Abstract
Persistent infections are frequently caused by dormant and biofilm-associated bacteria, which often display characteristically slow rates of growth. Antibiotics that require rapid cell growth may be ineffective against these organisms and thus fail to prevent reoccurring infections. In contrast to growth-based antimicrobial agents, membrane-targeting drugs effectively kill slow-growing bacteria. This communication introduces a potent antibiotic that disrupts bacterial membranes: (2-((3-(3,6-dichloro-9H-carbazol-9-yl)-2-hydroxypropyl)amino)-2-(hydroxymethyl)propane-1,3-diol), which we refer to as DCAP. DCAP is a broad-spectrum antibiotic that reduces the transmembrane potential of Gram-positive and Gram-negative bacteria and causes mislocalization of essential membrane-associated proteins, including MinD and FtsA. Importantly, DCAP kills nutrient-deprived microbes and sterilizes bacterial biofilms. DCAP is lethal against bacterial cells, has no effect on red blood cell membranes, and only decreases the viability of mammalian cells after ≥ 6 h. We conclude that membrane-active compounds are a promising solution for treating persistent infections. DCAP expands the limited number of compounds in this class of therapeutic small molecules and provides new opportunities for the development of potent broad-spectrum antimicrobial agents.
While the prevalence of multi-drug-resistant pathogens continues to rise1, the rate at which new clinical antimicrobials are introduced has declined significantly2. To add to this dismal picture of combating infectious diseases, the treatment of persistent infections has been complicated by the pathogen phenotypes3. Bacteria that grow very slowly are often associated with prolonged infections, and they are particularly tolerant of many of the clinically important classes of antibiotics that inhibit rapidly growing cells. For example, the β-lactam family of antibiotics inhibits enzymes involved in the synthesis of peptidoglycan, and is thus most effective at targeting microbes that grow rapidly and continuously synthesize new cell wall4,5. Relying on antibiotics that require fast metabolism creates long-term problems, as dormant bacteria may survive antibiotic treatments, become predisposed to developing drug resistance, and cause a relapse2.
An effective strategy for combating slow-growing bacteria is to target the lipid membrane3. Proteomic analyses have demonstrated that approximately one third of all proteins in bacteria are associated with membranes6. Peripheral and integral membrane proteins participate in various essential cellular processes, including: nutrient and waste transport, respiration, adhesion, mobility, cell-cell communication, and the transfer of genetic material3,6. Compounds that perturb these processes disrupt growth and the maintenance of cell homeostasis and may serve as potent therapeutic antimicrobial agents3,7.
Synthetic and naturally occurring small molecules that disrupt the bacterial membrane have been developed to treat persistent infections of mycobacterial and staphylococcal species3,8. This class of compounds exhibits multiple mechanisms of action, including: inhibiting specific enzymatic processes in the membrane, decreasing the transmembrane potential (ΔΨ), and increasing membrane permeability. The increase in permeability perturbs bacterial physiology and simultaneously facilitates the penetration of free radicals secreted by macrophages of the host immune system3.
The therapeutic benefit of membrane-active drugs has been demonstrated against dormant bacteria; however, there are no clear design rules for small molecules that are specific for bacterial versus eukaryotic membranes3. Many antibiotics in this class are ineffective against Gram-negative bacteria, presumably due to the outer membrane8. The identification of new broad-spectrum antibiotics that target bacterial membranes and the study of their mechanism of toxicity will provide an important step forward for this field.
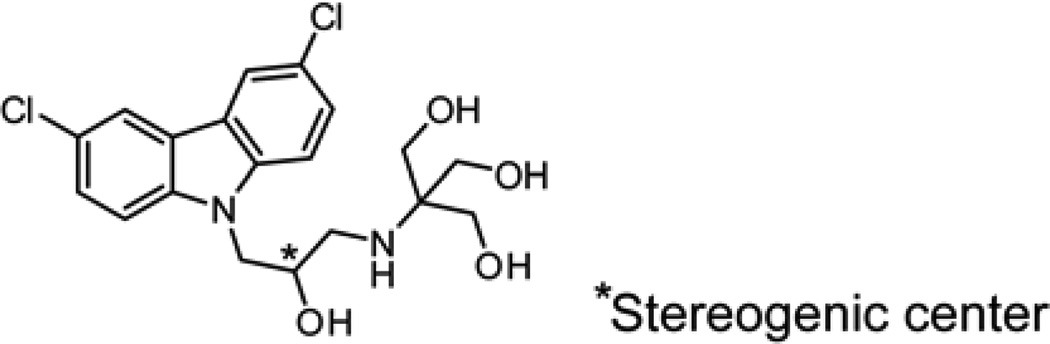
In this manuscript, we describe the discovery and characterization of a new compound that specifically targets the membranes of both Gram-positive and Gram-negative bacteria. For brevity, we refer to this compound, (2-((3-(3,6-dichloro-9H-carbazol-9-yl)-2-hydroxypropyl)amino)-2-(hydroxymethyl)propane-1,3-diol), as DCAP (Figure 1; its characterization is described in Figures S1 and S2). We identified DCAP via a high-throughput inhibitor screen of the in vitro activity of MipZ, which is an ATPase that regulates division site placement in Caulobacter crescentus9. Using a strain of C. crescentus in which MipZ was translationally fused to yellow fluorescent protein (YFP), we found that the treatment of cells with DCAP (20 µM) caused MipZ-YFP to mislocalize (Figure S3). At high concentrations of DCAP (≥ 75 µM), we observed cell lysis within minutes after treatment (Figure S4). This observation suggested to us that DCAP may not specifically inhibit MipZ in the cell but instead alter the properties of the cell envelope.
Figure 1.
Chemical structure of DCAP.
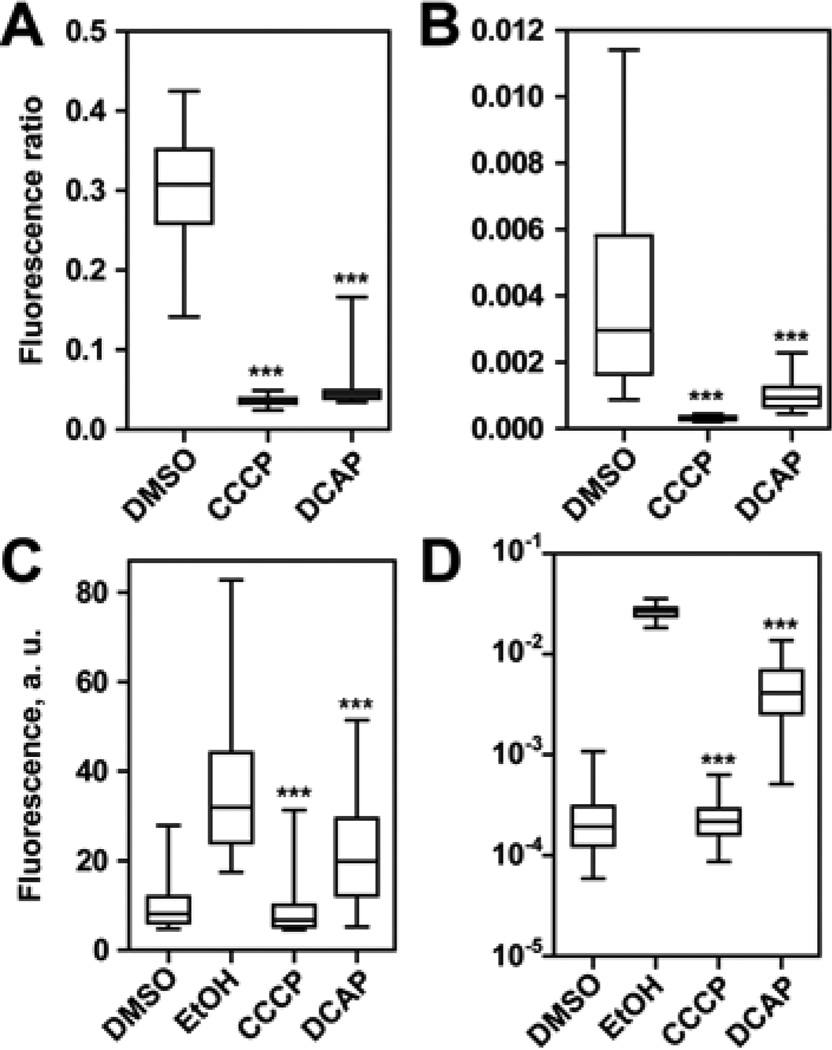
To test this hypothesis, we measured ΔΨ of two model bacteria, C. crescentus (Gram-negative) and Bacillus subtilis (Gram-positive), in the absence and presence of DCAP. As a positive control, we used carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Hydrophobic weak acids such as CCCP transport protons and other cations across the membrane and decrease ΔΨ10. CCCP dissolves into the lipid bilayer, and the acidic form associates with a cation near a leaflet of the membrane. The neutral complex moves to the other leaflet and dissociates to release the cation. After dissociation of the complex, the ionophore becomes available to bind to another cation and transport it across the membrane11. This increased permeability to ions dissipates ΔΨ. To visualize changes in the membrane potential, we used the fluorescent probe 3,3’-diethyloxacarbocyanine iodide (DiOC2). DiOC2 emits green fluorescence (λ=530 nm) in the monomeric form, and its fluorescence emission maximum is red-shifted (λ=576 nm) upon self-association12. Molecules of DiOC2 located inside cells reside either in the membrane or the cytoplasm. In the presence of a large ΔΨ, the number of positively-charged molecules of DiOC2 partitioned in the cytoplasm is greater than the number of molecules at the membrane13. The high local concentration in the cytoplasm causes DiOC2 to aggregate and increase the ratio of fluorescent light emitted at λ576/λ530. Conversely, λ576/λ530 decreases when ΔΨ is dissipated in bacteria12. Figure 2A and B illustrates λ576/λ530 for cells treated with small molecules. A significant decrease in λ576/λ530 was apparent after 20 min of treatment with CCCP and DCAP (p < 0.001) and indicated that ΔΨ dissipated rapidly. Antibiotics that do not target the bacterial membrane can decrease the potential over a long period of exposure (e.g. 3 – 4 h)14; however, the rapid action of DCAP suggests that the dissipation in ΔΨ is due to its direct effect on the inner membrane.
Figure 2.
(A, B) Measurement of ΔΨ using DiOC2. A) B. subtilis (n ≥ 8,964 cells). B) C. crescentus (n ≥ 158 cells). We calculated P-values by comparing data against the DMSO sample. (C, D) Measurement of membrane permeability using PI C) B. subtilis (n ≥ 2,546 cells). D) C. crescentus (n ≥ 35 cells). We calculated P-values by comparing data against the EtOH sample. In box plots, the top whisker represents 95%, the bottom whisker is 5%, the top of the box is 75%, the bottom of the box is 25%, and the line inside the box indicates the median of each sample population. Three asterisks (***) refers to p < 0.001.
Next we explored the mechanism of action of DCAP. One possibility is that it functions as an ionophore similar to CCCP. Alternatively, it may increase the general permeability of the membrane. To investigate the mechanism, we used propidium iodide (PI) to label the DNA of cells with compromised membranes12. As shown in Figure 2C and D, we observed that ethanol-treated cells were intensely labeled with PI, while the DMSO control sample was not. Treating cells with CCCP did not increase DNA labeling with PI; the fluorescence emission of these cells was similar to the DMSO sample. Addition of DCAP to cells increased the fluorescence of cells labeled with PI, although the intensity was significantly lower than the ethanol-treated cells (P < 0.001). These results suggest that DCAP has at least two mechanisms of antimicrobial action: it decreases ΔΨ by facilitating ion transport across the membrane and has a minor effect on the general permeability of the lipid bilayer. The bioactivity of DCAP may arise from its direct association with bacterial lipids or proteins in the membrane.
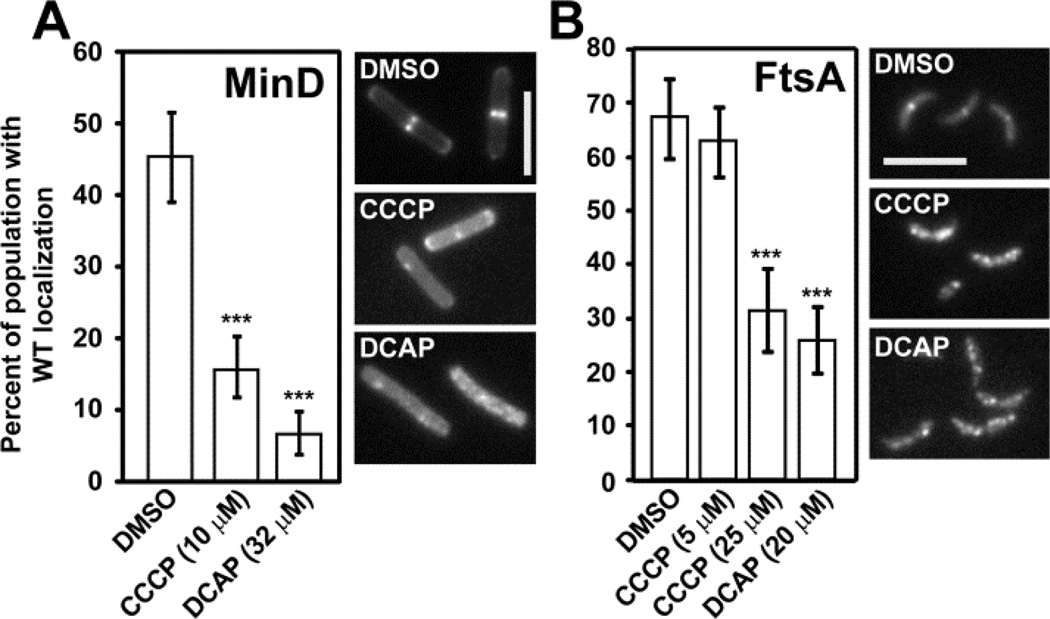
ΔΨ was identified recently as an important parameter for the in vivo localization of division proteins associated with the bacterial membrane, including MinD and FtsA15. We found that the treatment of B. subtilis cells with either CCCP or DCAP altered the localization of a fusion of green fluorescent protein to MinD (GFP-MinD) compared to DMSO-treated cells (Figure 3A). MinD localizes to the poles of B. subtilis cells and guides division plane formation at the mid-cell. As division progresses, MinD accumulates at the mid-cell and marks the sites of future cell poles15. Treating B. subtilis cells expressing GFP-MinD with CCCP increased diffuse fluorescence throughout the cell but had little effect on the location of the signal compared to the DMSO control (Figure S5A). In DCAP-treated cells, GFP-MinD mislocalized—the number of fluorescent foci increased in some cells while in others the fluorescence signal became more diffuse and was no longer concentrated at the poles (Figure S5A).
Figure 3.
Analysis of membrane protein localization in bacteria (p < 0.0001 = ***, compared to DMSO). Error bars represent the 95% confidence interval of percentages reported. Representative images for each sample are shown (scale bar, 5µm). (A) GFP-MinD in B. subtilis (n ≥ 252 cells) and (B) Venus-FtsA in C. crescentus (n ≥ 150 cells). The middle image shows cells from the treatment with 25 µM CCCP.
In addition to perturbing the localization of MinD in B. subtilis, CCCP and DCAP influenced the distribution of FtsA in C. crescentus. FtsA is a peripheral membrane protein that interacts with FtsZ and activates the recruitment of downstream division proteins16. FtsA resides at one pole (the pole opposite to the stalk) in non-dividing C. crescentus cells and is recruited to the mid-cell as division begins16. To study cells at this stage of division, we synchronized cells that express a fluorescent fusion of the protein Venus-FtsA and treated them with CCCP and DCAP. Treatment of C. crescentus cells with CCCP at its minimum inhibitory concentration (5 µM) did not alter the localization of FtsA; however, a higher concentration of CCCP (25 µM) had a significant effect on the position of FtsA. Most cells treated with DCAP exhibited multiple peaks of FtsA fluorescence (≥ 2) instead of a single peak — either at the pole or mid-cell — observed in untreated cells (Figure S5B). The observation that DCAP causes mislocalization of membrane proteins in B. subtilis and C. crescentus is consistent with the hypothesis that the compound decreases ΔΨ. The effect of DCAP was similar to the ionophore CCCP; however, DCAP causes more severe protein mislocalization at its MIC, which may arise from its influence on membrane permeability.
After confirming the membrane-targeting activity of DCAP, we tested the efficacy of DCAP against other bacterial species. Table S2 demonstrates that DCAP inhibits the growth of Escherichia coli, Pseudomonas aeruginosa, and other clinical pathogens. We observed that deleting one or more efflux pumps in E. coli and P. aeruginosa strains increased the sensitivity of cells to DCAP. Efflux pumps are active against a broad range of compounds and typically consist of three components: two transmembrane proteins, one in the inner and the other in the outer membrane, and a periplasmic protein that connects the two transmembrane components17. Deleting tolC in E. coli strain BW25113 eliminated a critical component of an efflux pump that is embedded in the outer membrane18 and reduced the MIC of DCAP by 4-fold. This result suggests that the activity of DCAP in Gram-negative bacteria is largely due to its effect on the inner membrane.
In addition to its activity against actively growing bacteria, DCAP kills cells in stationary phase (Table 1). We tested this property of DCAP against C. crescentus and S. aureus. We used S. aureus as a model Gram-positive bacterium rather than B. subtilis for these experiments, as B. subtilis can sporulate under starvation conditions and does not form robust biofilms on the plastic surfaces we used as substrates. To ensure that bacteria were deprived of nutrients, we grew the cells to late stationary phase and suspended them in isotonic solutions lacking amino acids or sugars. After treating cells with small molecules, we measured cell viability over time by plating culture aliquots on non-selective, solid growth media. We designated the minimum concentration of antibiotic required to completely eliminate colony formation as the minimum stationary-bactericidal concentration (MSC)19. To test whether membrane-targeting drugs have a greater efficiency in killing cells that grow slowly, we compared the MSC of membrane-active drugs (CCCP and DCAP) with ampicillin, which is only lethal to cells that are actively growing. We found that the MSC and MIC of DCAP for each organism were similar while the efficacy of ampicillin was significantly reduced for stationary cells of both organisms (Table 1). The MSC of ampicillin for S. aureus was 1000-fold higher than its MIC, while the MSC of ampicillin for C. crescentus was beyond the range of our measurements. CCCP inhibited the proliferation of C. crescentus cells regardless of their physiological status. However, the MSC of CCCP for S. aureus was > 300-fold higher than the MIC. This dramatic decrease in the effectiveness of CCCP in S. aureus may be due to changes in membrane composition as cells adjust their metabolism in nutrient-deprived conditions20. Overall, membrane-active CCCP and DCAP were more effective in killing stationary cells than ampicillin whose mechanism of action is more specific towards cells with robust growth.
Table 1.
Measurements of the minimum inhibitory concentration of growth (MIC), minimum stationary-bactericidal concentration (MSC), minimum biofilm inhibitory concentration (bMIC), and the minimum biofilm eradication concentration (MBEC).
| C. crescentus | S. aureus | |||||
|---|---|---|---|---|---|---|
| DCAPa | CCCPb | Ampc | DCAPa | CCCPb | Ampc | |
| MIC | 15 | 5 | 50 | 50 | 1.25 | 0.125 |
| MSC | 20 | 2.5 | >400 | 75 | >400 | 100 |
| bMIC | 20 | 5 | 100 | 100 | 2.5 | 0.8 |
| MBEC | 40 | 5 | >400 | >200 | 80 | >200 |
Concentration unit, µM.
Concentration unit, µg/mL
We found that membrane-active compounds are also efficient at eradicating biofilm-associated cells (Table 1). Biofilms are implicated in a wide range of human diseases, including cystic fibrosis and urinary tract infection, and are particularly recalcitrant to antibiotics21. The heterogeneity in the physiology of cells in biofilms makes it possible for the bacterial communities to persist in stressful conditions22. To determine the efficacy of antibiotics against biofilms, we adopted protocols for measuring the minimum biofilm inhibitory concentration (bMIC) and the minimum biofilm eradication concentration (MBEC)23. First, we formed biofilms on the surface of 96 individual plastic pins that protruded into the 96 wells of a microplate. The biofilms were exposed to compounds for 24 hrs, and we determined the lowest concentration of antibiotic that inhibited planktonic cell growth in the wells (bMIC). Since bMIC is a measurement of the rapid growth of freely suspended cells released from biofilms in the presence of antibiotics, the bMIC and MIC values did not differ significantly (Table 1).
After performing bMIC experiments, we transferred biofilms growing on the pins of the lid to nutrient media devoid of antibiotics to measure the minimum concentration of antibiotic that prevented planktonic growth from biofilms in antibiotic-free nutrient media (MBEC). MBEC indicates whether the exposure to antimicrobial agent used during the bMIC experiment sterilized biofilm-associated cells. MBEC values were generally larger than MICs and indicated an increased tolerance of stress exhibited by cells associated with biofilms. For C. crescentus, the MBEC values recapitulated the trend we observed in the MSC: CCCP and DCAP effectively eradicated biofilm cells while ampicillin was not cytotoxic at the highest concentration tested (400 µM). CCCP was the only effective antibiotic against S. aureus biofilms. Since CCCP was not as effective as DCAP at killing stationary S. aureus cells, we suspect that this variability in efficacy of membrane- active drugs is caused, in part, by changes in membrane composition (i.e. membrane proteins and lipid content) at different developmental stages of bacterial cells20. Despite the variations in efficacy, we conclude that the comparison of MIC, bMIC, MSC, and MBEC measurements for the three antibiotics support the hypothesis that membrane-active drugs eradicate slow-growing bacteria more effectively than antibiotics that rely on growth-dependent mechanisms.
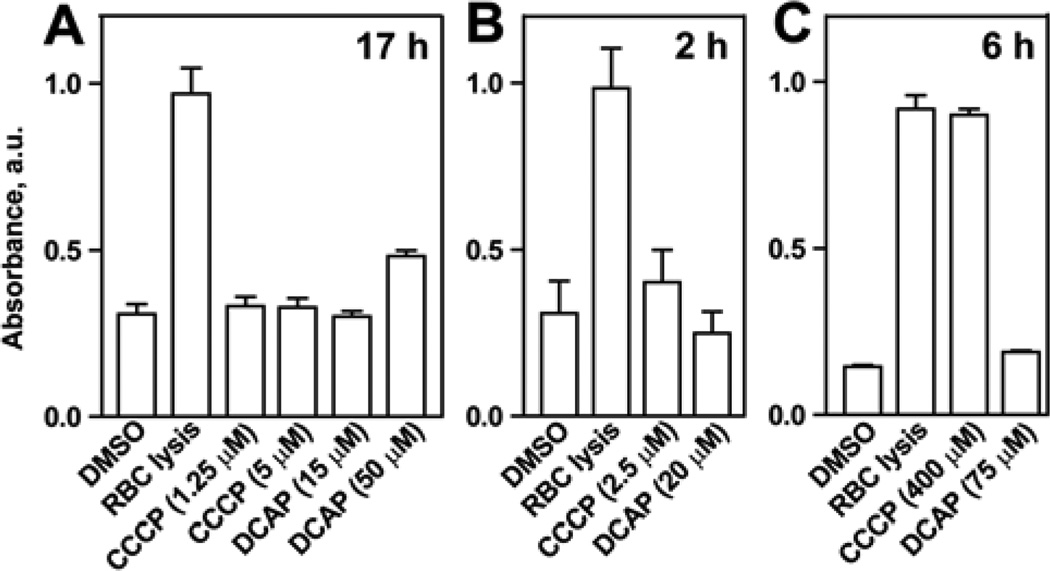
To test the toxicity of DCAP against mammalian membranes, we measured the hemolysis of rabbit red blood cells (RBC). We performed these experiments using conditions that closely mimicked the MIC and MSC assays. We treated RBCs with CCCP and DCAP at their MICs for the time periods used (17 h) to determine the MIC of C. crescentus and S. aureus. After incubation, we measured the absorbance of heme released from lysed RBCs. We found that the MIC concentrations of CCCP and DCAP did not significantly disrupt RBC membranes (Figure 4A) although higher concentrations of DCAP (i.e., 50 µM) were moderately toxic to RBCs.
Figure 4.
Rabbit red blood cell (RBC) hemolysis assay. Columns represent average values, and error bars represent the standard deviation of the mean for three replicates. Assay performed using the MIC conditions for C. crescentus and S. aureus (A), the MSC for C. crescentus (B), and the MSC for S. aureus (C).
We also reproduced the MSC assay conditions to measure toxicity against RBCs. First we determined the minimum time required to obtain the MSC for DCAP treatment of C. crescentus (2 h) and S. aureus (6 h; data not shown). Using these times, we performed the hemolysis assay and observed no significant toxicity of DCAP against RBCs (Figure 4B and C). In contrast, CCCP was toxic to RBCs at a high concentration (Figure 4C). These measurements indicate that DCAP does not appreciably perturb RBC membranes in conditions that are lethal to C. crescentus and S. aureus.
In addition to measuring the toxicity of membrane-active drugs on RBCs, we tested whether DCAP and CCCP dissipated ΔΨ of mitochondria in mammalian cells. We used human epithelial kidney (HEK) cells as a model tissue culture cell line. Similar to the bacterial assays, we used DiOC2 to probe the changes in ΔΨ in the presence of the compounds. As shown in Figure S6, we found that DCAP and CCCP slowly depolarized the mitochondrial ΔΨ in HEK cells. The efficiency of ΔΨ dissipation was lower in mitochondria compared to bacterial cells: treatment with DCAP and CCCP resulted in approximately 10-fold decrease in bacterial ΔΨ within 20 min (when comparing median values in Figure 1A and B) while the compounds reduced the mitochondrial ΔΨ by 2-fold (the first data point in Figure S6).
This eventual decrease of mitochondrial ΔΨ in HEK cells prompted us to investigate the viability of these cells when treated with DCAP and CCCP. We observed that the viability of HEK cells was minimally perturbed at short time intervals following treatment (2 h, Figure S7) while longer incubations decreased cell viability. As both CCCP and DCAP were similar in their ability to dissipate the mitochondrial ΔΨ in these cells, we speculate that the toxicity of DCAP on HEK cells may not be caused by its effect on the mitochondrial ΔΨ. We plan to investigate the basis of this toxicity of DCAP in future studies.
In summary, we report the discovery and characterization of a membrane-active antimicrobial agent, DCAP. DCAP kills bacteria by depolarizing ΔΨ and increasing membrane permeability. These activities of the compound disrupt the organization and integrity of the bacterial membrane and mislocalize essential membrane-associated proteins (e.g. MinD and FtsA). DCAP is inert to membranes of red blood cells at concentrations at which it is a potent antibacterial agent. However, DCAP slowly reduces the mitochondrial ΔΨ and becomes toxic to HEK cells. We plan to address the cytotoxicity of DCAP on mammalian cells by synthesizing and testing analogs. Furthermore, studies with DCAP analogs may provide insight into changes in the properties of membranes during the life cycle of bacteria and make it possible to correlate alterations in cell physiology with the vulnerability of cells to membrane-active drugs. Finally, studies of the structure-function relationship of DCAP and other broad-spectrum compounds may provide design rules for potent membrane-targeting drugs that kill bacterial cells specifically.
Supplementary Material
ACKNOWLEDGMENT
We thank Keith Poole for P. aeruginosa strains and Laura Kiessling for the human epithelial kidney cell line. D.B.W. and M.T. acknowledge the Human Frontiers Science Program (RGY0069/2008-C103). D.B.W also acknowledges the NIH (1DP2OD008735-01) and the Alfred P. Sloan research foundation for financial support of this research. M. T. also acknowledges the LOEWE Center for Synthetic Microbiology (SYNMIKRO).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Detailed experimental methods and materials used, DCAP characterization, and microscopy images. This material is available free of charge via the Internet at http://pubs.acs.org.
Authors declare no competing financial interest.
REFERENCES
- 1.Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, Lerner SA, Levy S, Lewis K, Lomovskaya O, Miller JH, Mobashery S, Piddock LJ, Projan S, Thomas CM, Tomasz A, Tulkens PM, Walsh TR, Watson JD, Witkowski J, Witte W, Wright G, Yeh P, Zgurskaya HI. Nat Rev Microbiol. 2011;9:894. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coates AR, Hu Y. Trends Pharmacol Sci. 2008;29:143. doi: 10.1016/j.tips.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Nat Rev Microbiol. 2011;9:62. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falconer SB, Czarny TL, Brown ED. Nat Chem Biol. 2011;7:415. doi: 10.1038/nchembio.590. [DOI] [PubMed] [Google Scholar]
- 5.Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Proc Natl Acad Sci U S A. 2009;106:21872. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poetsch A, Wolters D. Proteomics. 2008;8:4100. doi: 10.1002/pmic.200800273. [DOI] [PubMed] [Google Scholar]
- 7.Pathania R, Zlitni S, Barker C, Das R, Gerritsma DA, Lebert J, Awuah E, Melacini G, Capretta FA, Brown ED. Nat Chem Biol. 2009;5:849. doi: 10.1038/nchembio.221. [DOI] [PubMed] [Google Scholar]
- 8.Ganzle MG. Appl Microbiol Biotechnol. 2004;64:326. doi: 10.1007/s00253-003-1536-8. [DOI] [PubMed] [Google Scholar]
- 9.Thanbichler M, Shapiro L. Cell. 2006;126:147. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 10.Orlov VN, Antonenko YN, Bulychev AA, Yaguzhinsky LS. FEBS Lett. 1994;345:104. doi: 10.1016/0014-5793(94)00420-x. [DOI] [PubMed] [Google Scholar]
- 11.Terada H. Environ Health Perspect. 1990;87:213. doi: 10.1289/ehp.9087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro HM. Methods Mol Med. 2008;142:175. doi: 10.1007/978-1-59745-246-5_14. [DOI] [PubMed] [Google Scholar]
- 13.Waggoner A. J Membr Biol. 1976;27:317. doi: 10.1007/BF01869143. [DOI] [PubMed] [Google Scholar]
- 14.Novo DJ, Perlmutter NG, Hunt RH, Shapiro HM. Antimicrob Agents Chemother. 2000;44:827. doi: 10.1128/aac.44.4.827-834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strahl H, Hamoen LW. Proc Natl Acad Sci U S A. 2010;107:12281. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH, Shapiro L. Mol Microbiol. 2011;80:1680. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poole K. Clin Microbiol Infect. 2004;10:12. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 18.Sharff A, Fanutti C, Shi J, Calladine C, Luisi B. Eur J Biochem. 2001;268:5011. doi: 10.1046/j.0014-2956.2001.02442.x. [DOI] [PubMed] [Google Scholar]
- 19.Coates A, Hu Y, Bax R, Page C. Nat Rev Drug Discov. 2002;1:895. doi: 10.1038/nrd940. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YM, Rock CO. Nat Rev Microbiol. 2008;6:222. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 21.Parsek MR, Singh PK. Annu Rev Microbiol. 2003;57:677. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 22.Stewart PS, Franklin MJ. Nat Rev Microbiol. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 23.Ooi N, Miller K, Randall C, Rhys-Williams W, Love W, Chopra I. J Antimicrob Chemother. 2010;65:72. doi: 10.1093/jac/dkp409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.