Abstract
The banana weevil Cosmopolites sordidus (Germar) is one of a number of pests that attack banana crops. The use of the entomopathogenic fungus Beauveria bassiana as a biological control agent for this pest may contribute towards reducing the application of chemical insecticides on banana crops. In this study, the genetic variability of a collection of Brazilian isolates of B. bassiana was evaluated. Samples were obtained from various geographic regions of Brazil, and from different hosts of the Curculionidae family. Based on the DNA fingerprints generated by RAPD and AFLP, we found that 92 and 88 % of the loci were polymorphic, respectively. The B. bassiana isolates were attributed to two genotypic clusters based on the RAPD data, and to three genotypic clusters, when analyzed with AFLP. The nucleotide sequences of nuclear ribosomal DNA intergenic spacers confirmed that all isolates are in fact B. bassiana. Analysis of molecular variance showed that variability among the isolates was not correlated with geographic origin or hosts. A RAPD-specific marker for isolate CG 1024, which is highly virulent to C. sordidus, was cloned and sequenced. Based on the sequences obtained, specific PCR primers BbasCG1024F (5′-TGC GGC TGA GGA GGA CT-3′) and BbasCG1024R (5′-TGC GGC TGA GTG TAG AAC-3′) were designed for detecting and monitoring this isolate in the field.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-012-0292-9) contains supplementary material, which is available to authorized users.
Keywords: RAPD, AFLP, Strain-specific molecular marker, Environmental monitoring, Biological control
Introduction
Bananas (Musa sp.) are the most cultivated and consumed fruit worldwide. The cultivation of bananas has important economic and social impacts on various tropical countries, with crops grown by small, medium, and large-scale rural producers. In global terms, Brazil is the second largest producer after India.
The occurrence of pests in banana fields can reduce crop production, and affect fruit quality. Of all pests, Cosmopolites sordidus (Coleoptera: Curculionidae), commonly known as the banana weevil, remains the most important in Brazil. Damage is primarily caused by the larvae that tunnel in the rhizome and pseudostem of banana plants, consequently damaging the vascular system, interfering with nutrient uptake, and reducing plant stability. By damaging the rhizome, larvae may also cause indirect damage, since the tunnels allow pathogens to penetrate invaded sites [1]. As reviewed by Gold et al. [2], integrated pest management options for banana weevils comprise habitat management, host plant resistance, chemical control, and biological control.
The filamentous fungus Beauveria bassiana are well characterized in respect to pathogenicity to several insects and have been used as myco-biocontrol agents for biological control of agriculture pests worldwide (as reviewed by Sandhu et al. [3]) and it has been shown to be one of the most feasible agents of biological control against banana weevils [4]. The Beauveria genus is known to have broad genetic variability, facilitating the selection of fungal isolates with high virulence. In addition, this genus is adapted to climatic conditions of the regions in which it would be logically applied for the biological control of C. sordidus.
While Beauveria is morphologically distinguishable as a genus, species identification is difficult because of its structural simplicity, and the lack of distinctive features in phenotypic variation. Recently a global phylogenetic survey of the genus Beauveria, using both morphological and molecular methods was performed to assess diversity within the genus and to evaluated species concepts and their status taxonomic [5]. In this study B. bassiana and B. brongniartii type strains were proposed and six new species were described. According to the authors, extensive overlap in morphological characters limits their utility for species identification and only a minority of species can be positively identified on the basis of phenotype alone. In contrast, molecular data were effective for accurate diagnosis of all 12 Beauveria species.
Obviously if phenotypic characteristics are insufficient to differentiate the various Beauveria species, they are also unsuitable for monitoring specific strains used as biological control agents in fields [6]. The use of molecular tools has helped towards accurately identifying isolates, whether for patenting purposes or for monitoring the persistence and behavior of isolates after release into the environment (for overview see [7, 8]).
The purpose of our research was to analyze the genetic variability of a Brazilian collection of isolates of B. bassiana using RAPD and AFLP profiles, in order to develop a specific marker for the environmental monitoring of an isolate with high virulence to C. sordidus.
Materials and Methods
Establishing the Fungal Collection
A collection of B. bassiana isolates was established, with specimens originating from different geographic regions of Brazil, and collected from diverse hosts of the Curculionidae family. These isolates are available on the entomopathogenic fungi collection of the Universidade Estadual de Londrina (Londrina, Brazil) and Embrapa Cenargen (Brasília, Brazil) (Supplementary Material 1).
Extraction of Genomic DNA
Each isolate was grown in Pontecorvo’s liquid medium for 48 h at 28 °C [9], and the resulting mycelium was harvested by filtration. To extract the DNA, 0.5 g of frozen mycelium was ground to a fine powder in liquid nitrogen, and immediately transferred to a 2 ml microcentrifuge tube with 800 μl of DNA extraction buffer (100 mM Tris–HCl, pH 8.0; 25 mM EDTA, 1 % SDS, 25 mM NaCl) at 65 °C for 20 min. The suspension was deproteinized by extracting it once with an equal volume of phenol, and once with an equal volume of chloroform:isoamyalcohol (24:1). DNA was precipitated by adding two volumes of ice-cold ethanol and 10 % 3 M NaCl. The precipitate was collected by centrifuging, washed with 70 % ethanol, then dried and resuspended in TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA).
RAPD and AFLP Analyses
RAPD procedures were used as suggested by Fungaro et al. [10]. Primers OPX7, OPX13, OPE11, OPE14, and OPE15 (QIAGEN Operon) were used. Amplifications were carried out in a Mastercycler Gradient (Eppendorf, Hamburg, Germany). AFLP markers were amplified according to the protocol described by Munhoz et al. [11], which was modified from Vos et al. [12]. Briefly, 250 ng of genomic DNA from each isolate was double digested with EcoRI (Promega, Madison, WI) and MseI (NE Biolabs, Ipswich, MA) enzymes (6 U of each) in 20 μl of reaction solution with 1× One Phor-All buffer (Amersham Biosciences, Buckinghamshire, UK), 1× BSA (NE Biolabs, Ipswich, MA) and ultra-pure water q.s.p. for 4 h at 37 °C. Reactions were terminated by heat inactivation (65 °C for 20 min). All resulting fragments were ligated to adapter sequences by adding 1× T4 DNA ligase reaction buffer (NE Biolabs, Ipswich, MA), 0.5 μM of EcoRI adapter (denoted as E), 2.5 μM of MseI adapter (denoted as M), 67 U of T4 DNA ligase (NE Biolabs) and ultra-pure water q.s.p. (20 μl). Ligation reactions were incubated at 16 °C for 14 h and terminated as described above. Primers, which were complementary to the adapters with one, two or three selective nucleotides at the 3′ end, were used for selective amplification. The 20 μl PCR mix contained 1× reaction buffer, 0.25 μM of each primer, 0.25 mM of each dNTP, 1.5 mM MgCl2, 1 U of Go Taq Flexi (Promega, Madison, WI), and 5 μl of the adapter-ligated DNA. PCR conditions were as follows: 94 °C for 2 min, 35 cycles at 94 °C for 1 min, 56 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 5 min in a Gen Amp® PCR System 9700 thermocycler (Applied Biosystems, Carlsbad, CA). Samples (2 μl) of amplified products were denatured with 1× loading buffer (0.2 % each bromophenol blue and xylene cyanol, 10 mM EDTA, and 95 % formamide at pH 8.0) at 95 °C for 5 min before being were loaded onto the gels. AFLP bands were resolved in 5 % w/v denaturing polyacrylamide gels (19:1; acrylamide:bis-acrylamide) using Sequi-Gen GT electrophoresis apparatus (Bio-Rad, Hercules, CA) for 5 h at 80 W, and visualized by silver staining.
Amplification and Sequencing of the ITS1-5.8S-ITS2 Segment from Ribosomal DNA (rDNA)
The ITS1-5.8S-ITS2 DNA fragments of the UNI 40, IBCB 17, CG 138, CG 876, CG 475, CG 1024, and CG 890 isolates were amplified using the primer pairs ITS1 (forward) and ITS4 (reverse) as described by White et al. [13]. The amplicons were submitted to direct sequencing in both directions with a Big Dye Terminator Cycle Sequencing Standard kit Version 3.1 (Applied Biosystems, Foster City, CA, USA) under the following conditions: denaturation at 95 °C for 60 s, followed by 30 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 15 s, extension at 60 °C for 1.5 min, and a final extension at 55 °C for 3 min. A volume of HiDi formamide (10 μl) was added to the sequencing products, which were processed in an ABI 3500XL Genetic Analyser (Applied Biosystems, Foster City, CA, USA). The sequences obtained were aligned to sequences of 12 Beauveria species deposited in the NCBI database (http://www.ncbi.nlm.nih.gov/). The software package MEGA5 [14] was used to construct a neighbour joining tree [15].
Specific Primer Design
One of the RAPD products that was detected in the B. bassiana CG 1024 isolate, but was absent in all other isolates, was excised from the gel and purified with a GFX™ PCR DNA Kit and Gel Band Purification Kit (GE Healthcare, Piscataway, NJ). The resulting DNA was cloned using a TOPO TA Cloning Kit for sequencing (Invitrogen Life Technologies, Carlsbad, CA). The recombinant plasmid was purified with the CONCERT™ Rapid Plasmid Miniprep System (Invitrogen Life Technologies, Carlsbad, CA). The insert was sequenced using a Big Dye Terminator Cycle Sequencing Standard Kit Version 3.1 (Applied Biosystems, Foster City, CA, USA). Specific primers were designed using Gene Runner, version 3.05 (http://www.generunner.com), and PCR conditions were then optimized and tested for all isolates (n = 42) obtained from different geographic regions and hosts. Positive controls were prepared replacing the CG 1024-specific primer pair for rDNA primers ITS1 and ITS4 [13]. The reaction was submitted to a Mastercycler Gradient (Eppendorf, Hamburg, Germany) under the following conditions: 95 °C for 2 min, 35 cycles at 95 °C for 1 min, 62 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 5 min.
Statistical Treatment of RAPD and AFLP Data
Data were scored for the presence (1) or absence (0) of amplification products by means of indicator variables. This type of scoring was done for each locus across all 42 isolates of B. bassiana. Analysis of Molecular Variance (AMOVA) was used to estimate the components of variation directly from the molecular data with ARLEQUIN 2.0 software [16]. Total variance among the isolates was divided into variance between origins (geographical Brazilian states) and hosts, from which the isolates were collected. Each isolate was probabilistically assigned to a genetically distinct cluster based on Bayesian models using STRUCTURE 2.2.3 program [17], which is available online from Cornell University (http://cbsuapps.tc.cornell.edu/structure.aspx). The number of clusters (K) was set from 1 to 13, based on the number of geographical regions from which the isolates were collected. Ten interactions were performed for each K with the admixture model and correlated allele frequencies, using a burn-in period of 300,000 and 500,000 iterations based on the MCMC algorithm (Markov Chain Monte Carlo). The K statistics show a change in the probability of the log according to the number of K, as proposed by Evano et al. [18], while the ΔK is a prediction of the real number of clusters.
Efficiency of the Specific Marker in Detecting CG 1024-Isolate of B. bassiana in C. sordidus
The ability of the PCR-based marker to detect specifically the CG 1024-isolate in C. sordidus was tested. First, healthy insects were passed through a quarantine test to validate the absence of B. bassiana disease. These insects were then inoculated with two B. bassiana strains (CG 1024 and CG 1013, the latter as a negative control). Non-inoculated insects were also used as controls. Three insects from each treatment were immersed in a fungal suspension (1 × 109 conidia ml−1) for 90 s. Before DNA extraction, the insects were immersed in 1 % NaOCl solution for 3 min, and rinsed in three changes of sterile distilled water, to disinfect the insect surface (exoskeleton). Total genomic DNA was extracted at 2 and 4 day after inoculation using the standard phenol:chloroform:isoamyalcohol method. PCR assays were performed in a 25 μl-reaction mixture containing 10 ng of DNA and 0.2 μM of each specific primer-pair designed in the present study (BbasCG1024F and BbasCG1024R). Annealing conditions were 62 °C for 1 min. Amplicons were separated by agarose gel (1 %, v/v) electrophoresis.
Results and Discussion
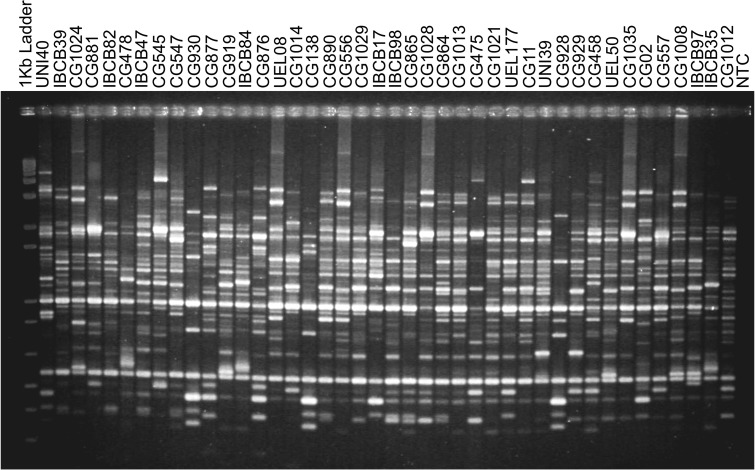
Amongst 50 RAPD loci generated with five random decamers, 92 % were polymorphic (Fig. 1). Accordingly, a high level of polymorphism was found using the AFLP technique. Out of 149 bands obtained from six primer combinations in the selective step, 131 (88 %) were polymorphic (Supplementary Material 2). Substantial correspondence between the clusters generated by RAPD and AFLP markers was observed.
Fig. 1.
Fingerprint generated by random amplified polymorphic DNA using the OPX13 primer; Lane M 1-kb molecular weight standard plus DNA Ladder, Lane NTC non-template control
When using the methodology implemented in the software STRUCTURE we found that the best K value determined according to the model described by Evano et al. [18] was two for RAPD data, and three for AFLP data. In other words, the isolates of B. bassiana were assigned to two and three genotypic clusters, depending on the molecular profile used. The assigning test revealed the participation of each isolate in a given cluster, with most isolates exhibiting Q-values close to 100 %. Just a few isolates exhibited low participation (Q < 44 %) in a second or a third cluster (Supplementary Material 3). By identifying isolates with similar band frequencies, this statistical analysis has assigned them to a number of clusters (K). Then, a value of cluster membership Q for all isolates was estimated. As Q-values were high, our results suggested a low level of gene flow between the isolates analyzed in the present study.
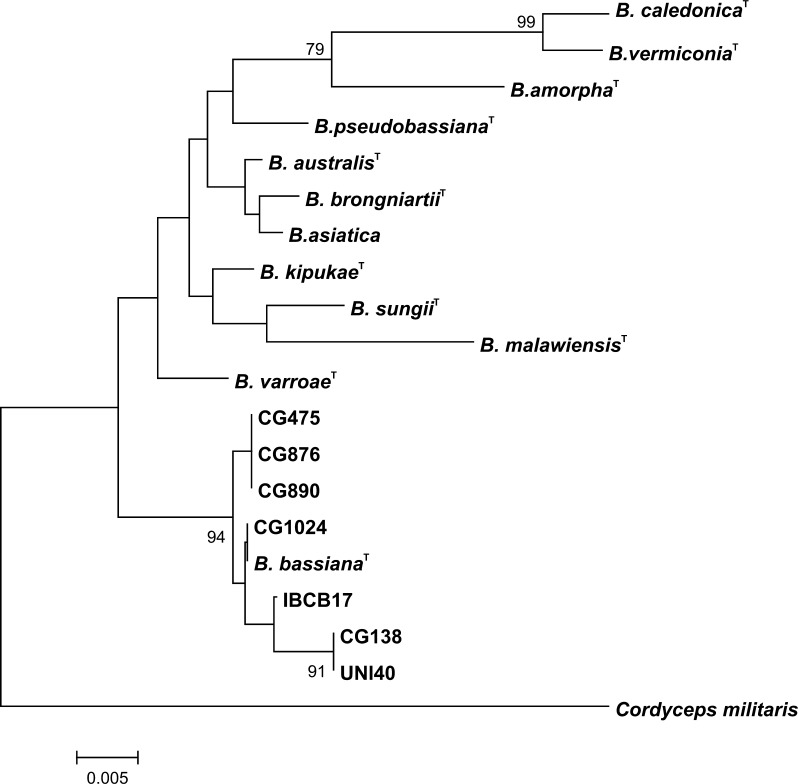
Since the genetic variations between genotypic clusters were high, representatives from each RAPD and AFLP group underwent nucleotide sequence analysis of rDNA-ITS region in order to confirm that all isolates are in fact B. bassiana. Alignment of the obtained sequences revealed six nucleotide variations among the strains. Despite these nucleotide variations, all of our isolates grouped together with an authentic culture of B. bassiana, which all together formed a group phylogenetically distant from other Beauveria species (Fig. 2). High levels of genetic variation within B. bassiana using multilocus markers have been also observed in several previous studies [7, 19–21].
Fig. 2.
Phylogenetic tree generated from sequences of the ITS1–5.8S–ITS2 region of isolates of B. bassiana using the CLUSTAL-W program and the neighbor-joining method. Accession numbers of the sequences used to generate the phylogenetic tree: B. bassiana (HQ880761), B. varroae (HQ880800), B. kipukae (HQ880803), B. brongniartii (HQ880782), B. australis (HQ880789), B. amorpha (AY532008), B. pseudobassiana (AY532022), B. sungii (AY531990), B. caledonica (AY532006), B. verniconia (AY532012), B. asiatica (AY532026), B. malawiensis (HQ880825)
The AMOVA results, which was carried out on the RAPDs and AFLPs separately, showed that variability among the isolates is not associated with geographical origin nor host, as the percentage of variation among populations was less than 0.72. Although some former studies have demonstrated some association between B. bassiana genetic groups and insect host, more recently was suggested that B. bassiana is a non-specialist entomopathogen, with no particular phylogenetic association concerning the specificity of the host, being the environmental factors the major selective forces for genotypic evolution in B. bassiana [7, 22, 23].
Amongst all 42 isolates of B. bassiana, the isolates CG 11, CG 138, CG 458, CG 475, CG 545, CG 547, CG 556, CG 864, CG 865, CG 876, CG 890, CG 919, CG 1013, CG 1024, CG 1028 were also presented in previous studies that evaluated their virulence against C. sordidus. Isolate CG 1024 was the most virulent, with a confirmed mortality of 96.7 % [24, 25]. Currently this isolate is being evaluated for its utility as a microbial control agent against the banana weevil under Brazilian field conditions, and the development of appropriate markers to distinguish CG 1024 from indigenous fungal strains will be essential.
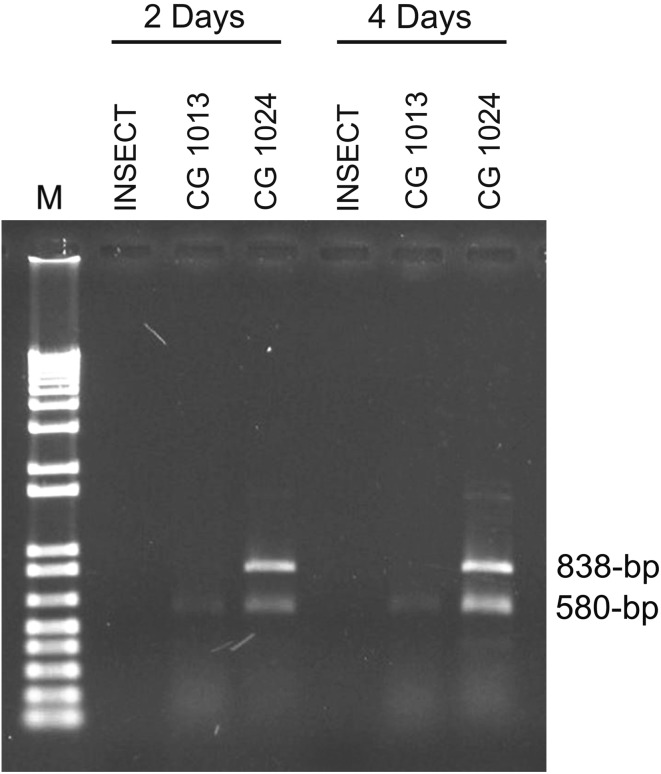
Comparison of all RAPD profiles allowed us to detect an amplicon (when using the OPE-14 primer) that was specific to isolate CG 1024. This DNA band, which was not revealed in the other isolates of B. bassiana, was cloned and sequenced. Then, the sequence obtained in this study was used for designing the primers BbasCG1024F (5′-TGC GGC TGA GGA GGA CT-3′) and BbasCG1024R (5′-TGC GGC TGA GTG TAG AAC-3′). Using the PCR conditions described in “Specific primer design” of the materials and methods section, the primer pair produced an amplicon of 838-bp, when the DNA mold of CG 1024 was used. No amplification product was observed in any other of the studied isolates (data not shown). It is promising that the specific primer-pair BbasCG1024838 was extremely efficient in detecting this fungal isolate in C. sordidus, even after the 2nd day of inoculation (Fig. 3). The CG 1024 isolate is expected to be released in the environment in the near future. The PCR assay here developed will be useful for monitoring the CG 1024 and to determine its effectiveness against C. sordidus.
Fig. 3.
PCR results using the 1024F838 and 1024R838 primers specific to the B. bassiana isolate CG 1024 2 and 4 days after inoculation on C. sordidus. Lane 1 PCR from DNA of C. sordidus non-inoculated with B. bassiana, lane 2 PCR from DNA of C. sordidus inoculated with B. bassiana isolate CG 1013, as an exclusivity control. Lane 3 PCR from DNA of C. sordidus inoculated with B. bassiana CG 1024. The 580-bp fragment corresponds to amplicon of the ITS1-5.8S-ITS2 region used to confirm the presence of PCR-compatible DNA and the 838-bp fragment is specific for CG1024 strain. Lane M 1-kb molecular weight standard plus DNA Ladder
Electronic supplementary material
Acknowledgments
This work was supported by grants and fellowships from the Brazilian institutions, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenadoria de Aperfeiçoamento de Pessoal do Ensino Superior (CAPES).
References
- 1.Akello J, Dubois T, Gold CS, Coyne D, Nakavuma J, Paparu P. Beauveria bassiana (Balsamo) Vuillemin as an endophyte in tissue culture banana (Musa spp.) J Invertebr Pathol. 2007;96:34–42. doi: 10.1016/j.jip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Gold CS, Pena JE, Karamura EB. Biology and integrated pest management for the banana weevil, Cosmopolites sordidus (Germar) (Coleoptera: Curculionidae) Int Pest Manag Rev. 2001;6:79–155. doi: 10.1023/A:1023330900707. [DOI] [Google Scholar]
- 3.Sandhu SS, Sharma AK, Beniwal V, Goel G, Batra P, Kumar A, Jaglan S, Sharma AK, Malhotra S (2012) Myco-biocontrol of insect pests: factors involved, mechanism, and regulation. J Pathog. doi:10.1155/2012/126819 [DOI] [PMC free article] [PubMed]
- 4.Oliveira FQ, Malaquias JB, Ferreira LL, Ribeiro TS, Pereira AIA. Efficacy of molasses and Beauveria bassiana (Balsamo) Vuill on the control of Cosmopolites sorditus Germar, 1824. Eng Ambient. 2010;7:127–132. [Google Scholar]
- 5.Rehner SA, Minnis AM, Sung GH, Luangsa-ard JJ, Devotto L, Humber RA. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia. 2011;103:1055–1073. doi: 10.3852/10-302. [DOI] [PubMed] [Google Scholar]
- 6.Castrillo LA, Vandenberg JD, Wraight SP. Strain-specific detection of introduced Beauveria bassiana in agricultural fields by use of sequence-characterized amplified region markers. J Invertebr Pathol. 2003;82:75–83. doi: 10.1016/S0022-2011(02)00190-8. [DOI] [PubMed] [Google Scholar]
- 7.Meyling NV, Lübeck M, Buckley EP, Eilenberg J, Rehner SA. Community composition, host range and genetic structure of the fungal entomopathogen Beauveria in adjoining agricultural and seminatural habitats. Mol Ecol. 2009;18:1282–1293. doi: 10.1111/j.1365-294X.2009.04095.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Fan M, Li Z, Butt TM. Molecular monitoring and evaluation of the application of the insect-pathogenic fungus Beauveria bassiana in southeast China. J Appl Microbiol. 2004;96:861–870. doi: 10.1111/j.1365-2672.2004.02215.x. [DOI] [PubMed] [Google Scholar]
- 9.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv Gen. 1953;5:141–238. doi: 10.1016/S0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 10.Fungaro MHP, Vieira MLC, Pizzirani-Kleiner AA, Azevedo JL. Diversity among soil and insect isolates of Metarhizium anisopliae var. anisopliae detected by RAPD. Lett Appl Microbiol. 1996;22:389–392. doi: 10.1111/j.1472-765X.1996.tb01186.x. [DOI] [Google Scholar]
- 11.Munhoz CF, Weiss B, Hanai LR, Zucchi MI, Fungaro MHP, Oliveira ALM, Monteiro-Vitorello CB, Vieira MLC. Genetic diversity and a PCR-based method for Xanthomonasaxonopodis detection in passion fruit. Phytopathology. 2011;101:416–424. doi: 10.1094/PHYTO-06-10-0169. [DOI] [PubMed] [Google Scholar]
- 12.Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White TJ, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 14.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Schneider S, Kueffer JM, Roessli D, Excoffier L. ARLEQUIN version 2.000. A software for population genetic data analysis. Switzerland: Genetic and Biometry Laboratory, University of Geneva; 2000. [Google Scholar]
- 17.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evano G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Shah FA, Patel N, Li ZZ, Butt TM. Molecular investigation on strain genetic relatedness and population structure of Beauveria bassiana. Environ Microbiol. 2003;10:908–915. doi: 10.1046/j.1462-2920.2003.00485.x. [DOI] [PubMed] [Google Scholar]
- 20.Castrillo LA, Bauer LS, Liu H, Griggs MH, Vandenberg JD. Beauveria bassiana (Ascomycota: Hypocreales) isolates associated with Agrilus planipennis (Coleoptera: Buprestidae) populations in Michigan. Biol Control. 2010;54:135–140. doi: 10.1016/j.biocontrol.2010.04.005. [DOI] [Google Scholar]
- 21.Meyling NV, Pilz C, Keller S, Widmer F, Enkerli J. Diversity of Beauveria spp. isolates from pollen beetles Meligethes aeneus in Switzerland. J Invertebr Pathol. 2012;109:76–82. doi: 10.1016/j.jip.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Ghikas DV, Kouvelis VN, Typas MA. Phylogenetic and biogeographic implications inferred by mitochondrial intergenic region analyses and ITS1-5.8S-ITS2 of the entomopathogenic fungi Beauveria bassiana and B. brongniartii. BMC Microbiol. 2010;10:174–189. doi: 10.1186/1471-2180-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garrido-Jurado I, Márquez M, Ortiz-Urquiza A, Santiago-Álvarez C, Iturriaga EA, Quesada-Moraga E, Monte E, Hermosa R. Genetic analyses place most Spanish isolates of Beauveria bassiana in a molecular group with word-wide distribution. BMC Microbiol. 2011;11:84–95. doi: 10.1186/1471-2180-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopes RB, Michereff-Filho M, Tigano MS, Neves PMOJ, López EL, Fancelli M, Silva JP. Virulence and horizontal transmission of selected Brazilian strains of Beauveria bassiana against Cosmopolites sordidus under laboratory conditions. B Insectol. 2011;64:201–208. [Google Scholar]
- 25.Dissertation. Lema EL (2010). Biological control of Cosmopolites sordidus (Germar) (Coleoptera: Dryophthoridae) with Beauveria bassiana (Bals.) Vuill. And population fluctuation in Ibiporã, Brazil. University of Londrina
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.