Background: DNAJA2 functions with Hsc70 to assist protein folding.
Results: An internal structure in DNAJA2, conserved in DNAJA1, is essential for folding activity and release of substrate triggered by the Hsc70 ATPase.
Conclusion: Substrate transfer from DNAJA2 to Hsc70 is a specific process fundamental to their folding mechanism.
Significance: The folding competence of Hsc70 may be determined by the DNAJs.
Keywords: ATPases, Chaperone Chaperonin, Heat Shock Protein, Protein Degradation, Protein Folding, DNAJ, Hsp40, Hsp70, J Domain
Abstract
DNAJA1 (DJA1/Hdj2) and DNAJA2 (DJA2) are the major J domain partners of human Hsp70/Hsc70 chaperones. Although they have overall similarity with the well characterized type I co-chaperones from yeast and bacteria, they are biologically distinct, and their functional mechanisms are poorly characterized. We identified DJA2-specific activities in luciferase folding and repression of human ether-a-go-go-related gene (HERG) trafficking that depended on its expression levels in cells. Mutations in different internal domains of DJA2 abolished these effects. Using purified proteins, we addressed the mechanistic defects. A mutant lacking the region between the zinc finger motifs (DJA2-Δm2) was able to bind substrate similar to wild type but was incapable of releasing substrate during its transfer to Hsc70. The equivalent mutation in DJA1 also abolished its substrate release. A DJA2 mutant (DJA-221), which had its C-terminal dimerization region replaced by that of DJA1, was inactive but retained its ability to release substrate. The release mechanism required the J domain and ATP hydrolysis by Hsc70, although the nucleotide dependence diverged between DJA2 and DJA1. Limited proteolysis suggested further conformational differences between the two wild-type co-chaperones and the mutants. Our results demonstrate an essential role of specific DJA domains in the folding mechanism of Hsc70.
Introduction
Chaperones of the Hsp70 family are highly conserved and have a central role in assisting polypeptide folding (1). The human cytosolic forms, constitutively expressed Hsc70 (HSPA8) and stress-induced Hsp70 (HSPA1A/B), also fulfill other functions, including selecting misfolded proteins for ubiquitin-dependent proteasomal degradation (2). The Hsp70 chaperones have a conserved ATPase cycle in which ATP hydrolysis to ADP results in binding of unfolded polypeptide substrate, and exchange of ADP for ATP causes release of substrate. The N-terminal nucleotide binding domain of an Hsp70 allosterically regulates the C-terminal substrate binding domain, in particular by interdomain contacts in the ATP state and by conformational changes during ATP hydrolysis, which are less well understood (3–8). The ATPase cycle of Hsp70 chaperones is further regulated by the DNAJ co-chaperones, which stimulate ATP hydrolysis, as well as a variety of nucleotide exchange factors (NEFs).4 Substrate folding is thought to require multiple cycles of binding and release by an Hsp70 (2, 3, 9).
DJA1 (DNAJA1/Hdj2) and DJA2 (DNAJA2) are the most abundant DNAJ co-chaperones of human Hsc70 (10–12). Both DJA1 and DJA2 share a domain architecture with the well characterized DnaJ from Escherichia coli and Ydj1 from Saccharomyces cerevisiae and are classified as type I DNAJs (2, 9). They have an N-terminal J domain, which like other conserved J domains binds Hsc70 to activate ATP hydrolysis and substrate binding by the chaperone. The ∼12-kDa J domain is connected by a partially conserved linker to an ∼34-kDa middle/C-terminal (MC) domain that contains two zinc finger motifs and a substrate-binding site (8, 10, 13, 14). Based on DnaJ and Ydj1, a model of function for the type I co-chaperones has been proposed in which a DNAJ binds substrate, contacts an Hsp70 through its J domain, and transfers substrate to the Hsp70 during ATP hydrolysis while dissociating from the complex (2, 3, 15, 16). This model is thought to apply to DJA1 and DJA2, at least in its broad outlines. We previously found that purified DJA2 and Hsc70 are sufficient for fast substrate folding. NEFs are not required, although they can enhance folding efficiency (13, 14). However, the mechanisms by which DJA2 binds and transfers substrate to Hsc70, and promotes substrate folding, have not been determined.
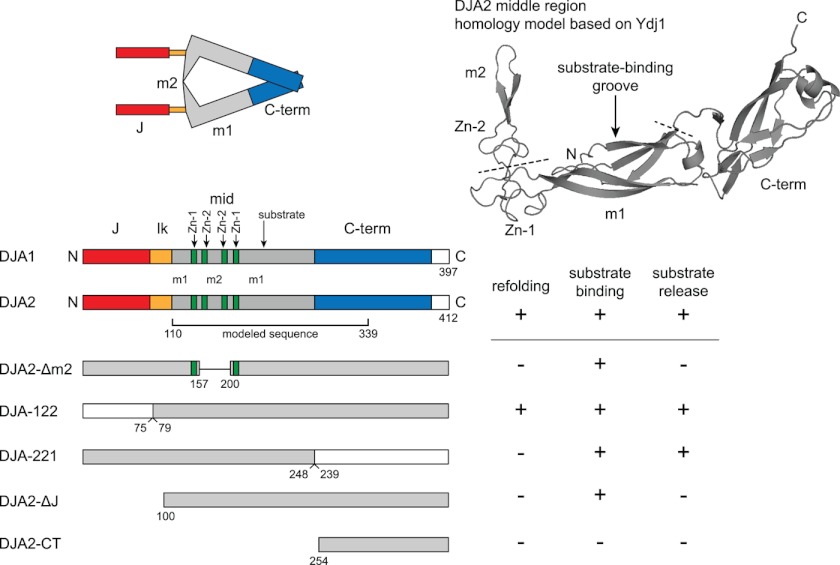
The function of DJA2 is expected to involve the structures within the MC domain. Studies with DnaJ and Ydj1, using mutations and chemical removal of the zinc ions, showed that the zinc finger motifs were necessary for function (16–18). The zinc-binding sites were suggested to contribute to substrate transfer to their Hsp70 partners (16). A crystallographic structure of a middle fragment of Ydj1 revealed three subdomains as follows: one with a putative substrate-binding site (middle 1 or m1); the zinc-binding sites forming a corner to another subdomain of unknown function (m2), and a C-terminal subdomain at the opposite end (Fig. 1 shows a homology model of DJA2 based on this structure) (19). Another crystallographic structure showed that the C-terminal subdomain extends into the homodimerization interface (20). The m1 subdomain can bind peptides, although additional substrate-binding sites are still possible (19, 21). Small angle x-ray scattering analyses of Ydj1 and DJA1 suggest the MC domains have a triangle shape with the m2 subdomains forming one side, close to the J domains and linkers (22, 23). DJA2 is expected to be similarly arranged (Fig. 1). The proximity of its m2 and J domains suggests they may functionally interact, possibly for substrate transfer to Hsc70, but to date experimental evidence is lacking.
FIGURE 1.
Domain architecture of DJA2. Top left, proposed arrangement of domains in DJA2 based on Ref. 2. Top right, a homology model of DJA2 was generated by SwissModel using the middle fragment of Ydj1 (1NLT) as template. The N and C termini, m1 and m2 subdomains, zinc-binding sites (Zn-1 and Zn-2), and partial C-terminal (C-term) dimerization region are marked. The positions of the Δm2 deletion and the C-terminal exchange with DJA1 are indicated by dashed lines. Bottom left, diagram of the domain arrangements in the DJA1 and DJA2 primary sequence, and mutants used in this study. The J domain (J), linker (lk), m1 and m2 sequences, zinc-binding sites (Zn-1 and Zn-2), and C-terminal dimerization sites are marked. Bottom right, summary of the results of this study.
Substrate transfer has recently been investigated for the endoplasmic reticulum (ER) lumenal co-chaperones p58 (DNAJC3) and ERdj3 (DNAJB11) and their partner Hsp70, BiP (HSPA5). ERdj3, although lacking the zinc-binding sites, is otherwise homologous to the type I co-chaperones. In contrast, p58 is conserved only in its J domain with a substrate binding region unrelated to other DNAJs, and it is classed as type III (24, 25). Dissociation of p58 from immobilized substrate required ATP and the J domain interaction with BiP. A p58 mutant with the J domain fused in a different location from the wild type could still release, but a BiP mutant defective in substrate binding could not (25). Dissociation of ERdj3 from immobilized substrate similarly required the J domain interaction with BiP. Moreover, formation of the ternary complex was not sufficient for substrate release from ERdj3, but this required stimulated ATP hydrolysis by BiP (24). Lack of substrate release from these DNAJs was associated with defects in their folding function in cells, suggesting that release is mechanistically important.
To gain further insight into the mechanistic requirements for substrate folding, we constructed a series of DJA2 mutants with disruptions in its internal structures. The competence of the mutants to support substrate folding was determined in cells and as pure proteins. We found that the function of DJA2 was abolished by deletion of its m2 subdomain and by exchanging its C-terminal dimerization region with that of DJA1. Using the pure proteins, we tested the binding of the DJA2 constructs to substrate and the release of the constructs from substrate upon addition of Hsc70 and ATP. The m2 deletion mutant was particularly impaired in its release from substrate. The nucleotide requirements for DJA2 release suggested a mechanism coupled to the Hsc70 ATPase cycle, and this coupling may differ for DJA1. Our results suggest a model of DJA2 function in which there is a coordination of the binding and release of substrate with different domains of DJA2 to promote folding.
EXPERIMENTAL PROCEDURES
Plasmids
pPROEX-HTa (Clontech) and pcDNA3.1 Myc-His (Invitrogen), encoding DJA2, DJA2-ΔJ, DJA1, DJA1-ΔJ, Hsc70, and Hsp110 in pPROEX-HTa, were as described previously (13, 14, 26). Hsp70 was amplified by PCR from pSV-Hsp70 (27) and inserted with an N-terminal FLAG tag into pcDNA3.1. Hsc70-K71M was amplified by PCR from pET-Hsc70K71M (28) and inserted into pPROEX-HTa. The pET plasmid encoding Myc-His-tagged firefly luciferase was as described previously (29). Plasmid pRK793 encoding tobacco etch virus protease (30) was obtained from Dr. Bhushan Nagar. Luciferase was amplified by PCR and inserted into pcDNA3.1 with a C-terminal HA tag. DNA purification kits were from Qiagen; restriction enzymes and other enzymes were from New England Biolabs; Expand long template PCR system was from Roche Applied Science; and QuikChange multisite-directed mutagenesis kit was from Stratagene. All constructs were verified by sequencing.
DJA2-Δm2 and DJA1-Δm2
The m2 fragment from residues 158 to 199 of DJA2 was removed by PCR and replaced with an EcoRI site encoding Glu-Phe. The corresponding fragment from residues 149 to 190 of DJA1 was similarly replaced with an XbaI site encoding Ser-Arg. The mutants were constructed in pPROEX-HTa and subcloned into pcDNA3.1 Myc-His as BamHI-NotI fragments.
DJA-221
The EcoRV site at amino acids Asp-299/Ile-300 of DJA1 was removed by silent mutation. EcoRV sites were reintroduced in DJA1 by silent mutation at Asp-238/Ile-239 and in DJA2 at Asp-247/Ile-248. The sequences encoding the N-terminal fragments were exchanged as BamHI-EcoRV fragments in pPROEX-HTa and subcloned into pcDNA3.1 as BamHI-NotI fragments.
DJA-122
A BamHI site was inserted into DJA1 by point mutation at Gly-75/Gly-76 mutating it to encode Gly-75/Ser-76. A BamHI site was inserted by silent mutation into DJA2 at Gly-78/Ser-79. DJA1 and DJA2 were re-inserted into pPROEX-HTa as SalI-NotI fragments to remove the BamHI site before the open reading frame. The C-terminal fragments were exchanged as BamHI-NotI fragments in pPROEX-HTa and subcloned into pcDNA3.1 as KpnI-NotI fragments.
DJA2-CT
The sequence encoding DJA2 from residue 254 to the C terminus was inserted into pcDNA3.1 as a BamHI-NotI fragment.
Protein Expression and Purification
All columns and bulk chromatography media were from GE Healthcare, except for HTP-gel hydroxyapatite from Bio-Rad. Rosetta 2 E. coli cells (Novagen) transformed with plasmid encoding the proteins were grown in TB medium at 37 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at OD (600 nm) around 1. Expression and purification of DJA1 and DJA2 and Hsc70 and Hsp110 were as described previously (13, 14), except that a Sephacryl S-300 26/60 high resolution column was used for the DJAs and a HiTrap Q column for Hsc70 and Hsp110. Hsc70-K71M was expressed and purified identically to Hsc70. The His tag was removed from Hsc70 by overnight incubation with purified His-tagged tobacco etch virus protease at 4 °C, followed by removal of the protease with nickel-Sepharose. For DJA2 mutants, purification was similar to DJA2, but expression time and temperature varied. DJA-221 was expressed at 37 °C for 1 h; DJA2-Δm2, DJA-122, and DJA2-ΔJ were at 30 °C for 1 h. Luciferase was expressed at 30 °C for 3 h, purified by nickel-Sepharose chromatography, and dialyzed overnight in buffer G containing 100 mm KOAc, 20 mm HEPES-KOH, pH 7.5, and 5 mm MgOAC2. Purified proteins were analyzed on SDS-PAGE for purification quality, and protein yield was determined by absorbance at 280 nm. Adenine nucleotide translocator (ANT) was purified from cow heart as described previously (13).
For limited proteolysis, 4 μm of purified DJAs in 500 mm NaCl, 20 mm HEPES-KOH, pH 7.5, and 5 mm MgOAc2 were treated for 10 min at 4 °C with various concentrations of trypsin or chymotrypsin as indicated. Digestion was stopped by adding 1 mm PMSF and Laemmli SDS-PAGE loading buffer. Samples were analyzed by SDS-PAGE and Coomassie staining or transferred onto PVDF and Ponceau staining. Edman degradation was performed by the Advanced Protein Technology Centre at the Hospital for Sick Children in Toronto.
For circular dichroism (CD) spectroscopy, DJA samples were desalted into buffer H containing 500 mm NaCl, 20 mm KH2PO4, pH 7.5, and 5 mm MgOAc2 using Micro Bio-Spin 6 chromatography columns (Bio-Rad). Measurements were performed on a Chirascan CD spectrometer with a 0.5-mm quartz cell. Protein concentration was 0.5 mg/ml in a total volume of 150 μl. CD wavelength scans were obtained at 1 nm step size and at 0.5-s time intervals. The signals obtained were in CD millidegrees and are represented as an average of three spectra obtained for each protein. To obtain melting curves, samples were measured in increasing concentrations of urea as indicated.
Refolding and ATPase Assays
Refolding of guanidine denatured luciferase with purified chaperones was as described previously (14). Final concentrations of proteins were 4 μm for Hsc70 and DJAs, 1 μm Hsp110 where indicated, and 5 nm luciferase.
For pulldown experiments, reactions were set up as for refolding except with 50 nm denatured luciferase and Hsc70 that had its His tag removed by tobacco etch virus protease digestion (14). The denatured luciferase was incubated with 4 μm wild-type or mutant DJA2 for 5 min, before addition of 4 μm Hsc70 and 2 mm ATP or ADP. At the indicated time points, reactions were stopped with 0.1 unit/μl apyrase, chilled to 4 °C, and diluted with buffer G containing 50 mm imidazole, 0.1% Triton X-100, and 1 mg/ml ovalbumin. Reactions were allowed to bind to nickel-Sepharose, washed with buffer G containing 50 mm imidazole and 0.1% Triton X-100, and eluted with Laemmli SDS-PAGE loading buffer.
Single-turnover ATPase experiments were based on a published procedure (31) with modifications. Purified Hsc70 was incubated with 5 μCi/ml [α-32P]ATP (PerkinElmer Life Sciences) for 5 min on ice. Hsc70-ATP complexes were separated from free ATP using Micro Bio-Spin 6 chromatography columns and flash-frozen until use. Purified DJAs (4 μm) were added on ice to Hsc70-ATP complexes (4 μm) in final conditions of buffer G containing 40 mm NaCl. The mixture was incubated at 30 °C to start Hsc70 ATPase activity. Aliquots were taken at various time points and inactivated with 40 mm EDTA and then loaded on polyethyleneimine cellulose thin layer chromatography plates (Fisher) and developed in 0.5 m LiCl and 0.5 m formic acid (14). The plates were exposed on a phosphorimager screen, and the ADP produced was quantified with a Typhoon scanner and ImageQuant software (GE Healthcare). Linear ATPase rates were calculated by regression (% ADP per min) with the amount of ADP produced over time.
ELISA Experiments
DJA2 Binding
0.5 μg of nontagged luciferase (Sigma) in 75 μl of PBS was immobilized on Maxisorp 96-well plates (Nunc) overnight at 4 °C. The following steps were performed at room temperature with a total volume of 75 μl. Unbound luciferase was washed three times with PBS containing 0.05% Tween 20 (PBST), and the immobilized luciferase was denatured with 8 m guanidinium chloride and 1 mm DTT in PBS for 15 min. Samples were washed with PBST three times, and blocking solution (PBS containing 0.5% BSA) was added and incubated for 45 min. Then wild-type or mutant DJA2 was added at various amounts in buffer H and allowed to bind for 45 min. Unbound DJA2 was removed, and samples were washed three times with PBST before primary antibody in the blocking solution was added for 45 min. The DJA2 and luciferase were decorated with antibodies against the His tag (Bioshop) and luciferase (Sigma), respectively. After washing three times with PBST, secondary antibody was added in PBST for 45 min. Goat anti-mouse IgG-conjugated HRP (Sigma) was used against the luciferase and His tag antibodies. After washing five times with PBST, Amplex Red reagent (Invitrogen) and hydrogen peroxide were prepared as described by the manufacturer and added onto the wells. The fluorescence signal was detected using a POLARstar BMG Labtech plate reader, with excitation at 544 nm and emission at 590 nm. Alternatively, Myc-His-tagged luciferase was used and decorated with antibodies against the Myc tag (clone 9E10, Covance), and DJA2 was decorated with specific polyclonal antibodies (13) and detected with goat anti-rabbit IgG-conjugated HRP (Jackson ImmunoResearch) with similar results as the first antibody set. Experiments with ANT were performed identically using 0.5 μg of ANT.
DJA2 Release and Hsc70 Binding
DJA2 binding reactions were carried out as above using 0.5 μg of Myc-His-tagged luciferase and 3 μg of wild-type or mutant DJA2 added for 45 min. Unbound DJA2 was removed, and samples were washed three times with PBST. 3 μg of Hsc70 in buffer G with 40 mm NaCl and 2 mm of the indicated nucleotides were added to wells with pre-bound DJAs as well as control wells with ATP and only denatured luciferase. Hsc70 was allowed to bind for 45 min at room temperature, and then unbound proteins were removed and samples washed three times with PBST. Primary and secondary antibody steps as well as detection of fluorescent signals were performed as above. For the primary antibodies we used anti-DJA2, anti-Hsc70 (StressMarq), and anti-Myc against luciferase. For the secondary antibodies we used goat anti-rabbit IgG-conjugated HRP against anti-DJA2 and goat anti-mouse IgG-conjugated HRP against anti-Myc and anti-Hsc70.
Luciferase Refolding in Cells
HeLa and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM), high glucose, and glutamine (Invitrogen) supplemented with 10% fetal bovine serum, 1 mm sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were cultured at 37 °C in 5% CO2. Plasmid transfections were carried out using Lipofectamine 2000 (Invitrogen).
Luciferase refolding assays were adapted from published procedures (12, 32). HEK293 cells were transfected with 2 μg of luciferase plasmid and 8 μg of DJA2 plasmid or control vector per 60-mm dish. One day after transfection, cells were treated with cycloheximide (Sigma) at 50 μg/ml to inhibit protein synthesis, transferred to 45 °C for 1 h, and then brought back to 37 °C for up to 2 h recovery. For proteasome inhibition, MG132 (Sigma) was added during the heat shock and recovery phases at 5 μg/ml. Cell samples, taken before heat shock and at 0, 1, and 2 h of recovery, were lysed on ice with PBS containing 1% Triton X-100, and the insoluble material was removed by centrifugation at 20,000 × g. The pellets were resuspended in PBS containing 0.1% Triton X-100, treated with 2 units of DNase I (New England Biolabs) at 37 °C for 30 min, and then denatured with 2% SDS. The total protein amounts in the supernatants and pellets were determined using the BCA protein assay (Pierce). Luciferase enzymatic activities in the supernatants were measured using the luciferase reporter assay kit (Promega), and activity values were normalized to total protein amounts. Equal amounts of protein were analyzed on immunoblots. Antibodies used were anti-FLAG-M2 (Sigma) to detect FLAG-tagged Hsp70, anti-HA (HA.11 clone 16B12, Covance) to detect HA-tagged luciferase, and anti-DJA1 and anti-DJA2 or anti-Myc to detect Myc-tagged DJA1 and DJA2. Detection was with ECL or ECL Plus reagent (GE Healthcare) visualized on film or for quantitation by a FluorChem HD2 digital camera and the FluorChem HD2 software (Alpha Innotech). For siRNA knockdowns, cells were transfected with 100 nm nonsilencing control duplex or duplex targeting DJA1 or DJA2 using DharmaFECT 1 (Dharmacon, ThermoScientific). The siRNAs targeted the sequences GTGAAGGACTGTAATCATA and CCACAAAGCTTTACATCTT (Dharmacon Custom siRNA Designer) in the 3′ UTRs of DJA1 and DJA2, respectively, and the nonsilencing duplex was the Dharmacon OnTarget Plus Control 1. Two days after siRNA transfection, cells were transfected with plasmids and tested for refolding as above.
For immunoprecipitations, HEK293 cells were transfected with the DJA2 plasmids (24 μg of DNA per 100-mm dish) using Lipofectamine 2000. One day after transfection, cells were lysed in PBS containing 1% Triton X-100; the lysates were adjusted to identical protein concentrations and volumes, and then anti-Myc and protein G-agarose fast flow (Upstate) were added to the lysates for incubation overnight at 4 °C. The beads were washed with PBS containing 0.1% Triton X-100 and eluted with Laemmli SDS-PAGE loading buffer.
HERG Trafficking
Assays for the trafficking of human ether-a-go-go-related gene (HERG) were based on previous work (26, 33, 34). HEK293 cells were transfected with 6 μg of HERG plasmid and 6 μg of DJA2 plasmid or control vector per 10-cm dish and distributed into 6-cm dishes following the transfection. Two days after transfection, the cells were starved for 1 h in DMEM lacking amino acids and serum and then labeled for 45 min with 100 μCi of [35S]methionine/cysteine labeling mix (PerkinElmer Life Sciences). The cells were washed with PBS, incubated with regular medium for the indicated times and then lysed, and immunoprecipitated as above except with antibody against HERG (Chemicon/Millipore). SDS-PAGE autoradiograms were analyzed by phosphorimager as above.
RESULTS
DJA2 Refolding Function
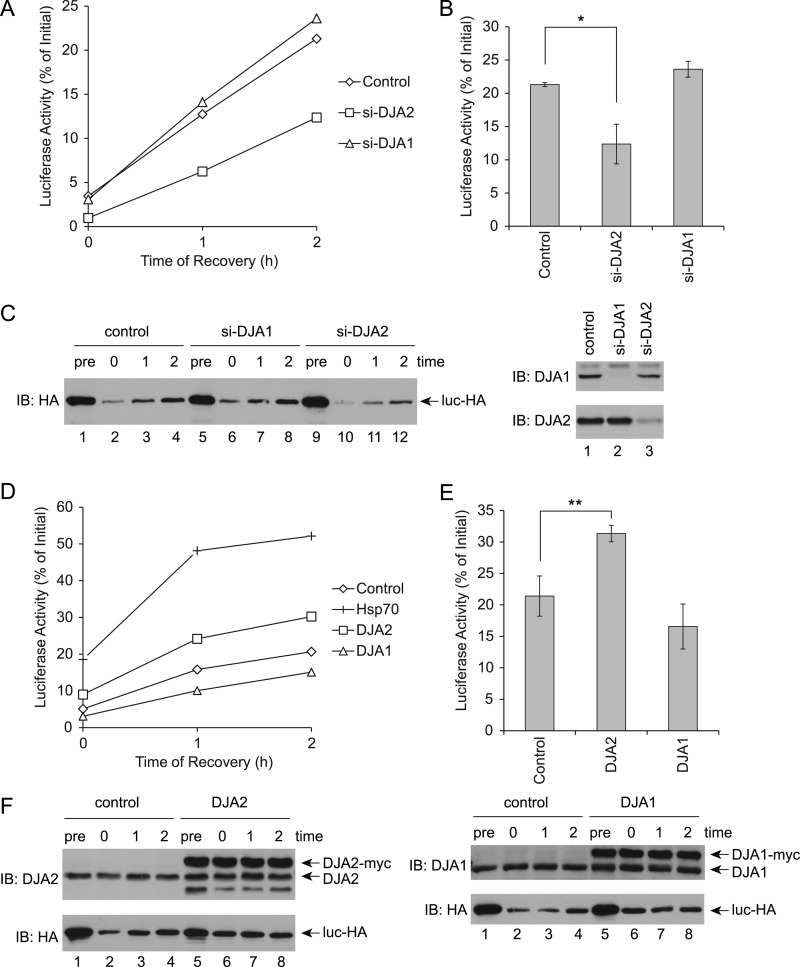
Our previous work showed that purified DJA2 was sufficient for efficient refolding of guanidine-denatured luciferase by Hsc70, whereas DJA1 was ineffective (13, 14). We now asked if this function of DJA2 was relevant in the cellular environment. An earlier assay for stress-inducible Hsp70 and DNAJB1 (32) was adapted to HEK293 cells. Transfected luciferase was denatured by a 45 °C heat shock for 1 h, and new protein synthesis was inhibited with cycloheximide. Luciferase refolding was initiated by allowing the cells to recover at 37 °C, and its enzymatic activity was monitored in cell lysates obtained at different time points. The heat shock reduced luciferase activity to ∼5% of the initial level before heat shock, and refolding reached its maximum at ∼20% by 2 h of recovery (Fig. 2, A and B). DJA1 and DJA2 levels were then depleted by siRNA knockdown, to below 25 and 40%, respectively, of levels in control cells (Fig. 2C). Consistent with our previous pure protein results, DJA2 knockdown significantly impaired refolding to ∼10% after 2 h, whereas DJA1 knockdown had no effect (Fig. 2, A and B). The levels of soluble luciferase in the Triton X-100-extracted lysates were also monitored by immunoblot. The amount of substrate was reduced by heat shock, due to either aggregation or degradation, but increased moderately over the 2-h recovery (Fig. 2C). The increase may be due to re-solubilization of aggregated material, as radiolabeling control experiments confirmed that cycloheximide inhibited translation to less than 1% (data not shown). Knockdown of DJA2 reduced somewhat the amount of soluble substrate during recovery (Fig. 2C).
FIGURE 2.
Refolding of denatured luciferase in cells. A, HEK293 cells were transfected with a control nonsilencing duplex, or siRNA duplexes targeting DJA2 (si-DJA2) or DJA1 (si-DJA1), and then transfected with luciferase. The cells were treated with cycloheximide and incubated at 45 °C for 1 h and then allowed to recover at 37 °C. Cells were lysed at the indicated times of recovery, and luciferase enzymatic activity in the lysates was measured. Luciferase activity during recovery was represented as a percentage of the initial activity before heat shock. B, average luciferase activities at 2 h of recovery. C, expression of luciferase and the DJAs detected by immunoblot (IB). The cell lysates either before heat shock (pre) or at 0, 1, and 2 h of recovery were probed with antibodies against the HA tag on luciferase. Lysates before heat shock were probed with antibodies against DJA2 or DJA1. D, HEK293 cells were transfected with luciferase and either control vector, Hsp70, DJA2, or DJA1. Luciferase refolding after heat shock was assayed as in A. Luciferase activities with the indicated transfections at different time points were plotted. E, average refolding at 2 h of recovery. F, expression of Myc-tagged DJA2 or DJA1 and luciferase at different time points. In this and all figures, error bars show standard deviations from the average of at least three independent experiments. ** denotes a difference with p < 0.01; * denotes a difference with p < 0.05.
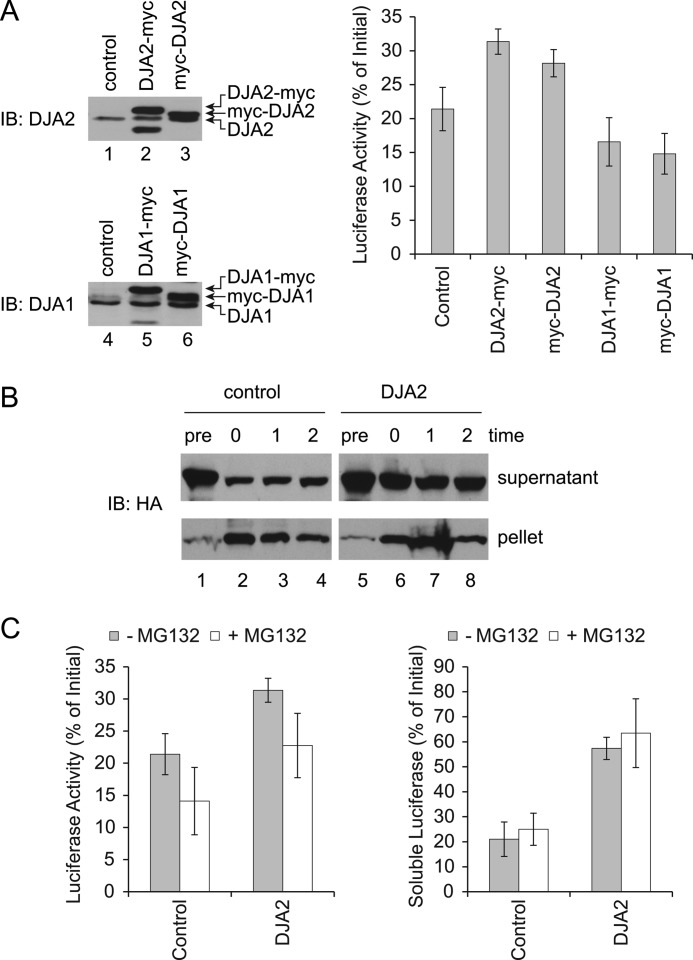
The function of transfected chaperones was then tested. As expected, transfection of Hsp70 increased the refolding yield to ∼50% of starting activity (Fig. 2, D and E). Hsc70 produced a similar although less pronounced effect, but it was found to express at 2- to 3-fold lower levels than Hsp70, suggesting it was functionally identical (data not shown). DJA2 also significantly increased the refolding of substrate to ∼30% (Fig. 2, D and E). In contrast, DJA1 had no significant effect. The expression of transfected DJA2 and DJA1 was at or somewhat above endogenous levels, and it remained unchanged during the experiment (Fig. 2F). Our C-terminally Myc-tagged DJA2 and DJA1 had their predicted farnesylation site blocked (10), and because farnesylation of Ydj1 is required for some of its functions (35, 36), we compared the refolding activity of the N-terminally Myc-tagged, farnesylated co-chaperones. We found that DJA2 was equally active in refolding with either an N- or C-terminal Myc tag, which suggests that the farnesylation was not important for this particular function. N-terminally Myc-tagged DJA1 remained nonfunctional (Fig. 3A). Our results suggested a refolding function specific to DJA2 compared with DJA1, which validates our previous in vitro experiments.
FIGURE 3.
A, refolding with Myc-tagged DJAs in cells. HEK293 cells were transfected with luciferase and N- or C-terminally tagged forms of DJA2 or DJA1, and luciferase refolding after heat shock was measured as in Fig. 2D. The expression levels of the indicated DJAs before heat shock were detected by immunoblot (IB). Luciferase activities at 2 h of recovery were measured. B, detection of soluble and insoluble luciferase in cell lysates. Luciferase refolding in HEK293 cells transfected with control vector or DJA2 was performed as in Fig. 2D. Cell lysates were extracted in Triton X-100 followed by centrifugation, and the amounts of HA-tagged luciferase in the supernatants and pellets were analyzed by immunoblot. C, refolding with proteasome inhibition. Luciferase refolding in HEK293 cells transfected with control vector or DJA2 was performed as in B, with or without MG132. Soluble HA-tagged luciferase was detected by immunoblot, and luciferase enzymatic activities were measured at 2 h of recovery. Quantitation of the HA signal at 2 h of recovery is shown.
DJA2 transfection reduced the loss of luciferase from the soluble fraction during heat shock, and the level of soluble luciferase then remained constant during recovery as its activity increased (Fig. 2F). DJA1 transfection also reduced the loss of soluble luciferase during heat shock and recovery, although luciferase activity was not increased compared with the control. We further analyzed the detergent-insoluble fraction from control and DJA2-transfected cells and, as expected, found an increase in aggregated substrate after the heat shock, with its levels persisting during the refolding phase (Fig. 3B). To test whether degradation of substrate was involved, the experiment was repeated with the proteasome inhibitor MG132. The amounts of soluble substrate were not affected by MG132, suggesting that loss of substrate during heat shock was due to aggregation rather than degradation. However, refolding was moderately decreased by MG132, with and without DJA2 transfection (Fig. 3C). This may be due to competition for the chaperones by other misfolded substrates that would normally be degraded. Overall, our results show an activity of both DJA2 and DJA1 in aggregation prevention. However, this activity was not sufficient for refolding, which was only promoted by DJA2.
Refolding Function of DJA2 Mutants
We constructed mutants to test the functional importance of different parts of DJA2. To identify structures within the MC region, a homology model was derived using the crystallographic structure of the highly similar Ydj1 central fragment as a template (Fig. 1) (19, 37). A mutant was constructed with deletions of the m2 subdomain and the second zinc-binding site (DJA2-Δm2), but the first zinc-binding site was left intact to avoid destabilization of the remaining protein (Fig. 1). We also constructed a mutant that replaced the C-terminal dimerization region of DJA2 with that of DJA1 (DJA-221). In other mutants, the J domain of DJA2 was replaced with that of DJA1 (DJA-122) or was completely deleted (DJA2-ΔJ). Because of the strong similarity (68%) between DJA2 and DJA1, the domain exchanges in DJA-122 and DJA-221 were not expected to be greatly disruptive. A final mutant contained only the C-terminal dimerization site (DJA2-CT).
The functions of the DJA2 mutants were then investigated in cells. Interestingly, DJA-122 appeared to be functional, whereas all of the other mutants did not promote refolding above the vector control (Fig. 4A). Although these results may have been expected for the DJA2-ΔJ and DJA2-CT mutants, the loss of function of DJA2-Δm2 and DJA-221 was notable. The overall pattern of soluble substrate before and after heat shock was observed to be similar to that of cells expressing wild-type DJA2, although the amounts remaining after heat shock varied between the mutants (Fig. 4B). The expression levels of all the mutants were comparable with or slightly above endogenous DJA2 (Fig. 4B). Like wild-type DJA2, the amounts of the mutants remained constant during the experiments (data not shown). Thus, within the MC domain, the m2 and C-terminal subdomains appear to be critical for DJA2 function.
FIGURE 4.
Refolding with DJA2 mutants in cells. A, HEK293 cells were transfected with luciferase and control vector or wild-type or mutant DJA2, and refolding after heat shock was measured as in Fig. 2D. Average luciferase activities at 2 h of recovery were plotted. Results with significant differences from those with wild-type DJA2 are marked (** denotes p < 0.01). B, expression of wild-type and mutant DJA2 was analyzed by immunoblot (IB). Endogenous DJA2 and DJA1 was visible in control transfected cells, and mutant DJA2 as bands with different migrations. The positions of molecular weight markers are shown on the left. Wild-type DJA2 produced a band corresponding to the full-length protein (black arrowhead) and a faster migrating band that appeared to be a truncated form (white arrowhead) detected by the antibody against DJA2. DJA2-Δm2 and DJA-122 also produced a full-length and truncated form, similarly marked. DJA-221 was detected by antibody raised against the DJA1 C terminus. Right, HA-tagged luciferase was detected at different time points.
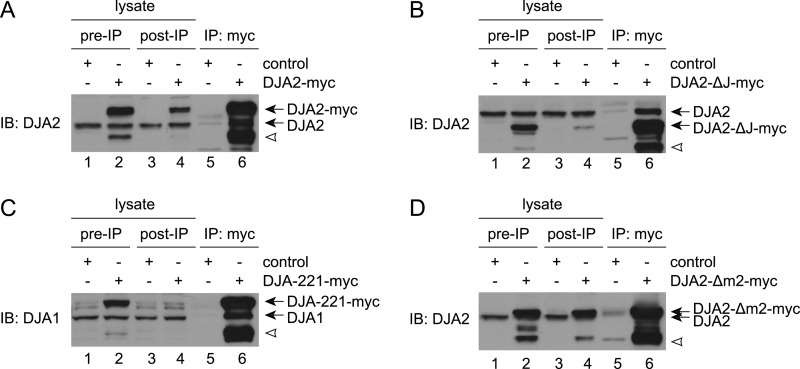
It was conceivable that the mutants were forming heterodimers with endogenous DJA2 or DJA1. We examined this possibility by immunoprecipitation of the transfected Myc-tagged DJA2 constructs and detection of co-precipitated endogenous DJA2 or DJA1. As hypothesized, endogenous DJA2 was co-precipitated with transfected DJA2 and DJA2-ΔJ, and DJA1 with transfected DJA-221 (Fig. 5, A–C). Endogenous DJA2 also appeared to co-precipitate with DJA2-Δm2, although the bands were less well separated (Fig. 5D). Although heterodimers could be partially active, the loss of function observed with DJA2-Δm2 and DJA2-ΔJ suggests that lack of a single m2 or J domain in DJA2 is detrimental. Similarly, DJA221 dimers with DJA1 appear to be less active than DJA2.
FIGURE 5.
A, co-immunoprecipitation of endogenous and transfected DJA2. HEK293 cells were transfected with control vector or Myc-tagged DJA2. Cells were lysed and immunoprecipitated (IP) with antibodies against the Myc tag (lanes 5 and 6). 10-μg samples of cell lysates before (lanes 1 and 2) and after (lanes 3 and 4) the immunoprecipitation reactions were also analyzed. DJA2 was detected by immunoblot (IB). The positions of endogenous and Myc-tagged DJA2 are marked, as well as a truncated form of Myc-tagged DJA2 (white arrowhead). B, co-IP of endogenous DJA2 with DJA2-ΔJ, as in A. C, co-IP of endogenous DJA1 with DJA-221, as in A except detected with antibodies against DJA1. D, co-IP of endogenous DJA2 with transfected DJA2-Δm2, as in A.
DJA2 Mutants and HERG Trafficking
We previously identified a function of DJA2 in ER-associated degradation of the HERG potassium channel by the ubiquitin-proteasome system. HERG is inserted into the ER and core-glycosylated (CG) during its synthesis and then modified to the fully glycosylated (FG) form upon trafficking through the Golgi to the cell surface. Like other secretory proteins, trafficking of HERG is thought to require its productive chaperone-dependent folding at the ER (33, 34, 38). In contrast to luciferase refolding, knockdown of DJA1 but not DJA2 reduced HERG trafficking. However, DJA2 overexpression also reduced trafficking in a proteasome-dependent manner and increased HERG ubiquitylation. A model was proposed in which DJA2 prolonged HERG association with Hsc70 leading to ubiquitylation by the Hsc70-associated E3 ligase CHIP (26).
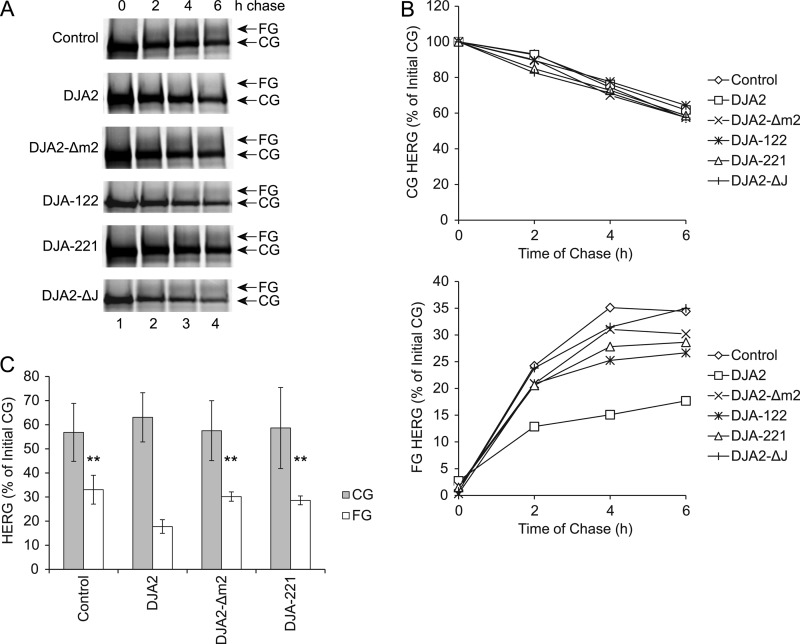
We asked whether our refolding-defective DJA2 mutants were still functional in inhibiting HERG trafficking by examining the kinetics of HERG trafficking and degradation with pulse-chase experiments. HEK293 cells were transfected with HERG and pulse-labeled, and the amounts of the HERG CG and FG forms were detected at different times of chase (Fig. 6A). As established previously, the amount of CG decreased over 6 h of chase to ∼60% of the starting amount, as the amount of FG increased to ∼35% of the initial CG amount (Fig. 6B). The rate of degradation was observed as a moderate reduction in the total amount of HERG (the sum of CG and FG amounts). Transfection of DJA2 significantly decreased the amount of FG to 15% of starting material, whereas the amount of CG was not greatly affected (Fig. 6, B and C). The clear reduction in total HERG was consistent with degradation induced by DJA2. Intriguingly, the mutants tested, DJA2-Δm2, DJA-122, DJA-221, and DJA2-ΔJ, were less effective than wild-type DJA2 in reducing trafficking and promoting degradation (Fig. 6, B and C). Thus, the m2 subdomain appears to be important for both the folding and degradation functions of DJA2, and exchange of the C-terminal region seems to also be disruptive. Exchange of the J domain was more detrimental for degradation than for folding.
FIGURE 6.
Impairment of HERG trafficking by DJA2 mutants. A, HEK293 cells were transfected with HERG and control vector or wild-type or mutant DJA2. Cells were pulse-labeled for 45 min and chased for the indicated times. At the time points, cell lysates were immunoprecipitated (IP) with antibody against HERG. The CG and FG forms of HERG were detected in autoradiograms. B, amounts of CG and FG HERG at different time points, represented as percentages of the initial CG amount at the 0-h time point. C, average CG and FG amounts at 6 h of chase. Results with significant differences from those with wild-type DJA2 are marked (** denotes p < 0.01).
In Vitro Function of DJA2 Mutants
To address the mechanistic defects in the mutants, DJA2-Δm2, DJA-221, DJA-122, and DJA2-ΔJ were purified and compared with wild-type DJA2 and DJA1. All of the proteins were isolated as dimers on gel filtration chromatography to eliminate aggregates. No changes in the overall secondary structure of the mutants compared with full-length protein were detected by CD spectroscopy, ruling out gross disruptions in protein structure (Fig. 7A). To further confirm the structural stability of DJA2-Δm2 and DJA-221, their urea denaturation curves were measured by CD spectroscopy. Both of the mutants had denaturation midpoints close to that of wild-type DJA2, from 5.6 to 6.1 m urea (Fig. 7B). Interestingly, wild-type DJA1 was somewhat less stable with its midpoint around 4.8 m urea. Overall, the mutants appeared to be properly folded, indicating that the subdomain boundaries were correctly identified, allowing analysis of subdomain function.
FIGURE 7.
A, CD spectra of wild-type DJA2, DJA1, and the indicated mutants at 0.5 mg/ml. Deconvolution suggested ∼12% α-helix and ∼38% β-sheet for all proteins. B, urea melting curves for DJA2, DJA1, and the indicated mutants determined by CD. Urea concentrations at the denaturation mid-points (CM) were calculated and marked above the graphs.
The mutants were compared with wild-type DJA2 in the ability to assist substrate folding by Hsc70. Luciferase was denatured with guanidine and diluted into reactions containing Hsc70, DJA2, and ATP, and its refolding at 30 °C was monitored over time by its enzymatic activity. As observed previously (13, 14), Hsc70 and DJA2 produced efficient refolding by 30 min, and the refolding yield at 60 min was set to 100% (Fig. 8A). Hsc70 alone was minimally functional with refolding reaching only ∼20%. The NEF Hsp110 increased refolding with DJA2, whereas DJA1 did not increase refolding above the control (Fig. 8, A and B) (14). Notably, both DJA2-Δm2 and DJA-221 were inactive, with less than 30% refolding compared with wild-type DJA2. The DJA-122 mutant was partially active with more than 60% refolding, although its refolding kinetic was slower than that of the wild type (Fig. 8, A and B). The negative control mutant DJA2-ΔJ was nonfunctional as expected (data not shown). These results were generally consistent with the functional defects observed in cells, most clearly for DJA2-Δm2 and DJA-221. Quantitative differences between the in vitro and in vivo results, such as the activity of DJA-122, were most likely due to the many differences in the experimental systems, including the means of denaturation and the presence of other chaperones. The results suggest that although the J domain may be somewhat interchangeable, the subdomains within the MC region of DJA2 may be more closely inter-related.
FIGURE 8.
Activity of purified DJA2 and mutants. A, refolding of denatured luciferase. Luciferase was denatured in guanidinium chloride and diluted into refolding reactions containing 4 μm Hsc70 alone (control) or with wild-type or mutant DJA2 or DJA1 (4 μm) and Hsp110 (1 μm). Luciferase enzymatic activity was monitored over time and represented as a percentage of the activity with Hsc70 and DJA2 at 60 min. B, average activities of luciferase at 60 min of refolding with Hsc70 alone (control) or wild-type or mutant DJA2. Significant differences between the indicated results are marked (** denotes p < 0.01; *, p < 0.05). C, binding of DJA2 to denatured luciferase detected by ELISA. 0.5 μg of luciferase was immobilized on microplates, and increasing amounts of wild-type or mutant DJA2 were allowed to bind and then detected by colorimetric ELISA quantitation. Background binding to wells without luciferase was subtracted.
Next, we examined the activity of the J domain in stimulating ATP hydrolysis by Hsc70. Single-turnover ATPase experiments were conducted to observe the hydrolysis step independently of nucleotide exchange. Complexes of Hsc70 with radiolabeled ATP were isolated; DJA2 or its mutants were added, and the production of ADP over time was monitored. Apparent hydrolysis rates were calculated from the linear slopes. Hsc70 alone had a basal low ATPase rate, which increased strongly upon addition of DJA2 (Table 1). The ATPase rates supported by DJA2-Δm2, DJA-122, and DJA-221 as well as wild-type DJA1 were not significantly different from that of DJA2 (Table 1), and as expected, DJA2-ΔJ was inactive. These results agreed with the relative independence of the J domain. However, the functional defect in DJA2-Δm2 and DJA-221 could not be due to loss of Hsc70 stimulation by their J domains, and they must have other causes.
TABLE 1.
Single-turnover ATPase activities of Hsc70 with DJA mutants
| Reaction | ATPase rate |
|---|---|
| % ADP/min ± S.D. | |
| Hsc70 | 0.47 ± 0.09 |
| Hsc70 + DJA2 | 3.06 ± 0.60 |
| Hsc70 + DJA2-Δm2 | 2.49 ± 0.32 |
| Hsc70 + DJA-122 | 2.27 ± 0.09 |
| Hsc70 + DJA-221 | 3.06 ± 0.53 |
| Hsc70 + DJA1 | 2.58 ± 0.20 |
| Hsc70 + DJA1-Δm2 | 2.72 ± 0.32 |
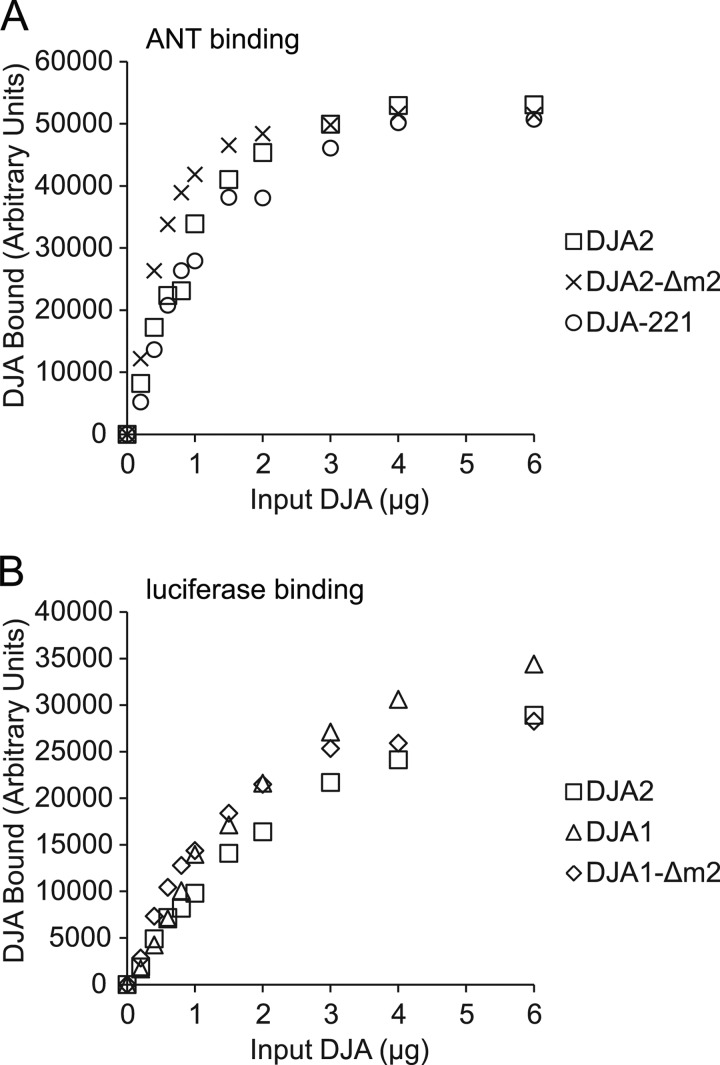
The MC domain of DJA2 contains a binding site for substrate (13), in agreement with the site identified in Ydj1. It was possible that the DJA2-Δm2 and DJA-221 mutants had defects in substrate binding, leading to their loss of function. We therefore established a microplate assay for substrate binding by DJA2. Luciferase was immobilized in the wells and denatured with guanidine. The denaturant was washed out, and different amounts of DJA2 from 0.2 to 6 μg were allowed to bind and then quantitatively detected by ELISA with specific antibodies. The amounts of DJA2 detected increased with input DJA2, approaching saturation at the highest amounts (Fig. 8C). As a positive control, the luciferase was separately detected with antibodies (data not shown). Also, no DJA2 binding was observed in negative control wells with no immobilized substrate (data not shown). Thus, the DJA2 detected represented its chaperone-substrate interaction. When the DJA2-Δm2, DJA-122, DJA-221, and DJA2-ΔJ were tested, they all bound to the substrate at levels comparable with wild-type DJA2 (Fig. 8C). These results were confirmed by testing the binding to another purified substrate, ANT (13). Again, the binding of DJA2-Δm2 and DJA-221 to ANT was similar to wild-type DJA2 (Fig. 9A). Together with the CD spectroscopy, these results were in agreement with natively folded MC domains in the mutants, with the substrate-binding site unaffected. Importantly, our results show that disrupted substrate binding could be ruled out as the basis for the functional defects of DJA2-Δm2 and DJA-221.
FIGURE 9.
A, binding of wild-type and mutant DJA2 to 0.5 μg of denatured ANT detected by ELISA and after subtraction of background binding, as in Fig. 8C. B, binding of wild-type and mutant DJA1 to denatured luciferase detected by ELISA as in Fig. 8C.
The m2 Subdomain Is Essential for Substrate Release
After substrate is bound by DJA2, it is thought to be transferred to Hsc70 upon J domain stimulation of the Hsc70 ATPase. To investigate this process, the ELISA experiments were extended to include Hsc70. The initial substrate binding step was repeated with an amount of DJA2 below saturation (3 μg), and in a second step, Hsc70 and ATP were added under conditions identical to the refolding reactions. After the second step, bound DJA2 and Hsc70 were detected separately. We found that Hsc70 and ATP reduced the amount of bound DJA2 to around 50% of starting DJA2, indicating that DJA2 was releasing the substrate (Fig. 10A). This level of release was in accord with that for ERdj3 with BiP in similar ELISA experiments (24). The amount of bound Hsc70 was then measured and expressed as a ratio of the amount bound in the absence of DJA2. As recently observed for DJA1 and Hsc70 using surface plasmon resonance (11), DJA2 partially decreased the amount of Hsc70 bound to substrate, although efficient Hsc70 binding was still observed at around 60% of maximum (Fig. 10B). Negative controls without substrate also showed no Hsc70 binding (data not shown). Therefore, Hsc70 binding to substrate and DJA2 release from substrate occurred in the same reaction.
FIGURE 10.
Substrate release by DJA2 and mutants. A, release of DJA2 from substrate. 3 μg of wild-type or mutant DJA2 was allowed to bind to immobilized denatured luciferase as in Fig. 8A and then incubated with 3 μg of Hsc70 and 2 mm ATP. The amount of DJA2 remaining bound after Hsc70 addition was detected by ELISA, quantified, and represented as a ratio to the starting amount of DJA2 bound before Hsc70 addition. Results with significant differences from those with wild-type DJA2 are marked (** denotes p < 0.01). B, substrate binding by Hsc70. The amounts of Hsc70 bound in the reactions in A were measured by ELISA detection. The amounts of Hsc70 bound were represented as a ratio to the maximum amount of Hsc70 bound in reactions without DJA2. C, substrate binding by DJA2 over time. Denatured nontagged luciferase was diluted into reactions containing His-tagged wild-type or mutant DJA2 as in Fig. 8A, and reactions were initiated by addition of nontagged Hsc70 and ATP. DJA2 was recovered by nickel-Sepharose pulldown at the indicated times. The amounts of DJA2 and luciferase bound were detected by immunoblot (IB). The amount of luciferase bound was represented as a percentage of the initial amount at the start of refolding. D, refolding reactions were performed as in C except with ADP, and amounts of luciferase bound by DJA2 at different time points were measured.
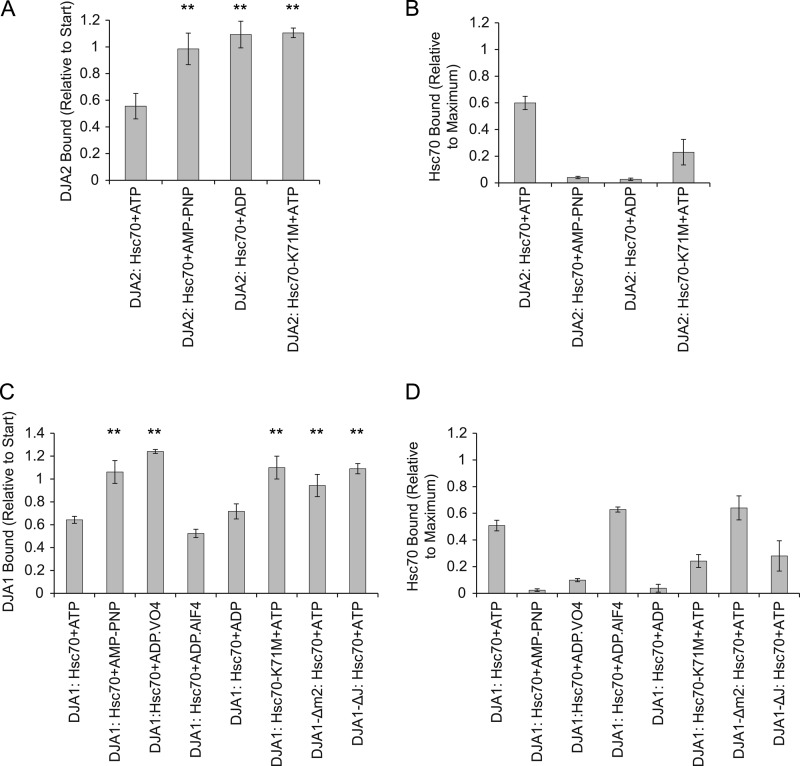
The DJA2 mutants were then tested for the ability to release from substrate. We found that DJA2-Δm2 was significantly impaired in substrate release, with more than 80% remaining bound (Fig. 10A). Notably, the amount of Hsc70 bound with DJA2-Δm2 was similar to that with wild-type DJA2 (Fig. 10B), which shows that substrate binding by Hsc70 is not sufficient for the release of DJA2. In contrast, DJA-122 and DJA-221 behaved like wild-type DJA2 in substrate release and Hsc70 binding to substrate, suggesting that a release defect is not an automatic result of DJA2 mutation. DJA2-ΔJ was unable to release substrate and led to a low level of Hsc70 substrate binding (Fig. 10, A and B), as predicted by the model of substrate transfer. The results with DJA2-Δm2 suggest that it can initiate Hsc70 binding to substrate but is impaired in releasing from substrate itself to complete the transfer reaction.
To further confirm the defect of DJA2-Δm2 in substrate release, the amount of luciferase bound to wild-type or mutant DJA2 was analyzed in solution under conditions similar to refolding reactions. The reactions were set up using His-tagged DJA2 or DJA2-Δm2 and nontagged denatured luciferase and initiated by addition of nontagged Hsc70 and ATP. At various times after Hsc70 addition, the DJA2 proteins were recovered with nickel-Sepharose, and bound luciferase was detected by immunoblot to examine the kinetics of release. In reactions with wild-type DJA2, luciferase binding was observed at 0 min, then decreased markedly by 5 min, and reached ∼20% of the initial amount by 15 min (Fig. 10C). With DJA2-Δm2, substrate binding at 0 min was also observed but remained relatively unchanged at 5 and 15 min (Fig. 10C). In negative control reactions lacking DJA2 and Hsc70, no luciferase was precipitated as expected (data not shown). As a further control, parallel reactions were set up containing ADP instead of ATP and analyzed the same way. Under these conditions without ATPase cycling by Hsc70, the substrate remained largely bound by both DJA2 and DJA2-Δm2 over 15 min (Fig. 10D). The persistent substrate binding of DJA2-Δm2 was consistent with a significant delay in the release of substrate and agreed with the defect observed in the ELISA experiments. Based on the combined results, we conclude that the loss of function of DJA2-Δm2 was most likely due to its defect in the release of substrate.
Nucleotide Dependence of Substrate Release
To analyze the DJA2 substrate release mechanism in more depth, we tested the requirement for nucleotides. When Hsc70 was added with the nonhydrolyzable ATP analog AMP-PNP, the amount of bound DJA2 remained at its starting level, indicating no release of substrate. No Hsc70 binding was observed with AMP-PNP (Fig. 11, A and B). When ADP was used instead, similar results were observed, with no substrate release of DJA2 and no binding of Hsc70 (Fig. 11, A and B). The Hsc70 binding data were consistent with its expected model of function. In the ATP-bound state, before hydrolysis, the Hsc70 substrate binding domain is considered to be open and unable to bind substrate. In the pre-formed ADP-bound state, it would be already closed and slow to bind subsequently added substrate. Thus, Hsc70 binding efficiency should be the highest when it converts between the ATP- and ADP-bound states. Interestingly, the results further suggested that ATP hydrolysis by Hsc70 is also necessary to induce substrate release from DJA2, as AMP-PNP and ADP were ineffective. To test this idea, the Hsc70-K71M mutant, which is defective in ATP hydrolysis, was used (28). In the presence of ATP, Hsc70-K71M was unable to induce DJA2 substrate release, although a low level of substrate binding was observed for the mutant Hsc70 (Fig. 11, A and B). Thus, Hsc70 ATPase activity was important for both Hsc70 substrate binding and DJA2 substrate release, in what appears to be a substrate transfer reaction. Moreover, this was consistent with the substrate release defect of DJA2-ΔJ observed above.
FIGURE 11.
Nucleotide dependence of substrate release. A, binding of DJA2 to substrate after reaction with Hsc70 or Hsc70-K71M and 2 mm of the indicated nucleotide was measured as in Fig. 10A. Results with significant differences from those with DJA2, Hsc70, and ATP are marked (** denotes p < 0.01). B, amounts of Hsc70 bound to substrate in the reactions in A were measured as in Fig. 10B. C, substrate release by DJA1. The binding of wild-type or mutant DJA1 to substrate after reactions with Hsc70 was measured. For the transition state analogs, 2 mm ADP and 20 mm Na3VO4 or 5 mm NaF and 0.5 mm AlCl3 were used. Results with significant differences from those with DJA1, Hsc70, and ATP are marked (** denotes p < 0.01). D, amounts of Hsc70 bound to substrate in the reactions in C were measured.
Because the J domain activity of DJA2-Δm2 was unaffected, the m2 subdomain may act downstream of the J domain in substrate release. We expected that the m2-dependent release mechanism should be conserved between DJA2 and DJA1, despite the poor refolding activity of DJA1. To address this, a DJA1-Δm2 mutant was constructed based on homology modeling. CD spectroscopy measurements of the purified mutant were essentially identical to the wild type (Fig. 7A). Both DJA1 and DJA1-Δm2 bound immobilized luciferase similarly to DJA2 in ELISA experiments (Fig. 9B). When Hsc70 and ATP were added to the bound DJA1, release of DJA1 from substrate and Hsc70 substrate binding was observed as for DJA2. Importantly, DJA1-Δm2 remained bound after Hsc70 was added, showing the same defect in substrate release as DJA2-Δm2, whereas Hsc70 substrate binding was again unaffected (Fig. 11, C and D). The mechanistic role of the m2 subdomain appeared to be the same for these type I co-chaperones.
To complete the comparison between DJA2 and DJA1, the nucleotide dependence of the DJA1 release reaction was tested. Similar to DJA2, Hsc70 with AMP-PNP could not support release of DJA1 from substrate or Hsc70 binding to substrate (Fig. 11, C and D). The negative control mutants Hsc70-K71M and DJA1-ΔJ were also inactive. When Hsc70 and ADP were used, ADP was surprisingly as effective as ATP in promoting substrate release of DJA1, although only a basal level of Hsc70 binding was observed (Fig. 11, C and D). This was an intriguing difference from DJA2, and it was explored further with ATP transition state analogues. When ADP and VO4− were used, no substrate release of DJA1 was observed and was accompanied by low levels of Hsc70 binding. In contrast, ADP and AlF4− caused efficient substrate release of DJA1 and substrate binding by Hsc70 comparable with reactions with ATP (Fig. 11, C and D). For different proteins, either ADP-VO4− or ADP-AlF4− may be a better mimic of ATPase transition states, and they may also represent early and late stages in the ATPase reaction (39, 40). Because ADP-AlF4− and ADP alone had similar effects with DJA1, we therefore postulated that the substrate release of DJA1 may be coupled to a late step in the ATPase reaction, perhaps the dissociation of free phosphate. Substrate release of DJA2 was not observed with ADP-AlF4− or ADP-VO4− (data not shown). Thus, the substrate release of DJA1 is distinct from that of DJA2, which may be coupled to the actual ATP hydrolysis step.
Conformation of the MC Regions
The mechanistic defect of DJA-221 was not related to its substrate release, which was similar to wild-type DJA2. Although there was no evidence that DJA-221 was misfolded, it was possible that the C-terminal exchange shifted the structure of the protein enough to impair function. To address this, DJA2 and DJA-221 were analyzed by limited proteolysis, and for comparison, DJA1 and DJA2-Δm2 were also examined. At 5 and 20 μg/ml trypsin, fragments of DJA2 were observed at around 33, 26, and 15 kDa (Fig. 12A). Protein sequencing revealed that the 33- and 27-kDa fragments of DJA2 had the same N termini at Gln-173, predicted to be at the tip of the m2 subdomain (Fig. 12C and Table 2). Taking into account the aberrant migration of DJA2 (the 46-kDa full-length protein migrated as a 54-kDa band), the 33-kDa fragment was projected to have a complete C terminus and the 26-kDa fragment a C-terminal end in the dimerization region. In digests of DJA-221, almost no 33-kDa fragment was observed, and a reduced amount of 26-kDa fragment was generated (Fig. 12A). Sequencing showed that they also started at Gln-173. This cleavage site was located in the part of DJA-221 that came from DJA2, suggesting that the C-terminal fusion caused some conformational changes throughout the rest of the MC domain, including the m2 region.
FIGURE 12.
Limited proteolysis of DJA2 mutants. A, wild-type or mutant DJA2 or DJA1 was incubated with the indicated concentrations of trypsin and analyzed by SDS-PAGE and total protein staining by Coomassie. The full-length proteins (black arrowhead) and proteolytic fragments corresponding to the J domain (white arrowhead) are marked. The major proteolytic fragments are marked with letters a–f. The positions of molecular weight markers (M) are shown on the right. B, DJAs were digested with chymotrypsin (chymo) and analyzed as in A. C, diagram of the primary sequences and fragments with N termini determined by protein sequencing and estimated C termini. Fragments correspond to those marked in A.
TABLE 2.
N-terminal sequences of proteolytic fragments
Residues are numbered according to the wild-type sequences
| Protein | Band | N-terminal sequence | Apparent mass |
|---|---|---|---|
| kDa | |||
| DJA1 | a | 101ERRGKN | 33 |
| DJA2 | b | 173QLAPGM | 33 |
| c | 173QLAPGM | 26 | |
| DJA2-Δm2 | d | 59NPEK | 28 |
| e | EFKD200 | 24 | |
| 213EVKI | 23 | ||
| DJA-221 | f | 173QLAPGM | 27 |
Digestion of DJA2-Δm2 with 5 and 20 μg/ml trypsin produced a 27-kDa fragment superficially similar to DJA2, with additional bands around 24 kDa being observed at 50 μg/ml trypsin (Fig. 12A). However, protein sequencing showed that the 27-kDa fragment had an N terminus within the J domain at Asn-59 and a predicted C terminus near the start of the dimerization region. The 24-kDa fragments had N termini at Lys-199 and Glu-213, after the segment removed by the mutation, and C termini in the dimerization site (Fig. 12C and Table 2; numbering refers to the wild-type sequence). Because the 27-kDa piece generated from DJA2-Δm2 was different from that observed for DJA2, there may be some rearrangement in the J domain and linker of DJA2-Δm2 relative to the wild type. However, the 24-kDa fragments resembled the wild-type fragments more closely.
When DJA1 was digested with trypsin, only the 33- and 15-kDa fragments were strongly present (Fig. 12A). Immunoblots identified the 15-kDa fragment of DJA1 as the J domain, and the similar fragment of DJA2 was presumably also its J domain. Interestingly, the 33-kDa fragment had its N terminus at Glu-101, the joining between the linker and MC domain, which was quite different from the 33-kDa fragment of DJA2 (Fig. 12C and Table 2). The lack of a 26-kDa fragment was somewhat surprising, as those trypsin cleavage sites are conserved between DJA1 and DJA2. The divergent profiles may be due to differences in accessibility of the tryptic sites or in the cooperative folding of the MC region. We investigated this further using limited proteolysis with chymotrypsin. When DJA2 was digested at 5 and 20 μg/ml chymotrypsin, a major fragment of 38 kDa was detected, corresponding to removal of only the J domain (Fig. 12B). Chymotrypsin digestion of DJA1 produced a fragment of 52 kDa, just shorter than the full-length, and no smaller fragments. Furthermore, treatment of DJA-221 resulted in almost complete digestion of the protein at 20 μg/ml chymotrypsin with no abundant stable fragments, whereas the profile of DJA-Δm2 was closer to wild-type DJA2. These results are consistent with some conformational difference between DJA2 and DJA1 and between DJA2 and DJA-221. Such differences may also be related to their folding activities.
DISCUSSION
The key features within DJA2 that we have determined to be necessary for its function are summarized in Fig. 1. The m2 subdomain is required for productive substrate release from DJA2, coupled to the hydrolysis of ATP by Hsc70. The J domain is necessary not only for activating Hsc70 but also for DJA2 substrate release. DJA1 has a similar m2-dependent release mechanism that may be coupled to a later stage in the Hsc70 ATPase reaction. Despite the strong homology between DJA1 and DJA2, they may be conformationally different, and structural changes in the C terminus affect the N-terminal regions important for function.
The in vitro data suggest a mechanism of substrate release from DJA2 that is critical for its function with Hsc70. Substrate release requires ATP hydrolysis by Hsc70, as AMP-PNP or the ATPase-inactive Hsc70-K71M mutants were ineffective. The J domain and the m2 subdomain are also necessary for substrate release, but their roles in the mechanism must be different. The J domain is the contact site for Hsc70 and most probably couples ATP hydrolysis in Hsc70 with DJA2. The m2 deletion impaired substrate release but had no effect on J domain activity or initial substrate binding by DJA2. Thus, the m2 subdomain may act downstream of the J domain in the release mechanism. It is possible that changes in Hsc70 during ATP hydrolysis are transmitted through the J domain and m2 to affect the substrate-binding site in DJA2. The same requirements for substrate release are found in DJA1, J and m2 regions and ATP hydrolysis, indicating that this is a conserved mechanism. Interestingly, the ability of DJA1 to release with ADP and a transition state analog suggests it has a subtle difference from DJA2 in coupling to Hsc70. This coupling and the structural variation suggested by the proteolytic profiles are the first experimental demonstrations of differences between DJA1 and DJA2 that may explain their functional divergence. There are likely other differences yet to be found.
The stretch of highest sequence similarity between DJA2 and DJA1 is in their J domains. The conserved surfaces may be interaction sites for other parts of the DJAs, by proximity the linker or the m2 subdomain. Two observations suggest that the linker in DJA2 contributes to function with its MC domain. First, in the limited proteolysis, the major tryptic cleavage was inside the m2 sequence, and the sequence N-terminal to this site may be degraded in one piece, consistent with it being a cooperatively folded unit. Second, we previously found that a chimeric mutant DJA1-2, having the J domain and linker of DJA1 and the MC domain of DJA2, was nonfunctional in Hsc70-mediated folding (14). In this study we found that DJA-122 retained some function with the J domain of DJA1 along with the linker and MC domain of DJA2. Thus, the linker and MC domain may be a functional as well as structural unit. It is possible that the linker and part of the MC domain, perhaps the m2 region, together form an interaction site for the J domain and the allosteric basis of the substrate release mechanism. Differences in the J domain, linker, and m2 arrangement in DJA1 may also underlie its distinct nucleotide dependence for release.
The proposed mechanism is in general agreement with the substrate release described for ERdj3 and p58 with BiP. Release from those co-chaperones required ATP hydrolysis by BiP and an active J domain able to stimulate the ATPase. Substrate release correlated with BiP binding to substrate, consistent with transfer of substrate to BiP. ATP binding without hydrolysis, or just the contact between J domain and BiP, was not sufficient for substrate release (24, 25). We now find the same features in the substrate release of DJA2 with Hsc70, and we suggest that these identify a mechanism conserved between the cytosolic and lumenal chaperone systems. Our results add further insight into the release mechanism. For ERdj3, a ternary complex with substrate and BiP was required for substrate release (24), and our results suggest that DJA2 has the same requirement. However, the substrate binding of both Hsc70 and DJA2-Δm2 was the same as for wild-type DJA2, and the J domain of the mutant was also fully active. Hsc70 substrate binding and ternary complex formation may be necessary, but are not sufficient, for DJA2 release. A p58 mutant with the J domain fused in a different location from that in the wild type could still release substrate, suggesting that local structures are not in principle required for release (25). Therefore, the m2-dependent release mechanism of DJA2 and DJA1 may be conserved only within type I co-chaperones. This idea is supported by studies with Ydj1 mutated in its zinc-binding sites at the base of the m2 subdomain. The mutations did not affect J domain and substrate binding activities but abolished refolding function with Hsp70, consistent with a defect in substrate transfer to Hsp70 (16). The m2 subdomain function in type I DNAJs may be generally conserved between cellular compartments and between species.
Our results suggest that the primary role of DJA2 in Hsc70-mediated folding may not be simply to initiate substrate binding by Hsc70 but to do so in a way that is productive. Indeed, Hsc70 bound the denatured substrate in the absence of DJA2, and the presence of DJA2 moderated Hsc70 binding. Similar results were reported in a study of Hsc70 and DJA1 (11), and we also observe Hsc70 substrate binding with DJA1. However, the functional defect of the DJA2-Δm2 mutant suggests that complete substrate transfer from DJA2 to Hsc70 is a critical factor for folding. We propose that the simultaneous, transient binding of both DJA2 and Hsc70 to substrate, followed by the correctly timed release of substrate from DJA2, are important to promote folding. It is possible that coordinated binding of DJA2 and Hsc70 to two sites on the substrate may act on its conformation, due to the allosteric changes in Hsc70 itself as part of its ATP hydrolysis reaction. Another possibility is that DJA2 directs Hsc70 to binding sites on the substrate that are more beneficial for its folding. In either case, DJA2 must release substrate appropriately for Hsc70 binding to be productive. Differences in the substrate release step may be one source of the functional differences between DJA2 and DJA1. These hypotheses remain to be tested in detail. Nevertheless, we note that DJA2-Δm2, with its defective substrate release, had the strongest impairment next to DJA2-ΔJ in the functions tested, luciferase refolding in cells and in vitro, and inhibition of HERG trafficking through degradation. The substrate transfer mechanism of DJA2 with Hsc70 may be essential for all functions of the DJA2 co-chaperone and, by extension, of DJA1 as well.
Our results in cells are consistent with distinct biological functions of DJA2 and DJA1. Knockdown of DJA1 but not DJA2 reduced HERG trafficking, whereas transfection of either one appeared to promote HERG degradation (26). Furthermore, knockdown of DJA1 reduced the clearance of misfolded mutant cystic fibrosis transmembrane regulator from the cell surface, and DJA2 knockdown did not (41). In contrast, DJA2 has been identified as an enhancer of G-protein-coupled signaling by the β2-adrenergic receptor (42). Although functions of DJA2 and DJA1 independent of Hsc70 are possible, it is generally thought that their main roles are as co-chaperones of Hsc70. The substrate release mechanisms of the DJAs coupled to the Hsc70 ATPase are likely to be important for their various activities, including processes distinct from folding. We now provide the first evidence of biochemical differences between DJA1 and DJA2, in their substrate release and apparent conformations, and there are likely to be further differences. Their molecular properties are likely the basis of their biological division of labor.
Acknowledgments
We thank Patrick Kim Chiaw for helpful discussions; Roxana Atanasiu for technical support, and the members of the Young laboratory.
This work was supported in part by the Canadian Institutes of Health Research.
- NEF
- nucleotide exchange factor
- ANT
- adenine nucleotide translocator
- CG
- core-glycosylated
- ER
- endoplasmic reticulum
- FG
- fully glycosylated
- HERG
- human ether-a-go-go related gene
- IP
- immunoprecipitation
- MC
- middle/C-terminal domain of DJA2
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 2. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery. J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang J., Prasad K., Lafer E. M., Sousa R. (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell 20, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daugaard M., Rohde M., Jäättelä M. (2007) The heat shock protein 70 family. Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581, 3702–3710 [DOI] [PubMed] [Google Scholar]
- 6. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertelsen E. B., Chang L., Gestwicki J. E., Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young J. C. (2010) Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qiu X. B., Shao Y. M., Miao S., Wang L. (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terada K., Mori M. (2000) Human DnaJ homologs dj2 and dj3 and bag-1 are positive cochaperones of hsc70. J. Biol. Chem. 275, 24728–24734 [DOI] [PubMed] [Google Scholar]
- 11. Terada K., Oike Y. (2010) Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. J. Biol. Chem. 285, 16789–16797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hageman J., van Waarde M. A., Zylicz A., Walerych D., Kampinga H. H. (2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 435, 127–142 [DOI] [PubMed] [Google Scholar]
- 13. Bhangoo M. K., Tzankov S., Fan A. C., Dejgaard K., Thomas D. Y., Young J. C. (2007) Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell 18, 3414–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tzankov S., Wong M. J., Shi K., Nassif C., Young J. C. (2008) Functional divergence between co-chaperones of Hsc70. J. Biol. Chem. 283, 27100–27109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson J. L., Craig E. A. (2001) An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 152, 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan C. Y., Ren H. Y., Lee P., Caplan A. J., Cyr D. M. (2005) The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J. Biol. Chem. 280, 695–702 [DOI] [PubMed] [Google Scholar]
- 17. Banecki B., Liberek K., Wall D., Wawrzynów A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. (1996) Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 271, 14840–14848 [DOI] [PubMed] [Google Scholar]
- 18. Linke K., Wolfram T., Bussemer J., Jakob U. (2003) The roles of the two zinc-binding sites in DnaJ. J. Biol. Chem. 278, 44457–44466 [DOI] [PubMed] [Google Scholar]
- 19. Li J., Qian X., Sha B. (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 20. Wu Y., Li J., Jin Z., Fu Z., Sha B. (2005) The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40. J. Mol. Biol. 346, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 21. Kota P., Summers D. W., Ren H. Y., Cyr D. M., Dokholyan N. V. (2009) Identification of a consensus motif in substrates bound by a type I Hsp40. Proc. Natl. Acad. Sci. U.S.A. 106, 11073–11078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borges J. C., Fischer H., Craievich A. F., Ramos C. H. (2005) Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 280, 13671–13681 [DOI] [PubMed] [Google Scholar]
- 23. Ramos C. H., Oliveira C. L., Fan C. Y., Torriani I. L., Cyr D. M. (2008) Conserved central domains control the quaternary structure of type I and type II Hsp40 molecular chaperones. J. Mol. Biol. 383, 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin Y., Awad W., Petrova K., Hendershot L. M. (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J. 27, 2873–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petrova K., Oyadomari S., Hendershot L. M., Ron D. (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 27, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker V. E., Wong M. J., Atanasiu R., Hantouche C., Young J. C., Shrier A. (2010) Hsp40 chaperones promote degradation of the HERG potassium channel. J. Biol. Chem. 285, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milarski K. L., Morimoto R. I. (1989) Mutational analysis of the human HSP70 protein. Distinct domains for nucleolar localization and adenosine triphosphate binding. J. Cell Biol. 109, 1947–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Brien M. C., Flaherty K. M., McKay D. B. (1996) Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis. J. Biol. Chem. 271, 15874–15878 [DOI] [PubMed] [Google Scholar]
- 29. Schneider C., Sepp-Lorenzino L., Nimmesgern E., Ouerfelli O., Danishefsky S., Rosen N., Hartl F. U. (1996) Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. U.S.A. 93, 14536–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 31. Fewell S. W., Smith C. M., Lyon M. A., Dumitrescu T. P., Wipf P., Day B. W., Brodsky J. L. (2004) Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 279, 51131–51140 [DOI] [PubMed] [Google Scholar]
- 32. Michels A. A., Kanon B., Konings A. W., Ohtsuka K., Bensaude O., Kampinga H. H. (1997) Hsp70 and Hsp40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J. Biol. Chem. 272, 33283–33289 [DOI] [PubMed] [Google Scholar]
- 33. Akhavan A., Atanasiu R., Noguchi T., Han W., Holder N., Shrier A. (2005) Identification of the cyclic nucleotide binding domain as a conserved determinant of ion-channel cell-surface localization. J. Cell Sci. 118, 2803–2812 [DOI] [PubMed] [Google Scholar]
- 34. Walker V. E., Atanasiu R., Lam H., Shrier A. (2007) Co-chaperone FKBP38 promotes HERG trafficking. J. Biol. Chem. 282, 23509–23516 [DOI] [PubMed] [Google Scholar]
- 35. Flom G. A., Lemieszek M., Fortunato E. A., Johnson J. L. (2008) Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell 19, 5249–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Summers D. W., Douglas P. M., Ren H. Y., Cyr D. M. (2009) The type I Hsp40 Ydj1 utilizes a farnesyl moiety and zinc finger-like region to suppress prion toxicity. J. Biol. Chem. 284, 3628–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 38. Ficker E., Dennis A. T., Wang L., Brown A. M. (2003) Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ. Res. 92, e87–100 [DOI] [PubMed] [Google Scholar]
- 39. Smith C. A., Rayment I. (1996) X-ray structure of the magnesium(II)·ADP·vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry 35, 5404–5417 [DOI] [PubMed] [Google Scholar]
- 40. Inobe T., Kikushima K., Makio T., Arai M., Kuwajima K. (2003) The allosteric transition of GroEL induced by metal fluoride-ADP complexes. J. Mol. Biol. 329, 121–134 [DOI] [PubMed] [Google Scholar]
- 41. Okiyoneda T., Barrière H., Bagdány M., Rabeh W. M., Du K., Höhfeld J., Young J. C., Lukacs G. L. (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosales-Hernandez A., Beck K. E., Zhao X., Braun A. P., Braun J. E. (2009) RDJ2 (DNAJA2) chaperones neural G protein signaling pathways. Cell Stress Chaperones 14, 71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]