Abstract
Background:
It has been a challenge to determine breast cancer clonality accurately. The aim of the present study was to assess methods using formalin-fixed paraffin-embedded (FFPE) tissue to differentiate new primary tumours from true recurrences that are associated with poorer prognoses and often require more aggressive treatment.
Methods:
We investigated the novel method of analysing gene alterations of mitochondrial DNA D-loop region (GAMDDL) and compared it with the conventional method of analysing the X-chromosome-linked human androgen receptor (HUMARA). The FFPE sections of primary and secondary breast cancers, the non-neoplastic mammary gland, and lymph nodes were examined.
Results:
Informative rates for HUMARA, GAMDDL, and combined analyses were 42.1%, 76.9%, and 89.5%, respectively. All of the 10 contralateral breast cancers were determined to be non-clonal. In contrast, 3 out of 8 (37.5%) of the ipsilateral secondary tumours shared a clonal origin with the primary tumour and were classified as true recurrences, whereas 4 out of 8 (50%) were classified as new primary tumours.
Conclusion:
GAMDDL analysis represents a novel and useful molecular method for examining the precise cell lineages of primary and secondary tumours, and was more accurate than HUMARA in determining clonality.
Keywords: breast cancer, mitochondria DNA, cell lineage, clonality, D-loop
Breast cancer is a systemic disease that may take years to manifest, and can recur even 20 years after treatment for primary tumours (Karrison et al, 1999). Because the patients with secondary new primary tumours are associated with more favourable prognosis than those with recurrent tumours (Haffty et al, 1996), it is important to determine whether a secondary breast tumour is a true recurrent tumour, or is a new primary tumour. Despite advances in the methods, it remains difficult to assess tumour clonality accurately. In order to treat secondary tumours appropriately, methods that allow a retrospective analysis of primary tumour samples are needed to evaluate long-term recurrence.
Previous studies have demonstrated that a molecular analysis is more useful than clinicopathological characterisations to define tumour clonality. One well-known molecular method is an analysis of polymorphisms in X-chromosome-linked genes, such as glucose-6-phosphate dehydrogenase (Linder and Gartler, 1965), phosphoglycerate kinase (Vogelstein et al, 1985), and the human androgen receptor (HUMARA) (Mashal et al, 1993; Noguchi et al, 1995). However, because this method is based on the random inactivation of somatic X chromosomes by methylation, a process known as lyonisation (Lyon, 1961), it is only useful for analysing samples from female patients. Another molecular method compares loss of heterozygosity (LOH) patterns in chromosomal regions associated with breast cancer susceptibility by analysing polymorphic microsatellite markers (Kollias et al, 2000; Goldstein et al, 2005a). However, the complexity associated with comparing LOH patterns in a large panel of genes renders this assay impractical for large cohorts.
Recently, a unique approach for analysing gene alterations of the mitochondrial DNA (mtDNA) D-loop region (GAMDDL) was applied to investigate lobular carcinoma in situ as a precursor lesion, or a risk factor, of invasive lobular carcinoma (Morandi et al, 2006; Aulmann et al, 2008). The mtDNA is encoded separately from nuclear DNA, and by virtue of its proximity to reactive oxygen species and lack of protective histones, mtDNA accumulates mutations at much higher rates than nuclear DNA (Chatterjee et al, 2011). These high mutation rates in mtDNA have been observed for many carcinomas, including breast, lung, and head and neck cancers (Lee et al, 2010; Yu, 2011). The D-loop of mtDNA appears particularly to be susceptible to mutation, and several insertions, deletions, and point mutations have been identified in this region in breast cancer tissues (Lee et al, 2010). Although the D-loop represents a non-coding region of mtDNA, the D-loop is essential for mtDNA transcription because it contains promoters and an origin of replication of mtDNA (Shadel, 2008).
The maternal mode of inheritance for mtDNA (Giles et al, 1980) and high variability in sequence offers the opportunity to be a more sensitive method (Salk and Horwitz, 2010). To estimate the accuracy of this novel approach for assessing clonality, we studied GAMDDL in primary and secondary tumours from both ipsilateral and contralateral breast tissue, and compared the results with the conventional approach of HUMARA analysis.
Materials and methods
Patients and tissue preparation
Among the 558 patients who underwent breast conservative surgery at the Tokai University Hospital between January 1991 and December 2004, 17 patients with secondary ipsilateral breast tumours and 14 patients with contralateral breast tumours were identified during the follow-up period. Of these patients, 8 and 11, respectively, had formalin-fixed paraffin-embedded (FFPE) sections of normal tissue (lymph node), a non-neoplastic mammary gland, a primary tumour, and secondary tumour available.
The location of breast cancers was indicated as quadrant A: medial upper, B: medial under, C: lateral upper, D: lateral under, and E: around nipple. All samples were pathologically examined according to the World Health Organisation classification system (Tavassoli and Devilee, 2003) and the Scarff–Bloom–Richardson grading system (Elston and Ellis, 1991). Immunohistochemical intrinsic subtypes for these samples were also defined as: Luminal A, oestrogen receptor (ER) and/or progesterone receptor (PgR) positive and HER2 negative; Luminal B, ER and/or PgR positive and HER2 positive; HER2, ER negative and HER2 positive; triple negative, ER, PgR and HER2 negative; and basal-like subtype, ER, PgR and HER2 negative, cytokeratin5/6 and/or EGFR positive (Carey et al, 2006).
The study design was approved by an institutional ethics committee, and the patients were informed of the privacy policy of the study.
Laser microdissection and extraction of DNA
The FFPE sections (5 μm) were mounted on PEN membrane-covered slides (Membrane Slide 1.0 PEN; Carl Zeiss MicroImaging, Jena, Germany). Sections were deparaffinised, stained with hematoxylin and eosin, dehydrated with 99.5% ethanol, air-dried for 20 min, and stored in a desiccators. Breast cancer tissues were microdissected using a PALM MicroBeam (Carl Zeiss MicroImaging). Carcinoma cell nests were captured using a solid-state 355-nm UV laser and transferred to microcentrifuge tubes. The average number of cells captured was approximately 2 000 per case (1 mm2). All captured tissue sections were incubated for 16 h at 56 °C with 25 μl tissue lysis buffer (QIAamp DNA Micro Kit; Qiagen K.K., Tokyo, Japan) containing 10 μl proteinase K (20 mg ml−1). Carrier RNA was added to the sample to improve DNA affinity and isolated by Qiagen spin column. DNA was eluted in 30 μl DNAse/RNAse-free distilled water (GibcoBRL, Grand Island, NY, USA) and stored at −20 °C.
The HUMARA analysis
To determine the methylation patterns of HUMARA in patient tissues, DNA samples (13 μl) of the lymph node were incubated at 37 °C with or without 18 U of HpaII (Toyobo, Osaka, Japan) in a total volume of 30 μl. After 16 h, samples were deactivated at 90 °C for 5 min.
PCR primers were designed to amplify fragments of androgen receptor (AR) exon 1 that included the HpaII site and the short tandem repeat of the variable CAG region as follows: AR1 forward (5′-TGTGGGCCTCTACGATG-3′) and reverse (5′-TCCAAGACCTACCGA-3′) (product size, 238–298 bp); and for nested amplification, AR2 forward (5′-CCGAGGAGCTTTCCAGAATC-3′) and reverse (5′-TACGATGGGCTTGGGGAGAA-3′) (product size, 215–273 bp) (Wu et al, 2003). For AR1 amplification, digested or nondigested DNA (8 μl) was amplified in a total volume of 20 μl containing 10 μl AmpliTaq Gold PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and 20 μℳ primers. The PCR amplification was performed with an initial step of 95 °C for 10 min, followed by 28 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 30 s, and a final extension step at 72 °C for 10 min. Amplification was performed using a Mastercycler ep gradient S (Eppendorf, Hamburg, Germany). For AR2 amplification, digested or undigested DNA (2 μl) was amplified in a total volume of 20 μl containing 10 μl AmpliTaq Gold PCR Master Mix and 4 pmol each PCR primer. PCR amplification was performed as described above, except 38 cycles were used. PCR products were separated on 5–20% acrylamide gradient gels (Tris-Borate, EDTA, e-PAGEL Precast Gels; ATTO Corp., Tokyo, Japan) and were stained with ethidium bromide.
Samples were further analysed if undigested, control non-neoplastic lymph-node DNA yielded two distinct alleles. As a negative control, samples were incubated without methylation-specific HpaII under the same condition.
The GAMDDL analysis
The mtDNA D-loop encompasses positions 16045–60 (MITOMAP, www.mitomap.org) (Anderson et al, 1981). Following DNA extraction from FFPE tissue, the D-loop region was PCR amplified as two overlapping fragments, DL1 and DL2. Primers for DL1 (16045–16347) include forward (5′-CCACCCAAGTATTGACTCACCCATCAA-3′) and reverse (5′-ATTTGACTGTAATGTGCTATGTACGGTA-3′)(product size, 302 bp). Primers for DL2 (16216–60) included forward (5′-CTTCAACTATCACACATCAACTGCAACT-3′) and reverse (5′-CATGGAGAGCTCCCGTGAGTGGTTAAT-3′) (product size, 395 bp). PCR reactions were performed in 20 μl containing 4 pmol of each primer, 200 μℳ dNTPs, 4 μl PrimeSTAR GXL Buffer with Mg2+ (Takara Bio, Shiga, Japan), and 0.5 U PrimeSTAR GXL DNA Polymerase (Takara Bio). Samples were amplified at 95 °C for 10 s, 58 °C for 15 s, and 72 °C for 60 s. PCR products were separated on a 2% agarose gel and purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). Purified products were sequenced using a 3100 Genetic Analyzer (Applied Biosystems).
The sequence of amplified products (567 bp) was compared against a comprehensive human mitochondrial databank to estimate relative phylogenetic distances from normal mammary tissue. Phylogenetic clusters were constructed using the neighbour-joining method of MEGA4 (http://www.megasoftware.net/index.html) (Tamura et al, 2007) and were analysed using the unweighted pair group method with arithmetic mean (UPMEGA) method (Sokal and Michener 1958).
Interpretation by the UPMEGA method was verified by comparing the incidence of genetic alterations detected in the examined tissues. A statistical analysis was also performed using Mann–Whitney’s U-test. The incidence of genetic alteration in the mtDNA D-loop was calculated as a percentage; x bp (the number of altered bases compared with the normal mammary gland around the primary tumour)/567 bp (the number of bases examined in D-loop region) × 100.
Results
Clinical and pathological characteristics of secondary tumours
Clinicopathological characteristics of the ipsilateral and contralateral secondary tumours analysed are summarised in Tables 1 and 2, respectively. Pathological characteristics, such as immunohistochemical intrinsic subtype and histological or nuclear grade, were found to be similar between primary and secondary ipsilateral tumours (Table 1). Despite variations in the distance from the margin and time to detection, 7 out of 8 (87.5%) ipsilateral secondary tumours were clinically characterised as recurrent because they were adjacent to a primary tumour. Secondary tumours in contralateral breast were detected at the same time (e.g., case 13) or 7–67 months after the primary tumour was treated. In 3 out of 11 (27.3%) secondary contralateral tumours, their intrinsic subtypes and grading differed from those of the primary tumour (e.g., cases 10, 12, and 15; Table 2).
Table 1. Clinicopathological characteristics of cases involving secondary ipsilateral breast tumours.
|
Clinical characteristics
|
Pathological characteristics
|
Molecular characteristics
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
First tumour
|
Second tumour
|
First tumour | Second tumour | HUMARA | GAMDDL | |||||||
| Case | Age | pTNM | Margin | Adjuvant therapy | Quadrant | Quadrant | Time to second tumour (M) | Outcome | IIS, HG/NG | IIS, HG/NG | First/second tumours | First/second tumours |
| Clinically recurrent: | ||||||||||||
| 1 | 43 | T1Nx | 5 mm | Right D | Right ECA | 39 | Alive | Luminal A, HGII | Luminal A, HGII | NP | Identical | |
| 2 | 47 | T1N0M0 | 5 mm | CMF, Tam | Right C | Right CA | 37 | Alive | Luminal A, HGIII | Luminal A, HGIII | Same allele | Identical |
| 3 | 45 | T1N0M0 | 12 mm | Right BD | Right D | 12 | Alive | Luminal A, HGI | Luminal A, HGI | Same allele | Identical | |
| 4 | 51 | T1N0M0 | 4 mm | IR, CMF | Left C | Left EBD | 15 | Alive | HER2, HGIII | HER2, HGIII | Same allele | No mutation |
| 5 | 26 | T2N2M0 | 10 mm | CMF, Gos, T | Left C | Left CD | 36 | Dead | Luminal B, HGIII | Luminal B, HGIII | Same allele | Different mutation |
| 6 | 49 | T1N0M0 | 5 mm | IR | Left D | Left BD | 19 | Alive | TNBC, HGII | TNBC, HGIII | Same allele | Different mutation |
| 7 | 43 | T1N0M0 | < 1 mm | Left A | Left AC | 45 | Alive | BLBC, HGII | TNBC, HGII | Different allele | Different mutation | |
| Clinically new primary: | ||||||||||||
| 8 | 66 | T1N0M0 | 9 mm | IR, Tam | Right B | Right C | 41 | Dead | Luminal A, HGII | Luminal A, HGIII | Different allele | No mutation |
Abbreviations: BLBC=basal-like breast cancer; CMF=CPM+methotrexate+5-FU; HG=histological grade I, II, and III; HUMARA=human androgen receptor; GAMDDL=gene alterations of mitochondrial DNA D-loop region; Gos=goserelin; IR=irradiation; IIS=immunohistochemical intrinsic subtype; M=months; NP=no polymorphism; Quadrant A=medial upper; Quadrant B=medial under; Quadrant C=lateral upper; Quadrant D=lateral under; Quadrant E=around nipple; T=trastuzumab; Tam=tamoxifen; TNBC=triple-negative breast cancer.
Table 2. Clinicopathological characteristics of cases involving secondary contralateral breast tumours.
|
Clinical characteristics
|
Pathological characteristics
|
Molecular characteristics
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
First tumour
|
Second tumour
|
First tumour | Second tumour | HUMARA | GAMDDL | |||||||
| Case | Location | pTNM | Adjuvant Therapy | Loation | pTNM | Time to 2nd tumour (M) | Adjuvant Therapy | Outcome | IIS, HG/NG | IIS, HG/NG | First/second tumours | First/second tumours |
| 9 | Left | T1micN0 | Right | Tis | 7 | AI | Alive | Luminal A, NG2 | Luminal A, NG2 | NP | No mutation | |
| 10 | Right | T1N0 | Tam | Left | T1micN0 | 24 | Alive | BLBC, HGI | HER2, NG3 | NP | Different mutation | |
| 11 | Left | T1N0 | Tam | Right | T1N0 | 51 | Alive | Luminal A, HGI | Luminal A, HGI | Same allele | Different mutation | |
| 12 | Left | T1N0 | IR, Tam | Right | T1micN0 | 34 | AI | Alive | Luminal A, HGI | TNBC, NG3 | Same allele | Different mutation |
| 13 | Right | T1N0 | Left | Tis | 0 | Alive | Luminal A, HGI | Luminal A, NG2 | Same allele | Different mutation | ||
| 14 | Right | T1N0 | Tam | Left | T1mic | 67 | Alivea | Luminal A, HGI | Luminal A, NG1 | Different allele | NE | |
| 15 | Right | T1N1 | Tam | Left | T1mic | 14 | Alive | Luminal A, HGII | TNBC, NG3 | Different allele | NE | |
| 16 | Right | T1N1 | Left | T1N1 | 47 | CMF | Alive | BLBC, HGIII | BLBC, HGIII | Different allele | NE | |
| 17 | Right | T2N1 | Left | T1 | 18 | Alive | Luminal A, HGI | Luminal A, HGI | Different allele | NE | ||
| 18 | Right | T2N0 | Left | T2N0 | 36 | IR | Alive | Luminal A, HGII | Luminal A, HGIII | Different allele | NE | |
| 19 | Right | T1N0 | Left | T1 | 28 | IR, CMF | Alive | Luminal A, HGI | Luminal A, HGII | Different allele | NE | |
With lung metastasis.
Abbreviations: AI=aromatase inhibitor; BLBC=basal-like breast cancer; CMF=CPM+methotrexate+5-FU; GAMDDL=gene alterations of mitochondrial DNA D-loop region; HG=histological grade I, II, and III; HUMARA=human androgen receptor; IR=irradiation; IIS=immunohistochemical intrinsic subtype; M=months; NE=not examined; NG=nuclear grade 1, 2, and 3; NP=no polymorphism; Tam=tamoxifen; TNBC=triple-negative breast cancer.
The HUMARA analysis of clonality
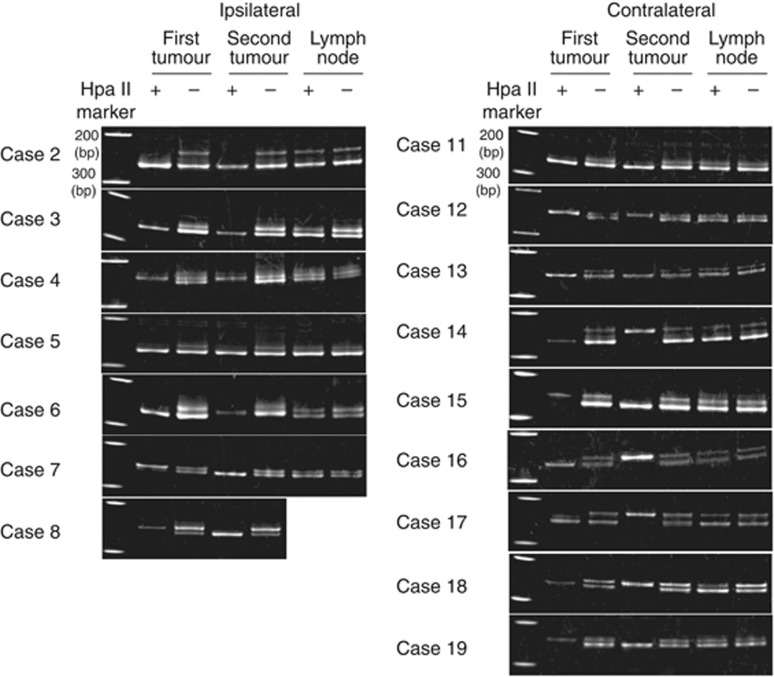
Two distinct HUMARA alleles were detected in normal lymph-node tissue in 16 out of 19 (84.2%) cases. Of these patients, seven had ipsilateral tumours and nine had contralateral tumours (Figure 1). For cases 7 and 8 of the former group, and cases 14–19 of the latter group, the location of the methylated allele in these secondary tumours differed from the primary tumours, thereby leading to the classification of these secondary tumours as non-clonal (Figure 1). For case 7, this classification was in contrast with the classification assigned based on clinicopathological characteristics, whereas the classification of case 8 was consistent with its clinicopathological characteristics. The informative rate of this method was 8 out of 19 (42.1% Table 3).
Figure 1.
Determination of tumour clonality by HUMARA. DNA isolated from primary tumours, secondary tumours, and non-neoplastic lymph-node tissues from each patient were incubated with (+) methylation-specific HpaII and were amplified with primers for exon 1 of AR. As a negative control, samples were incubated without methylation-specific HpaII under the same conditions. A methylated AR gene was found on different alleles in cases 7 and 8 (in ipsilateral tumour samples) and cases 14–19 (in contralateral tumour samples).
Table 3. Summary of molecular analysis results.
| HUMARA | No polymorphism | Same allele | Different allele | Total | Informative rate (%) |
| Ipsilateral | 1 | 5 | 2 | 8 | 8/19 (42.1%) |
| Contralateral | 2 | 3 | 6 | 11 | |
| 3 | 8 | 8 | 19 | ||
| GAMDDL | No mutation | Identical mutation | Different mutation | Total | 10/13 (76.9%) |
| Ipsilateral | 2 | 3 | 3 | 8 | |
| Contralateral | 1 | 0 | 4 | 5 | |
| 3 | 3 | 7 | 13 | ||
| HUMARA and GAMDDL | Not determined | True recurrence | New primary | Total | 17/19 (89.5%) |
| Ipsilateral | 1 | 3 | 4 | 8 | |
| Contralateral | 1 | 0 | 10 | 11 | |
| 2 | 3 | 14 | 19 |
Abbreviations: GAMDDL=gene alteration of mitochondrial DNA D-loop region; HUMARA=human androgen receptor.
Informative cases are presented in shadowed boxes.
The GAMDDL analysis of clonality in non-neoplastic tissue
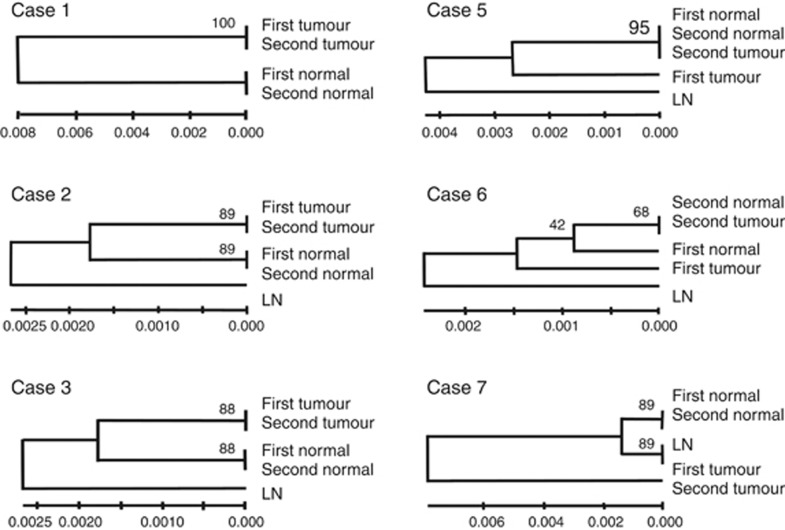
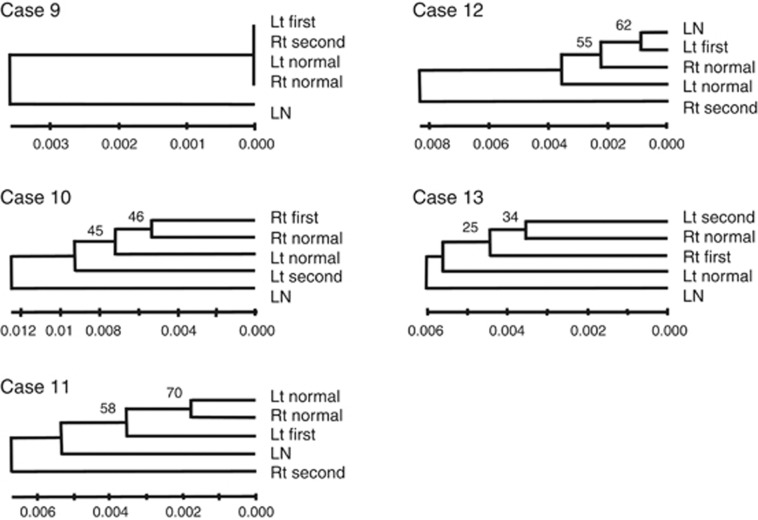
The GAMDDL analysis revealed that sequences of the mtDNA D-loop region isolated from lymph-node tissues differed from the sequences obtained from mammary gland tissues in all cases (Figures 2 and 3). In addition, sequences obtained from left vs right non-neoplastic mammary glands differed in all cases except case 9, where the DNA sequences were identical (Figure 3). In contrast, sequences from non-neoplastic mammary glands surrounding primary and secondary ipsilateral tumours were identical in 5 out of 6 cases (83.3%), with exceptional case 6 (Figure 2).
Figure 2.
Phylogenetic clustering of mtDNA D-loop regions from primary and secondary ipsilateral tumours. The relative phylogenetic distances between lymph node (LN), non-neoplastic mammary gland surrounding the primary (first normal) and secondary tumour (second normal), primary tumour (first tumour), and ipsilateral secondary tumour (second tumour) were determined using the neighbour-joining method. Scale bars represent length where 0.001 indicates that one altered base is present in 1000 bp. Bootstrap value represents the expected reproducibility of clustering. Gene alterations were not observed in any of the sequences obtained from cases 4 and 8 (not shown).
Figure 3.
Phylogenetic clustering of mtDNA D-loop regions in primary and secondary contralateral tumours. The relative phylogenetic distances between lymph node (LN), non-neoplastic mammary gland surrounding the primary (first normal) and secondary tumour (second normal), primary tumour (first tumour), and contralateral secondary tumour (second tumour) were determined using the neighbour-joining method. Scale bars represent length where 0.001 indicates that one altered base is present in 1000 bp. Bootstrap values represent the expected reproducibility of clustering. Mutations were not observed in any of the sequences obtained from case 9 (data not shown).
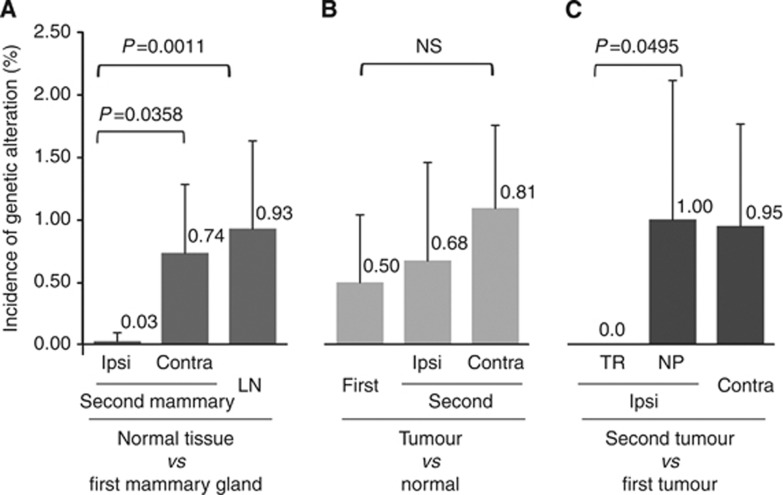
The incidence of gene alterations in the ipsilateral mammary gland around a second tumour was significantly lower (0.03%±0.07) than that of the contralateral mammary gland (0.74%±0.55, P=0.0358) and lymph nodes (0.93%±0.71, P=0.0011) (Figure 4).
Figure 4.
The incidence of genetic alterations in normal tissue vs primary and secondary tumour tissues. The proportion of number of altered bases to the number of bases examined in the D-loop region (567 bp) is shown as the incidence of genetic alteration (%). (A) The incidence of genetic alterations detected in the secondary mammary gland (ipsilateral and contralateral) and lymph-node tissue, in comparison with the primary mammary gland, is presented. (B) The incidence of genetic alterations detected in a tumour, in comparison with the non-neoplastic mammary gland, is presented. (C) The incidence of genetic alterations in secondary tumours, in comparison with, a primary tumour is presented. The incidences were compared and statistically analysed using the Mann–Whitney test. contra=contralateral; Ipsi=ipsilateral; LN=lymph node.
The GAMDDL analysis of clonality in ipsilateral breast tumours
Genetic alterations that were detected in the D-loop region (position 16045–16569, 1–60) of the samples obtained from ipsilateral breast tumours are summarised in Supplementary Table. GAMDLL analysis revealed that identical mutations were present in the sequences from primary and secondary ipsilateral tumours in 3 out of 8 (37.5%) cases (e.g., cases 1, 2, and 3). Therefore, these secondary tumours were classified as true recurrences (Table 1). In contrast, in cases 5, 6, and 7, GAMDDL analysis revealed unique mutations that were present in the primary and secondary tumours. There were no mutations in the D-loop in primary or secondary tumour samples of cases 4 and 8.
In case 5, two nucleotides (16261 and 16362) were found to be altered in the primary tumour tissue (Supplementary Table, Supplementary Figure 2). However, the sequence obtained from a secondary tumour that was detected 36 months later was found to be identical to the non-neoplastic tissues isolated both times. In case 6, the primary tumour sequence was altered at position 9 (Supplementary Table, Supplementary Figure 2), and 19 months later, the non-neoplastic mammary gland acquired one nucleotide mutation at position 16362. The sequence of the secondary tumour was identical to the non-neoplastic tissue. In case 7, multiple mutations were identified in the primary tumour, and these differed from the mutations identified in the secondary tumour. Therefore, cases 5, 6, and 7 were classified as novel primary tumours.
The incidence of gene alterations in the tumours classified as true recurrence (0.0 (%)) were significantly lower than tumours classified as novel primary tumours by UPMEGA method (1.00%±1.12, P=0.0495) (Figure 4).
The GAMDDL analysis of clonality in contralateral breast tumours
In 4 out of 5 (80%) cases involving secondary contralateral breast tumours, for which clonality could not determined by HUMARA, secondary tumours were found to be non-clonal compared with the primary tumours (Figure 3). In case 9, there were no differences in the mtDNA isolated from tumours or non-neoplastic tissues, therefore clonality could not be established.
The incidence of gene alterations was not significant between the tumour classified as novel primary tumours by the UPMEGA method (1.00%±1.12) and contralateral tumours (0.95%±0.82) (Figure 4).
Combined analysis
The informative rate of HUMARA analysis was 42.1% (8 out of 19), and 76.9% (10 out of 13) for GAMDDL analysis. When results from both methods were combined, the informative rate increased to 89.5% (Table 3). Based on the combined results, 37.5% of the ipsilateral breast tumours were characterised as true recurrent tumours, whereas 50% and 100% of ipsilateral and contralateral tumours, respectively, were characterised as non-clonal (Table 3).
Discussion
Accurate assessment of tumour clonality is important for determining patient prognosis and for identifying the most appropriate treatment options for secondary tumours. In particular, patients with recurrent tumours have a less favourable prognosis and may require a more aggressive treatment than patients with new primary tumours. However, the methods currently utilised to assess the clonality of secondary breast tumours are not consistently reliable. Based on the direct comparison of GAMDDL and HUMARA analyses performed in this study, GAMDDL analysis was found to be a more informative method for determining the clonality of secondary breast cancers.
Recently, the biological significance of mtDNA as related to aging and cancer has been the spotlight of numerous investigations (Lee et al, 2010). The mtDNA mutations, found in most cancers, have received particular attention as potential molecular markers of cancer status (Salk & Horwitz, 2010). Human mtDNA, which replicates independently of nuclear DNA, consists of 16579 nucleotides encoding mitochondrial proteins and RNAs for mitochondrial protein synthesis. Each mammalian cell contains thousands of mitochondria containing many mitochondrial nucleoids, and each mitochondrial nucleoid contains 2–10 copies of mtDNA (Shadel, 2008). The clonal inheritance of mtDNA suggests analysis of mtDNA mutations as an attractive candidate method for determining the clonality of secondary tumours. In breast cancers, mtDNA copy numbers are often decreased (Mambo et al, 2005; Tseng et al, 2006; Yu et al, 2007; Fan et al, 2009). Other alterations in nucleotide sequences have been reported in breast cancer tissue, including a large-scale deletion (4977 bp) of positions 8470–13 447 (Radpour et al, 2009; Shen et al, 2010). Deletions of various sizes and accumulations of gene alterations have been reported to occur during the aging process, possibly contributing to mitochondrial dysfunction that leads to carcinogenesis.
Numerous alterations have been documented in the nucleotide sequences of mitochondrial D-loops in many cancers, including breast cancer. Tan et al (2002) reported that 14 out of 19 (73.7%) breast cancers samples analysed had at least one somatic mtDNA mutation. Moreover, 81.5% of these mutations were detected in the non-coding D-loop region compared with 3.7% in the rRNA region. Similarly, Zhu et al (2005) reported that somatic mtDNA mutations were present in 14 out of 15 (94%) breast cancer tissues analysed with a seven-fold higher mutation rate detected in the D-loop region compared with the coding region. In the present study, high rate of GAMDDL was detected in 10 out of 13 (76.9%) samples.
The LOH (Goldstein et al, 2005a, 2005b; Vicini et al, 2007) and CGH analysis (Teixeira et al, 2004) found that 75% and 76% of ipsilateral secondary tumours were clonally related to the primary tumours, respectively. However, in the current study, only 37.5% of ipsilateral secondary tumours were confirmed as true recurrence, and a higher rate (50%) of ipsilateral tumours were found to be clonally distinct from the primary tumours. A possible explanation for this higher rate is that GAMDDL is a precise method to detect differences between cancer cells that have been harvested from a very restricted area (approximately 2 000 cells in 1 mm2). Correspondingly, if clonal heterogeneity is present in primary tumours, only a small proportion of the cancer cells analysed could be recurrent, and as a result, a higher primary rate is obtained. Cytogenetic studies by Teixeira and Heim (2011) support this speculation. For example, Teixeira et al (2001, 2002) have provided evidence regarding polyclonal carcinogenesis where a considerable population of cells from a clone distinct from the primary clone were detected in a breast cancer tissue based on an analysis of cytogenetic alterations and comparative genomic hybridisation. Therefore, four pathways for clonal evolution of a neoplastic cell population were proposed. Briefly, (1) initial monoclonality is retained, (2) monoclonal tumorigenesis with additional aberrations lead to secondary clonal heterogeneity, (3) polyclonal tumorigenesis is followed by a reduction in genomic complexity, or (4) polyclonal tumorigenesis occurs with additional cytogenetic changes (Teixeira and Heim, 2011). Considering the heterogeneity due to polyclonal carcinogenesis, analysis using microdissection might increase the new primary rate. To clarify the clonality in breast cancer with heterogeneity, GAMDDL would be further investigated, including the comparative study to CGH analysis with hierarchical clustering methodologies.
Although informative, previous studies have not conducted a systematic examination of GAMDDL for breast cancer tissues and surrounding non-neoplastic mammary gland tissue. Therefore, we also analysed the sequences of mtDNA D-loop regions in non-neoplastic mammary glands and lymph nodes from individuals with secondary breast tumours. Only for case 6 was the analysis of non-neoplastic ipsilateral breast tissue obtained at the time of primary and secondary tumour incidence, and D-loop sequence alterations were also identified. Moreover, patient age and time to second tumour were not particularly higher for case 6 than the other cases. One possibility for the genetic alterations detected is the irradiation the patient underwent for the treatment of remnant mammary tissue that remained following the primary surgery. In contrast, differential D-loop sequences were identified in contralateral non-neoplastic mammary gland tissues in all but one case. Furthermore, a comparison of D-loop sequences from lymph nodes and mammary glands revealed nucleotide alterations in all cases. These results suggest that the cell origins of lymph nodes, left mammary glands, and right mammary glands differ even in the embryonic period.
Overall, informative rates of 42.1% and 76.9% were obtained for HUMARA and GAMDDL analyses, respectively, in the present study. In comparison, the informative rate for LOH has been reported to range from 15.8 to 32.4% (Kollias et al, 2000). It appears that GAMDDL analysis is superior to other molecular methods for establishing clonality, and is useful for determining clonality of primary and secondary tumours, even if the primary tumours are genetically heterogeneous.
Acknowledgments
We thank Ms Ai Kakuhira in the Department of Pathology at the Nihon University School of Medicine for her technical support. This work was partially supported by a Grant-in-Aid for Scientific Research (C) (no. 21590387) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290(5806): 457–465 [DOI] [PubMed] [Google Scholar]
- Aulmann S, Penzel R, Longerich T, Funke B, Schirmacher P, Sinn HP (2008) Clonality of lobular carcinoma in situ (LCIS) and metachronous invasive breast cancer. Breast Cancer Res Treat 107(3): 331–335 [DOI] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295(21): 2492–2502 [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Dasgupta S, Sidransky D (2011) Mitochondrial subversion in cancer. Cancer Prev Res (Phila) 4(5): 638–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5): 403–410 [DOI] [PubMed] [Google Scholar]
- Fan AX, Radpour R, Haghighi MM, Kohler C, Xia P, Hahn S, Holzgreve W, Zhong XY (2009) Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J Cancer Res Clin Oncol 135(8): 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC (1980) Maternal inheritance of human mitochondrial DNA. Proc Nat Acad Sci USA 77(11): 6715–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein NS, Vicini FA, Hunter S, Odish E, Forbes S, Kestin LL (2005a) Molecular clonality relationships in initial carcinomas, ipsilateral breast failures, and distant metastases in patients treated with breast-conserving therapy: evidence suggesting that some distant metastases are derived from ipsilateral breast failures and that metastases can metastasize. Am J Clin Pathol 124(1): 49–57 [DOI] [PubMed] [Google Scholar]
- Goldstein NS, Vicini FA, Hunter S, Odish E, Forbes S, Kraus D, Kestin LL (2005b) Molecular clonality determination of ipsilateral recurrence of invasive breast carcinomas after breast-conserving therapy: comparison with clinical and biologic factors. Am J Clin Pathol 123(5): 679–689 [PubMed] [Google Scholar]
- Haffty BG, Reiss M, Beinfield M, Fischer D, Ward B, McKhann C (1996) Ipsilateral breast tumor recurrence as a predictor of distant disease: implications for systemic therapy at the time of local relapse. J Clin Oncol 14(1): 52–57 [DOI] [PubMed] [Google Scholar]
- Karrison TG, Ferguson DJ, Meier P (1999) Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 91(1): 80–85 [DOI] [PubMed] [Google Scholar]
- Kollias J, Man S, Marafie M, Carpenter K, Pinder S, Ellis IO, Blamey RW, Cross G, Brook JD (2000) Loss of heterozygosity in bilateral breast cancer. Breast Cancer Res Treat 64(3): 241–251 [DOI] [PubMed] [Google Scholar]
- Lee HC, Chang CM, Chi CW (2010) Somatic mutations of mitochondrial DNA in aging and cancer progression. Ageing Res Rev 9(Suppl 1): S47–S58 [DOI] [PubMed] [Google Scholar]
- Linder D, Gartler SM (1965) Glucose-6-phosphate dehydrogenase mosaicism: utilization as a cell marker in the study of leiomyomas. Science 150(3692): 67–69 [DOI] [PubMed] [Google Scholar]
- Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, Sukumar S, Sidransky D (2005) Tumor-specific changes in mtDNA content in human cancer. Int J Cancer 116(6): 920–924 [DOI] [PubMed] [Google Scholar]
- Mashal RD, Lester SC, Sklar J (1993) Clonal analysis by study of X chromosome inactivation in formalin-fixed paraffin-embedded tissue. Cancer Res 53(19): 4676–4679 [PubMed] [Google Scholar]
- Morandi L, Marucci G, Foschini MP, Cattani MG, Pession A, Riva C, Eusebi V (2006) Genetic similarities and differences between lobular in situ neoplasia (LN) and invasive lobular carcinoma of the breast. Virchows Arch 449(1): 14–23 [DOI] [PubMed] [Google Scholar]
- Noguchi S, Yokouchi H, Aihara T, Motomura K, Inaji H, Imaoka S, Koyama H (1995) Progression of fibroadenoma to phyllodes tumor demonstrated by clonal analysis. Cancer 76(10): 1779–1785 [DOI] [PubMed] [Google Scholar]
- Radpour R, Fan AX, Kohler C, Holzgreve W, Zhong XY (2009) Current understanding of mitochondrial DNA in breast cancer. Breast J 15(5): 505–509 [DOI] [PubMed] [Google Scholar]
- Salk JJ, Horwitz MS (2010) Passenger mutations as a marker of clonal cell lineages in emerging neoplasia. Sem Cancer Biol 20(5): 294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS (2008) Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol 172(6): 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Fang H, Chen T, He J, Zhang M, Wei X, Xin Y, Jiang Y, Ding Z, Ji J, Lu J, Bai Y (2010) Evaluating mitochondrial DNA in cancer occurrence and development. Ann N Y Acad Sci 1201: 26–33 [DOI] [PubMed] [Google Scholar]
- Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Univ Kansas Sci Bull 38: 1409–1438 [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8): 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tan DJ, Bai RK, Wong LJ (2002) Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res 62(4): 972–976 [PubMed] [Google Scholar]
- Tavassoli A, Devilee P (eds) (2003) Pathology & Genetics Tumours of the Breast and Female Genital Organs. International Agency for Research on Cancer Press: Lyon [Google Scholar]
- Teixeira MR, Heim S (2011) Cytogenetic analysis of tumor clonality. Adv Cancer Res 112: 127–149 [DOI] [PubMed] [Google Scholar]
- Teixeira MR, Pandis N, Heim S (2002) Cytogenetic clues to breast carcinogenesis. Genes Chromosomes Cancer 33(1): 1–16 [DOI] [PubMed] [Google Scholar]
- Teixeira MR, Ribeiro FR, Torres L, Pandis N, Andersen JA, Lothe RA, Heim S (2004) Assessment of clonal relationships in ipsilateral and bilateral multiple breast carcinomas by comparative genomic hybridisation and hierarchical clustering analysis. Br J Cancer 91(4): 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MR, Tsarouha H, Kraggerud SM, Pandis N, Dimitriadis E, Andersen JA, Lothe RA, Heim S (2001) Evaluation of breast cancer polyclonality by combined chromosome banding and comparative genomic hybridization analysis. Neoplasia 3(3): 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC (2006) Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 45(7): 629–638 [DOI] [PubMed] [Google Scholar]
- Vicini FA, Antonucci JV, Goldstein N, Wallace M, Kestin L, Krauss D, Kunzmann J, Gilbert S, Schell S (2007) The use of molecular assays to establish definitively the clonality of ipsilateral breast tumor recurrences and patterns of in-breast failure in patients with early-stage breast cancer treated with breast-conserving therapy. Cancer 109(7): 1264–1272 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Feinberg AP (1985) Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science 227(4687): 642–645 [DOI] [PubMed] [Google Scholar]
- Wu Y, Basir Z, Kajdacsy-Balla A, Strawn E, Macias V, Montgomery K, Guo SW (2003) Resolution of clonal origins for endometriotic lesions using laser capture microdissection and the human androgen receptor (HUMARA) assay. Fertil Steril 79(Suppl 1): 710–717 [DOI] [PubMed] [Google Scholar]
- Yu M (2011) Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci 89(3-4): 65–71 [DOI] [PubMed] [Google Scholar]
- Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, Zhang N, Hao X, Niu R (2007) Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB life 59(7): 450–457 [DOI] [PubMed] [Google Scholar]
- Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER (2005) Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis 26(1): 145–152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.