Background: Glucose-responsive ChREBP binds 14-3-3 and localizes cytosolically during starvation.
Results: ChREBP interacts with 14-3-3 through its α2 helix in a novel phosphorylation-independent conformation.
Conclusion: 14-3-3 binds to ChREBP with high affinity to stabilize ChREBP in the cytosolic compartment under low glucose conditions.
Significance: ChREBP·14-3-3 structure is the first step toward understanding the mechanism by which ChREBP responds to glucose levels.
Keywords: Carbohydrate Metabolism, Glucose Metabolism, Signal Transduction, Transcription Factors, X-ray Crystallography, Fatty Acid Synthesis, Lipogenesis
Abstract
Carbohydrate response element-binding protein (ChREBP) is an insulin-independent, glucose-responsive transcription factor that is expressed at high levels in liver hepatocytes where it plays a critical role in converting excess carbohydrates to fat for storage. In response to fluctuating glucose levels, hepatic ChREBP activity is regulated in large part by nucleocytoplasmic shuttling of ChREBP protein via interactions with 14-3-3 proteins. The N-terminal ChREBP regulatory region is necessary and sufficient for glucose-responsive ChREBP nuclear import and export. Here, we report the crystal structure of a complex of 14-3-3β bound to the N-terminal regulatory region of ChREBP at 2.4 Å resolution. The crystal structure revealed that the α2 helix of ChREBP (residues 117–137) adopts a well defined α-helical conformation and binds 14-3-3 in a phosphorylation-independent manner that is different from all previously characterized 14-3-3 and target protein-binding modes. ChREBP α2 interacts with 14-3-3 through both electrostatic and van der Waals interactions, and the binding is partially mediated by a free sulfate or phosphate. Structure-based mutagenesis and binding assays indicated that disrupting the observed 14-3-3 and ChREBP α2 interface resulted in a loss of complex formation, thus validating the novel protein interaction mode in the 14-3-3β·ChREBP α2 complex.
Introduction
Liver is the primary site of de novo fatty acid and triglyceride synthesis from excess carbohydrate. These fats can be stored locally or released by the liver in various forms and taken up by adipocytes for long term storage. ChREBP,3 discovered in this laboratory (1), is the master regulator controlling the conversion of carbohydrate to fat in liver and acts independently of insulin signaling. ChREBP is a large transcription factor of 96 kDa containing several functional domains, including two nuclear export signals, NES1 and NES2, an extended bipartite nuclear localization signal, a proline-rich region, and a DNA binding bHLH/ZIP domain (Fig. 1A) (1–3). ChREBP senses and responds to fluctuating levels of glucose and glucose metabolites with altered intracellular localization, DNA binding, and transcriptional activity (4); the mechanisms, however, are not yet completely elucidated. ChREBP and the insulin-responsive transcription factor steroid-response element-binding protein (SREBP)-1c independently induce hepatic lipogenic enzyme genes in response to elevated blood glucose and insulin levels, respectively (5, 6). However, ChREBP but not SREBP-1c induces liver pyruvate kinase and multiple additional enzymes in hepatic glycolytic pathways that provide substrate for fatty acid synthesis (5, 7–11) Thus, ChREBP plays a critical role in tightly coupling enhanced glycolysis and de novo fatty acid synthesis following ingestion of high carbohydrate meals.
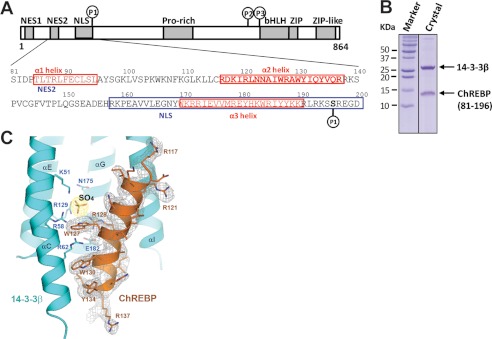
FIGURE 1.
Domain organization and the sequence of the N-terminal glucose-responsive regulatory region of ChREBP. A, residues 81–196 of the N-terminal regulatory region, including the predicted α1, α2, and α3 helices, the nuclear export signal 2 (NES2), nuclear localization signal (NLS), and Ser-196 phosphorylation site, were found in a complex with 14-3-3β. B, SDS-PAGE analysis of 14-3-3β·ChREBP crystals revealed the presence of both 14-3-3β and ChREBP fragment. C, simulated annealing omit map of ChREBP α2 helix bound to 14-3-3β and the interface SO4 molecule. The maps are contoured at 1.0σ for ChREBP and 2.0σ for the sulfate.
ChREBP activity is regulated predominantly through multiple post-translational modifications of which phosphorylation and dephosphorylation are the best characterized (4, 12). Acetylation and O-GlcNAcylation have also been reported (13–15). Phosphorylation by cAMP-dependent protein kinase (PKA) and dephosphorylation mediated by a xylulose 5-phosphate-activated protein phosphatase, PP2ABδC, of one or more sites on the ChREBP N-terminal region regulate nucleocytoplasmic transport of ChREBP (2, 3, 16–20). When circulating blood glucose levels are low and glucagon is high, phosphorylated and inactive ChREBP is mainly localized to the cytoplasm in liver (16). As the glucose level increases after a meal, dephosphorylated and active ChREBP translocates to the nucleus where it dimerizes with Max-like protein X (Mlx) and binds to glucose-responsive elements within the promoters of liver pyruvate kinase and all genes involved in hepatic lipogenesis.
14-3-3 proteins are a family of regulatory proteins acting primarily as phosphoserine/phosphothreonine-binding modules and regulate the functions of a large number of different substrate proteins (22–25). 14-3-3 proteins play a dual role in the regulation of ChREBP activity, affecting both nuclear export and import of ChREBP. 14-3-3 interacts directly with phosphorylated (p)-ChREBP (inactive) under low glucose conditions, and as a result, ChREBP is retained in the cytosol or exported out of the nucleus (19, 20). 14-3-3 binding to phosphorylated ChREBP inhibits its interaction with importin-α, which is required for nuclear import of ChREBP. It has been shown that 14-3-3 binds to two sites on ChREBP, a high affinity primary binding site encompassing the α2 helix (residues 117–137) (3, 26) and a secondary binding site overlapping the importin-α-binding site (residues 158–190) ( 3). Here, we report the co-expression and co-crystallization of the 14-3-3β and ChREBP N-terminal regulatory region (residues 1–250) and the crystal structure determination of the complex at 2.4 Å resolution. Our results show that ChREBP binds to 14-3-3β through the primary binding site, the α2 helix. A free sulfate or phosphate ion is found at the interface between ChREBP and 14-3-3 and likely contributes to the tight binding of the two proteins. The 14-3-3β·ChREBP complex represents a novel interaction mode of a 14-3-3 protein with its target protein and provides new insights into the mechanism by which 14-3-3 regulates ChREBP function.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The full-length mouse 14-3-3β and the N-terminal region of ChREBP (residues 1–250) containing two phosphoserine mimetic mutations, S140D and S196D, were cloned into the pETDuet vector (Novagen). The ChREBP fragment was inserted into the first multiple cloning site (MCS1) between EcoRI and HindIII restriction sites, and 14-3-3β was cloned into the MCS2 between the NdeI and XhoI sites. The resultant plasmid was transformed into BL21-CodonPlus (DE3) Escherichia coli cells and grown at 32 °C until absorption at 600 nm reached 0.6. Protein expression was induced with the addition of isopropyl β-d-thiogalactopyranoside (Sigma) to a final concentration of 0.1 mm, and cells were left to grow overnight at 16 °C. Cells were harvested and resuspended in 50 mm NaH2PO4, 300 mm NaCl, 1 mm β-mercaptoethanol, 1 mm benzamidine, 10 μg/ml leupeptin (Roche Applied Science), and 5 mm imidazole (Buffer A). Clarified cell lysate was loaded onto a nickel-nitrilotriacetic acid affinity column (GE Healthcare) pre-equilibrated with Buffer A. After washing with Buffer A containing 20 mm imidazole, the bound protein was eluted with 200 mm imidazole. The resulting protein complex was treated with thrombin (Hematologic Technologies, Inc.) and further purified using a Superdex 200 gel filtration column (GE Healthcare) equilibrated with 20 mm HEPES-KOH, pH 7.5, 150 mm NaCl, and 1 mm DTT and then concentrated to 20 mg/ml for crystallization.
Crystallization, Data Collection, and Structure Determination
Crystals of the 14-3-3β·ChREBP complex were grown using the hanging drop vapor diffusion technique. A 1.5-μl protein at a concentration of 20 mg/ml was mixed with 1.5 μl of well solution containing 100 mm sodium citrate, pH 6.5, 200 mm sodium/potassium tartrate, and 2.2 m ammonium sulfate and equilibrated against the well at 20 °C. Crystals grew to a maximum size of 0.2 × 0.05 × 0.05 mm3 after 3 days and were cryoprotected by dipping the crystals into 2.2 m lithium sulfate and flash-frozen in liquid nitrogen. Diffraction data were collected at beamline 19-ID, Advanced Photon Source, Argonne National Laboratory, Argonne, IL. Data were processed using the program HKL2000 (27). Initial phases were obtained by the molecular replacement method using the programs PHASER (28). A human 14-3-3β structure (Protein Data Bank code 2BQ0) (22) was used as a starting model. Extra density from ChREBP was observed from the map generated by PHASER, and the model of ChREBP with side chains unambiguously assigned was manually built into the density using COOT (29). Structure refinements were performed using phenix.refine (30). The crystal data and final refinement statistics are summarized in Table 1. The coordinates of 14-3-3β·ChREBP have been deposited in the Protein Data Bank (Protein Data Bank code 4GNT) (31).
TABLE 1.
Summary of crystallographic data collection and refinement statistics
| 14-3-3β·ChREBP | |
|---|---|
| Data collection | |
| Space group | P4222 |
| Cell dimensions | |
| a, b, c | 115.9, 115.9, 59.5 Å |
| α, β, γ | 90, 90, 90° |
| Resolution | 50.0 to 2.40 Å (2.49 to 2.40 Å) |
| Rsym or Rmerge | 0.078 (0.818) |
| I/σI | 27.1 (1.95) |
| Completeness () | 96.1% (94.7%) |
| Redundancy | 5.9 (5.8) |
| Refinement | |
| Resolution | 33.7 to 2.4 Å |
| No. of reflections | 14,627 |
| Rwork/Rfree | 0.177/0.228 |
| No. of atoms | |
| Protein | 2077 |
| Water | 71 |
| Inorganic | 20 |
| B-factors | |
| Protein | 73.4 |
| Water | 72.5 |
| Inorganic | 123.9 |
| Root mean square deviations | |
| Bond lengths | 0.010 Å |
| Bond angles | 0.678° |
Mutagenesis and Pull-down Assays
Site-specific mutations were generated using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. Mutant 14-3-3β was constructed in plasmid pET21 and expressed in BL21 codon plus (DE3). FLAG-tagged full-length ChREBP or the indicated ChREBP mutants were expressed in HEK293T cells using the mammalian expression vector pExchange5A (Stratagene) (20). Cells containing overexpressed ChREBP or ChREBP mutants were then lysed in a buffer containing 10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5 mm EDTA, 10 mm sodium fluoride, and 1% Nonidet P-40. Proteins were purified by incubating with M2 beads bearing anti-FLAG antibodies (Sigma) in the binding buffer containing 20 mm HEPES-KOH, pH 7.3, 110 mm potassium acetate, 2 mm magnesium acetate, 5 mm sodium acetate, 0.5 mm EGTA, 0.01% Nonidet P-40, and 0.5% BSA for 1 h at 4 °C with gentle rocking. For analysis of mutant 14-3-3β binding to wild-type ChREBP, the beads with bound FLAG-ChREBP were incubated with the purified 14-3-3β mutants in 500 μl of the binding buffer for 1.5 h at 4 °C with gentle rocking. The beads were then washed three times with the same binding buffer without BSA followed by elution with SDS-PAGE sample buffer. The eluted samples were subjected to SDS-PAGE (10%) and transferred to Trans-Blot nitrocellulose membranes (Whatman) for Western blotting.
The following primary antibodies were used to detect 14-3-3 proteins and FLAG-ChREBP, respectively, pan-14-3-3 (Santa Cruz Biotechnology) and anti-FLAG (Sigma). Horseradish peroxidase-conjugated anti-rabbit or mouse IgG (Zymed Laboratories Inc.) was used as the secondary antibody, and bound proteins were visualized with the ECL Western blotting detection system (Pierce).
Isothermal Titration Calorimetry (ITC)
ITC experiments were carried out using a VP-ITC microcalorimeter (MicroCal/GE Healthcare). His-tagged human wild-type (WT) and mutant (R62A and E182A) 14-3-3β were dialyzed overnight into a buffer containing 20 mm HEPES, pH 7.5, and 1 mm EDTA. All titration experiments were performed in the same dialysis buffer. A synthetic ChREBP peptide of residues 117–140(S140P) containing the α2 helix and phosphorylated Ser(P)-140 was used in the binding assay. The peptide at 300 μm concentration was injected in 12-μl increments into the reaction cell containing 1.4 ml of 30 μm WT or mutant 14-3-3β proteins. The binding isotherm derived from heat changes was used to calculate the dissociation constant Kd and enthalpy (ΔH0) values with the ORIGIN version 7.0 software (MicroCal/GE Healthcare). The concentrations for WT and mutant 14-3-3 proteins were determined by using the colorimetric Coomassie Blue protein reagent.
RESULTS
14-3-3β Forms a Stable Complex with the N-terminal Regulatory Region of ChREBP
To understand the interactions between 14-3-3 and ChREBP, and how 14-3-3 regulates the function of ChREBP, we reconstituted a complex of 14-3-3β and ChREBP N-terminal regulatory region (residues 1–250, see Fig. 1) by co-expressing and co-purifying the two proteins. The N-terminal region of ChREBP binds tightly and stably to 14-3-3β, as demonstrated by the presence of both ChREBP and 14-3-3β in the same fractions after several steps of purification, including Ni2+ affinity purification, thrombin treatment, and size exclusion chromatography (data not shown). Both proteins were also present in the crystals (Fig. 1B). Notably, the ChREBP band on SDS-PAGE had a molecular mass of ∼14 kDa, smaller than what was expected for the 250 amino acids encoded in the construct. N-terminal sequencing and mass spectral analysis revealed that the ChREBP fragment in the crystal had a mass of 13,977 kDa and consisted of residues from 81 to 196, a region encompassing all three previously predicted α helices, α1, α2, and α3, the NES2 and the nuclear localization signal regulatory elements (Fig. 1A). However, after the complex structure was determined and refined, only residues 117–137 of ChREBP, containing the α2 helix, are structured and visible in the electron density map (Fig. 1C). Apparently, the regions outside α2 are flexible or disordered in the crystal. This observation is consistent with the lack of a well defined structured domain in this region of ChREBP despite the presence of three predicted α helices.
Interactions between ChREBP α2 Helix and 14-3-3β
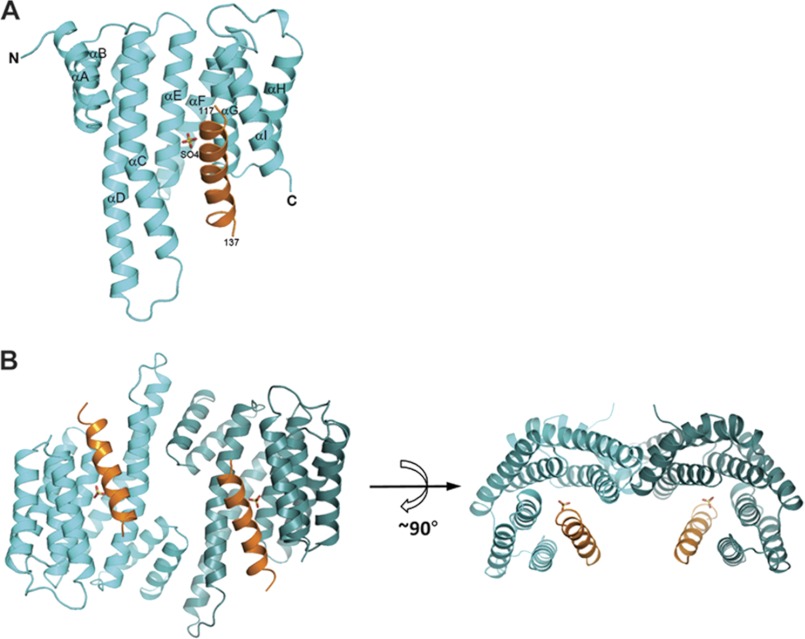
The overall conformations of the 14-3-3β protein in the complex with ChREBP is similar to those reported previously (22) and contains nine α helices (from αA to αI) arranged into a right-handed superhelical bundle with the conserved peptide-binding groove located at the concave face of the protein (Fig. 2). Previously the structures of apo-14-3-3β and its complex with non-phosphorylated Pseudomonas aeruginosa exoenzyme S (ExoS) peptide have been reported (22). The root mean square deviations between Cα atoms of these 14-3-3β proteins and the 14-3-3β bound to ChREBP presented here are only ∼0.4 Å, indicating that they adopt essentially identical conformation. In the crystal structure of 14-3-3β·ChREBP complex, the α2 helix of ChREBP is wedged between the third (αC) and the last (αI) helices of 14-3-3β (Fig. 2) and oriented roughly parallel to both αC and αI helices. The 14-3-3β in complex with ChREBP retained its dimer conformation, and the two 14-3-3β monomers in the dimer are related by a strict crystallographic 2-fold symmetry (Fig. 2B).
FIGURE 2.
Overall structure of 14-3-3β·ChREBP complex. A, ribbon diagram of a monomer of the 14-3-3β (cyan) complexed with the α2 helix of ChREBP (orange). The SO4 molecule is shown as sticks. B, two orthogonal views of 14-3-3β dimer in complex to ChREBP α2.
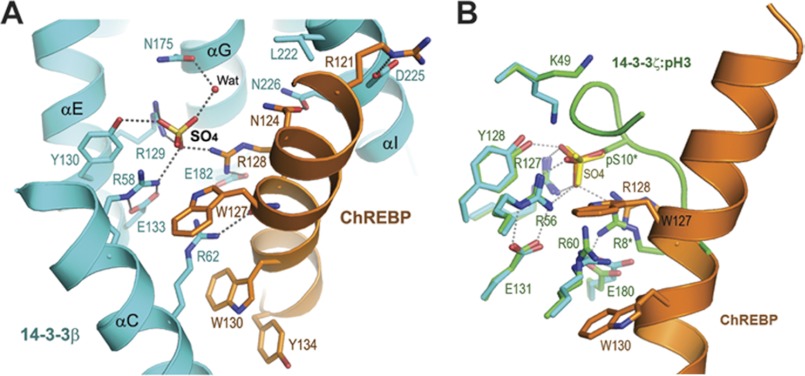
A close inspection of the interface between ChREBP and 14-3-3β revealed the presence of a free sulfate (SO4) molecule mediating some of the interactions between the two proteins (Fig. 3A). This sulfate molecule interacts directly with Arg-58, Arg-129, and Tyr-130 of 14-3-3β, the so-called Arg-Arg-Tyr triad important for phosphopeptide binding (22), and indirectly with Asn-175 through a water molecule (Fig. 3A); it also interacts directly with Arg-128 of ChREBP. Although it is equally likely that a phosphate molecule can bind to the same site, we assigned a sulfate at this position because of the presence of 2.2 m ammonium sulfate as the crystallization agent. In addition to these sulfate-mediated or potentially phosphate-mediated interactions, there are a number of direct contacts between ChREBP and 14-3-3β, including hydrogen bonds between Asn-124, Trp-127, and Arg-128 of ChREBP to Asn-226, Arg-58, and Glu-182 of 14-3-3β, respectively. A number of prominent van der Waals contacts are also observed between the two proteins. In particular, Arg-62 of 14-3-3β intercalates between two tryptophan side chains of ChREBP (Trp-127 and Trp-130) (Fig. 3A), and the side chain of Glu-182 of 14-3-3β stacks against Tyr-131 of ChREBP. As a result, about 35% of surface area of ChREBP α2 helix is buried upon forming the complex with 14-3-3β (916 Å2 contact area out of a total of 2673 Å2). Importantly, the structural analysis indicates that residue Ser-140, which has been mutated from Ser to Asp in the construct to mimic phosphoserine, is not involved in the interaction with 14-3-3.
FIGURE 3.
Involvement of a free sulfate at the interface between 14-3-3β and ChREBP α2. A, detailed interactions between 14-3-3β (cyan) and ChREBP α2 (orange). Residues involved in the intermolecular contacts are shown as sticks. A water molecule (WAT) is shown as a small red sphere. Hydrogen bonds are shown as dotted lines. B, comparison of the 14-3-3β·ChREBP interface with that between 14-3-3ξ and a phosphorylated histone H3 peptide (pH 3, in green). Residues of 14-3-3ξ are labeled.
Structure-based Mutagenesis Analysis of 14-3-3β·ChREBP Interface
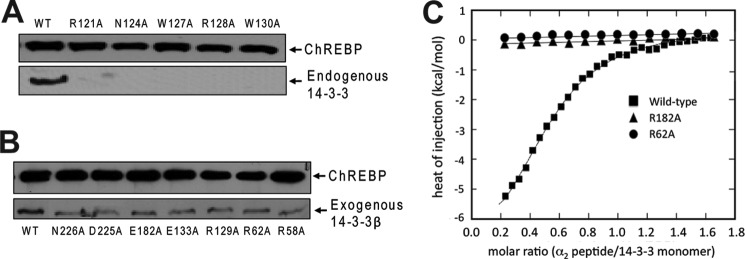
To validate the observed interface between 14-3-3 and ChREBP, we made a series of site-directed mutations on ChREBP α2 aimed to disrupt its interactions with 14-3-3. These include R121A, N124A, W127A, R128A, and W130A of ChREBP. Full-length FLAG-tagged ChREBP containing these mutations were expressed in HEK293 cells and purified by anti-FLAG affinity gel. Any endogenous 14-3-3 proteins associated with ChREBP are expected to be co-purified. As shown in Fig. 4A, a substantial amount of endogenous 14-3-3 proteins co-purified with wild-type (WT) ChREBP, whereas mutation of any one of these ChREBP residues abolished its binding to endogenous 14-3-3.
FIGURE 4.
Mutagenesis and binding analysis of ChREBP and 14-3-3β. A, pull-down assays were performed to assess the binding between WT and mutant ChREBP and endogenous 14-3-3 proteins. Full-length FLAG-tagged ChREBP proteins were expressed in HEK293T cells and purified via the anti-FLAG resin. The association of 14-3-3 proteins with ChREBP was detected via Western blotting using anti-14-3-3 pan-antibody. B, pull-down assays of the binding between the WT ChREBP and exogenously expressed WT and mutant 14-3-3β. C, ITC experiments were performed to measure the binding constant between a ChREBP peptide containing the α2 helix (residues 117–140) to the WT and two 14-3-3β mutants, R62A and E182A. The isotherm titrations from three data sets were analyzed with ORIGIN version 7.0 software package (MicroCal/GE Healthcare). They correspond to wild-type (black squares), R62A (black circles), E182A (black triangles), respectively.
We also made a series of mutations on 14-3-3β at its interface with ChREBP. These include R58A, R62A, R129A, E133A, E182A, D225A, and N226A. In an in vitro pull-down assay, binding of all purified 14-3-3β mutants to full-length wild-type ChREBP was decreased compared with wild-type 14-3-3β, although not abolished (Fig. 4B). We further examined the binding affinity of 14-3-3β R62A and E182A mutants to ChREBP using ITC assays. Arg-62 and Glu-182 of 14-3-3β directly interact with the ChREBP α2 helix. In particular, Arg-62 has never been shown previously to directly coordinate binding in any 14-3-3·target protein complex. Results from the ITC experiments showed that WT 14-3-3β binds a ChREBP α2 peptide tightly with a Kd of 2.8 μm (Fig. 4C). Both R62A and E182A mutant 14-3-3βs failed to bind the α2 peptide. These data support the important roles of residues Arg-62 and Glu-182 of 14-3-3β in binding ChREBP as observed in the crystal structure.
Comparison of 14-3-3·ChREBP Interface to that of 14-3-3·Phosphopeptides
Because the majority of 14-3-3 binding partners are phosphorylated peptides, we compared the ChREBP-binding site on 14-3-3β with that of a phosphoserine peptide as represented by the complex of 14-3-3ζ and histone H3(Ser(P)-10) (Fig. 3B) (32). The conformations of the 14-3-3 proteins in the two complexes are similar, with a root mean square deviation between the Cα atoms of the two proteins being 0.5 Å. Not unexpectedly, the sulfate in the 14-3-3β·ChREBP structure superimposes exactly on the phosphoryl group of the Ser(P)-10 of H3. All the residues on 14-3-3β that interact with the sulfate or phosphate group are also conserved in 14-3-3ζ (Fig. 3B). Additionally, the side chain of Arg-8 of histone H3, which is two amino acids upstream of the phosphorylated Ser(P)-10 and conserved in the phosphorylation-dependent 14-3-3-binding motifs, superimposes well with Arg-128 of ChREBP. In both complexes, these Arg residues provide additional interactions to the bound phosphate or sulfate group. Other than these shared interactions with the phosphate/sulfate group, ChREBP α2 interacts with 14-3-3 in a fundamentally different manner from those 14-3-3-bound phosphopeptides. ChREBP α2 adopts an extended well defined α-helical conformation with several bulky aromatic residues, Trp-127, Trp-130, and Tyr-131, interacting intimately with 14-3-3 at the edge of the conserved peptide-binding groove between αC and αI. In particular, Arg-62 of 14-3-3β is closely involved in these interactions, sandwiched between the side chains of Trp-127 and Trp-130 (Fig. 3A). Arg-62 of 14-3-3 has not been shown previously to directly bind any phosphopeptide or the nonphosphorylated ExoS peptide.
Comparison of 14-3-3·ChREBP Interface with That of 14-3-3·ExoS
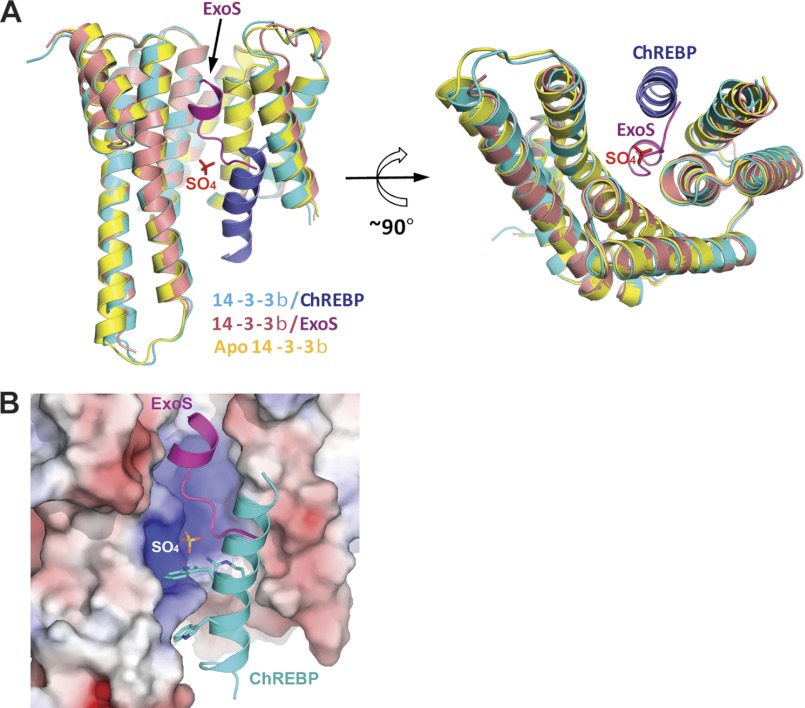
So far the only other available structure of a 14-3-3 protein bound to a nonphosphorylated peptide is that complexed to ExoS (22, 33). Comparing 14-3-3·ChREBP and 14-3-3·ExoS complex structures showed that the binding mode of ChREBP α2 helix to 14-3-3 is completely different from that of ExoS peptide (Fig. 5). The binding sequence of ChREBP α2, 121RLNNAIWRAWY131 (the underlined residues are those in contact with 14-3-3β), is very different from that of the ExoS peptide bound to 14-3-3 (421GLLDALDLASK431). In 14-3-3·ExoS complex structures, the bound ExoS peptide contains a single α-helical turn and binds to a hydrophobic patch at one side of the conserved peptide-binding groove of 14-3-3 mostly through hydrophobic interactions and does not involve Arg-58 or Arg-129 of the conserved Arg-Arg-Tyr triad (22). ChREBP α2, in contrast, binds to the positively charged patch of the peptide-binding groove of 14-3-3 through a sulfate/phosphate molecule (Fig. 5B), and to the residues on αC and αI helices at the edge of the groove. Thus, ChREBP α2 and ExoS represent two distinct and largely nonoverlapping phosphorylation-independent protein/peptide recognition modes by 14-3-3.
FIGURE 5.
Comparison of 14-3-3β·ChREBP with 14-3-3β·ExoS revealed two distinct nonphosphorylated peptide-binding modes. A, two orthogonal views of the superposition of 14-3-3β (cyan)·ChREBP (blue) complex with 14-3-3β (pink)·ExoS (magenta) complex. The apo-14-3-3β (yellow, Protein Data Bank code 2BQ0) in closed conformation is also superimposed. The sulfate in the complex of 14-3-3β·ChREBP is shown as sticks. B, ExoS peptide and ChREBP α2 bind to the nonoverlapping sites at the conserved peptide-binding groove of 14-3-3β. The molecular surface of 14-3-3β is shown and colored by electrostatic potentials. Negative potential is colored red and positive potential blue.
DISCUSSION
14-3-3 proteins bind to a wide variety of target proteins in response to activation of diverse cell signaling pathways, predominantly through recognition of phosphoserine or phosphothreonine sites with loosely conserved motifs RSXp(S/T)XP (mode 1) and RXXXp(S/T)XP (mode 2) (34, 35). There are a few exceptions in which target proteins bind to 14-3-3 independent of phosphorylation state. These include exoenzyme S (36), p190RhoGEF (37), R18 peptide inhibitor (38), as well as ChREBP. Two distinct 14-3-3-binding sites in the N-terminal regulatory region of ChREBP have been identified (16, 20, 26) as follows: a primary binding site located around α2 helix (residues 116–135) and a secondary binding site near the predicted α3 helix (residues 170–190) potentially covering a cAMP-dependent protein kinase (PKA) phosphorylation site at Ser-196. The N-terminal regulatory region of ChREBP is responsible for the glucose-regulated intracellular localization of full-length ChREBP, and in the absence of the C terminus acts as an independent glucose-responsive protein in nuclear export and import (16, 20). Previous reports have shown that 14-3-3 binding plays a vital role in glucose-regulated subcellular localization, thereby regulating the first essential step for transcriptional activation of ChREBP in response to glucose. However, how 14-3-3 interacts with ChREBP was not known. In this work, we took an unbiased approach to co-express and co-purify 14-3-3β with the entire N-terminal regulatory domain of ChREBP and determined the structure of the complex. We showed that 14-3-3β binds stably to a 115-residue fragment (residues 81–196) of ChREBP and that the complex is formed primarily through interactions with the α2 helix in a novel phosphorylation-independent manner. Several unique features set ChREBP α2 apart from all previously characterized 14-3-3 target protein-binding modes. First, the entire 21-residue peptide of ChREBP adopts a well defined helical conformation and is packed against two 14-3-3 helices (αC and αI) at the edge of the conserved peptide-binding groove. Second, a free sulfate or phosphate molecule mediates the interactions between ChREBP α2 and the positively charged patch of the binding groove on 14-3-3 involving the conserved Arg-Arg-Tyr triad. Third, several large aromatic side chains of α2 are intimately involved in both hydrogen bonding and van der Waals stacking interactions with 14-3-3. These observations provide insight into the structural basis for the high affinity binding of ChREBP α2 to 14-3-3β. In particular, the free sulfate/phosphate molecule appears to play a crucial role in this interaction between ChREBP and 14-3-3. Therefore, although 14-3-3 may bind to ChREBP constitutively independent of the phosphorylation state of ChREBP, this binding could be affected by the levels of phosphate or sulfate in cells. Conceivably, other metabolites may also be able to affect the interactions between 14-3-3 and ChREBP α2, which could provide an additional regulatory mechanism of ChREBP function, as discussed below.
The regulation of ChREBP in response to changing levels of glucose is complex, and it likely involves multiple steps. Previous studies have suggested that interactions of ChREBP with importin-α and 14-3-3 proteins are regulated by ChREBP phosphorylation and represent important regulatory mechanisms of glucose-responsive activation and inactivation of ChREBP (3, 19). Importantly, 14-3-3 was shown to inhibit importin-α binding to ChREBP. Careful characterization of the ChREBP importin-α-binding site identified it as an extended classical bipartite nuclear localization signal that minimally encompasses ChREBP residues 158–190 (3). This overlaps with the proposed secondary 14-3-3-binding site on ChREBP covering approximately residues 170–200. However, due to the lower affinity of 14-3-3 binding to this secondary site, and perhaps the proteolytic cleavage immediately after residue 196 during purification, the binding of 14-3-3 to the secondary site was not observed in the current structure. Different approaches will be needed to determine the structure of 14-3-3 bound to the secondary binding site of ChREBP.
In this study, the finding that 14-3-3 binds to ChREBP α2 with high affinity independent of phosphorylation highlighted a role for 14-3-3 in regulating ChREBP protein stability. ChREBP can be degraded by the ubiquitin-proteasome pathway in hepatocytes (15), and 14-3-3 binding to the N-terminal domains of ChREBP may act as a stabilizing factor preventing ChREBP from proteolytic degradation and thus help to maintain a relatively stable pool of ChREBP in the cytosol (16, 20, 21.). In support of this notion, the full-length and the N-terminal domains of ChREBP are extremely labile and susceptible to proteolysis when expressed in bacterial cells that do not express 14-3-3 proteins. However, ChREBP can be overexpressed and is stable in mammalian cells that do express 14-3-3. Moreover, during purification of ChREBP from liver or overexpressed ChREBP from mammalian cells, a significant amount of endogenous 14-3-3 protein remains bound to ChREBP. Results from the studies using ChREBP mutants with disruptions of either the α2- or α3-binding sites indicate that the majority of this endogenous 14-3-3 is bound to the primary α2 site (3, 20). Notably, the tight binding between ChREBP α2 and 14-3-3β is partially mediated by SO4 or PO4 as observed in the crystal structure. It is tempting to speculate that the interaction between ChREBP α2 and 14-3-3 could also be affected by other anionic metabolites in cells, which may affect the stability of ChREBP. The regulation of ChREBP protein turnover is an area that has not been extensively investigated so far. Knowledge of the structural interactions between ChREBP and 14-3-3 is critical for understanding glucose regulation of ChREBP function. The crystal structure of 14-3-3 bound to the primary binding site of ChREBP represents the first step toward that goal.
Acknowledgments
We thank Yuh Min Chook for helpful discussions. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK063948 from NIDDK. This work was also supported by a Veterans Affairs merit review grant, Welch Foundation Grant I-1720 (to K. U.), and American Heart Association Grant 10GRNT4310090 (to H. Z.).
The atomic coordinates and structure factors (code 4GNT) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ChREBP
- carbohydrate response element-binding protein
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. (2001) A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. U.S.A. 98, 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fukasawa M., Ge Q., Wynn R. M., Ishii S., Uyeda K. (2010) Coordinate regulation/localization of the carbohydrate-responsive binding protein (ChREBP) by two nuclear export signal sites. Discovery of a new leucine-rich nuclear export signal site. Biochem. Biophys. Res. Commun. 391, 1166–1169 [DOI] [PubMed] [Google Scholar]
- 3. Ge Q., Nakagawa T., Wynn R. M., Chook Y. M., Miller B. C., Uyeda K. (2011) Importin-α protein binding to a nuclear localization signal of carbohydrate response element-binding protein (ChREBP). J. Biol. Chem. 286, 28119–28127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uyeda K., Repa J. J. (2006) Carbohydrate response element-binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 4, 107–110 [DOI] [PubMed] [Google Scholar]
- 5. Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. (2004) Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. U.S.A. 101, 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horton J. D., Bashmakov Y., Shimomura I., Shimano H. (1998) Regulation of sterol regulatory element-binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. U.S.A. 95, 5987–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma L., Tsatsos N. G., Towle H. C. (2005) Direct role of ChREBP·Mlx in regulating hepatic glucose-responsive genes. J. Biol. Chem. 280, 12019–12027 [DOI] [PubMed] [Google Scholar]
- 8. Towle H. C. (2005) Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol. Metab. 16, 489–494 [DOI] [PubMed] [Google Scholar]
- 9. Iizuka K., Miller B., Uyeda K. (2006) Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am. J. Physiol. Endocrinol. Metab. 291, E358–E364 [DOI] [PubMed] [Google Scholar]
- 10. Denechaud P. D., Bossard P., Lobaccaro J. M., Millatt L., Staels B., Girard J., Postic C. (2008) ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J. Clin. Invest. 118, 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeong Y. S., Kim D., Lee Y. S., Kim H. J., Han J. Y., Im S. S., Chong H. K., Kwon J. K., Cho Y. H., Kim W. K., Osborne T. F., Horton J. D., Jun H. S., Ahn Y. H., Ahn S. M., Cha J. Y. (2011) Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE 6, e22544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Havula E., Hietakangas V. (2012) Glucose sensing by ChREBP/MondoA-Mlx transcription factors. Semin. Cell Dev. Biol. 23, 640–647 [DOI] [PubMed] [Google Scholar]
- 13. Bricambert J., Miranda J., Benhamed F., Girard J., Postic C., Dentin R. (2010) Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J. Clin. Invest. 120, 4316–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sakiyama H., Fujiwara N., Noguchi T., Eguchi H., Yoshihara D., Uyeda K., Suzuki K. (2010) The role of O-linked GlcNAc modification on the glucose response of ChREBP. Biochem. Biophys. Res. Commun. 402, 784–789 [DOI] [PubMed] [Google Scholar]
- 15. Guinez C., Filhoulaud G., Rayah-Benhamed F., Marmier S., Dubuquoy C., Dentin R., Moldes M., Burnol A. F., Yang X., Lefebvre T., Girard J., Postic C. (2011) O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60, 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. (2001) Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element-binding protein. Proc. Natl. Acad. Sci. U.S.A. 98, 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawaguchi T., Osatomi K., Yamashita H., Kabashima T., Uyeda K. (2002) Mechanism for fatty acid “sparing” effect on glucose-induced transcription. Regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J. Biol. Chem. 277, 3829–3835 [DOI] [PubMed] [Google Scholar]
- 18. Kabashima T., Kawaguchi T., Wadzinski B. E., Uyeda K. (2003) Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc. Natl. Acad. Sci. U.S.A. 100, 5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merla G., Howald C., Antonarakis S. E., Reymond A. (2004) The subcellular localization of the ChoRE-binding protein, encoded by the Williams-Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum. Mol. Genet. 13, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 20. Sakiyama H., Wynn R. M., Lee W. R., Fukasawa M., Mizuguchi H., Gardner K. H., Repa J. J., Uyeda K. (2008) Regulation of nuclear import/export of carbohydrate response element-binding protein (ChREBP). Interaction of an α-helix of ChREBP with the 14-3-3 proteins and regulation by phosphorylation. J. Biol. Chem. 283, 24899–24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Postic C., Dentin R., Denechaud P. D., Girard J. (2007) ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 22. Yang X., Lee W. H., Sobott F., Papagrigoriou E., Robinson C. V., Grossmann J. G., Sundström M., Doyle D. A., Elkins J. M. (2006) Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Obsil T., Obsilova V. (2011) Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672 [DOI] [PubMed] [Google Scholar]
- 24. Kleppe R., Martinez A., Døskeland S. O., Haavik J. (2011) The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell Dev. Biol. 22, 713–719 [DOI] [PubMed] [Google Scholar]
- 25. Obsilová V., Silhan J., Boura E., Teisinger J., Obsil T. (2008) 14-3-3 proteins: a family of versatile molecular regulators. Physiol. Res. 57, S11–S21 [DOI] [PubMed] [Google Scholar]
- 26. Li M. V., Chen W., Poungvarin N., Imamura M., Chan L. (2008) Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3. Mol. Endocrinol. 22, 1658–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L. C. (2005) Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol. Cell 20, 199–211 [DOI] [PubMed] [Google Scholar]
- 33. Ottmann C., Yasmin L., Weyand M., Veesenmeyer J. L., Diaz M. H., Palmer R. H., Francis M. S., Hauser A. R., Wittinghofer A., Hallberg B. (2007) Phosphorylation-independent interaction between 14-3-3 and exoenzyme S. From structure to pathogenesis. EMBO J. 26, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. (1997) The structural basis for 14-3-3·phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 35. Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., Yaffe M. B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153–166 [DOI] [PubMed] [Google Scholar]
- 36. Masters S. C., Pederson K. J., Zhang L., Barbieri J. T., Fu H. (1999) Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38, 5216–5221 [DOI] [PubMed] [Google Scholar]
- 37. Zhai J., Lin H., Shamim M., Schlaepfer W. W., Cañete-Soler R. (2001) Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 276, 41318–41324 [DOI] [PubMed] [Google Scholar]
- 38. Wang B., Yang H., Liu Y. C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999) Isolation of high affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 [DOI] [PubMed] [Google Scholar]