Table 1). The 99 % of Good’s coverage indicate the level of coverage of the 16S rRNA sequences identified in these groups represents majority of bacterial sequences present in the tested sample. The phylogenetic classification of the sequences from the sediment sample, at the phylum level is shown in Fig. 2a. The sequences represented eight phyla, where the majority of the sequences were assigned to the phylum firmicutes (89.4 %), followed by proteobacteria (8.1 %) and unclassified phyla (2.4 %). Planctomycetes, actinobacteria, chloroflexi, bacteroidetes, verucomicrobium, gemmatimonadetes were the other phyla identified with relatively low abundance (<1 %). A total of 429 sequence reads (2.4 %) were from unclassified phyla, representing the possibility of the presence of novel unidentified group of microorganism in the pelagic sediment of Andaman Sea.
Fig. 1.
Rarefaction curves representing the richness of the pyrosequencing reads with distance values (dissimilarity level) of 0 (unique), 3 (0.03) and 5 % (0.05). The vertical axis shows the number of OTUs that would be expected to be found after sampling the number of tags or sequences shown on the horizontal axis
Table 1.
Phylotype coverage and diversity estimation of the pyrosequencing analysis
| Distance units | Reads | Coveragea | OTUsb | Chao1 | ACE | Shannon | Evennessc |
|---|---|---|---|---|---|---|---|
| Unique | 18,206 | 99 % | 2,403 | 4,904 | 5,340 | 4.09 | 0.525 |
| 0.03 | 18,206 | 99 % | 305 | 372 | 380 | 3.04 | 0.532 |
| 0.05 | 18,206 | 99 % | 143 | 167 | 183 | 2.35 | 0.474 |
aThe coverage percentages (Good), richness estimators (ACE, Chao1) and diversity indices (Shannon and evenness) were calculated by Good’s method and RDP
bThe operational taxonomic units (OTU) were defined with 0, 3 and 5 % dissimilarity using the RDP classifier
cThe Shannon index of evenness was calculated with the formula E = H/ln(S), where H is the Shannon diversity index and S is the total number of sequences in that group
Fig. 2.
Phylogenetic classification obtained by classifying the metagenomic dataset against the RDP database using the RDP classifier. a Percentage abundance of the phylum, firmicutes representing 89 % of the sample, while proteobacteria 8 %, Unclassified bacteria 2.5 % and other phylum share the remaining 0.5 %. b Heatmap representing the relative abundance of 20 bacterial genera detected in the sample
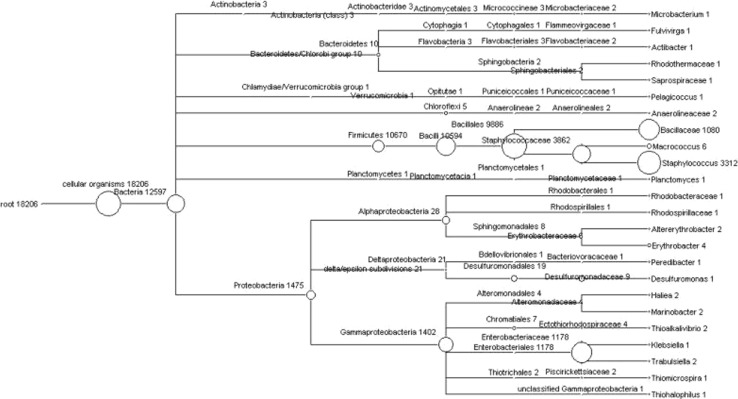
The phylogenetic tree represents the BLAST analysis of the 16S rRNA sequence against the SILVA database. The tree was represented at genus level and the species level classification was obtained by creating a mapping file using the ARB-software [22]. At the genus level, 21 different genera were found in the sample (Figs. 2b, 3). Of these, the genera Bacillus and Staphylococcus roughly constituted about 75 % of the total sequences. The genus Escherichia represented 9 % and another 2 % of the sample constituted by other genera such as Microbacterium, Fulvivirgo, Actibacter, Pelagicoccus, Macrococcus, Planctomycetes, Altererythrobacter, Erythrobacter, Pelobacter, Desulfuromonas, Hailea, Marinobacter, Thioalkalivibrio, Klebsiella, Trabulsiela, Thiomicrospira and Thiohalophilus. Approximately, 14.5 % of the total sequence reads could not be assigned to specific genera.
Fig. 3.
Phylogenetic diversity of 18,206 reads of the Andaman Sea sample computed by MEGAN, the classification for the sequence reads were obtained by using Ribosomal Database Project Classifier tool against the RDP database. Each circle represents bacterial taxa in the RDP taxonomy database and the size of the circle represents the number of reads assigned to the particular taxon
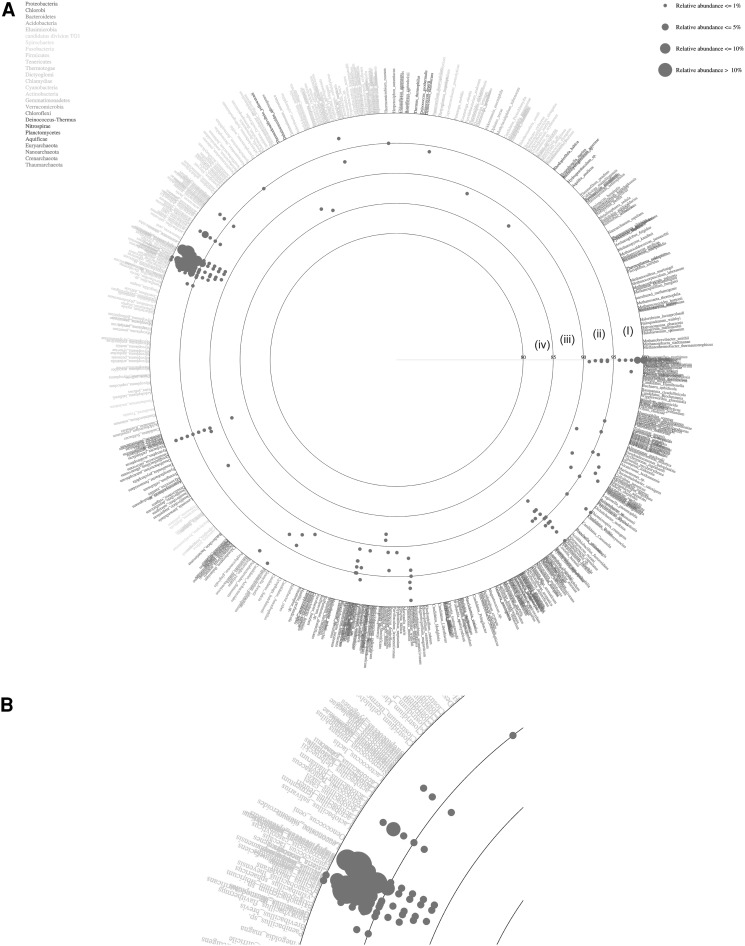
The total BLAST score was calculated against each sequence of the reference database (http://mg.bio.titech.ac.jp/vitcomic/) to identify the nearest relative of the sample sequence in order to classify at species level (Fig. 4). In Fig. 4, each species name in the reference database is placed in circles with ordered phylogenetic relatedness. Physical distances between nearest species in the plot indicate genetic distances of 16S rRNA genes between them. The circles indicate the boundaries of BLAST average similarities (inner most circle starting at 80 %, followed by 85, 90, 95 and 100 % identity to the database sequence). Each dot represents average similarities of each sequence against the nearest relative species in the reference dataset. The size of these dots indicates the relative abundance of sequences in the sample. The VITCOMIC plot contains four categories of dot size that indicate the relative abundance of the sample sequence. The reads represented by the largest dot (Fig. 4b) i.e., >10 % of relative abundance were B. licheniformis, M. caseolyticus and Staphylococcus epidermidis. The sequences represented by dots with >5 % similarity were B. cytotoxicus, B. weihenstephanensis and Shigella flexneri. In addition to these major species, each of B. halodurans, S. aureus, B. anthracis, B. pumilus, B. subtilis, and Lactobacillus sakei were represented by more than 1 % of the total reads. E. litoralis, Thioalkalivibrio sp., B. thuringiensis, Geobacillus kaustophilus, Methylacidiphilum infernorum, Methylococcus capsulatus, Oceanobacillus iheyensis, Pelobacter carbinolicus, Thermomicrobium roseum and T. crunogena were represented by less than 1 % of the total reads (data not shown).
Fig. 4.
a Vitcomic map representing the species diversity of the sample, based on the comparison of the reads against the NCBI non-redundant database using the blastn tool. Large circles indicate boundaries of BLAST average similarities, inner most circle (iv) 80–85 %, followed by (iii) 85–90 %, (ii) 90–95 %, (i) 95–100 % similarity of the database sequence. b High-resolution view of the region containing the predominant phylum firmicutes. The Larger size dots indicate the relative abundance of the particular taxon is more than 10 % of the sample sequence
Discussion
An attempt was made to analyze the bacterial diversity and bacterial community structure of the deep sea sediment of Andaman Sea through bTEFAP pyrosequencing analysis. The accuracy of bacterial diversity of the particular environment is dependent on the size and number of sequences used for analysis. The length of the sequences also strongly affects the phylogenetic affiliation [23–25]. The accuracy of the taxonomic assignment of the microorganisms also depends on the targeted 16S rRNA gene sequence as the required length varies to different hypervariable regions. Different hypervariable regions have different efficacies with respect to species calls in different genera [26]. The V6 hypervariable region is widely used as it has been reported to have good discriminating power. In this study, we have used the V6–V9 variable region with an average read length of 390 bp, which is long enough to classify the reads up to the genus level.
In this study, the obtained sequences and the observed OTUs represented a near saturation of rarefaction curves. In addition, Good’s coverage revealed that 99 % of the phylotypes identified in the sediment sample could represent majority of the sequences present in the sample. A total 305 OTUs representing the members of eight different phyla were identified. Firmicutes was the most predominant phylum in the sample, in agreement with the bacterial diversity reported from the subsea floor sediment of Andaman Sea [24]. In contrast, gammaproteobaceria was reported to be the dominant phylum in the marine sediment of China Sea and Arctic Ocean [27–29]. In this study, the proteobacteria was represented by only 8 % of the total reads, whereas firmicutes represented more than 89 %. Microbial communities in deep-sea is dependent on variables such as substrate availability and type, biogeochemistry, nutrient input, productivity and hydrological conditions of the regional scale [30]. Thus, the microbial diversity is impacted by temporal and spatial dimensions of deep-sea [31].
The genus Bacillus was most abundant in the sediment sample. The occurrence and distribution of the genus Bacillus in marine environment has been extensively studied. The genus Bacillus consists of 222 recognized species distributed widely across many aquatic habitats including marine sediments [32–34]. Bacillus subtilis, B. pumilus, B. licheniformis, and B. cereus were reported to be the common inhabitants of Pacific Ocean habitat [35]. Similarly, marine Bacillus sp. have been isolated from shallow marine sediments of Scripps pier, California [36] and from coastal marine sediments in San Diego area [37]. The genus Bacillus is capable of producing and secreting many industrially important enzymes and antibiotics. The next abundant genus in the sample was Staphylococcus, which might possibly be transported from the surface regions. The sequence reads also showed affiliation to a number of bacteria specifically isolated from the deep sea sediments, such as A. sediminis, E. litoralis and Thiohalospira sp., etc. Actibacter sediminis strain JC2129T was isolated from tidal flat sediment of Dongmak on Ganghwa Island, South Korea [11]. Erythrobacter are gram-negative facultative photoheterotrophs, metabolising organic carbon when available, but are capable of photosynthetic light utilization when organic carbon is scarce. They are globally distributed in the euphotic zone and only recently recognized as a component of the marine microbial community that appears to be critical for the cycling of both organic and inorganic carbon in the ocean. E. litoralis is a halotolerant aerobic phototroph, an abundant marine organism that has been little studied to date [38]. The genus Thiohalospira of the family Ectothiorhodospiraceae is of special interest because, unlike other purple sulfur bacteria, these organisms oxidize H2S and produce Sº outside of the cell but also because some species are extremely halophilic and are among the most halophilic of all known prokaryotes [39]. Other bacterial genera such as Microbacterium, Fulvivirgo, Macrococcus, Pelagicoccus, Planctomycetes, Desulfuromonas, Hailea, Marinobacter, Thioalkalivibrio and Thiomicrospira were represented with lower abundance. Interestingly, we have found a number of phylogenetic lineages of unclassified genera that belongs to the phyla proteobacteria, acidobacteria and firmicutes. Altogether 44.5 % of sequence reads could not be classified up to the species level indicating the possibility for the presence of novel uncultivated or unidentified bacterial species.
Acknowledgments
The authors thankfully acknowledge funding from Department of Biotechnology, New Delhi (No. BT/PR-10486/BCE/08/657/2008). The authors also thank Mr. Ravi, vessel manager, NIOT for his support. Authors also acknowledge the central facilities, CAS, CEGS, NRCBS and IPLS at MKU.
References
- 1.Polymenakou PN, Lampadariou N, Mandalakis M, Tselepides A. Phylogenetic diversity of sediment bacteria from the Southern Cretan margin, Eastern Mediterranean Sea. Syst Appl Microbiol. 2009;32:17–26. doi: 10.1016/j.syapm.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Surajit D, Lyla PS, Ajmal Khan S. Marine microbial diversity and ecology: importance and future perspectives. Curr Sci. 2006;90:1325–1337. [Google Scholar]
- 3.Azam F. Oceanography: microbial control of oceanic carbon flux:the plot thickens. Science. 1998;280:694–696. doi: 10.1126/science.280.5364.694. [DOI] [Google Scholar]
- 4.Wu H, Yatao Guo Y, Wang G, Dai S, Li X. Composition of bacterial communities in deep-sea sediments from the South China Sea, the Andaman Sea and the Indian Ocean. Afr J Microbiol Res. 2011;5:5273–5283. [Google Scholar]
- 5.Biddle JF, SylvanJB Brazelton WJ, Tully BJ, Edwards KJ, Moyer CL, Heidelberg JF, Nelson WC. Prospects for the study of evolution in the deep biosphere. Frontiers Microbiol. 2012 doi: 10.3389/fmicb.2011.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 7.Dini-Andreote F, Andreote FD, Araújo WL, Trevors JT, Elsas JD. Bacterial genomes: habitat specificity and uncharted organisms. Microb Ecol. 2012;64:1–7. doi: 10.1007/s00248-012-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards RA, Rodriguez-Brio B, Wegley L, et al. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roesch LF, Fulthope PR, Riva A, Casella G, Hadwin AK, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 11.Kim BS, Kim BK, Lee JH, Kim M, Lim YW, Chun J. Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J Microbiol. 2008;46:357–363. doi: 10.1007/s12275-008-0071-9. [DOI] [PubMed] [Google Scholar]
- 12.Keijser BJF, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucl Acid Res. 2008;36:120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Brunns MA, Tiedje JM. DNA recovery from soil of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James CA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43–58. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zajec N, Stres B, Avgustin G. Distinct approaches for the detection and removal of chimeric 16S rRNA sequences can significantly affect the outcome of between-site comparisons. Aquat Microb Ecol. 2012;66:13–21. doi: 10.3354/ame01510. [DOI] [Google Scholar]
- 18.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): sequences and tools for high- throughput rRNA analysis. Nucl Acids Res. 2004;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011 doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori H, Maruyama F, Kurokawa K. SVofItTwareOMI: visualization tool for taxonomic compositions of microbial communities based on 16S rRNA gene sequences. BMC Bioinformatics. 2010;11:332. doi: 10.1186/1471-2105-11-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biol Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra S, Stark M, Huson DH. Analysis of 16S rRNA environmental sequences using MEGAN. BMC Genomics. 2011;21:17. doi: 10.1186/1471-2164-12-S3-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering and region on the analysis of 16s rRna gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briggs BR, Inagaki F, Morono Y, Futagami T, Huguet C, Rosell-Mele A, Lorenson TD, Colwell FS. Bacterial dominance in subseafloor sediments characterized by methane hydrates. FEMS Microbiol Ecol. 2012;81:88–98. doi: 10.1111/j.1574-6941.2012.01311.x. [DOI] [PubMed] [Google Scholar]
- 25.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tekere M, Prinsloo A, Olivier J, Jonker N, Venter S. An evaluation of the bacterial diversity at Tshipise, Mphephu and Sagole hot water springs, Limpopo Province, South Africa. Afr J Microbiol Res. 2012;6:4993–5004. [Google Scholar]
- 27.Bowman JP, McCuaig RD. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol. 2003;69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M, Wang P, Wang F, Xiao X. Microbial diversity at a deep-sea station of the Pacific Nodule Province. Biodiversity Conserv. 2005;14:3363–3380. doi: 10.1007/s10531-004-0544-z. [DOI] [Google Scholar]
- 29.Ravenschlag K, Sahm K, Amann R. Quantitative molecular analysis of the microbial community in marine arctic sediments (svalbard) Appl Environ Microbiol. 2000;67:387–395. doi: 10.1128/AEM.67.1.387-395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin LA, Etter RJ, Rex MA, Gooday AJ, Smith CR, Pineda J, Stuart CT, Hessler RR, Pawson D. Environmental influences on regional deep-sea species diversity. Annu Rev Ecol Syst. 2001;132:51–93. doi: 10.1146/annurev.ecolsys.32.081501.114002. [DOI] [Google Scholar]
- 31.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arretia JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siefert JL, Larios-Sanz M, Nakamura LK, Slepecky RA, Paul JH, Moore ER, Fox GE, Jurtshuk P. Phylogeny of marine Bacillus isolates from the Gulf of Mexico. Curr Microbiol. 2000;41:84–88. doi: 10.1007/s002840010098. [DOI] [PubMed] [Google Scholar]
- 33.Miranda CA, Martins OB, Clementino MM. Species-level identification of Bacillus strains isolated from marine sediments by conventional biochemical, 16S rRNA gene sequencing and inter-tRNA gene sequence lengths analysis. Antonie Van Leeuwenhock. 2008;93:297–304. doi: 10.1007/s10482-007-9204-0. [DOI] [PubMed] [Google Scholar]
- 34.Jang SK, Zhang W, Qian PY. Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J Microbiol Methods. 2009;77:48–57. doi: 10.1016/j.mimet.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Ivanova EP, Vysotskii MV, Svetashev VI, Nedashkovskaya OI, Gorshkova NM, Mikhailov VV, Yumoto N, Shigeri Y, Taguchi T, Yoshikawa S. Characterization of Bacillus strains of marine origin. Int Microbiol. 1999;2:267–271. [PubMed] [Google Scholar]
- 36.Nealson KH, Ford J. Surface enhancement of bacterial manganese oxidation: implications for aquatic environments. Geo Microbiol J. 1980;2:21–37. [Google Scholar]
- 37.Tebo BM, Ghiorse WC, Waasbergen LG, Sering PL, Caspi R (1997) Bacterially-mediated mineral formation: insights into manganese(II) oxidation from molecular genetic and biochemical studies. In: Banfield JF, Nealson KH (eds) Geomicrobiology: interactions between microbes and minerals, Washington, DC. Mineral Soc Am 35:225–266
- 38.Oh HM, Giovannoni SJ, Ferriera S, Johnson J, Cho JC. Complete genome sequence of Erythrobacter litoralis HTCC2594. J Bacteriol. 2009;191:2419–2420. doi: 10.1128/JB.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorokin DY, Tourova TP, Muyzer G, Kuenen GJ. Thiohalospira halophila gen. nov., sp. nov., and T. alkaliphila sp. nov., novel obligately chemolithoautotrophic, halophilic, sulfur-oxidizing gammaproteobacteria from hypersaline habitats. Int J Syst Evol Microbiol. 2008;58:1685–1692. doi: 10.1099/ijs.0.65654-0. [DOI] [PubMed] [Google Scholar]