Background: hTRF2 and hRap1 prevent non-homologous end joining and exist as a complex at human telomeres.
Results: The TRF2-Rap1 complex has high specificity for telomeres and a higher affinity for 3′ telomeric ends than hTRF2.
Conclusion: hRap1 regulates the DNA binding characteristics of hTRF2.
Significance: This is the first evidence of hRap1 interacting with DNA and altering the affinity of hTRF2 for telomeric DNA.
Keywords: DNA Binding Protein, DNA Damage, DNA Physical Chemistry, Electron Microscopy (EM), Telomeres, TRF2, hRap1
Abstract
The TRF2-Rap1 complex suppresses non-homologous end joining and interacts with DNAPK-C to prevent end joining. We previously demonstrated that hTRF2 is a double strand telomere binding protein that forms t-loops in vitro and recognizes three- and four-way junctions independent of DNA sequence. How the DNA binding characteristics of hTRF2 to DNA is altered in the presence of hRap1 however is not known. Here we utilized EM and quantitative gel retardation to characterize the DNA binding properties of hRap1 and the TRF2-Rap1 complex. Both gel filtration chromatography and mass analysis from two-dimensional projections showed that the TRF2-Rap1 complex exists in solution and binds to DNA as a complex consisting of four monomers each of hRap1 and hTRF2. EM revealed for the first time that hRap1 binds to DNA templates in the absence of hTRF2 with a preference for double strand-single strand junctions in a sequence independent manner. When hTRF2 and hRap1 are in a complex, its affinity for ds telomeric sequences is 2-fold higher than TRF2 alone and more than 10-fold higher for telomeric 3′ ends. This suggests that as hTRF2 recruits hRap1 to telomeric sequences, hRap1 alters the affinity of hTRF2 and its binding preference on telomeric DNA. Moreover, the TRF2-Rap1 complex has higher ability to re-model telomeric DNA than either component alone. This finding underlies the importance of complex formation between hRap1 and hTRF2 for telomere function and end protection.
Introduction
Eukaryotic chromosomes ends are protected by a combination of looped or fold-back structures and proteins that generate protective telomere-specific complexes (1, 2). In higher eukaryotes, a set of six telomere-specific proteins termed the shelterins has been identified, which are central to telomere maintenance (3). In addition to the shelterins, the reverse transcriptase telomerase and many general DNA replication and repair factors complete the repertoire of proteins protecting telomere ends (4–6). TRF1 and TRF2 bind directly to duplex telomeric repeats through their myb domains, whereas the other shelterins are suggested to scaffold onto these two proteins (7, 8). TRF2 appears to be most central to these events. In vitro TRF2 is able to generate t-loop structures in which the 3′ single strand (ss)2 overhang of the telomere invades the internal double strand (ds) telomere regions to generate a large duplex loop (1, 9). Furthermore, TRF2 is able to bind another shelterin factor, Rap1, as well as numerous DNA replication and repair factors including Ku 70, Apollo, MRN complex, WRN, Fen1, ORC1, PARP1, and PARP2 presumably to tether these factors near the chromosome end (10, 11). The complex of TRF2 and Rap1 prevents NHEJ and works with DNA-PK to suppress joining of telomere ends and thus must be central to telomere maintenance and architecture (12, 13).
The importance of TRF2 has been documented by studies involving overexpression of dominant negative alleles and more recently inducible knock-outs of TRF2 (14, 15). These studies show that TRF2 suppresses NHEJ and prevents chromosome end-to-end fusions (14, 15). Additionally, down-regulation or loss of TRF2 activates the ATM kinase pathway, induces the formation of DNA damage induced foci, up-regulates p53, and arrests cells at the G1/S checkpoint (16–18). The interaction of TRF2 with telomeric DNA has been examined in detail in vitro in this and other laboratories (9, 19, 20). The protein contains a single myb domain as well as a dimerization domain and a highly basic N terminus (7). Because two myb domains are needed for stable DNA binding, the homodimer of TRF2 represents the minimal oligomer required for stable binding to duplex telomeric DNA (21). TRF2 however is also able to bind to DNA in a non-sequence but structure-specific manner, mediated by the basic N terminus (19). These studies described first in this laboratory showed that telomeric DNA is highly prone to replication fork slippage, and thus, the binding of TRF2 to slipped structures may be key in stopping further fork slippage and preventing the generation of deleterious structures (19). Moreover, TRF2 prevents NHEJ at non-telomeric sites through its basic domain (22).
hRap1 was first identified by yeast two hybrid analysis with hTRF2 as a bait (10). hRap1 binds directly to hTRF2 through its RCT domain and pulldown experiments in human cell extracts have identified stable complexes of hTRF2 and hRap1 (2, 10). hRap1 is more abundant in the cell than hTRF2, and immunodepletion of hRap1 results in a reduction of TRF2 levels (23). In contrast, lowering TRF2 with shRNA resulted in only a moderate change in the total Rap1 levels due to its overabundance (24). This may indicate that in contrast to mouse Rap1, hRap1 does not require hTRF2 for its stability in the cell. It was suggested that hRap1 does not bind DNA directly and is recruited to human telomeres by its binding partner hTRF2 (10). However, as observed by CHIP analysis, hTRF2 and hRap1 have distinct and overlapping binding sites along the chromosome (25, 26). Furthermore, mammalian Rap1 is found at non-telomeric loci in addition to telomeres (25). Some of the non-telomeric sites have consensus (TTAGGG)2 motifs, whereas others do not (25). The localization of hRap1 to regions with TTAGGG sequences could be through its interaction with TRF2, but how it localizes to non-telomeric sites is not clear (26).
Budding yeast Rap1 plays a role in transcription, telomere length regulation, and end capping. Rap1 is the most conserved shelterin component among different species (27), and recently, hRap1 was shown to have similar functions to its yeast counterpart. hRap1 regulates the expression level of genes that are in close proximity to its binding sites and regulates telomere length through its BRCT domain (11, 26). Moreover, Rap1 alone suppresses NHEJ in human extracts and homologous recombination in mouse cells (23, 28). However, significantly less is known about the function and role of Rap1 in human or mouse cells in the suppression of the DNA damage response.
Our previous in vitro studies (9, 19) of the interaction of TRF2 with telomeric DNA as well as Holiday junctions and replication forks have led to a better understanding of its role in telomere protection. To further understand the role of hRap1 alone or bound to hTRF2, it will be critical to determine how the binding of hRap1 to hTRF2 augments or modifies its binding to telomeric DNA and to unusual DNA structures. In this study, we characterized the DNA binding ability of hRap1 to different model DNA templates in the absence of hTRF2 and then tested how the binding preference and affinity is altered when hRap1 is in a complex with hTRF2.
EXPERIMENTAL PROCEDURES
Preparation of DNA Molecules
Preparation of Linear Telomeric Templates
The pRST5 plasmid contains ∼575 bp of ds human telomeric DNA (9). A plasmid containing ∼1.1-kb telomeric repeats, pOST6, was prepared from pRST5 with expansive cloning as described in Stansel et al. (9). The minichromosome template consists of two model telomeres joined at their non-telomeric ends so that ds telomeric DNA with 3′ ss overhangs are present at both ends. The minichromosome templates were prepared similarly to the model telomeres (9) by treating pRST5 with BsmBI and NotI followed by 3′ overhang ligation with a 1:10 molar ratio of linear ds template to a 124-nt oligonucleotide consisting of 5′-TTAGGG-3′ repeats (IDT, Coralville, IA). The NotI digestion product has 5′-GGCC-3′ ends that facilitate dimerization at the nontelomeric ends upon ligation. Blunt end DNA molecules were obtained by treating BsmBI digested pOST6 with S1 nuclease as described in the manufacturer's protocol (Fermentas, Inc., Glen Burnie, MD). To generate a template with a telomeric 5′ overhang, BsmBI digested pOST6 was treated with ExoIII on ice for 5 min per the manufacturer's instructions (New England Biolabs). A nontelomeric template with a 3′ overhang was generated by treating EcoRI-digested pGLGAP, which does not contain telomeric repeats (29), with T7 exonuclease at room temperature for 40 s followed by incubation on ice for 5 min (New England Biolabs).
Preparation of Stalled Replication Forks and Holliday Junctions
To generate a circular ds DNA with a 400-nt single strand tail, pGLGAP plasmid was nicked using Nb.BbvCI (New England Biolabs), and the nicked strand was displaced by incubation with the Klenow fragment (exo-) of DNA polymerase I in the presence of dNTPs except for dCTP (29). To make the tail ds with different gap sizes at the fork, primers were annealed to the ss tail and incubated with Klenow (exo-) polymerase in the absence of dGTP for 30 min at 37 °C (30). Preparation of telomeric and nontelomeric Holliday junctions were done as described (31).
Preparation of DNA for EMSA
pRST1 plasmid, which contains a 154-bp ds telomeric insert, was digested with BsmBI and HindIII and the telomeric insert isolated by gel electrophoresis (Qiagen, Germantown, MD). The purified insert was used as a ds telomeric template for EMSA after the 5′ ends were radiolabeled with [γ-32P]ATP in a standard T4 polynucleotide kinase reaction as described by the manufacturer (New England Biolabs). To generate a radiolabeled telomeric template with a 3′ overhang, the gel isolated duplex DNA was ligated to a G-rich oligonucleotide with eight telomeric repeats after the oligonucleotide was phosphorylated with [γ-32P]ATP from its 5′ end as described above. A nontelomeric template with a 3′ overhang was prepared from two oligonucleotides with sequences: 5′-ATAGCTAGACATAGACCTAGGATTCCGTAGCTAGCACTGGCATACTGCTAGATCGCGATACTGGTCACTAGCTAGGCTACAGTCCTGACG-3′ and 5′-CGCGATCTAGCAGTATGCCAGTGCTAGCTACGGAATCCTAGGTCTATGTCTAGCTAT-3′ (MWG Operon, Huntsville, AL). Only oligonucleotide 1 was phosphorylated at its 5′ end with [γ-32P]ATP as described by the manufacturer with T4 polynucleotide kinase (New England Biolabs) and annealed to oligonucleotide 2 with a gradual cool down from 90 °C.
Purification of Proteins
The full-length hRap1 and hTIN2 genes were purchased from Open Biosystems (Fisher Scientific, Pittsburg, PA) and the full-length hTRF2 gene was a gift of Dr. Christopher Counter. Each gene was cloned into the pFastBacHtA plasmid. The full-length NH2-terminal His6-tagged hTRF2 was purified with a TalonTM metal affinity resin (Clontech, Palo Alto, CA) from Sf21 extracts as described (32) and stored in 20 mm Hepes (pH 7.5), 300 mm NaCl, 20% glycerol, 3 mm MgCl2, 1 mm DTT at −80 °C. The full-length NH2-terminal His6-tagged hRap1 was purified with nickel-nitrilotriacetic acid chromatography (Qiagen) from Hi-5 cell extracts (10) and stored in 50 mm NaPO4 (pH 7.5), 300 mm NaCl, 20% glycerol, 8 mm β-mercaptoethanol. Similarly, NH2-terminal His6-tagged hTIN2 was purified with nickel-nitrilotriacetic acid chromatography (Qiagen) from Sf21 extracts (33) and stored at −80 °C in 50 mm NaPO4, 150 mm NaCl, 8 mm β-mercaptoethanol, and 20% glycerol.
TRF2-Rap1 Complex Formation
TRF2-Rap1 complexes were formed by incubating 100 μg of hTRF2 and 100 μg of hRap1 on ice for 30 min in 10 mm Hepes (pH 7.5), 25 mm NaPO4, 300 mm NaCl, 4 mm β-mercaptoethanol, 1,5 mm MgCl2, 0.5 mm DTT, and 20% glycerol, followed by size exclusion chromatography on Sepharose 6 (GE Healthcare) using an elution buffer of 20 mm Hepes (pH 7.5), 300 mm NaCl, 20% glycerol, 3 mm MgCl2, and 8 mm β-mercaptoethanol. Fractions collected from the Sepharose 6 size exclusion column (GE Healthcare) were stored at −80 °C. To determine the elution profile of the individual components, 100 μg of NH2-terminal His6-tagged hTRF2 or NH2-terminal His6-tagged hRap1 proteins were passed through the Sepharose 6 size exclusion column (GE Healthcare) in a buffer of 300 mm NaCl, 20 mm Hepes (pH 8.75), and 8 mm β-mercaptoethanol.
EM Analysis
Tungsten Shadowcasting
DNA-protein complexes in the binding reaction mixture were cross-linked with 0.6% (w/v) gluteraldehyde for 5 min at room temperature and passed through 2-ml size exclusion columns with A5M beads (Agarose Bead Technologies) pre-equilibrated with 0.01 mm Tris-HCl (pH 7.6) and 0.1 mm EDTA. The cross-linked complexes were mixed with a buffer containing 2.5 mm spermidine, adsorbed to glow-charged carbon foil grids for 3 min and dehydrated with a series of water/ethanol washes, air-dried, and rotary shadowcast with tungsten at 1 × 10−6 torr as described (34). An FEI Tecnai 12 instrument (Hillsboro, OR) equipped with a Gatan Orius CCD camera (Gatan, Pleasanton, CA) at 40 kV was used to capture the images using Digital Micrograph software. Images for publication were arranged and contrast optimized using Adobe Photoshop CS5 (Adobe Systems, San Jose, CA). At least 100 molecules were scored in sequence as they were encountered at the EM, and statistical analysis was done using Student's t test (Graphpad Software, Inc., La Jolla, CA).
Negative Staining and Mass Analysis
To generate the TRF2-Rap1 complex for negative staining, hTRF2 (200 ng) and hRap1 (200 ng) were incubated on ice for 30 min and then diluted to 20 ng/μl in 20 mm Hepes (pH 8.75) and 100 mm NaCl. To negative stain individual proteins, hRap1 or hTRF2 was diluted to 20 ng/μl as above and stained with 2% (w/v) uranyl acetate in water. Samples were examined in a Philips CM12 TEM at 80 kV, and images were captured on a Gatan First Light high sensitivity CCD camera (Gatan, Pleasanton, CA). Negative stained images (1500–2000) for each protein were analyzed using NIH ImageJ software (Bethesda, MD) for mass determination as described previously in numerous papers from this laboratory (31, 35). Ferritin was used as a size standard for these experiments.
Mass Analysis of Proteins Bound to Holliday Junctions
DNA-protein complexes were tungsten shadowcast as described above and a size standard (ferritin) was prepared side-by-side for each experiment (31). Images were captured as described above (Tecnai 12, Hillsboro, OR). For each protein ∼ 100 images were analyzed with NIH ImageJ software to determine the area distribution.
Electrophoretic Mobility Shift Assays
DNA binding reactions were done at room temperature for 20 min in 20 mm Tris (pH 8.0) at different protein concentrations and 5 nm DNA template. Reactions were quenched with 2.5 mm EDTA, 5% glycerol, 0.25 mg of bromphenol blue and 0.25 mg of xylene cyanol and loaded onto a 4% native polyacrylamide gel. The dried gel was analyzed by autoradiography and imaged with a Typhoon 9400 phosphoimager (Amersham Biosciences). Graphpad prism software was used for the nonlinear regression analysis to determine Kd values (GraphPad Software, Inc., La Jolla, CA). To verify that binding is specific to hRap1, a His-tagged antibody was used in the binding reactions as described above and the antibody amount in the reaction was determined according to manufacturer's protocol (ABGENT, San Diego, CA).
RESULTS
hRap1 Binds to Minichromosomes, Holliday Junctions, and Replication Forks
The DNA binding ability of hRap1 has been previously examined using gel shift assays employing short (72 bp) ds human telomeric DNA or ss and ds yeast telomeric DNA (10). By this approach, hRap1 did not show significant binding to telomeric DNA. However, short DNAs do not recapitulate the full telomere architecture, and thus, we used EM to examine the DNA binding ability of hRap1 using DNA templates in the range of 0.7 to 7 kb, including the minichromosomes, Holliday Junctions and replication forks illustrated in Fig. 1, A–C. The minichromosome template mimics a human chromosome with a 6-kb segment of plasmid DNA flanked by ∼575 bp of repeating 5′-TTAGGG-3′ duplex repeats at both ends terminating in 120-nt, 3′-TTAGGG overhangs (Fig. 1A). We also used a Holliday junction (HJ) with 175-bp intersecting arms and a similar size telomeric HJ with two TTAGGG repeats at the intersection site (Fig. 1B) (36). Finally, a replication fork was created from a 3.4-kb duplex circle that contains a ∼400-bp displaced arm with different gap sizes at the fork (30), which enables us to test the ability of proteins to bind to junction sites through structure recognition as these replication forks do not contain telomere repeats (Fig. 1C).
FIGURE 1.
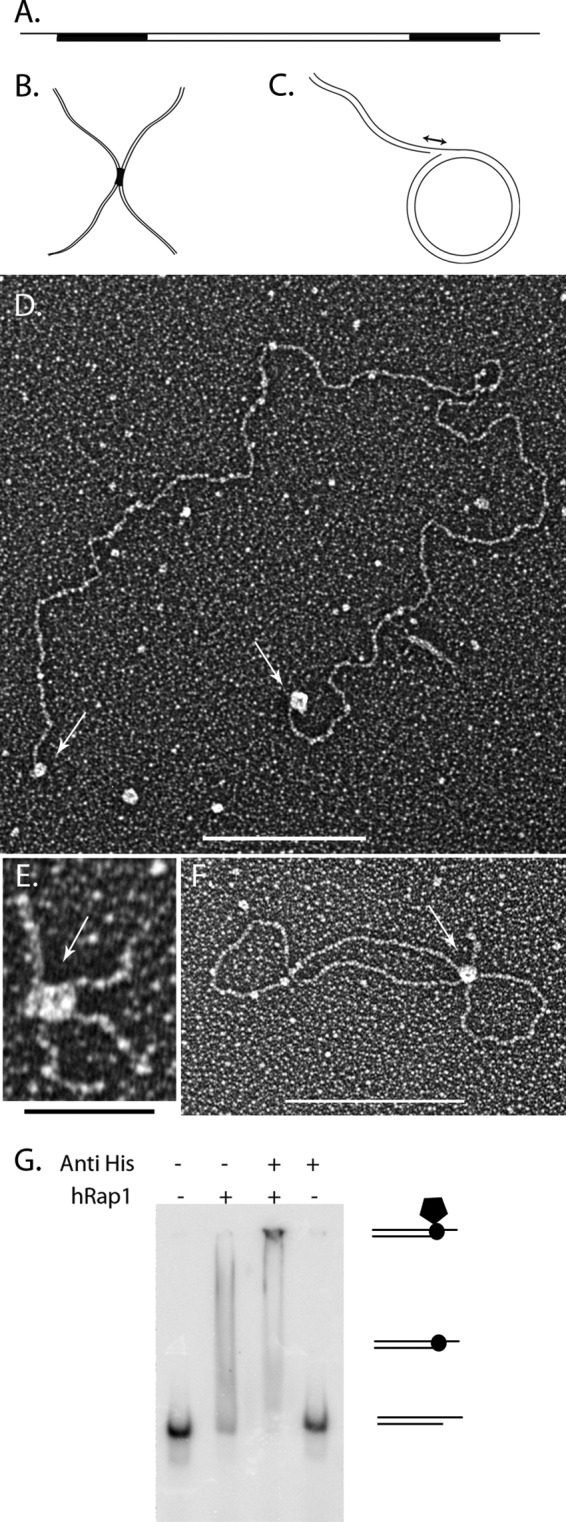
hRap1 binds to model DNA templates. Schematic representations of the minichromosome (A), replication fork with a 25-nt gap (B), and Holliday junction templates (C). Black regions correspond to the telomeric DNA. Tungsten shadowcast images of hRap1 illustrate protein bound to the minichromosome (D), replication fork (E), and telomeric Holliday junction template (F). Data are shown in reverse contrast. Scale bar in D and F are 100 nm and is 50 nm in E. Shown is EMSA with 4% PAGE demonstrates hRap1 bound to the nontelomeric template with a 3′ overhang in the presence and absence of anti-His6 antibody (G).
Human Rap1 was purified from insect cells. To examine the binding preference of each protein on the different DNA templates, sub-saturating ratios of protein:DNA were used to gain evenly sized particles on the DNA and to avoid a significant number of aggregates consisting of multiple DNAs bound together by protein. To do this, different protein:DNA ratios were tested, and the ones that yielded roughly one-half of the DNA bound by protein, while <10% was in aggregates, were selected. The optimal binding for each protein was observed at 14:1 and 26:1 protein tetramers:DNA molar ratios for hTRF2 and hRap1, respectively. hTIN2 interacts with hTRF2 (37) but has not been shown to interact with telomeric DNA directly, whereas the behavior of hTRF2 with similar templates has been studied previously (19, 37). Thus, hTIN2 and hTRF2 were used as controls in these assays. Supplemental Fig. 1 shows the binding of hTRF2 to the minichromosomes (supplemental Fig. 1A), a replication fork with a 25-nt gap (Gap25) (supplemental Fig. 1B), and telomeric HJs (supplemental Fig. 1C). In each case, hTRF2 bound to the three- and four-way junctions confirming previous findings (19). Under the same binding conditions even with as many as 568 hTIN2 protein monomers to DNA, TIN2 did not bind to the minichromosome (data not shown). hRap1 localized to the telomeric ends of the minichromosomes (Fig. 1D), to the crossover of the telomeric HJ (Fig. 1E), and to the fork junction of Gap25 DNA (Fig. 1F). In each case, the protein-bound DNA species showed a uniformly sized and shaped protein particle bound. hRap1 has an N-terminal histidine tag and to confirm that the binding activity was specific to hRap1, we did an EMSA with an anti-His tag antibody. The DNA template used was 57-nt-long with a 33-nt 3′ overhang and did not contain any telomeric sequences (see “Experimental Procedures”). The lack of a shift with antibody alone but the presence of a supershift with the antibody and hRap1 confirmed the binding activity to be specific to hRap1 (Fig. 1G). These data suggest that hRap1 has the ability to directly interact with DNA.
hRap1 Binds to ds-ss Junctions Independent of Sequence but Prefers 3′ Overhang Structures over 5′ Overhangs
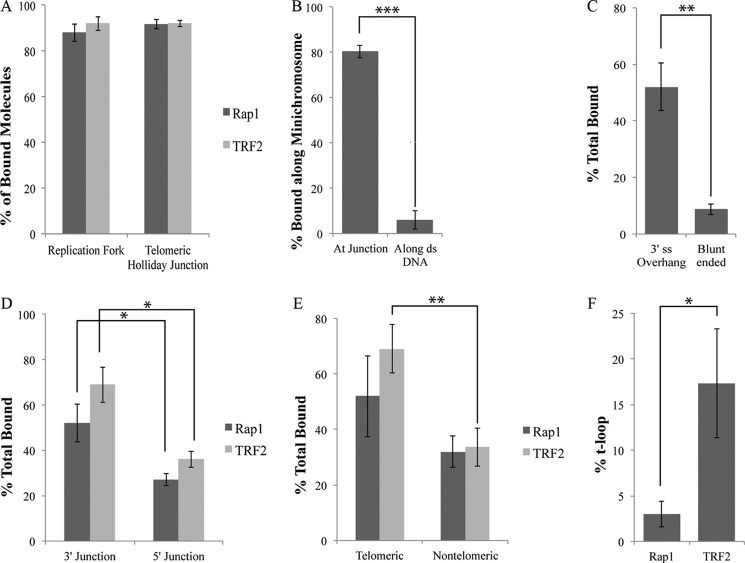
We previously showed that hTRF2 binds to replication forks, HJs, and ds-ss overhangs independent of sequence (19). Because hRap1 bound to the same DNA templates, we wished to quantify binding of hRap1 with different DNA templates. On replication forks and Holliday junctions, the majority of hRap1 localized to the junction site (Fig. 2A) at comparable levels with hTRF2, confirming that hRap1 recognizes three- and four-way junctions. Scoring the distribution of hRap1 molecules along the minichromosomes, we found that hRap1 localizes to the termini of the minichromosomes containing the 3′ ds-ss junction site with a ∼13-fold greater preference over the ds telomeric DNA (Fig. 2B). When the 3′ ss overhang was removed, the total hRap1-binding dropped to background levels, confirming that hRap1 specifically recognizes the ds-ss junctions of minichromosomes (Fig. 2C).
FIGURE 2.
hRap1 recognizes the 3′ ds-ss junction structures independent of sequence. A illustrates that both hRap1 and TRF2 have a strong and similar preference for binding to the ss-ds junction at replication fork with a 25-nt gap and the crossover at the HJ. B illustrates the strong preference for hRap1 binding to the end of the minichromosome containing a 3′ ss extension as contrasted to binding internally along the ds telomeric segment. C showed the strong preference for hRAP1 binding to the minichromosome containing a 3′ ss extension as contrasted to the same but blunt-ended DNA. D shows that both hRap1 and TRF2 prefer to bind at the ss-ds junction of DNAs with a 3′ overhang as contrasted to a 5′ overhang. E compares the binding to DNAs containing 3′ overhangs joined to either telomeric ds segments or non-telomeric. F compares the t-loop formation percentages of hRap1 and hTRF2 on the minichromosome. Each binding experiment was done in triplicate, and at least 100 molecules were counted. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We wanted to determine whether localization of hRap1 to minichromosomes is sequence- or structure-dependent. Human telomeres engaged in homologous recombination-dependent telomere maintenance have 5′ C-rich overhangs (38). To further probe the structure specificity of hRap1 binding, we used a linear template containing a 1.1-kb telomeric tract at one end and then digested it with a 3′ to 5′ exonuclease to generate 5′ tails up to 340 nt in length (as seen by EM). hRap1 was found to preferentially bind to DNA with 3′ overhangs over 5′ ends as does hTRF2 (Fig. 2D). To examine the sequence specificity of hRap1 binding at junction sites, a linear 3.5-kb nontelomeric plasmid DNA was digested with a 5′ to 3′ exonuclease to generate 3′ overhangs of an average of ∼560 nts. hRap1 localized to the 3′ ends of the telomeric and nontelomeric linear templates equally with no statistical difference, whereas hTRF2 showed a preference for telomeric DNA with 3′ overhangs (Fig. 2E). There was no significant effect of the presence of telomeric sequences at HJs on hRap1 binding (supplemental Fig. 2). These findings suggest that hRap1 has a strong preference for 3′ ends on linear templates and has the ability to interact with the ds-ss junction sites independent of their sequence, in contrast to hTRF2. The 3′ G-rich overhang at telomeric ends is a requirement for the formation of a t-loop (1, 9), and thus, we addressed the t-loop forming ability of hRap1. Despite binding to junction sites, hRap1 formed t-loops 5-fold less well when compared with hTRF2, suggesting that this is not a strong activity for hRap1 alone (Fig. 2F).
hTRF2-Rap1 Complex Is Formed from Four Molecules of hRap1 and Four Molecules of hTRF2
The oligomeric states of proteins can influence their binding characteristics. hTRF2 and hRap1 exist in a complex at human telomeres (2, 39), and although it has been demonstrated that they exist in a 1:1 ratio (39), their oligomeric state is unknown. Therefore, we analyzed the mass of hRap1, hTRF2, and the TRF2-Rap1 complexes in the absence of DNA by EM. The TRF2-Rap1 complex was formed from the purified proteins (see “Experimental Procedures”). In negative-stained fields of hTRF2 (Fig. 3A), Rap1 (Fig. 3B), and the TRF2-Rap1 complex (Fig. 3C), a variety of particle sizes and shapes were observed for all three, but in each case, there was a predominant particle, and there was a clear size difference between hTRF2, hRap1, and the TRF2-Rap1 complex.
FIGURE 3.
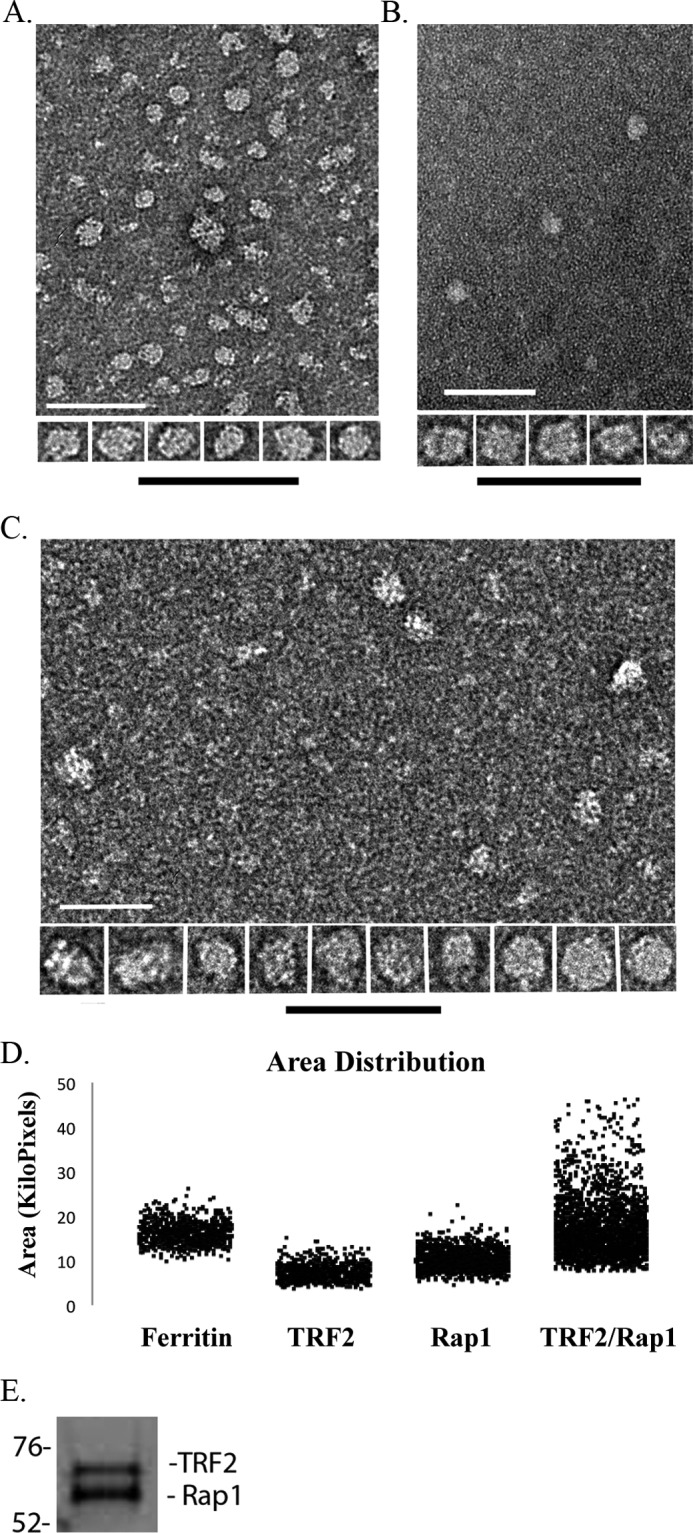
Mass and oligomeric state analysis of hRap1, hTRF2, and the hRap1-TRF2 complex. Representative negative stained images of hTRF2 (A), hRap1 (B), and the TRF2-Rap1 complex (C) are shown as fields and an array of selected single particles at higher magnification. Bars are equivalent to 200 nm. Area distributions of single protein particles and ferritin as a size standard were calculated from two-dimensional projections of the EM images (D). The TRF2-Rap1 complex was separated using 10% SDS-PAGE and stained with Coomassie Orange to determine the ratio of hRap1 to hTRF2 (E).
To obtain estimates for the mass of the predominant species in each preparation, we compared their projected areas in the negative-stained images to that of ferritin as carried out in previous studies (31). The projected area distributions of the TRF2-Rap1 complex showed a wider range than that of hTRF2 or hRap1 (Fig. 3D) and presents a good example of a positively skewed Gaussian distribution (supplemental Fig. 3D), which could result, for example, from a cylindrical shape for the TRF2-Rap1 complex rather than spherical shape as observed in hTRF2 and hRap1 where the size distribution would be much more narrow. Analysis of 715–2135 particles (see “Experimental Procedures”) yielded an estimated mass of 136 ± 0.4 kDa for hTRF2, 221 ± 0.3 kDa for hRap1, and 496 ± 1.0 kDa for the TRF2-Rap1 complex (Table 1).
TABLE 1.
Size analysis of hRap1, hTRF2, and the TRF2-Rap1 complex in solution
The estimated mass values obtained from the statistical analysis of the area distributions derived from negative staining as in Fig. 3 and the size exclusion chromatography (supplemental Fig. 4) are shown with the corresponding oligomeric states.
| Calculated mass | Expected mass | Oligomeric state | |
|---|---|---|---|
| kDa | kDa | ||
| Size exclusion chromatography | |||
| hTRF2 | 121 | 140 | Dimer |
| hRap1 | 285 | 240 | Tetramer |
| TRF2-Rap1 | 514 | 520 | TRF2 and Rap1 tetramers |
| Negative staining | |||
| hRap1 | 221 | 240 | Tetramer |
| TRF2-Rap1 | 496 | 520 | TRF2 and Rap1 tetramers |
| hTRF2 | 136 | 140 | Dimer |
Size exclusion chromatography was used to confirm the EM results. The TRF2-Rap1 complex (see “Experimental Procedures”), hRap1, and hTRF2 were passed through a Sepharose S6 matrix, and each fraction was analyzed by SDS-PAGE followed by Coomassie Blue or Coomassie Orange staining (supplemental Fig. 4, A and B). Based on parallel filtration of mass standards using the same column, hTRF2 eluted in a single peak equivalent to a mass of 121 kDa, and hRap1 eluted in a wider peak with a mass of 285 kDa. When hTRF2 was in a complex with hRap1 there was a significant shift of the peak in the elution profiles. The co-complex was found to elute as a wide peak with a mass of ∼496–514 kDa likely due to the high glycerol concentration of the column buffer (supplemental Fig. 4C). To determine the TRF2 to Rap1 ratio in the complex, SDS gel electrophoresis of the TRF2-Rap1 complex was carried out and the gel stained with a quantitative dye, Coomassie Orange. Analysis using NIH ImageJ software revealed that hTRF2 and hRap1 are in 1:1.17 ratio (Fig. 3E), confirming previous findings (39).
In summary, similar masses were obtained from the EM and gel filtration: 121 to 136 kDa for hTRF2, 221 to 285 kDa for hRap1, and 496 to 514 kDa for the TRF2-Rap1 complex. The mass estimates derived by EM and gel filtration are in good agreement, but in the future, other methods may be applied to further confirm these findings. To deduce the oligomeric state corresponding to these masses, we used published protein values, determined by SDS-PAGE, as a reference (hTRF2 ∼ 70 kDa, hRap1 ∼ 60 kDa) (39). The calculated masses based on the DNA sequence do not take into account post-translational modifications, and these can change the mass and shape of the protein and thus their mobility on size exclusion chromatography. Indeed, hTRF2 is known to migrate at a much higher apparent mass in gel filtration than its calculated mass and one explanation offered is its flexible structure due to the presence of the long unstructured linker domain (21). The values equate to a dimer for TRF2 and a tetramer for hRap1. Even though various combinations are possible because hRap1 exists as a tetramer and hTRF2 is stable as a dimer in solution, and they exist in 1:1 ratio, we conclude that the TRF2-Rap1 complex consists of a 4:4 complex consisting of one tetramer of hRap1 and either two dimers of hTRF2 or a tetramer of both proteins (Table 1).
TRF2-Rap1 Complex Recognizes Three- and Four-way Junctions and Binds to DNA in as a 4:4 Complex
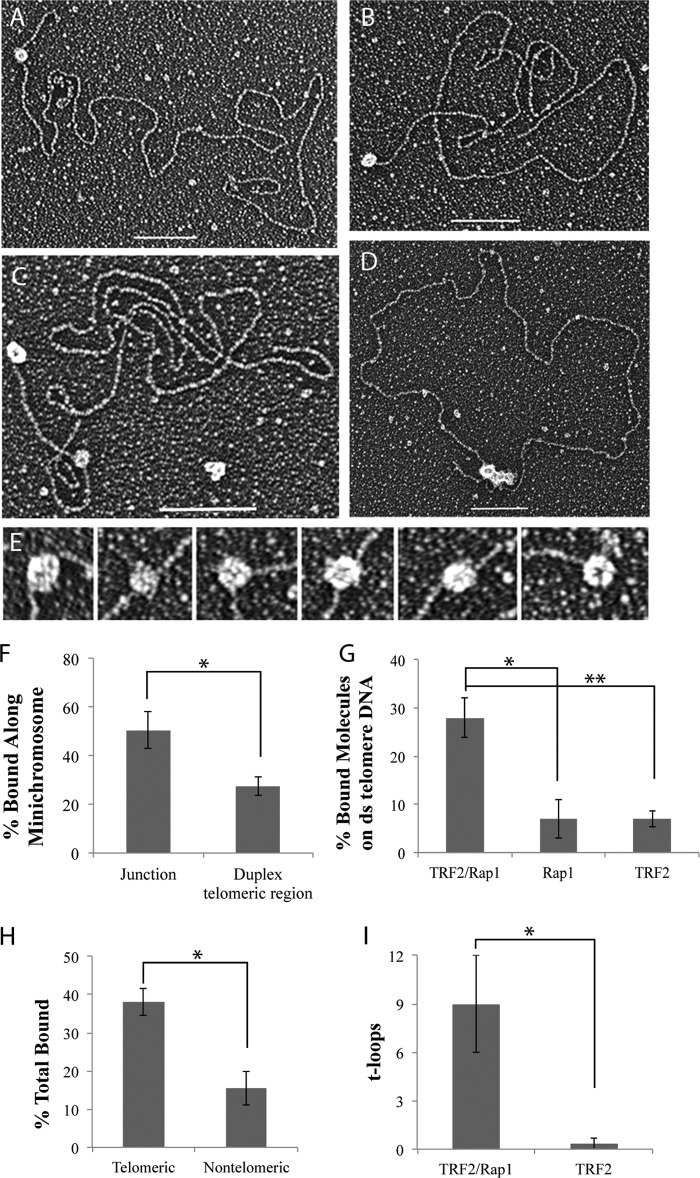
hRap1 and hTRF2 are present at human telomeres and have been identified in cell extracts in complex with each other. However, little is known about the binding preference of this complex on different templates or whether the affinity of the complex is different from its components. To examine the binding preference of the TRF2-Rap1 complex, we used the same model junction templates described above and found that a molar ratio of 7:1 TRF2-Rap1 complex to DNA provided optimal binding for EM studies (Fig. 4 and Fig. 5). The TRF2-Rap1 complex bound to the Gap25 and telomeric HJ DNAs in a manner similar to hRap1 and hTRF2 alone (Fig. 4, A and B), and localized specifically to the junction of gap25 and telomeric HJ DNAs (Fig. 4C). No significant difference in binding or preference was observed on the telomeric HJ versus nontelomeric HJs (Fig. 4D). These results suggest that the 4:4 complex of TRF2-Rap1 can also recognize three- and four-way junctions independent of sequences.
FIGURE 4.
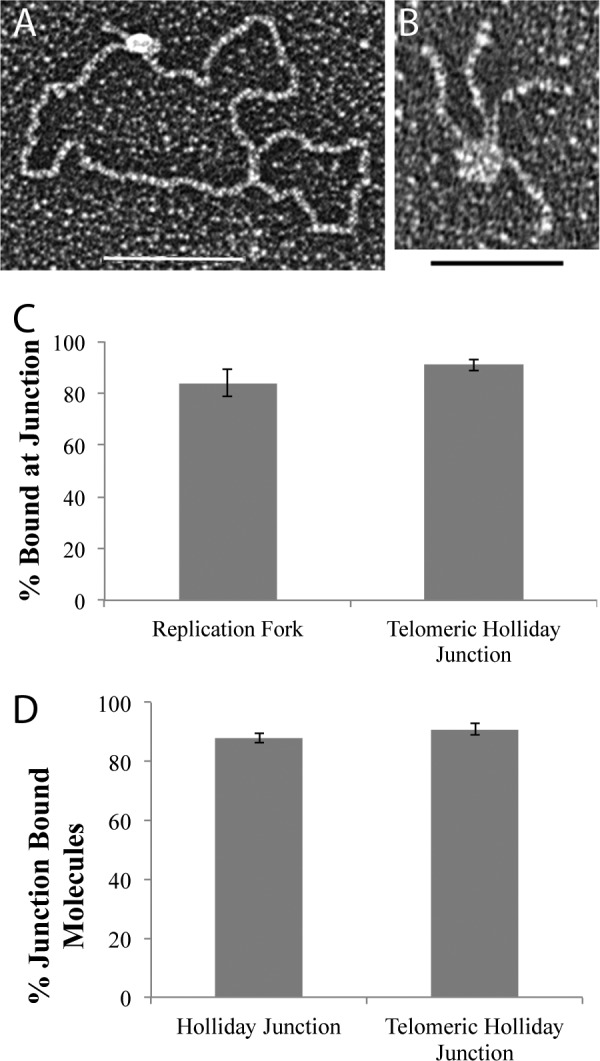
The hTRF2-Rap1 complex recognizes replication forks and Holliday junctions. The TRF2-Rap1 complex localizes to the junction site of the replication fork (A) and the telomeric HJ DNA (B). Junction preference of the protein bound molecules on the DNA templates is in C, whereas D represents the sequence preference of the hTRF2-Rap1 complex on the HJ DNAs. Each EM binding reaction was done in triplicate, and 100 molecules each were counted. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 5.
The binding properties of the TRF2-Rap1 complex along the minichromosome. Tungsten shadowcast images of the TRF2-Rap1 complex on the minichromosome illustrate the protein bound to the ds-ss junction and/or to the ds telomeric DNA (A–C). An example of the TRF2-Rap1 complex bringing the ends of the minichromosome together to form a circle is depicted in D. Bars in A–D are equivalent to 100 nm. In high magnification images, the ds telomeric DNA appears to pass through the TRF2-Rap1 complex (E). Analysis of the binding preference of the TRF2-Rap1 complex along the minichromosome DNA is in F, showing significant binding to the internal duplex telomeric sequences. G further compares the binding percentages of hRap1, hTRF2, and the complex to the ds telomeric DNA segments of the minichromosomes. The effect of DNA sequence on the binding of the TRF2-Rap1 is shown in H. I demonstrates the comparison of the t-loops formed by the TRF2-Rap1 complex and hTRF2 at 10 nm protein concentration. Corresponding p values are shown by the different number of asterisks on each graph as described in the legend to Fig. 2.
When mass analysis was carried out on tungsten shadow cast proteins bound to telomeric HJ DNA (Table 2) (corresponding Gaussian distributions are in supplemental Fig. 5), as described above, the masses obtained were 276 kDa for hRap1 and 476 kDa for the TRF2-Rap1 complex, which were consistent with the negative staining and gel filtration values. The only exception was that the estimated mass for hTRF2 was 315 kDa, arguing that it is a tetramer when it is bound to DNA, consistent with the observation of Fouche et al. (19). Therefore, hTRF2 and hRap1 bind to telomeric HJ as tetramers, whereas the TRF2-Rap1 complex binds to telomeric HJs as a 4:4 complex of each protein.
TABLE 2.
Mass and oligomeric state of hRap1, hTRF2, and the TRF2-Rap1 complex on telomeric Holliday junctions
Mass, mean area, and the oligomeric state of each protein were obtained from the two-dimensional projection analysis of the tungsten shadowcast images as in Figs. 1 and 4. Masses were calculated based on using ferritin as a size standard. Area distributions with Gaussian fits are shown in supplemental Fig. 5.
| Bound to telomeric Holliday junction | No. of molecules counted | Mean area | Calculated mass | 95% Confidence Interval +/− | No. of oligomers |
|---|---|---|---|---|---|
| pixel | kDa | kDa | |||
| TRF2 | 168 | 13,771 | 315 | 1.6 | Tetramer |
| Rap1 | 87 | 10,472 | 256 | 2.1 | Tetramer |
| TRF2-Rap1 complex | 119 | 15,823 | 475 | 3.3 | Tetramer of TRF2 and Rap1 |
TRF2-Rap1 Complex Has Higher Specificity for Telomeric DNA and Junction Structures than hTRF2 or hRap1 Alone
Incubation of the TRF2-Rap1 complex with the minichromosome template revealed binding along the internal duplex telomeric region (Fig. 5A), at the ends with the ds-ss junction (Fig. 5B), or both (Fig. 5C). Because the duplex telomeric DNA constitutes one-twelfth of the total DNA length from each end of the minichromosome molecule, we were able to verify binding at the ds telomeric regions by measuring the length of the DNA from its end to the protein-binding site. When protein molecules were examined in detail from 60 examples, it appeared that the ds DNA passed through the TRF2-Rap1 complex as contrasted to binding on one side (Fig. 5E). On the minichromosomes, the major preference for binding was to the ds-ss junction site (Fig. 5F). However, approximately one-third of the bound complexes were localized to the internal duplex telomeric DNA, which is a significantly higher value than the percentage of hRap1 or hTRF2 molecules alone bound to the ds telomeric DNA on the minichromosome where the binding was at background levels for both proteins (Fig. 5G). These data suggest that in addition to binding to the DNA junction, the TRF2-Rap1 complex has a second binding site available to it on the duplex telomeric DNA; the complex binds more readily to this region than hTRF2 and hRap1 alone.
To examine the influence of sequence on TRF2-Rap1 complex binding, we used the linear nontelomeric template with a 3′ overhang used for hRap1 binding reactions (described above). Under the same reaction conditions, the amount of the TRF2-Rap1 complex bound to the nontelomeric template was 2.5-fold less than to the minichromosomes (Fig. 5H). On the minichromosome, the majority of the TRF2-Rap1 complexes were observed at the ds-ss junction site while on the nontelomeric linear template, no preferential binding was observed (data not shown). These observations reveal that the TRF2-Rap1 complex has high specificity for telomeric sequences with a major preference being the ds-ss junction site at the end of the telomere.
The TRF2-Rap1 complex formed a new structure with the minichromosome template in which both ends of the DNA were joined by the protein complex in a circle: 9% of the DNA was observed in this form (Fig. 5D). Neither hRap1 nor hTRF2 alone exhibited this property (data not shown). Intermolecular bridges between two or more DNA molecules were present but at very low levels, likely reflecting the low DNA concentration used in the binding reaction. As the TRF2-Rap1 complex concentration was increased, the amount of circles, which might be a different form of t-loops, increased. When the TRF2-Rap1 complex concentration was titrated down to a lower level where hTRF2 no longer makes t-loops, the percentage of DNAs arranged into t-loops by the TRF2-Rap1 complex was 9% (Fig. 5I). This finding suggests that the TRF2-Rap1 complex has higher capacity to re-model telomeric DNA than TRF2 or Rap1 alone.
Kd Values for DNA Binding Confirm the EM Observations
The higher specificity of the TRF2-Rap1 complex binding to telomeric sequences, observed by EM, could be due to a change in the substrate affinity when TRF2 and Rap1 form the 4:4 complex. To further explore this, we determined the Kd values of the two individual proteins and the complex on telomeric and nontelomeric DNA templates. A telomeric duplex DNA template of 154 bp with or without a 3′ 54-mer G-rich overhang and a nontelomeric template with a 57-bp ds region that has a 33-nt 3′ overhang were prepared, incubated with TRF2, Rap1, or the TRF2-Rap1 complex in similar binding conditions to the EM assays and quantified as described under “Experimental Procedures.” Table 3 summarizes the Kd values obtained for each template and protein and the Hill coefficients determined for the telomeric 3′ overhang DNA. Typical EMSA gels are shown in supplemental Fig. 6, and graphs are shown in supplemental Fig. 7. Hill coefficients on the model telomere template are 2.4 ± 0.4 for hRap1, 5.3 ± 1.3 for hTRF2, and 3.3 ± 0.6 for the complex. All values are >1.5, showing that each protein has positive cooperativity of binding. hRap1 has the lowest affinity for ds telomeric DNA (Kd of 265 ± 57 nm) and has very similar affinity for telomeric and nontelomeric 3′ overhang structures (119 ± 12 nm and 161 ± 25.3 nm). These data are consistent with the EM observations and confirm that hRap1 prefers ds-ss junction sites irrespective of sequence. hTRF2 binds three times tighter to ds telomeric regions than hRap1, and both hRap1 and hTRF2 have similar affinities for nontelomeric junctions (161 ± 25.3 and 133 ± 16.36 nm). The Kd of hTRF2 on ds telomeric DNA is lower than the previously published value of 180 nm (40). The major reason for the tighter binding we observed could be the higher pH of the binding buffer, which inhibited TRF2 aggregation in solution. When telomeric sequences are present, the affinity of hTRF2 for junction sites is nearly two times that of hRap1. Consistent with EM, hTRF2 prefers telomeric junction sites over nontelomeric, whereas hRap1 has a slight preference for telomeric junction sites.
TABLE 3.
Affinity of hRap1, hTRF2, and the TRF2-Rap1 complex on telomeric and nontelomeric DNA templates
The dissociation constants of hRap1, hTRF2, and the TRF2-Rap1 complex on the telomeric and nontelomeric DNA templates with a 3′ overhang or on the duplex telomere DNA are presented derived from electrophoretic experiments shown in the supplemental Figs. 6 and 7. The Hill coefficient values of each protein on the model telomeric DNA are shown with the error values.
| Hill coefficient |
KD |
|||
|---|---|---|---|---|
| Telomeric 3′ overhang | Duplex telomere template | Nontelomeric 3′ overhang | ||
| nm | nm | nm | ||
| hRap1 | 2.4 ± 0.4 | 118.9 ± 12.1 | 265.0 ± 57.2 | 161.3 ± 25.3 |
| hTRF2 | 5.3 ± 1.3 | 64.7 ± 3.4 | 85.5 ± 11.6 | 133.0 ± 16.4 |
| hTRF2-hRap1 | 3.3 ± 0.6 | 5.8 ± 0.3 | 37.0 ± 9.1 | No binding |
When both proteins were present in a 4:4 complex, we observed a great increase in the affinity for telomeric DNA. The binding affinity increased a minimum of 2-fold on ds telomeric DNA and ∼10-fold on the model telomere structure containing a 3′ overhang. The TRF2-Rap1 complex has a 5-fold higher affinity for 3′ ds-ss junctions than ds telomeric regions. The TRF2-Rap1 complex cannot bind to the nontelomeric 3′ overhang structures even if the molarity of protein used in the reaction was up to three times the molarity in other binding reactions. The significant increase in affinity of the TRF2-Rap1 complex for internal duplex telomeric regions and for telomere 3′ overhang structures explains the dual binding of the TRF2-Rap1 complex along the minichromosome.
DISCUSSION
To understand the role of hRap1 at telomeres, we studied the DNA binding characteristics of hRap1 and the TRF2-Rap1 complex on different DNA templates that mimic telomeric structures. In this study, we demonstrated that hRap1 directly interacts with ds-ss DNA junctions in the absence of hTRF2. The specificity of hRap1 for junction sites is structure specific rather than sequence-specific. The dissociation rates of hRap1 and hTRF2 at nontelomeric 3′ ds-ss junction sites are similar in value, but in the presence of adjoining duplex telomeric DNA, hTRF2 shows tighter binding. Each protein binds to DNA as a tetramer and with positive cooperativity. The TRF2-Rap1 complex is ∼500 kDa and consists of four molecules each of TRF2 and Rap1. When hRap1 and hTRF2 form a complex, the binding specificity for telomeric DNA increases significantly. As the affinity for ds telomeric DNA increases by more than 2-fold, the affinity for the 3′ telomeric ds-ss junction sites increases by more than 10-fold. Interestingly, the affinity of the TRF2-Rap1 complex for linear nontelomeric DNA with a 3′ overhang is lost, indicating that upon complex formation, there is a significant change in substrate specificity.
In this work, we found that hRap1 directly binds to DNA and has a preference for ds-ss junction sites. However, it was previously reported that hRap1 does not bind to DNA on its own but rather needs hTRF2 for binding (10). The reason for this contradiction is most likely the different sets of DNA templates employed. Li et al. (10) tested the DNA binding ability of hRap1 using a 72-bp DNA template with ds human telomeric repeats or ds and ss yeast telomeric DNA templates and did not observe binding upon addition of hRap1. They suggested that the lack of binding was due to the neutral character of the myb domain. We used much longer DNA templates and ones with ds-ss junction structures or with three- and four-way junctions. We observed that hRap1 does not bind ds telomeric DNA but has high preference for ds-ss junctions. Moreover, hRap1 has similar Kd values for telomeric and nontelomeric ds-ss junctions, demonstrating that its myb domain does not directly bind to telomeric duplex DNA, and NMR studies revealed that the myb domain does not contain charged residues on the surface (41), in contrast to the myb domain of hTRF2, which is positively charged (40). On the other hand, the affinity of hTRF2 for ds-ss ends increases in the presence of telomeric sequences, which could reflect the presence of its positively charged myb domain. Thus, hRap1 appears to recognize ds-ss junction structures in the absence of hTRF2 and without a sequence preference.
Previously, Li et al. (10) failed to detect a change in the dissociation rate of hTRF2 when TRF2 is in complex with hRap1. With a similar template, we observed a 2-fold increase in binding affinity for ds telomeric DNA and 10-fold increase in binding affinity for 3′ telomeric ds-ss ends. The reason of the difference may lie in the assay conditions. In their assay, hTRF2 and hRap1 were added to the ds telomeric DNA sequentially, and DNA was already bound by hTRF2 when hRap1 was added. However, in our assays, the TRF2-Rap1 complex was formed first. Consistent with their finding (10), we did not observe any positive cooperativity of binding on a duplex telomere template. In contrast, the TRF2-Rap1 complex shows positive cooperativity of binding in the presence of a 3′ overhang structure and the affinity of binding increases by 10-fold compared with TRF2 alone.
We observed that both hRap1 and hTRF2 have the ability to bind to nontelomeric junction sites with similar affinities, raising the question what domain is responsible for the DNA binding ability of hRap1. We previously showed that the N-terminal basic domain facilitates the binding of hTRF2 to junction structures independent of sequence through a 16-amino acid stretch with eight positive charges (19). Similarly, an 18-amino acid stretch with five positive charges is present at the N-terminal BRCT domain of hRap1 (10). This is consistent with the finding of a single BRCT domain involved in the DNA binding of TopBP1 (42), XRCC1 (43) and replication factor C (44) to ds-ss junctions. Thus, the BRCT domain of hRap1 may account for its binding to these ds-ss junctions. Alteration of telomere length upon deletion of the BRCT domain in Rap1 underlines the importance of this domain for its function (11, 45). In the future, further analysis with deletion mutants should help elucidate the role of the BRCT domain of hRap1, including how the binding of hRap1 to DNA is affected by its interaction with hTRF2 and how the binding properties of hTRF2 to DNA are affected when it is complexed with hRap1. It is always possible that the effect of hRap1 on hTRF2 is dependent on its DNA binding activity and that the increase in its affinity for telomeric DNA may be due to allosteric interaction between hRap1 and hTRF2. Moreover, the DNA binding activity of hRap1 may be important for non-telomeric functions at internal sites on the chromosomes but at the telomere requires hTRF2 as a partner.
hRap1 binds to junction sites and also modulates the binding of hTRF2 along linear telomeric DNA by facilitating a positive cooperativity of binding and by increasing its affinity for telomeric sequences. Moreover, we found that the duplex telomeric region is required for binding of the complex because even if the 3′ overhang structure was present, the TRF2-Rap1 complex could not bind when duplex telomeric sequences were absent. Indeed, the TRF2-Rap1 complex bound to the ds telomeric DNA in the absence of a 3′ ds-ss junction with a ∼2-fold higher affinity than the individual components. The observation that one-fourth of the TRF2-Rap1 complexes localized within the ds telomeric region and three-fourths of the TRF2-Rap1 complexes were at the ds-ss junction site (or both at the junction site and the ds telomeric DNA) indicate that once the 3′ overhang is available, the TRF2-Rap1 complexes can slide and until it reaches the ds-ss junction where it binds tighter. These observations lead to the following model.
During telomere extension, the newly synthesized DNA will be exposed and needs to be coated with histones and/or shelterin components that bind to the ds telomeric DNA. The 3′ overhang is generated by the end processing machinery later in S phase and thus should not be available as a binding site (46). We propose that (Fig. 6) TRF2 and Rap1 bind to the ds telomeric DNA as a complex due to the higher affinity of the complex for ds telomeric DNA than either individual component. Once the 3′ overhang is formed, the TRF2-Rap1 complex slides to the junction site, and it remains due to its ∼ 6-fold lower Kd value for the junction over duplex telomeric DNA and thus covers the open telomere end. On long telomeres, the TRF2-Rap1 complex could initiate t-loop formation and prevent DDR or as the telomere length becomes shorter (to a level where the t-loops cannot be formed) then with its very high affinity for the 3′ ds-ss telomeric end, the TRF2-Rap1 complex could suppress the unnecessary DDR by blocking the end and making it less accessible to DNA repair proteins. Indeed, the finding that very short telomeres without t-loops exist stably in vivo (47), that the TRF2-Rap1 complex can prevent NHEJ of telomeres with less than 10 repeats and in the absence of Rap1, can only be compensated by significantly high amounts of hTRF2 to facilitate end protection (12, 48) provide support to this model. Depending on what parameter one measures, the loss of Rap1 in cells may be considered to be important for end protection or not (12, 28). This illustrates importance of further work that will better link end protection in vivo with the physical complexes and structures formed at telomere by the shelterin.
FIGURE 6.
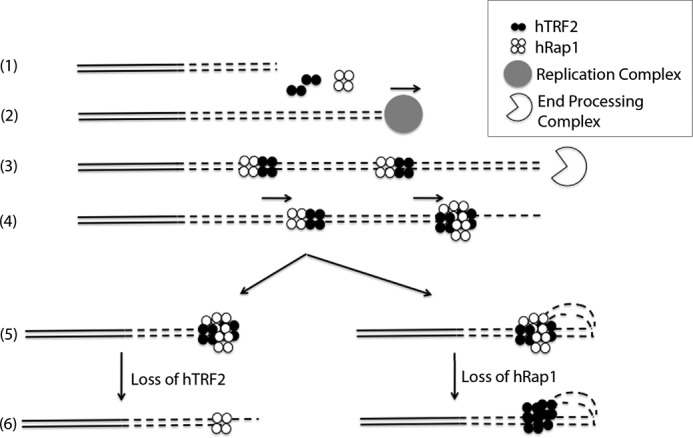
Model for the TRF2-Rap1 complex loading onto and protecting the telomere. (1), the nontelomeric and the telomeric DNA are presented in solid lines and in dashed lines (2), respectively. The new telomeric DNA is synthesized in S phase (3). hTRF2 and hRap1 bind to the newly synthesized ds telomeric DNA as a complex and then end resection occurs forming the 3′ overhang (4). The TRF2-Rap1 complexes slide along the ds telomeric DNA toward the newly formed 3′ overhang (5). End protection by the TRF2-Rap1 complex occurs through t-loop formation or simply by blocking the exposed end (6). In the absence of hTRF2, hRap1 binds to the junction site to prevent NHEJ (or HDR in mice) and in the absence of hRap1, hTRF2 prevents DDR by forming t-loops.
Acknowledgments
We thank Dr. Christopher Counter for the hTRF2 gene, Dr. Sarah Compton for the purification of hRap1, and Drs. Oya Bermek and Sezgin Ozgur for advice during the preparation of this work. We further thank Dr. Thomas Traut for help in analyzing enzymatic reactions and Kavon Javaherian for help in data collection.
This work was supported by National Institutes of Health Grants GM31819 and ES3773 (to J. D. G.).

This article contains supplemental Figs. 1–7.
- ss
- single-stranded
- hRap1
- human Rap1
- nt
- nucleotides
- ds
- double-stranded
- NHEJ
- non-homologous end joining
- HJ
- Holliday junction
- Gap25
- replication fork with a 25-nt gap.
REFERENCES
- 1. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 2. Liu D., O'Connor M. S., Qin J., Songyang Z. (2004) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- 3. Palm W., de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 4. Hsu H. L., Gilley D., Galande S. A., Hande M. P., Allen B., Kim S. H., Li G. C., Campisi J., Kohwi-Shigematsu T., Chen D. J. (2000) Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14, 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., de Lange T. (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 [DOI] [PubMed] [Google Scholar]
- 6. Tarsounas M., Muñoz P., Claas A., Smiraldo P. G., Pittman D. L., Blasco M. A., West S. C. (2004) Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117, 337–347 [DOI] [PubMed] [Google Scholar]
- 7. Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 [DOI] [PubMed] [Google Scholar]
- 8. Bianchi A., Stansel R. M., Fairall L., Griffith J. D., Rhodes D., de Lange T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 18, 5735–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stansel R. M., de Lange T., Griffith J. D. (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li B., Oestreich S., de Lange T. (2000) Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483 [DOI] [PubMed] [Google Scholar]
- 11. O'Connor M. S., Safari A., Liu D., Qin J., Songyang Z. (2004) The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 279, 28585–28591 [DOI] [PubMed] [Google Scholar]
- 12. Bae N. S., Baumann P. (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol. Cell 26, 323–334 [DOI] [PubMed] [Google Scholar]
- 13. Bombarde O., Boby C., Gomez D., Frit P., Giraud-Panis M. J., Gilson E., Salles B., Calsou P. (2010) TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J. 29, 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Steensel B., Smogorzewska A., de Lange T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- 15. Konishi A., de Lange T. (2008) Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 22, 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Celli G. B., de Lange T. (2005) DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7, 712–718 [DOI] [PubMed] [Google Scholar]
- 17. Denchi E. L., de Lange T. (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 [DOI] [PubMed] [Google Scholar]
- 18. d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 [DOI] [PubMed] [Google Scholar]
- 19. Fouché N., Cesare A. J., Willcox S., Ozgür S., Compton S. A., Griffith J. D. (2006) The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem. 281, 37486–37495 [DOI] [PubMed] [Google Scholar]
- 20. Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 21. Court R., Chapman L., Fairall L., Rhodes D. (2005) How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 6, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mao Z., Seluanov A., Jiang Y., Gorbunova V. (2007) TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 104, 13068–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sarthy J., Bae N. S., Scrafford J., Baumann P. (2009) Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 28, 3390–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takai K. K., Hooper S., Blackwood S., Gandhi R., de Lange T. (2010) In vivo stoichiometry of shelterin components. J. Biol. Chem. 285, 1457–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martínez P., Blasco M. A. (2011) Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 11, 161–176 [DOI] [PubMed] [Google Scholar]
- 26. Yang D., Xiong Y., Kim H., He Q., Li Y., Chen R., Songyang Z. (2011) Human telomeric proteins occupy selective interstitial sites. Cell Res. 21, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y., Rai R., Zhou Z. R., Kanoh J., Ribeyre C., Yang Y., Zheng H., Damay P., Wang F., Tsujii H., Hiraoka Y., Shore D., Hu H. Y., Chang S., Lei M. (2011) A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 18, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sfeir A., Kabir S., van Overbeek M., Celli G. B., de Lange T. (2011) Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian D., Griffith J. D. (2005) p53 Monitors replication fork regression by binding to “chickenfoot” intermediates. J. Biol. Chem. 280, 42568–42572 [DOI] [PubMed] [Google Scholar]
- 30. Compton S. A., Ozgür S., Griffith J. D. (2010) Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. J. Biol. Chem. 285, 13349–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D. (2008) The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 283, 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bianchi A., Smith S., Chong L., Elias P., de Lange T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S. H., Kaminker P., Campisi J. (1999) TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Griffith J. D., Christiansen G. (1978) Electron microscope visualization of chromatin and other DNA-protein complexes. Ann. Rev. Biophys. Bioeng. 7, 19–35 [DOI] [PubMed] [Google Scholar]
- 35. Griffith J. D., Makhov A., Zawel L., Reinberg D. (1995) Visualization of TBP oligomers binding and bending the HIV-1 and adeno promoters. J. Mol. Biol. 246, 576–584 [DOI] [PubMed] [Google Scholar]
- 36. Lee S., Cavallo L., Griffith J. (1997) Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem. 272, 7532–7539 [DOI] [PubMed] [Google Scholar]
- 37. Ye J. Z., Donigian J. R., van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., de Lange T. (2004) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279, 47264–47271 [DOI] [PubMed] [Google Scholar]
- 38. Oganesian L., Karlseder J. (2011) Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell 42, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu X. D., Küster B., Mann M., Petrini J. H., de Lange T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25, 347–352 [DOI] [PubMed] [Google Scholar]
- 40. Hanaoka S., Nagadoi A., Nishimura Y. (2005) Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 14, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanaoka S., Nagadoi A., Yoshimura S., Aimoto S., Li B., de Lange T., Nishimura Y. (2001) NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 312, 167–175 [DOI] [PubMed] [Google Scholar]
- 42. Yamane K., Tsuruo T. (1999) Conserved BRCT regions of TopBP1 and of the tumor suppressor BRCA1 bind strand breaks and termini of DNA. Oncogene 18, 5194–5203 [DOI] [PubMed] [Google Scholar]
- 43. Yamane K., Katayama E., Tsuruo T. (2000) The BRCT regions of tumor suppressor BRCA1 and of XRCC1 show DNA end binding activity with a multimerizing feature. Biochem. Biophys. Res. Commun. 279, 678–684 [DOI] [PubMed] [Google Scholar]
- 44. Kobayashi M., Figaroa F., Meeuwenoord N., Jansen L. E., Siegal G. (2006) Characterization of the DNA binding and structural properties of the BRCT region of human replication factor C p140 subunit. J. Biol. Chem. 281, 4308–4317 [DOI] [PubMed] [Google Scholar]
- 45. Li B., de Lange T. (2003) Rap1 affects the length and heterogeneity of human telomeres. Mol. Bio. Cell 14, 5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chow T. T., Zhao Y., Mak S. S., Shay J. W., Wright W. E. (2012) Early and late steps in telomere overhang processing in normal human cells: the position of the final RNA primer drives telomere shortening. Genes Dev. 26, 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Capper R., Britt-Compton B., Tankimanova M., Rowson J., Letsolo B., Man S., Haughton M., Baird D. M. (2007) The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 21, 2495–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu L., Blackburn E. H. (2007) Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]