Background: Telomeric repeat-containing RNA has recently been found in mammalian cells.
Results: Oligonucleotide models of telomeric DNA and RNA form a hybrid G-quadruplex structure.
Conclusion: We suggest a model system for understanding the structure and function of human telomeres.
Significance: Our finding provides valuable information for understanding the structure and function of human telomeric DNA and RNA.
Keywords: DNA, DNA Structure, Nucleic Acid Chemistry, RNA Structure, Telomeres, G-quadruplex
Abstract
Telomeric repeat-containing RNA, a non-coding RNA molecule, has recently been found in mammalian cells. The detailed structural features and functions of the telomeric RNA at human chromosome ends remain unclear, although this RNA molecule may be a key component of the telomere machinery. In this study, using model human telomeric DNA and RNA sequences, we demonstrated that human telomeric RNA and DNA oligonucleotides form a DNA-RNA G-quadruplex. We next employed chemistry-based oligonucleotide probes to mimic the naturally formed telomeric DNA-RNA G-quadruplexes in living cells, suggesting that the process of DNA-RNA G-quadruplex formation with oligonucleotide models of telomeric DNA and RNA could occur in cells. Furthermore, we investigated the possible roles of this DNA-RNA G-quadruplex. The formation of the DNA-RNA G-quadruplex causes a significant increase in the clonogenic capacity of cells and has an effect on inhibition of cellular senescence. Here, we have used a model system to provide evidence about the formation of G-quadruplex structures involving telomeric DNA and RNA sequences that have the potential to provide a protective capping structure for telomere ends.
Introduction
Telomeres play an important role in genome stability and in cell growth by protecting chromosomal ends (1). These protective functions of telomeres are thought to be due to telomere-specific DNA conformations (1). In humans, telomeric DNA consists of a duplex region, composed of TTAGGG repeats, ending in a shorter G-rich single-stranded overhang. Previous studies have suggested that human telomeric DNA may exist in multiple states such as G-quadruplexes or T-loops (2–5). For example, we, along with two other groups, determined the topology of human telomeric G-quadruplex in K+ solution (6–8). Our recent studies also demonstrated that telomeric overhang DNA forms a higher-order DNA structure containing consecutive G-quadruplexes (9). Recently, it has been suggested that the protective function of the telomere may depend on its structures (10). However, how these structures provide this protection is not yet well known.
Telomeres have long been considered to be transcriptionally silent. However, recent studies have demonstrated that telomeric DNA is transcribed into telomeric repeat-containing RNA (referred to as TERRA2) in mammalian cells (11, 12). These telomeric RNA molecules were detected in various human and rodent cell lines, containing mainly UUAGGG repeats of heterogeneous length. These findings raise the important question of how telomeric RNA is specifically associated with telomeric DNA in terms of chromosome end protection. The existence of telomeric RNA may reveal a new level of protection of chromosome ends that could provide new insights into fundamental biological processes such as cancer and aging (11–14). Determining the structure and function of TERRA RNA will be essential for understanding telomere biology and telomere-related diseases. Using NMR or x-ray crystallography, we and other groups have demonstrated that human telomere RNA forms G-quadruplex structures (15–19). More recently, we found that human telomere RNA forms a G-quadruplex structure in living cells by employing a light-switching probe (20).
In this study, we first investigated the possible association between telomeric RNA and telomeric DNA. Multiple methods such as CD, NMR, MALDI-TOF MS, and DMS footprinting were used to provide several complementary lines of evidence for intermolecular G-quadruplex formation by telomeric RNA and DNA oligonucleotides. Employing two fluorophore-labeled probes to mimic the DNA-RNA G-quadruplex in living cells, we found that oligonucleotide models of telomeric DNA and RNA can colocalize and form a DNA-RNA G-quadruplex in living cells. We further designed and synthesized a “light-up” probe to mimic the naturally formed structure in living cells. A profluorophore probe that resulted in a strong fluorescence by eliminating its fluorescence quenching was used to detect DNA-RNA G-quadruplex structures. Using this probe, we found that the process of DNA-RNA G-quadruplex formation by oligonucleotide models of telomeric DNA and RNA could occur in cells.
Next, we investigated the possible roles of the DNA-RNA G-quadruplex in telomere protection. We found that the human telomeric DNA-RNA G-quadruplex causes a significant increase in the clonogenic capacity of cells when exposed to an oligonucleotide (T-oligo) homologous to the telomeric 3′ single-stranded overhang to activate DNA damage signals and induce senescence in cells. Furthermore, we demonstrated that the DNA-RNA G-quadruplex inhibits chromosome end fusions and has an effect on the inhibition of cellular senescence using the inhibition of telomeric repeat binding factor (TRF 2) by inducing a dominant negative allele of TRF 2 (TRF 2ΔBΔM). These results suggest that the formation of the DNA-RNA G-quadruplex by telomeric RNA and DNA contributes to telomere end protection, providing valuable information for understanding the structure and function of human telomere DNA and RNA.
EXPERIMENTAL PROCEDURES
DMS Footprinting
50 μm of FAM-labeled DNA-1 with or without 50 μm telomeric RNA-1 in 140 mm KCl was denatured at 95 °C and annealed with cooling 0.5 °C/min to 4 °C. 10 μl of 0.5% dimethyl sulfate was added to 40 μl of 2.5 μm annealed oligonucleotides in 140 mm KCl solution for 1–25 min on ice. Each reaction was quenched with addition of 13 μl of stop solution (1.5 m sodium acetate, 1 m β-mercaptoethanol, and 1 μg/μl calf thymus DNA). After two ethanol precipitations, oligonucleotides were cleaved with 1 m piperidine at 95 °C for 30 min. The cleaved products were dissolved in loading buffer and analyzed by 20% denaturing PAGE, followed by visualization with GelStar (Lonza). FLA-3000 (Fujifilm) was used for detection.
CD Measurements and Analysis of CD Melting Profile
CD spectra were measured using a Jasco model J-725 CD spectrophotometer. The spectra were recorded using a 1-cm path-length cell. In CD melting studies, diluted samples were equilibrated at room temperature for several hours to obtain equilibrium spectra. The melting curves were obtained by monitoring a 260-nm CD band. Solutions for CD spectra were prepared as 0.3-ml samples at 10 μm strand concentration in the presence of 100 mm KCl or NaCl and 5 mm HEPES buffer (pH 7.0). CD spectra of telomeric DNA (DNA-2) in the presence of KCl or NaCl were examined. Then, upon addition of telomeric RNA (RNA-1) to telomeric DNA (DNA-2), the solutions of CD spectra were investigated.
MALDI-TOF MS
The MALDI matrix was 3-hydroxypicolinic acid, 50:50 acetonitrile/H2O, ammonium iron citrate. 1 μl of sample (0.5 mm DNA-RNA complex oligo, 200 mm KCl) was mixed with 1 μl of matrix solution. A spot of 1 μl of the sample-matrix mixture was placed on a stainless steel (384-well) MALDI target plate and allowed to air dry at room temperature. The MALDI-TOF MS spectrum was measured using a matrix-assisted laser desorption/ionization time of flight mass spectrometer on an Autoflex III Smartbeam mass spectrometer (positive mode).
NMR Experiments
NMR experiments were performed on a Bruker DRX-600 spectrometer. An RNA-1 (0.1 mm) and DNA-2 (0.1 mm) complex oligo was dissolved in 0.5 ml of 90% H2O/10% D2O, 10 mm Na-phosphate (pH 6.8), and 200 mm KCl.
Fluorescent Measurements
Fluorescent spectra were measured using a Jasco model FP-6500 spectrofluorometer. The spectra were recorded using a 1-cm path-length cell. For each sample, at least two spectrum scans were accumulated over a wavelength range from 300–650 nm. In photography experiments, UV irradiation of 365 nm was achieved with a UV Spot Light Source (Hamamatsu Photonics, 200 W) and UV-D36C filter (Asahi Technoglass).
Synthesis of 7-Azidocoumarin-labeled Oligonucleotide
Preparation of 7-azidocoumarin is described in the supplemental material. To incorporate the azido group at the 5′ end of C6-aminoalkyl oligonucleotide, 30 nmol of the oligonucleotide in 300 μl of 0.5 m Na2CO3/NaHCO3 buffer (pH 9.2) was incubated for 12 h at room temperature with 10 μmol of succinimidyl-7-azidocoumarin 6 in 80 μl of dimethyl sulfoxide. The crude oligonucleotide was purified by reverse-phase HPLC (MALDI-TOF MS: calculated, 4136.79; found, 4137.64).
Click Ligation of Oligonucleotides
To 50 μl of Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)-methyl]amine ligand (0.7 mm as final concentration), sodium ascorbate (1 mm), and CuSO4·5H2O (100 μm) were added sequentially to prepare the “Cu catalyst.” 7-azidocoumarin-contained DNA-3 (1 μm) and alkyne-contained RNA-2 (1 μm) were added to the Cu catalyst solution, and the reaction mixture was kept at room temperature for 2 h. After completion of the reaction, click-ligated oligonucleotide was purified by RP-HPLC and analyzed by MALDI-TOF mass spectrometry (calculated, 8215.24; found, 8217.43).
Fluorescence Microscopy
HeLa cells (3 × 104) were seeded in a 35-mm dish for fluorescence microscopy experiments. Cultures were incubated at 37 °C and 5% CO2 in DMEM containing 10% FBS and antibiotics. For transfection, probes (0.4 μm) were diluted with DMEM without 10% FBS and antibiotics. Lipofectamine 2000 reagent (Invitrogen) (10 μl) was activated in DMEM and antibiotics by equilibration for 10 min at room temperature. The probes and activated Lipofectamine were mixed together, and the lipid complexes were incubated at 37 °C for 20 min. The lipid complexes were directly added to a 35-mm dish containing HeLa cells and mixed gently by rocking after incubation at 37 °C for 16 h. Then the Cu catalyst was added and incubated at 37 °C for 3 h. For imaging the probes, the excitation and absorbance filters were 360/40 and 470/40 nm, respectively.
Clonogenic Assay
A clonogenic assay was performed as described previously (21). Cells were treated with DNA-2 (T-oligo), RNA-1 (telomeric RNA oligo), DNA-2/RNA-1 (DNA-RNA complex oligo), or diluent for 1 week and then trypsinized and counted. Each oligo concentration is 40 μm. Three hundred cells were seeded into 60-mm culture dishes in triplicate and then incubated in complete medium for 2 weeks with medium changes twice per week. Subsequently, the cells were fixed for 5 min in 100% methanol. The methanol was then removed, and the culture dishes were rinsed briefly with water. Then the cell colonies were stained for 10 min in 4% (w/v) methylene blue solution in PBS, washed once again with water, and then counted.
β-Galactosidase Staining (SA-β-Gal) Assay
β-Galactosidase staining (SA-β-Gal) assay was performed as described previously (22). The cells were washed in PBS, fixed for 15 min at room temperature in 4% formaldehyde in PBS, washed, and incubated at 37 °C with cell staining working solution of the β-galactosidase staining kit (Mirus). Staining was evident in 16 h.
Cell Growth
HT-1080 and HeLa cells were grown at 37 °C and 5% CO2 in DMEM (1.5 ml) containing 10% FBS and antibiotics (penicillin and streptomycin). FuGENE HD transfection reagent (Roche) was used for transfection. pWZL Hygro-TRF2ΔBΔM with transfection reagent was transfected in a ratio of 1:3 (w/w) to 90% confluent cells in a 24-well plate at 37 °C and 5% CO2 for 16 h. Then the cells were seeded in a 24-well plate with or without 40 μm of telomeric RNA (RNA-1). Hygromycin B was added for selection. After 7 days of selection, each cells were seeded in duplicate at concentration of 1 × 104 cells/ml/well in a 24-well plate. Living cells were counted using a Countess automated cell counter (Invitrogen) every other day. HT1080 cells expressing TRF2ΔBΔM in treatment with or without telomeric RNA-1 (40 μm) were incubated.
RESULTS
G-quadruplex Formation by Human Telomeric DNA and RNA Oligonucleotides as a Model System to Study the Relationship between Telomeric DNA and RNA
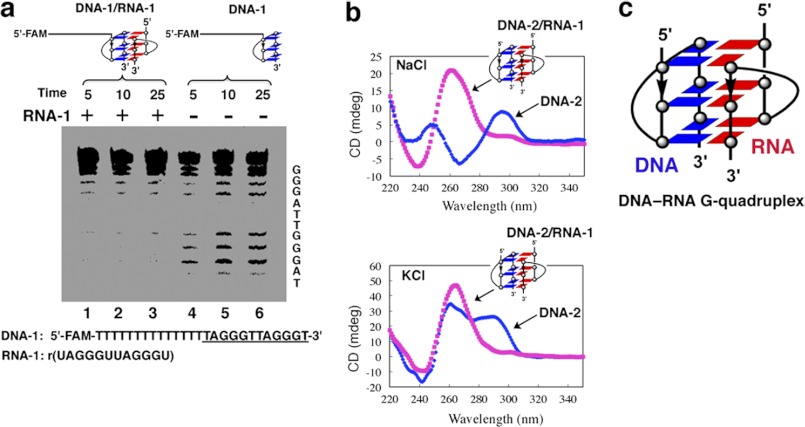
Telomeric RNAs were found to be localized with telomeric DNA (11, 12), suggesting a possible association between telomeric RNA and telomeric DNA. We assume that this association of telomeric RNA with telomeric DNA may be involved in the formation of an intermolecular G-quadruplex structure. To test this hypothesis, we performed a DMS footprinting assay. DMS footprinting has been used to identify G residues involved in the formation of a G-quadruplex (23). The N7 position of each G involved in the formation of a G-quadruplex is protected against methylation by DMS through Hoogsteen bonding (23). The cleavage pattern of G residues is used to probe G-quadruplex formation. In the absence of telomeric RNA (RNA-1), DNA-1, a sequence containing telomeric DNA repeats, was cleaved randomly at every G residue, suggesting an unstructured form (Fig. 1a, lanes 4–6). However, in the presence of the telomere RNA sequence RNA-1, G residues were either partially or fully protected from piperidine cleavage (Fig. 1a, lanes 1–3). The DMS footprinting pattern suggests that oligonucleotide models of telomeric DNA and RNA can form an intermolecular DNA-RNA G-quadruplex. We noted that the dominant CD band of DNA-1 at 280 nm was shown as the characteristic of the single-stranded structure, consistent with the results of the DMS footprinting assay and melting temperature (Tm) curve analysis (supplemental Fig. S1) (24–26).
FIGURE 1.
DNA-RNA G-quadruplex formation by telomeric DNA and RNA oligonucleotides. a, DMS footprinting assay for analysis of G-quadruplex formation. A representative gel electropherogram of a FAM-labeled oligonucleotide (DNA-1) in the presence or absence of telomeric RNA (RNA-1) is shown. The RNA-1 contains two repeats of the human telomeric RNA sequence r(UAGGGUUAGGGU). Lanes 1, 2, and 3 show DMS footprinting in the presence of telomeric RNA-1. Lanes 3, 4, and 5 show DMS footprinting in the absence of telomeric RNA-1. Reaction times: 5, 10, and 25 min. b, CD spectrometry. Upper panel, d(TAGGGTTAGGGT) DNA-2 with or without RNA-1 in the presence of 100 mm NaCl at 25 °C. Bottom panel, DNA-2 with or without RNA-1 in the presence of 100 mm KCl at 25 °C. c, schematic structure of an intermolecular DNA-RNA G-quadruplex formed by human telomeric DNA and RNA. The blue and red boxes show dG of DNA and rG of RNA.
To define the structural features of the DNA-RNA G-quadruplex, we examined the conformation of human telomeric DNA-RNA complex using CD spectroscopy. The CD spectrum of DNA-2 in the presence of Na+ showed a positive peak at 290 nm and a negative peak at 265 nm, which is a characteristic CD signature of an antiparallel G-quadruplex structure (Fig. 1b). Upon the addition of telomeric RNA (RNA-1) to the telomeric DNA (DNA-2), the peak at 290 nm disappeared, whereas a strong positive peak at 260 nm appeared, suggesting that a parallel G-quadruplex was formed by oligonucleotide models of telomeric DNA and RNA (Fig. 1b). Similarly, CD spectrum analysis showed that a parallel G-quadruplex was formed when equal amounts of telomeric RNA were added to the telomeric DNA in the presence of K+, when the peak at 290 nm disappeared, whereas the intensity of the 260-nm peak increased (Fig. 1b) (27, 28). According to CD melting experiments, the Tm value of the intermolecular G-quadruplex in 100 mm KCl solution was 65 °C (supplemental Fig. S2), indicating high thermodynamic stability.
Recent advances in MS have made it a successful technique for investigating noncovalent intermolecular complexes of DNA, RNA, and PNA-DNA hybrids (29, 30). To further characterize the DNA-RNA G-quadruplex structure, MALDI-TOF MS was used to directly observe the G-quadruplex formation. We observed peaks near m/z (7591.5 + nK+), corresponding to the sum of the molecular weights (MW) of DNA-2 and RNA-1 of the DNA-RNA G-quadruplex (the sum of MW = 3892.3 (DNA-2 MW) + 3700.2 (RNA-1 M W)) (supplemental Fig. S3), suggesting that the intermolecular G-quadruplex remained stable even in the gas phase (30).
G-quadruplex structures display characteristic resonances for the imino protons, whose presence may be used to confirm the presence of the G-quadruplex structure in solution (31, 32). We investigated the structure of the RNA-1 and DNA-2 complex using NMR. In the imino proton region of the 600 MHz 1H NMR spectrum in the presence of K+, peaks that were assignable to the guanine imino protons of the G-quadruplex structure were observed at 11.0–12.0 ppm (supplemental Fig. S4) (31, 32). These signals corresponded to the G residues of the G-quadruplex, indicating that RNA-1 and DNA-2 form a DNA-RNA G-quadruplex (Fig. 1c).
Mimicking the Naturally Formed Telomeric DNA-RNA G-quadruplexes in Living Cells Using Oligonucleotide Models of Telomeric DNA and RNA
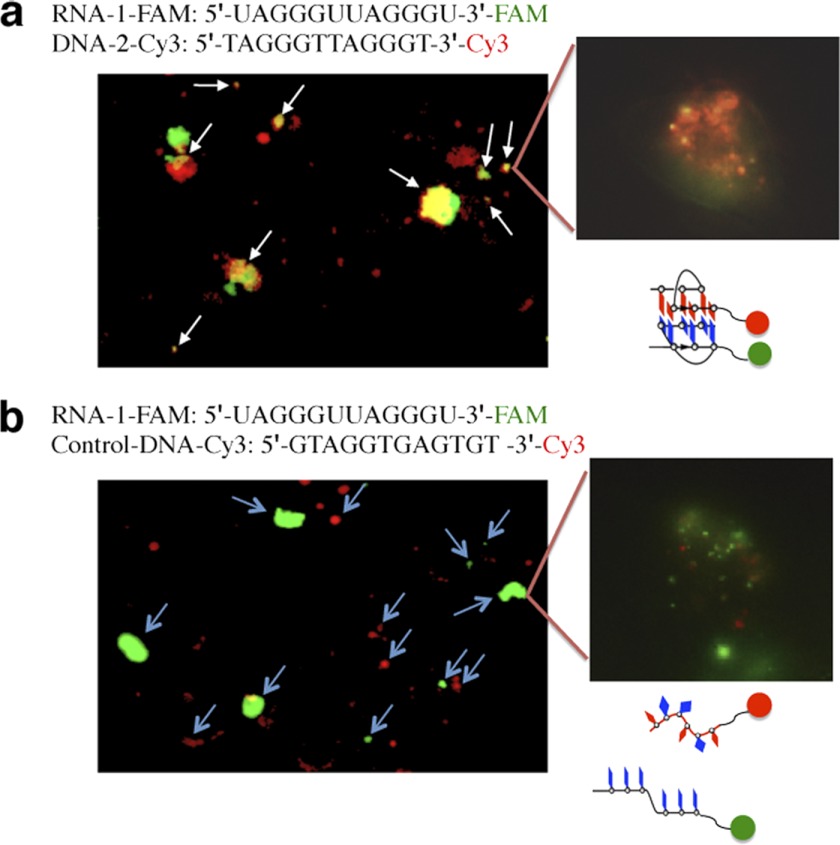
As the naturally formed telomeric G-quadruplex structures are difficult to probe in living cells, we next employed a model system using telomeric RNA and DNA oligonucleotides to mimic the naturally formed telomeric DNA-RNA G-quadruplexes in living cells. Therefore, we prepared three oligonucleotide probes with fluorophores at their 3′ terminus to investigate whether an intermolecular G-quadruplex is present in living cells (33). For the DNA-2-Cy3 and control-DNA-Cy3 probes, the Cy3 fluorophore was incorporated into the 3′ end of the G-rich sequence 5′-TAGGGTTAGGGT-3′ (DNA-2) and the 3′ end of random-sequence 5′-GTAGGTGAGTGT-3′ (control DNA, non-telomeric sequence), respectively. For the RNA-1-FAM probe, the FAM fluorophore was incorporated into the 3′ end of the G-rich sequence 5′-UAGGGUUAGGGU-3′ (RNA-1). When two fluorophore-labeled probes are in their free form without G-quadruplex formation, both fluorophore molecules are spatially separated, and two colors can be observed. Formation of the G-quadruplex brings the fluorescent molecules at the 3′ ends into close proximity and produces a color change. We observed colocalization of the fluorescent signals (orange) when the FAM-labeled probe RNA-1-FAM (green) and the Cy3-labeled probe DNA-2-Cy3 (red) were transfected into living cells (Fig. 2a). This colocalization was not observed when the random probe control-DNA-Cy3 was used (Fig. 2b). These observations suggest that oligonucleotide models of telomeric DNA and RNA can colocalize and form a DNA-RNA G-quadruplex in cells. This is consistent with recent results showing that a G-quadruplex dimer is formed in living cells (20).
FIGURE 2.
Fluorescence microscopy images of the intermolecular G-quadruplex formed by oligonucleotide models of telomeric DNA and RNA in living cells. a, RNA-1-FAM signals are shown in green, and DNA-2-Cy3 signals are shown in red. The colocalization of the two signals generates orange signals (arrows). b, for RNA-1-FAM and control-DNA-Cy3, there is no colocalization of fluorescent signals. RNA-1-FAM, the FAM fluorophore is located at the 3′ end of the G-rich sequence 5′-UAGGGUUAGGGU-3′ (RNA-1). DNA-2-Cy3, the Cy3 fluorophore is located at the 3′ end of the G-rich sequence 5′-TAGGGTTAGGGT-3′ (DNA-2). Control-DNA-Cy3, the Cy3 fluorophore is located at the 3′ end of the random sequence 5′-GTAGGTGAGTGT-3′ (control DNA).
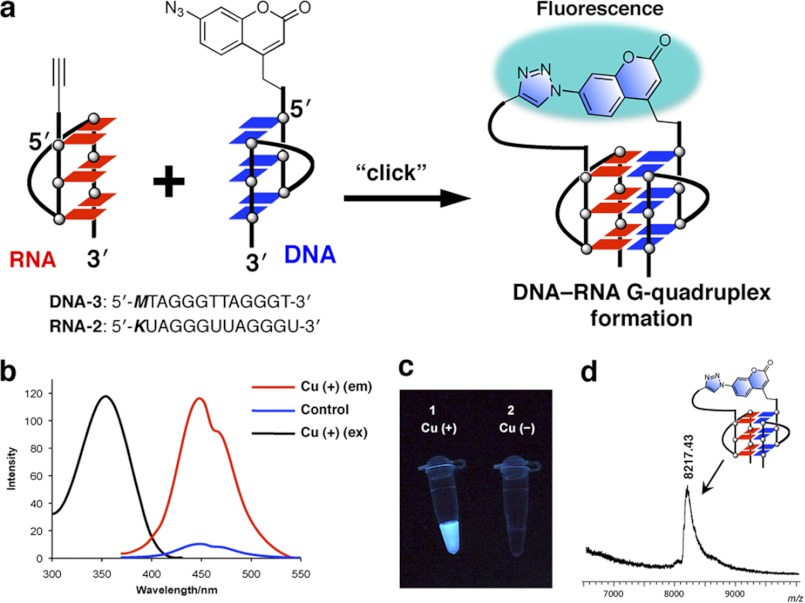
To further prove DNA-RNA hybrid G-quadruplex formation using oligonucleotide models of telomeric DNA and RNA in living cells, we developed a light-up reporter strategy to probe for the telomeric DNA-RNA G-quadruplex. We designed and synthesized a probe (DNA-3) having the two-repeat 12-mer human telomere DNA and azidocoumarin moiety at its 5′ terminus and another probe with a 5′ alkyne at the 5′ end of a 12-mer RNA (RNA-2) (Fig. 3a) (supplemental schemes 1–3). Nonfluorescent azidocoumarin was chosen as a profluorophore. The profluorophore has no fluorescence because of the quenching effect of the electron-rich α-nitrogen of the azido group (34, 35). Substitution at the 7-position of coumarin dyes has a significant influence on their fluorescence properties. In fact, formation of a triazole ring at its 7-position by azide-alkyne cycloaddition will eliminate quenching, resulting in a strong fluorescence. The coumarin dye as a profluorophore can produce a strong fluorescence when a triazole ring is formed by azide-alkyne cycloaddition (Fig. 3a) (35–37). When the coumarin dye-labeled probe is free in the solution (or in the cell) without G-quadruplex formation, two molecules (azidocoumarin and alkyne at the 5′ end) are separated spatially, and no fluorescence is observed. Formation of the G-quadruplex brings the two molecules at the 5′ ends into close proximity, allowing the azide-alkyne cycloaddition reaction to produce a strong fluorescence by the formation of a triazole ring (Fig. 3a). The emission color serves as a means to rapidly probe DNA-RNA G-quadruplex formation. The advantage of the structure dependence of an emission production is that the cycloaddition reaction can strongly provide evidence for G-quadruplex formation. A strong interaction by G-quadruplex formation is the only effective way to induce the cycloaddition reaction. The simple molecule colocalization and decoy effects of the oligomers are unable to promote the click reaction by bringing the 5′ alkyne and 5′ azidocoumarin reaction partners into close proximity to one another.
FIGURE 3.
A light-up reporter strategy to probe DNA-RNA hybrid G-quadruplex formation. a, 7-azidocoumarin as a profluorophore was incorporated into the 5′ terminus of 12-mer human telomere DNA (DNA-3, 5′-MTAGGGTTAGGGT-3′). The profluorophore containing azide moiety is fluorescent inactive. G-quadruplex formation by the DNA and RNA (RNA-2, 5′-KUAGGGUUAGGGU-3′) promotes a click reaction to trigger the fluorescence signals by formation of a triazole ring. This can be used as a reporter to probe DNA-RNA G-quadruplex formation. b, fluorescence spectra for oligonucleotide probes. Cu (+) (em), with Cu catalyst for emission spectrum; control, using the 12-mer RNA-3 (5′-KUUUUUUUUUUUU-3′) as a control; Cu (+) (ex), excitation spectrum (350 nm). c, fluorescence image of probes without and with Cu catalyst after illumination with a UV lamp (365 nm). d, MALDI-TOF MS spectrum of the click reaction product of DNA-3 and RNA-2. After completion of the reaction, the click-ligated oligonucleotide was purified by RP-HPLC and analyzed by MALDI-TOF mass spectrometry.
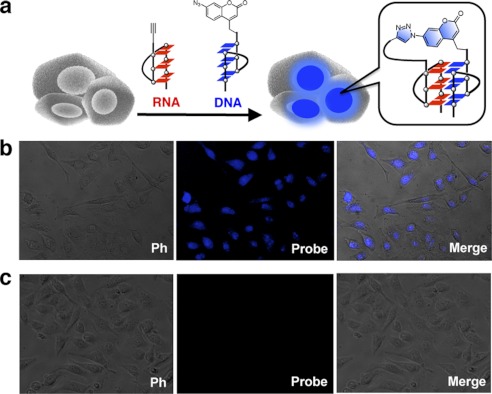
We found that the formation of G-quadruplex between the 12-mer azidocoumarin-contained DNA-3 and the 5′ alkyne 12-mer RNA (RNA-2) activated the weakly fluorescent coumarin to give an intensely fluorescent triazole product by the cycloaddition reaction (Fig. 3, b and c). A clear blue color was observed by the naked eye with the addition of the Cu catalyst (Fig. 3c). The fluorescence spectrum in the presence of the Cu catalyst exhibits an emission band at 450 nm whereas, on using a 12-mer RNA-3 as a control (without telomeric sequence), no emission was observed (Fig. 3b). The resulting product of the click reaction was characterized by MALDI-TOF MS, revealing that the product is the azido-alkyne cycloaddition product (M-H+ calculated, 8215.2; found, 8217.4) (Fig. 3d). To document the direct evidence of the presence of DNA-RNA G-quadruplexes in living cells, oligonucleotide probes were applied to living cells (Fig. 4a). We incubated human HeLa cells with the probes and visualized the live cells by fluorescence microscopy. The cycloaddition reaction was started by adding new cell medium containing the Cu catalyst. We directly observed the cells by using a fluorescent microscope without fixation and washing. We clearly observed the blue fluorescence from the catalyst-treated cells (Fig. 4b). Negative cells that were untreated with the Cu catalyst remained virtually nonfluorescent (Fig. 4c). Taken together, these observations suggest that the process of DNA-RNA G-quadruplex formation using oligonucleotide models of telomeric DNA and RNA could occur in cells, consistent with the results of our previous study using a click chemistry approach, and showing that a DNA-RNA G-quadruplex structure was formed in solution using human telomeric DNA and RNA oligonucleotides (38, 39).
FIGURE 4.
Schematic for detection of DNA-RNA G-quadruplex formation in living cells using oligonucleotide probes. a, G-quadruplex formation in living cells will induce the fluorescence signals. b and c, fluorescence microscopy images of live cells with Cu catalyst (b) and without Cu catalyst (c). ph, phase-contrast imaging.
We next prepared a FRET molecular beacon to investigate telomeric DNA-RNA G-quadruplex formation by natural telomeric RNA and beacon DNA in living cells. The molecular beacon containing telomere DNA sequence is designed to form a stem-loop hairpin structure in the absence of a target, quenching the fluorophore reporter FAM (supplemental Fig. S5). The formation of a G-quadruplex with a natural telomeric RNA target causes the hairpin to open, separating the fluorophore and the quencher (DABCYL) and restoring the fluorescence. To detect DNA-RNA G-quadruplex formation, the molecular beacon was delivered into HeLa cells. The resulting fluorescence signal was visualized 6 h after delivery, as shown in Fig. 6S. The imaging result was consistent with the click chemistry approach, suggesting that a DNA-RNA G-quadruplex could form using telomeric RNA and artificial DNA oligomers in cells.
FIGURE 6.
Growth curve analysis of HT1080 cells expressing TRF2ΔBΔM in treatment with or without telomeric RNA oligo (RNA-1) and diluent alone. a, after 3 weeks, cells treated with telomeric RNA-1 proliferated at almost the same rate as diluent control cells. Data are mean ± S.E. of three experiments. b, effect of human telomeric RNA on the TRF2ΔBΔM-induced senescent morphology and SA-β-Gal expression. TRF2ΔBΔM-expressing cells were treated with telomeric RNA-1, diluent, or control RNA (non-telomeric sequence). The resulting cells were assessed for SA-β-Gal activity 15 days later.
Protective Effect of DNA-RNA G-quadruplex on the Clonogenic Capacity of T-oligo-treated Cells
Having demonstrated that oligonucleotide models of telomeric DNA and RNA can form a DNA-RNA G-quadruplex structure, we next investigated the possible roles of the DNA-RNA G-quadruplex in telomere protection. Recent evidence suggests that exposure to oligonucleotides (T-oligos) homologous to the telomere 3′ telomere overhang activated the p53 pathway and induced senescence in cells (21, 40). The T-oligos are believed to mimic exposure of the 3′ telomeric overhang DNA (uncapping the telomere) and activate DNA damage responses, which induce a transient cell cycle arrest or apoptosis (21, 40). To investigate the function of telomeric RNA, we induced telomere uncapping by treatment with the T-oligo and used a clonogenic assay to examine the protective effect of the DNA-RNA G-quadruplex. HT1080 cells were treated with a T-oligo (DNA-2), a telomeric RNA oligo (RNA-1), a DNA-RNA oligo complex containing telomere DNA and RNA (DNA-2/RNA-1) (two oligos with homologous sequence from the DNA-RNA G-quadruplex), or diluent left alone for 1 week (Fig. 5). To enhance cellular uptake of the DNA fragment and mimic T-oligos, DNA-2 with a homologous telomeric sequence was used instead of DNA-1. An equal number of cells were seeded in 60-mm culture dishes in triplicate and then incubated in a complete medium for 2 weeks, and the medium was changed twice a week. The dishes were then stained with methylene blue (Fig. 5a). The clonogenic capacity of cells pretreated with T-oligo (DNA-2) was almost completely suppressed (8.7 ± 1.5% of diluent-treated control; p < 0.01; Fig. 5, a and b), consistent with previous results showing that the addition of T-oligo caused cells to undergo apoptosis (21, 40). Surprisingly, in contrast, we found that the treatment of cells with the DNA-RNA oligo complex (DNA-2/RNA-1) caused a significant increase in the clonogenic capacity (85.1 ± 9.8% of diluent-treated control; Fig. 5, a and b), similar to that observed with telomeric RNA alone or diluent alone. Enhanced clonogenicity is known to accompany the inhibition of SA-β-Gal activity (40). Thus, we next examined whether the DNA-RNA G-quadruplex could inhibit SA-β-Gal activity induced by exposure to T-oligo. HT1080 cells were treated with diluent, DNA-2, control DNA (non-telomeric sequence), DNA-RNA oligo complex, or DNA/control RNA oligo complex (DNA-2/control RNA; control RNA was a non-telomeric sequence RNA) as a control for 1 week, and SA-β-Gal activity was then assessed (Fig. 5c). The cells treated with the DNA-2/RNA-1 did not exhibit senescent morphology, and SA-β-Gal activity was almost completely abolished (Fig. 5c), similar to diluent- and control DNA-treated cells. In a parallel experiment, cells treated with the DNA-2/control RNA showed increased SA-β-Gal activity and exhibited senescent morphology similar to DNA-2 (Fig. 5c). These findings suggest that the DNA-RNA G-quadruplex formation induced a protective response regarding the survival and proliferation of cells.
FIGURE 5.
Effect of human telomeric RNA on clonogenic capacity of T-oligo-treated cells. a, the exogenous T-oligo has been suggested to mimic the uncapped dysfunctional endogenous telomere overhang, inducing DNA damage responses and a senescent phenotype, reducing the clonogenic capacity of the cells. Cells were treated with DNA-2 (T-oligo), RNA-1 (telomeric RNA oligo), DNA-2/RNA-1 (DNA-RNA oligo complex), or diluent. The cells were then incubated and assayed (clonogenic assay). Colonies of cells were stained with methylene blue. The appearance of the stained dishes showed that the number of colonies in cells treated with DNA-RNA oligo complex markedly increased compared with DNA-2 alone. b, quantification of clonogenic capacity. Colonies in triplicate cultures, shown above, were counted and plotted as a percentage of the diluent-treated control. Error bars represent mean ± S.D. c, effect of human telomeric RNA on SA-β-Gal activity in T-oligo-treated cells. Cells were treated with diluent, DNA-2 (T-oligo), control DNA (non-telomeric sequence), DNA-2/RNA-1 (DNA-RNA oligo complex), or DNA-2/control RNA, control RNA r(GUAGGUGAGUGU) (non-telomeric sequence). The resulting cells were stained and assayed for SA-β-Gal activity.
Protective Effect of the DNA-RNA G-quadruplex in TRF2ΔBΔM-expressing Cells
To further reveal the function of the DNA-RNA G-quadruplex, we used a dominant-negative mutant of TRF2, TRF2ΔBΔM, which lacks the NH2-terminal basic domain and the COOH-terminal Myb domain. Overexpression of TRF2ΔBΔM results in the rapid induction of senescence in human fibroblasts (41, 42). Cells in TRF2ΔBΔM-induced senescence display a reduction in the G-rich single-stranded overhang, leading to the disruption of the “capping” structure of telomeres (42). Consistent with the effect of TRF2ΔBΔM, TRF2ΔBΔM-expressing HT1080 human fibrosarcoma cells ceased to proliferate, whereas control cells continued to grow (Fig. 6a). In contrast, we found that cell proliferation was significantly augmented at almost the same rate as that observed in the diluent control-treated cells, when the cells were treated with a telomeric RNA oligo (RNA-1) (Fig. 6a). To further understand this protective function, we examined whether telomeric RNA could inhibit SA-β-Gal activity. SA-β-Gal activity has been used as an indicator of cellular senescence (21). In contrast to cells expressing TRF2ΔBΔM alone, cells treated with the telomeric RNA oligo did not exhibit senescent morphology and showed significantly decreased SA-β-Gal activity, similar to that observed in the diluent-treated cells (Fig. 6b). We also noted that cells treated with a control RNA (non-telomeric sequence) showed increased SA-β-Gal activity and exhibited senescent morphology, indicating that the mutated RNA had no effect on the inhibition of cellular senescence (Fig. 6b). Together, these results suggest that human telomeric RNA may provide a protective effect for telomere ends by forming a DNA-RNA G-quadruplex with telomeric DNA.
DISCUSSION
The discovery of telomere RNA molecules opens new doors to a better understanding of the essential biological role of telomeres. There is a clear need to revisit the structural and functional mechanisms of telomeres accompanying telomere RNA participation. Our findings provide evidence that human telomeric RNA contributes to telomere end protection. It is noted that the system described here potentially mimics a naturally formed structure in cells. Human telomere structures are difficult to probe in cells using antibodies because the concentration of G-quadruplexes in humans is too low to be detected in the presence of only a few dozen chromosome ends (92 telomeres/cell in G1 of the cell cycle and twice as many in G2). In a previous study, Schaffitzel et al. (43) suggested that Stylonychia lemnae telomeric DNA forms an antiparallel G-quadruplex in vivo by using a single-chain antibody, in which ∼2 × 108 telomeres are present in one macronucleus. Human telomeric G-quadruplex structures are difficult to probe in living cells, and thus far, direct evidence for the existence of human telomeric DNA and RNA G-quadruplexes in cells has not yet been obtained. In this study, to overcome these limitations, we employed a model system of telomeric RNA and DNA oligonucleotides to mimic the naturally formed telomeric DNA-RNA G-quadruplexes in living cells, providing suggestive evidence for the natural existence of this structure.
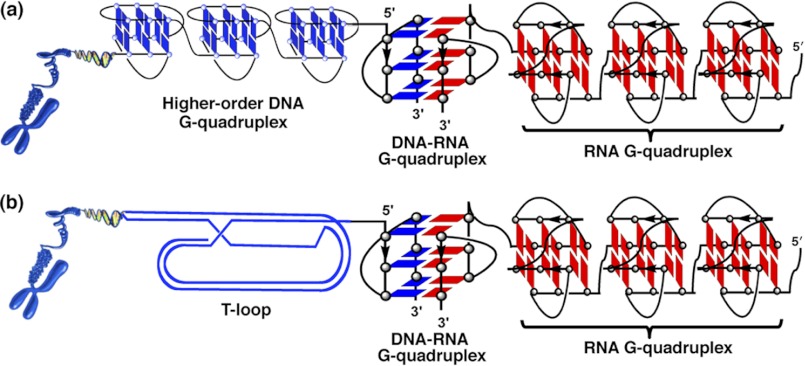
The formation of an intermolecular G-quadruplex by oligonucleotide models of telomeric DNA and RNA provides a protective effect. A simple explanation for the protective action of telomeric RNA involves a steric interference model in which a higher-order structure formed by the DNA-RNA complex may avoid exposure of single-stranded telomeric DNA and block the access of key regulatory molecules to the single-stranded DNA. Recently, we demonstrated that long telomeric overhang DNA consecutively forms G-quadruplexes (9). Long telomeric RNAs were also suggested to form a higher-order telomeric G-quadruplex structure (16–18). The telomeric DNA-RNA G-quadruplex may be involved in a superhelix structure formed by human telomeric DNA and RNA and provide a protective effect (Fig. 7).
FIGURE 7.
Schematic superhelix structure of human telomeric G-quadruplexes formed by telomere DNA and RNA. a, a higher-order telomeric DNA G-quadruplex or telomeric DNA-RNA hybrid G-quadruplex, or telomeric RNA G-quadruplex may be involved in the structure. b, a t-loop, telomeric DNA-RNA hybrid G-quadruplex, or telomeric RNA G-quadruplex may be involved in the structure.
Some proteins have been suggested to be associated with telomeric RNA. A shelterin component, such as TRF2, was found to recruit telomeric RNA to telomeric DNA (44). Overexpression of TRF2ΔBΔM, which caused the loss of TRF 2 from telomeres, may lead to failure in recruiting telomeric RNA to telomeric DNA. This is consistent with our observations that the addition of telomeric RNA oligo to TRF2ΔBΔM-expressing cells inhibits the senescent morphology and fusion events. Several telomeric DNA-binding proteins have been found to be associated with telomere G-quadruplex. Rap1 is known to promote G-quadruplex formation (45). Stm1p aids in telomere protection and can bind a G-quadruplex (46). POT1 promotes G-quadruplex unfolding (47, 48). Recent studies demonstrated that telomere RNA promotes POT1 binding to telomeric single-stranded DNA by removing hnRNPA1 (49), suggesting that telomeric RNA is involved in telomere capping to preserve genomic integrity, consistent with our results. Whether these proteins or other factors interact with the telomeric DNA-RNA G-quadruplex remains to be discovered. In addition, telomere DNA, telomere RNA, and related proteins may form a TDRP body (telomeric DNA, RNA, and proteins are referred to as TDRP (50).
Acknowledgments
We thank Titia de Lange for providing the TRF2ΔBΔM plasmid.
This work was partially supported by Ministry of Education, Science, Sports, Culture, and Technology, Japan Grant-in-aid for Scientific Research (B) 22350071. This work was also supported by grants from the Takeda Science Foundation, the Naito Foundation and Mochida Memorial Foundation for Medical and Pharmaceutical Research.

This article contains supplemental Figs. S1–S6 and Schemes 1–3.
- TERRA
- telomeric repeat-containing RNA
- DMS
- dimethyl sulfate
- SA-β-Gal
- senescence-associated β-galactosidase.
REFERENCES
- 1. Blackburn E. H. (2001) Switching and signaling at the telomere. Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 2. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y., Patel D. J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1, 263–282 [DOI] [PubMed] [Google Scholar]
- 4. Parkinson G. N., Lee M. P., Neidle S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. (2004) T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5, 323–329 [DOI] [PubMed] [Google Scholar]
- 6. Xu Y., Noguchi Y., Sugiyama H. (2006) The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 14, 5584–5591 [DOI] [PubMed] [Google Scholar]
- 7. Luu K. N., Phan A. T., Kuryavyi V., Lacroix L., Patel D. J. (2006) Structure of the human telomere in K+ solution. An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 128, 9963–9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambrus A., Chen D., Dai J., Bialis T., Jones R. A., Yang D. (2006) Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 34, 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Y., Ishizuka T., Kurabayashi K., Komiyama M. (2009) Consecutive formation of G-quadruplexes in human telomeric-overhang DNA. A protective capping structure for telomere ends. Angew. Chem. Int. Ed. Engl. 48, 7833–7836 [DOI] [PubMed] [Google Scholar]
- 10. Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 [DOI] [PubMed] [Google Scholar]
- 11. Dai X., Huang C., Bhusari A., Sampathi S., Schubert K., Chai W. (2010) Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection. EMBO J. 29, 2788–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schoeftner S., Blasco M. A. (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 [DOI] [PubMed] [Google Scholar]
- 13. Luke B., Lingner J. (2009) TERRA. Telomeric repeat-containing RNA. EMBO J. 28, 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schoeftner S., Blasco M. A. (2009) A 'higher order' of telomere regulation. Telomere heterochromatin and telomeric RNAs. EMBO J. 28, 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu Y., Kaminaga K., Komiyama M. (2008) G-quadruplex formation by human telomeric repeats-containing RNA in Na+ solution. J. Am. Chem. Soc. 130, 11179–11184 [DOI] [PubMed] [Google Scholar]
- 16. Martadinata H., Phan A. T. (2009) Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 131, 2570–2578 [DOI] [PubMed] [Google Scholar]
- 17. Randall A., Griffith J. D. (2009) Structure of long telomeric RNA transcripts. The G-rich RNA forms a compact repeating structure containing G-quartets. J. Biol. Chem. 284, 13980–13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collie G. W., Haider S. M., Neidle S., Parkinson G. N. (2010) A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 38, 5569–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Y., Ishizuka T., Kimura T., Komiyama M. (2010) A U-tetrad stabilizes human telomeric RNA G-quadruplex structure. J. Am. Chem. Soc. 132, 7231–7233 [DOI] [PubMed] [Google Scholar]
- 20. Xu Y., Suzuki Y., Ito K., Komiyama M. (2010) Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 14579–14584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O. (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li G. Z., Eller M. S., Hanna K., Gilchrest B. A. (2004) Signaling pathway requirements for induction of senescence by telomere homolog oligonucleotides. Exp. Cell Res. 301, 189–200 [DOI] [PubMed] [Google Scholar]
- 23. Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kypr J., Kejnovská I., Renciuk D., Vorlícková M. (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clark C. L., Cecil P. K., Singh D., Gray D. M. (1997) CD, absorption and thermodynamic analysis of repeating dinucleotide DNA, RNA and hybrid duplexes [d/r(AC)]12.[d/r(GT/U)]12 and the influence of phosphorothioate substitution. Nucleic Acids Res. 25, 4098–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vorlicková M., Kejnovská I., Kovanda J., Kypr J. (1998) Conformational properties of DNA strands containing guanine-adenine and thymine-adenine repeats. Nucleic Acids Res. 26, 1509–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S. (2006) Quadruplex DNA. Sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumari S., Bugaut A., Huppert J. L., Balasubramanian S. (2007) An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3, 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krishnan-Ghosh Y., Stephens E., Balasubramanian S. (2004) A PNA4 quadruplex. J. Am. Chem. Soc. 126, 5944–5945 [DOI] [PubMed] [Google Scholar]
- 30. Rueda M., Luque F. J., Orozco M. (2006) G-quadruplexes can maintain their structure in the gas phase. J. Am. Chem. Soc. 128, 3608–3619 [DOI] [PubMed] [Google Scholar]
- 31. Phan A. T., Patel D. J. (2003) Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution. Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 125, 15021–15027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu Y. (2011) Chemistry in human telomere biology. Structure, function and targeting of telomere DNA/RNA. Chem. Soc. Rev. 40, 2719–2740 [DOI] [PubMed] [Google Scholar]
- 33. Sedoris K. C., Thomas S. D., Clarkson C. R., Muench D., Islam A., Singh R., Miller D. M. (2012) Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol. Cancer Ther. 11, 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sivakumar K., Xie F., Cash B. M., Long S., Barnhill H. N., Wang Q. (2004) A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org. Lett. 6, 4603–4606 [DOI] [PubMed] [Google Scholar]
- 35. Zhou Z., Fahrni C. (2004) A fluorogenic probe for the copper(I)-catalyzed azide-alkyne ligation reaction. Modulation of the fluorescence emission via 3(n,pi)-1(pi,pi) inversion. J. Am. Chem. Soc. 126, 8862–8863 [DOI] [PubMed] [Google Scholar]
- 36. Beatty K. E., Xie F., Wang Q., Tirrell D. A. (2005) Selective dye-labeling of newly synthesized proteins in bacterial cells. J. Am. Chem. Soc. 127, 14150–14151 [DOI] [PubMed] [Google Scholar]
- 37. Beatty K. E., Liu J. C., Xie F., Dieterich D. C., Schuman E. M., Wang Q., Tirrell D. A. (2006) Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew. Chem. Int. Ed. Engl. 45, 7364–7367 [DOI] [PubMed] [Google Scholar]
- 38. Xu Y., Suzuki Y., Komiyama M. (2009) Click chemistry for the identification of G-quadruplex structures. Discovery of a DNA-RNA G-quadruplex. Angew. Chem., Int. Ed. Engl. 48, 3281–3284 [DOI] [PubMed] [Google Scholar]
- 39. El-Sagheer A. H., Brown T. (2010) Click chemistry with DNA. Chem. Soc. Rev. 39, 1388–1405 [DOI] [PubMed] [Google Scholar]
- 40. Li G. Z., Eller M. S., Firoozabadi R., Gilchrest B. A. (2003) Evidence that exposure of the telomere 3′ overhang sequence induces senescence. Proc. Natl. Acad. Sci. U.S.A. 100, 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Steensel B., Smogorzewska A., de Lange T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- 42. Karlseder J., Smogorzewska A., de Lange T. (2002) Senescence induced by altered telomere state, not telomere loss. Science 295, 2446–2449 [DOI] [PubMed] [Google Scholar]
- 43. Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H. J., Plückthun A. (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 98, 8572–8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P. M. (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giraldo R., Rhodes D. (1994) The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 13, 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hayashi N., Murakami S. (2002) STM1, a gene which encodes a guanine quadruplex binding protein, interacts with CDC13 in Saccharomyces cerevisiae. Mol. Genet. Genomics 267, 806–813 [DOI] [PubMed] [Google Scholar]
- 47. Wang H., Nora G. J., Ghodke H., Opresko P. L. (2011) Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem. 286, 7479–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaug A. J., Podell E. R., Cech T. R. (2005) Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. U.S.A. 102, 10864–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Flynn R. L., Centore R. C., O'Sullivan R. J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L. (2011) TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu Y., Komiyama M. (2012) Structure, function and targeting of human telomere RNA. Methods 57, 100–105 [DOI] [PubMed] [Google Scholar]