Background: Interaction between lysyl-tRNA synthetase and HIV-1 Gag is critical for tRNALys primer packaging and virus replication.
Results: Mutation of residues of dimerization helix 7 abolishes hLysRS packaging into virions, reduces binding to HIV-1 Gag, and affects the synthetase dimerization state and aminoacylation activity.
Conclusion: LysRS dimer interface residues interact with HIV-1 Gag.
Significance: Mapping host cell-viral protein interaction will aid in development of novel antiviral agents.
Keywords: Aminoacyl tRNA Synthetase, Biophysics, Fluorescence, HIV-1, Protein-Protein Interactions, Gag, Capsid, Capsid C-terminal Domain, Lysyl-tRNA Synthetase
Abstract
The primer for reverse transcription in human immunodeficiency virus type 1, human tRNALys,3, is selectively packaged into the virion along with tRNALys1,2. Human lysyl-tRNA synthetase (hLysRS), the only cellular factor known to interact specifically with all three tRNALys isoacceptors, is also selectively packaged into HIV-1. We have previously defined a tRNALys packaging complex that includes the tRNALys isoacceptors, LysRS, HIV-1 Gag, GagPol, and viral RNA. Numerous studies support the hypothesis that during tRNALys packaging, a Gag·GagPol complex interacts with a tRNALys·LysRS complex, with Gag interacting specifically with the catalytic domain of LysRS, and GagPol interacting with both Gag and tRNALys. In this work, we have identified residues along one face of the motif 1 dimerization helix (H7) of hLysRS that are critical for packaging of the synthetase into virions. Mutation of these residues affects binding to Gag in vitro, as well as the oligomerization state and aminoacylation activity of the synthetase. Taken together, these data suggest that H7 of LysRS has a dual function. In its canonical role it maintains the synthetase dimer interface, whereas in its function in tRNA primer recruitment, it bridges interactions with HIV-1 Gag.
Introduction
Upon infection of the host cell by HIV-1, the viral RNA genome is copied into a double-stranded cDNA by the viral enzyme reverse transcriptase. Host cellular tRNALys,3 is required to initiate the first step of reverse transcription in the viral life cycle (1). The resultant viral DNA is translocated into the nucleus of the infected cell where it integrates into the host cell DNA and codes for viral RNA and proteins. The mature viral structure includes glycosylated envelope proteins and proteins resulting from processing of the large precursor proteins Gag and GagPol (2). Both Gag and GagPol are translated from the same full-length viral RNA, which is also packaged into assembling virions where it serves as the genomic RNA. During maturation, Gag is cleaved by HIV-1 protease to yield matrix, capsid (CA),2 nucleocapsid, and p6 proteins. Protease, reverse transcriptase, and integrase are enzymes produced as a result of GagPol processing. During the assembly step of the viral life cycle, Gag, GagPol, viral RNA, and specific cellular components are selectively packaged into the virion for initiating subsequent infectious cycles (3).
Host-encoded tRNALys,3, which serves as the primer for reverse transcription, is selectively packaged into HIV-1, along with the other major human tRNALys isoacceptors (tRNALys1,2). The packaging of tRNALys requires GagPol (4) and human lysyl-tRNA synthetase (hLysRS), which is also selectively packaged into HIV-1 (5). Human LysRS, the enzyme that aminoacylates tRNALys, is the only known cellular factor that specifically recognizes all tRNALys species. The selective packaging of tRNALys isoacceptors into HIV-1 raised the possibility that LysRS also participates in viral tRNA packaging. Indeed, overexpression of hLysRS increases tRNALys packaging into HIV-1 particles (6), and siRNA knockdown of LysRS decreases the tRNALys amounts incorporated (7). Thus, LysRS is the limiting factor for tRNALys packaging (6). Increasing the concentration of viral tRNALys,3 in HIV-1 by overexpressing exogenous tRNALys,3 also results in increased annealing of the tRNA onto the primer binding site and enhanced viral infectivity (6). Furthermore, packaging of tRNALys isoacceptors requires binding to LysRS (8), but aminoacylation of the tRNA is not required (9).
Human LysRS is a class II synthetase forming a closely related subgroup (known as IIb) with aspartyl- and asparaginyl-tRNA synthetases (10, 11). Class II synthetases are generally functional dimers or tetramers, and their active sites consist of an antiparallel β-sheet structure and three highly degenerate consensus sequences (motifs 1, 2, and 3). Motif 1, which is comprised of α-helices 5, 6, and 7 (H7) and part of a β-sheet (β6), constitutes the dimer interface, whereas motif 2 and 3 together constitute the aminoacylation active site (12–14). Although an α2 homodimer is the functional oligomerization state for aminoacylation, hLysRS crystallized as an α2α2 tetramer (14). LysRS is also part of the high molecular weight multisynthetase complex (MSC) present in higher eukaryotes (15). Within the MSC, LysRS specifically interacts with the scaffold protein p38/AIMP2 and is present in a unique α2β1:β1α2 orientation, which is designed to control both retention and mobilization of LysRS from the MSC (16).
The source of LysRS packaged into HIV-1 is still unclear. One report suggests that the packaged viral LysRS is of host mitochondrial origin (17). However, earlier studies suggested that packaged LysRS does not appear to originate from any of its identified steady-state cellular compartments, which include the cytoplasmic MSC, nuclei, mitochondria, or cell membrane. Instead, newly synthesized cytoplasmic LysRS has been shown to interact with HIV-1 Gag before entering any of these compartments upon viral infection (18). More recently, it has been suggested that interactions also occur between LysRS and the Pol domain of the GagPol precursor (19).
The packaging of hLysRS into HIV-1 is specific; of nine aminoacyl-tRNA synthetases and three additional components of the mammalian MSC tested, only hLysRS has been shown to be packaged (18). The domains critical for interaction between LysRS and Gag have been identified to include the motif 1 domain of LysRS and the C-terminal domain (CTD) of CA (20). Deletions that extend into HIV-1 CA-CTD or LysRS motif 1 H7 abolish the interaction between Gag and hLysRS in vitro. Furthermore, deletion of motif 1 eliminates LysRS packaging into Gag viral-like particles (20). Interestingly, both of these primarily helical regions (motif 1 of LysRS and HIV-1 CA-CTD) are critical for homodimerization of the individual proteins. We have previously investigated the interaction between Gag and LysRS by fluorescence anisotropy (FA) measurements and gel chromatography (21). An apparent equilibrium binding constant (Kd) of 310 nm was measured for the Gag/LysRS interaction, and CA alone binds to LysRS with a similar affinity (∼400 nm) as full-length Gag. Gag and LysRS variants containing point mutations in their dimerization motifs that effectively eliminated homodimerization still interacted in vitro, suggesting that dimerization of each protein per se is not required for the interaction. Furthermore, nuclear magnetic resonance and mutagenesis studies mapped the CA residues critical for the interaction to the helix 4 region of CA-CTD (22). More recently, an ab initio energy minimized “bridging monomer” model of the HIV-1 CA-CTD·LysRS·tRNALys ternary complex has been proposed, which is also consistent with an interaction between helix 4 of CA-CTD and the H7 region of LysRS (23). In addition, circular dichroism experiments along with in silico studies also support this helix 4/H7 interaction (24).
Although the CA-CTD residues involved in LysRS interaction are known, amino acids in the motif 1 region of LysRS that are involved in interaction with HIV-1 Gag have not been mapped. In this work, we carried out both cell-based and in vitro studies aimed at fine mapping of the critical H7 residues. Analyses of truncated LysRS constructs along with alanine-scanning mutagenesis experiments demonstrate the importance of H7 residues along one face of the dimerization helix in packaging of LysRS into HIV-1 virions. LysRS variants with single and double amino acid changes in H7 were purified and subjected to biochemical and biophysical in vitro characterization to determine binding affinity, oligomeric state, and aminoacylation ability. Changes that reduced or eliminated LysRS packaging into HIV-1 particles were strongly correlated with defects in binding to HIV-1 Gag/CA-CTD, LysRS dimerization, and aminoacylation activity. Taken together, these studies reveal a dual role for specific motif 1 residues of hLysRS in modulating the dimerization state of the synthetase and packaging in HIV-1.
EXPERIMENTAL PROCEDURES
Cell-based Analysis
Truncated variants of the gene encoding hLysRS were constructed by PCR using primers listed in the supplemental Methods and inserted into the EcoRI and XhoI cloning sites of plasmid pcDNA3.1 as previously described (25). For preparation of V5 epitope-tagged hLysRS containing double point mutations, the sense and antisense oligonucleotides (listed in the supplemental Methods) were first purified by polyacrylamide gel electrophoresis. Alanine scanning mutagenesis of hLysRS H7 was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Plasmids encoding hLysRS variants along with HIV-1 BH10 proviral DNA were then transfected into human HEK-293T cells (CRL-11268; ATCC) using Lipofectamin 2000 (Invitrogen), and cell and viral lysates were subjected to Western blot analysis using antibodies for V5 epitope, CAp24, and β-actin as previously described (25).
Protein Purification and Labeling
WT and histidine-tagged mutant LysRS proteins were derived from plasmid pM368 (21). Alanine scanning mutagenesis of hLysRS was also performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutations were confirmed by sequencing the entire gene. For purification, the following proteins were overexpressed in Escherichia coli and purified according to previously published procedures: WT and variant hLysRS (21), CA (21), monomeric CA-CTD variant containing two point changes to Ala at Trp-184 and Met-185 (WM CA-CTD) (21, 22), and HIV-1 Gag lacking the p6 domain (GagΔp6) (26). Protein concentrations were estimated using the Bradford assay (Bio-Rad). HIV-1 GagΔp6 was labeled with Texas Red-X, succinimidyl ester (Molecular Probes) following the manufacturer's protocol, as previously described (27). Briefly, 100 μm protein was incubated with Texas Red-X dye freshly dissolved in anhydrous dimethyl sulfoxide at a 10:1 dye:protein ratio for 60 min at room temperature in 150 mm NaCl, 40 mm HEPES, pH 7.5. The reaction was quenched by addition of 5 μl of 1 m Tris-HCl, pH 8.5, and unreacted dye was removed by passing the reaction mixture through a column assembly containing the purification resin provided by the manufacturer. Covalent labeling and complete removal of free dye were confirmed by visualizing the fluorescence on a denaturing polyacrylamide gel. The final labeling stoichiometry was determined by measuring the absorbance at 280 and 595 nm and using the following excitation coefficients: ∈280 = 63,090 m−1 cm−1 (GagΔp6) and ∈595 = 80,000 m−1 cm−1 (Texas Red-X). The labeling stoichiometry was estimated to be 0.7:1 GagΔp6:fluorophore. Labeling of WT LysRS with fluorescein was performed as previously described (21). Biotinylation (EZ-Link Sulfo-NHS-LC-LC-Biotin; Thermo Scientific) of WM CA-CTD was performed by incubating protein (250 μm) with 20-fold molar excess of biotin on ice for 2 h. Excess nonreacted biotin reagent was removed by overnight dialysis according to the manufacturer's protocol (Thermo Scientific).
Binding Measurements
Fluorescence Anisotropy Measurements
Apparent Kd values were determined by measuring the FA of 100-nm Texas Red-labeled GagΔp6 as a function of increasing concentrations of hLysRS. The labeled protein was incubated in amber tubes with varying amounts of the target protein for 1 h at room temperature in binding buffer (40 mm HEPES, pH 7.5, 150 mm NaCl, and 2 mm DTT). All measurements were made on a Spectramax M5 plate reader (Molecular Devices). The wavelengths for monitoring excitation (Ex), emission (Em), and the emission cutoffs (Co) for fluorescein and Texas Red were as follows: fluorescein, Ex = 494 nm, Em = 518 nm, and Co = 515 nm; and Texas Red, Ex = 585 nm, Em = 620 nm, and Co = 610 nm. Slit widths of 5 nm were used in all experiments. Data analysis was performed as previously described by fitting the data to a 1:1 binding model with a correction for changes in fluorophore intensity caused by protein binding (OriginPro 8 SRO) (27).
ELISA
The binding affinity between hLysRS proteins and HIV-1 GagΔp6 (or Biotin-WM CA-CTD) was measured by coating 96-Microwell polysorp plates (Nunc Maxisorp; Thermo Scientific) overnight using either 100 μl of 100 nm GagΔp6 or 100 μl of 4 μg/ml neutravidin at 4 °C. The wells were washed three times with 250 μl of buffer A (50 mm Tris-HCl, 150 mm NaCl, pH 7.4) followed by a 2-h blocking step using a 3% BSA solution dissolved in buffer A. WM CA-CTD was biotin-labeled, and 100 nm Biotin-WM CA-CTD was bound to the neutravidin-coated plates. Varying amounts of His-tagged LysRS variants (diluted in 3% BSA) were then applied, and plates were incubated at room temperature for 1 h. The wells were washed five times with 250 μl of ice-cold buffer B (150 μl of buffer A and 100 μl of Tween 20) followed by the addition of 100 μl of ice-cold anti-His-horseradish peroxidase antibody (Immunology Consultant Laboratory, Inc.) (1:5000 dilution) followed by incubation for 45 min on ice. The wells were again washed with 250 μl of ice-cold buffer B prior to the addition of 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Scientific). The reaction was incubated in the dark at room temperature for 30 min before the addition of 100 μl of 2 m H2SO4 to quench the reaction. The product absorbance was monitored at 450 nm using a Spectramax M5 plate reader. Data analysis was performed by fitting the data to the modified Hill equation for calculation of the apparent Kd in OriginPro 8 SRO,
where y is the calculated absorbance at 450 nm, Amin is the minimum absorbance, Amax is the highest absorbance, x is the titrated protein concentration, and n is the Hill coefficient.
tRNALys,3 Preparation and Aminoacylation Activity
Unmodified WT tRNALys,3 was prepared from FokI-digested plasmid pLYSF119 by in vitro transcription using T7 RNA polymerase as described previously (21). An extinction coefficient of 604,000 m−1 cm−1 was used to determine the concentration of tRNALys,3. Aminoacylation assays were conducted as described previously (21). To determine the initial rate of aminoacylation, assays were performed using 10 and 20 nm hLysRS (WT and mutants, respectively) and 0.5 μm tRNALys,3. Under these conditions, the initial rate is proportional to kcat/Km (28).
Dynamic Light Scattering
Dynamic light scattering (DLS) measurements were performed on a Malvern Instruments Zeta Sizer Nano-ZS apparatus. LysRS proteins were first exchanged into 50 mm NaHPO4, 150 mm NaCl, pH 7.5. The samples were then centrifuged for 5 min at 10,000 × g in a table top microcentrifuge to remove any aggregated protein particles. The collected supernatant was then filtered through a 0.22-μm PVDF syringe filter prior to analysis. Protein samples (60 μl at 20 μm) were put in a quartz cuvette, and the data were collected at 25 °C.
Gel Chromatography
The oligomeric states of WT and variant LysRS (F244A/I245A and I254D/R255A) were monitored by size exclusion chromatography using a HiLoad 16/60 Superdex 200 column (GE Healthcare) attached to a GE AKTApurifier INV-907 fast performance liquid chromatography system. The mobile phase was 50 mm NaPO4, pH 7, 150 mm NaCl, and 10 mm β-mercaptoethanol. The column was calibrated using molecular mass markers ranging from 13.7 to 232 kDa (Amersham Biosciences). A 50 μm solution of protein (100 μl) was loaded onto the column, and absorbance at 280 nm was monitored.
RESULTS
LysRS H7 Residues Are Essential for Packaging into HIV-1
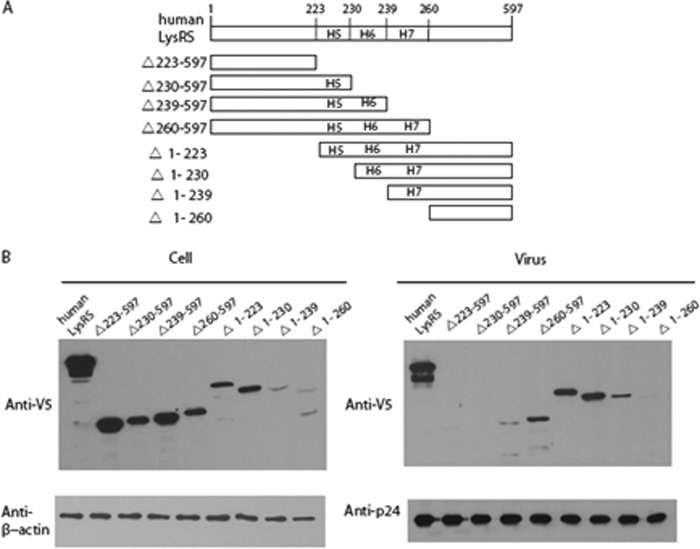
To map the motif 1 region of LysRS critical for its packaging into virions, eight truncation constructs of LysRS were constructed with deletions extending into H5, H6, and H7 (Fig. 1A). The DNAs coding for each of the V5-tagged truncated LysRS variants together with HIV-1 (BH10) were co-transfected into HEK-293T cells. Expression of LysRS variants observed in the cellular fraction probed with anti-V5 and anti-β-actin antibodies is shown in Fig. 1B (left panel). Viral lysates were probed with anti-V5 and anti-CAp24 antibodies (Fig. 1B, right panel). Only LysRS constructs that retained H7 (Δ260–597, Δ1–223, Δ1–230, and Δ1–239) were packaged to a significant extent into HIV-1. On the other hand, packaging was eliminated or drastically reduced for constructs that lacked H7 (Δ223–597, Δ230–597, Δ239–597, and Δ1–260), thereby suggesting a critical role for H7 in packaging of LysRS into HIV-1 (Fig. 1B, right panel).
FIGURE 1.
Effect of LysRS truncation on packaging into HIV-1. HEK-293T cells were co-transfected with plasmids coding for BH10 and wild-type or mutant hLysRS, and Western blots of cell and viral lysates were probed for either cellular hLysRS and β-actin (B, left panel) or viral LysRS and CAp24 (B, right panel). A, scheme showing hLysRS truncation constructs. B, Western blots of cellular (left panel) and viral lysates (right panel).
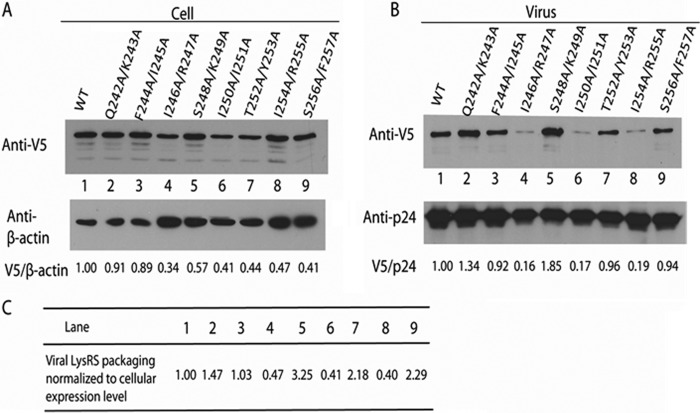
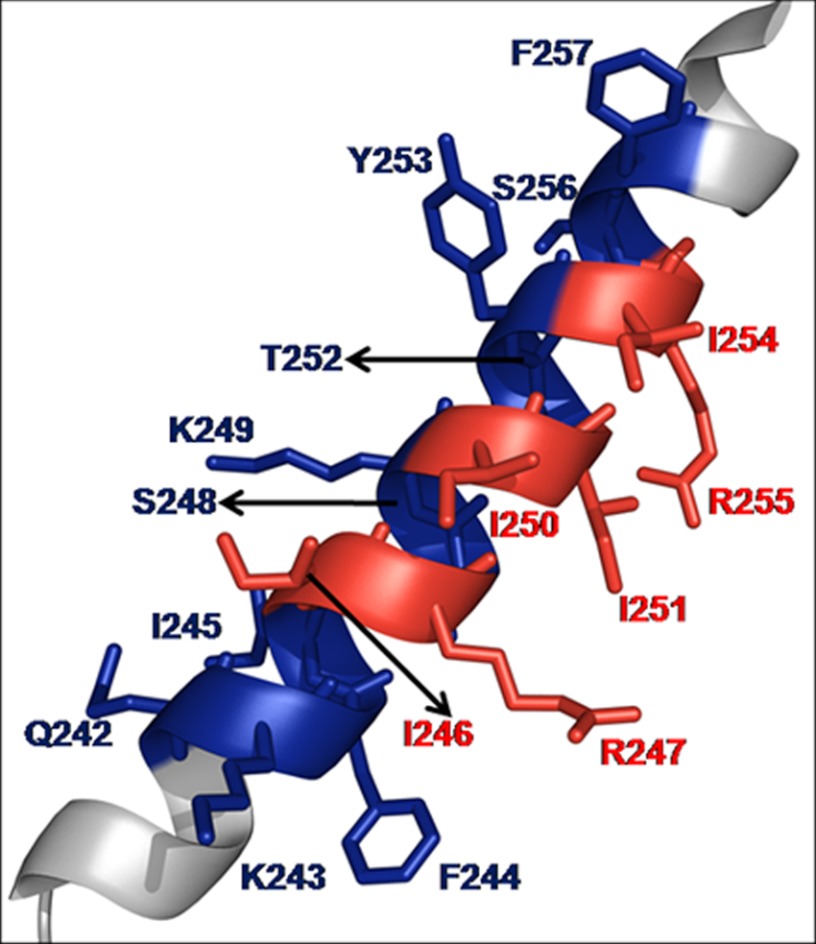
To map specific residues within H7 of hLysRS responsible for packaging, Ala scanning mutagenesis of H7 residues was carried out. There are six turns in H7, and eight double mutant variants were prepared to probe the role of each in packaging. HEK-293T cells were co-transfected with plasmids coding for BH10 and wild-type or mutant hLysRS, and Western blots of cell and viral lysates were probed as described above. As shown in Fig. 2B, three mutants displayed >80% reduction in viral hLysRS packaging: I246A/R247A (lane 4), I250A/I251A (lane 6), and I254A/R255A (lane 8). After taking into account the variable cell expression of exogenous LysRS seen in Fig. 2A, these three H7 variants still display >50% reduction in LysRS packaging into virus (listed in Fig. 2C). Another variant displayed WT levels of packaging (F244A/I245A, lane 3), and interestingly, four variants displayed increased levels of packaging relative to WT hLysRS (Q242A/K243A, lane 2; S248A/K249A, lane 5; T252A/Y253A, lane 7; and S256A/F257A, lane 9). In three independent experiments, whereas the pattern of exogenous LysRS expression in the cell varied, the pattern of incorporation of mutant LysRS into virions remained similar to that listed in Fig. 2C. Fig. 3 shows the location of the mutated residues on H7. Interestingly, the second residue of each of the double mutations that resulted in decreased packaging is located on the same face of H7 and in a similar location of each turn (Fig. 3, Arg-247, Ile-251, and Arg-255).
FIGURE 2.
Effect of H7 double mutations on LysRS packaging into HIV-1. HEK-293T cells were co-transfected with plasmids coding for BH10 and wild-type or mutant hLysRS, and Western blots of cell and viral lysates were probed for either cellular hLysRS and β-actin (A) or viral LysRS and CAp24 (B). Lanes 1, WT; lanes 2, Q242A/K243A; lanes 3, F244A/I245A; lanes 4, I246A/R247A; lanes 5, S248A/K249A; lanes 6, I250A/I251A; lanes 7, T252A/Y253A; lanes 8, I254A/R255A; lanes 9, S256A/F257A. In C, the viral LysRS packaged is normalized to the cellular expression of hLysRS by dividing V5/p24 by V5/β-actin.
FIGURE 3.
Structure of hLysRS H7 (Protein Data Bank code 3bju) highlighting residues probed in this study. Mutation of the residues shown in red resulted in decreased packaging of LysRS into HIV-1. Mutation of the residues in blue did not adversely affect packaging of LysRS.
LysRS Double Mutants That Are Defective in Packaging into HIV-1 Are Also Defective in Binding to GagΔp6 and CA-CTD
We next probed the role of the hLysRS H7 residues determined to be important for LysRS packaging for their ability to bind viral proteins. To quantify the binding affinity between HIV-1 GagΔp6 and LysRS variants, we selected five variants for analysis based on the results shown in Fig. 2. Three variants that showed a reduction in LysRS packaging (I246A/R247A, I250A/I251A, and I254A/R255A), one that had no effect on packaging (F244A/I245A), and one that showed the largest increase in packaged LysRS (S248A/K249A) were selected for analysis. Based on the crystal structure of hLysRS (14), four pairs of selected residues had one side chain in the pair pointing in the same direction (see Fig. 3, Phe-244, Arg-247, Ile-251, and Arg-255), whereas the side chains of residues Ser-248 and Lys-249 of the final pair of residues selected for in vitro analysis both point in the opposite direction relative to Phe-244, Arg-247, Ile-251, and Arg-255. In addition to Ala substitutions at all positions, we made additional variants wherein we substituted the hydrophobic Ile residue, as well as Ser, with a hydrophilic Asp residue to ensure greater disruption of potential interactions. Both double and single point mutations were constructed as summarized in Table 1 and supplemental Table S1.
TABLE 1.
Comparison of the GagΔp6 and WM CA-CTD binding affinity, oligomeric state, and aminoacylation activity of WT and mutant hLysRS
| LysRS variant |
Kd |
Dynamic light scatteringc |
Relative aminoacylation activity kcat/Kmd | ||
|---|---|---|---|---|---|
| Fluorescence anisotropya GagΔp6 | ELISAb biotin-WM-CA-CTD | d(H) | Volume | ||
| μm | nm | % | |||
| WT | 0.56 ± 0.08 | 0.62 ± 0.04 | 10.4 | 96.8 | 1 |
| F244A | 0.7 ± 0.1 | 0.8 ± 0.15 | 10.4 | 98.6 | 0.9 |
| I245A | 0.62 ± 0.05 | 0.73 ± 0.1 | 10.5 | 94.8 | 1 |
| F244A/I245A | 0.52 ± 0.07 | 0.7 ± 0.04 | 10.2 | 97.8 | 0.9 |
| S248D | 0.53 ± 0.09 | 0.56 ± 0.07 | 10.3 | 99.5 | 1 |
| K249A | 0.59 ± 0.07 | 0.74 ± 0.2 | 10.4 | 99.2 | 0.9 |
| S248D/K249A | 0.62 ± 0.1 | 0.66 ± 0.1 | 10.1 | 97.6 | 1 |
| I246D | 2.6 ± 1.1 | 1.2 ± 0.4 | 9.2 | 99.8 | 0.6 |
| R247A | 1.5 ± 0.1 | 1.8 ± 0.6 | 8.3 | 62.7 | 0.3 |
| 17.2 | 35.8 | ||||
| I246D/R247A | 2.5 ± 0.5 | 3.6 ± 0.9 | 8.9 | 99.9 | 0.2 |
| I250D | 1.8 ± 0.5 | 1.8 ± 0.4 | 19.9 | 57.7 | 0.4 |
| 8.5 | 34.6 | ||||
| I251D | 2.6 ± 0.3 | 1.5 ± 0.5 | 8.7 | 99.9 | 0.4 |
| I250D/I251D | 2.7 ± 0.7 | 3.8 ± 0.6 | 8.1 | 99.9 | 0.2 |
| I254D | 2.1 ± 0.5 | 1.6 ± 0.3 | 8.8 | 99.9 | 0.4 |
| R255A | 1.9 ± 0.1 | 3.4 ± 0.9 | 8.9 | 98.4 | 0.4 |
| I254D/R255A | 5.1 ± 2.3 | 5.5 ± 1.8 | 8.6 | 99.8 | 0.3 |
a Measurements were performed using 100 nm Texas Red-labeled GagΔp6 in the presence of binding buffer and varying amounts of hLysRS proteins as described under “Experimental Procedures.”
b Measurements were performed by immobilizing 100 nm biotin-WM CA-CTD on neutravidin coated 96-well microtiter plate.
c The polydispersity index was between 0.2 and 0.6 for all of the samples. d(H) is the hydrodynamic diameter, and the volume denotes the relative amounts of the predominant species.
d Initial rates of aminoacylation are proportional to kcat/Km under the conditions used here, and all values are relative to WT hLysRS, which was set to 1. With the exception of the DLS measurements, all the results are the averages of three trials.
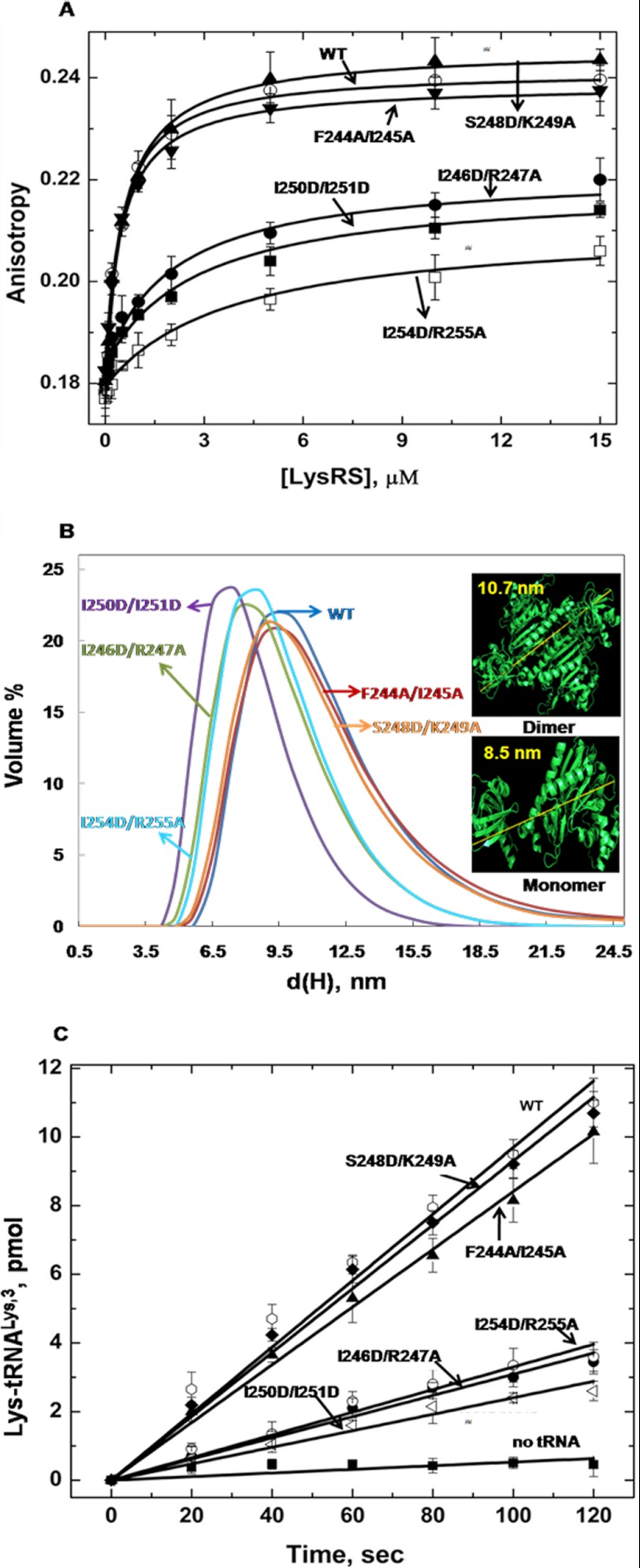
FA binding assays were performed to determine the binding affinity of WT and variant hLysRS proteins for HIV-1 GagΔp6, and WM CA-CTD. As shown in Fig. 4A, LysRS double mutants at positions that were important for viral packaging (I246D/R247A, I250D/I251D, and I254D/R255A) also showed a ∼ 5–10-fold reduction in binding to GagΔp6 (Table 1). The control variants F244A/I245A (Kd = 0.52 μm) and S248D/K249A (Kd = 0.62 μm) bound with affinities comparable with that of the WT protein (Kd = 0.56 μm). When changes to only Ala were made, the double mutants also displayed an ∼3-fold reduction in binding to GagΔp6 (supplemental Table S1), which was somewhat less severe compared with the Asp containing double mutants (Table 1). To confirm the binding results, we also employed ELISA to measure the binding of a subset of the LysRS mutants to biotinylated WM CA-CTD. As shown in Table 1, F244A/I245A and S248D/K249A hLysRS once again bound with a similar affinity as the WT protein, but variants with changes in residues critical for LysRS packaging showed a 5–9-fold weaker binding affinity, consistent with the FA results. Single point mutations of these residues generally showed 3–5-fold reduced binding. We also tested the binding of hLysRS double mutants to GagΔp6 by ELISA and obtained very similar results to FA studies (data not shown). Taken together, these results confirm the importance of three critical pairs of H7 residues (Ile-246/Arg-247, Ile-250/Ile-251, and Ile-254/Arg-255) for binding to HIV-1 GagΔp6. Furthermore, these results suggest that a 5-fold reduction in binding affinity is sufficient to significantly reduce packaging of hLysRS into virions.
FIGURE 4.
A, FA binding assay to determine the affinity between 100 nm GagΔp6 and hLysRS variants. B, size distribution of hLysRS proteins as measured by dynamic light scattering, where d(H) is the hydrodynamic diameter. The crystal structure (Protein Data Bank code 3bju) of hLysRS was used to estimate the expected diameter of a dimer (10.7 nm) and monomer (8.5 nm) (inset). C, comparison of initial rates of aminoacylation of human tRNALys,3 by WT and variant hLysRS proteins.
Like other class II synthetases, LysRS is a functional dimer in its canonical role in tRNA aminoacylation, and mutations that affect dimerization do not affect tRNA binding but do reduce enzymatic activity (21, 23). Because the mutations we made that affected LysRS packaging and binding to HIV-1 Gag were in the H7 dimerization domain, it was important to determine the effect of these changes on the oligomeric state of the protein and on its enzymatic activity.
LysRS Mutants Defective in Binding to GagΔp6 Are Shifted toward Monomeric State
DLS measurements were carried out to examine the effect of mutations on the monomer/dimer equilibrium of hLysRS. The published crystal structure (Protein Data Bank code 3bju) of LysRS (14) was first used to measure the largest end to end distance of the dimeric (∼10.7 nm) and monomeric (∼8.5 nm) proteins (Fig. 4B, inset). As shown in Fig. 4B, DLS measurements of the WT protein along with the control double mutants, F244A/I245A and S248D/K249A showed an equilibrium distribution toward the dimeric species. In contrast, the variants that were defective in packaging and bound more weakly to GagΔp6 and CA-CTD showed an equilibrium shift toward the monomeric form of the protein. These results are summarized in Table 1 and are consistent with gel chromatography experiments performed using WT, F244A/I245A, and I254D/R255A hLysRS (data not shown). The I250D and R247A variants showed the presence of a population with a hydrodynamic diameter distribution of >10.7 nm, suggesting the formation of a higher order complex. A second population corresponding to a monomer was also present in this case (Table 1). Overall, these results suggest that mutating key residues in H7 of LysRS significantly perturbs the monomer-dimer equilibrium, with a correlation between residues important for LysRS packaging into HIV-1 and a shift toward the monomeric form of the protein.
LysRS Variants with a Mutated Dimer Interface Are Also Defective in Aminoacylation
To determine the effect of the H7 changes on enzymatic activity, we performed aminoacylation assays using in vitro transcribed cognate human tRNALys,3. The hLysRS variants shifted toward a monomeric state (I246D/R247A, I250D/I251D, and I254D/R255A) showed a 3–4-fold reduced kcat/Km relative to the WT protein or to control proteins F244A/I245A and S248D/K249A (Fig. 4C and Table 1). A slightly lower, ∼2-fold reduction in aminoacylation activity was measured for variants with single point mutations at residues Ile-246, Arg-247, Ile-250, Ile-251, Ile-254, and Arg-255 (Table 1 and supplemental Table S1). Thus, mutations in H7 that perturb the oligomeric state also affect charging of cognate tRNALys,3, as expected. Although the charging activity was reduced, the decreases were modest, suggesting that the shift to monomer was likely incomplete.
DISCUSSION
In this work, we mapped residues in the H7 dimerization domain of hLysRS that contribute to packaging of the synthetase-tRNA complex into HIV-1. Mutating three pairs of residues along primarily one face of the helix reduced the packaging of hLysRS into HIV-1, as well as binding to GagΔp6 and CA-CTD in vitro. These mutations were also effective in disrupting the dimeric interface of the protein, which was evident by DLS measurements and reduced aminoacylation activity. HIV-1 Gag and hLysRS are both capable of homodimer formation, as well as higher order oligomerization (14, 29). However, previous work showed that homodimerization of these proteins is not important for their interaction, because a monomeric triple mutant variant still bound with an affinity within 2-fold of that of WT LysRS (21). Moreover, size exclusion chromatography using the WT proteins provided evidence for exclusively heterodimer formation and not a higher order complex (21). Recently, it was shown that upon insertion of a flavodoxin domain in a region proximal to motif 1, the resulting monomeric LysRS species retained its ability to interact with CA-CTD (23). Using available data, computational approaches were used to generate a model of the CA-CTD/LysRS interaction (23). Consistent with this model, our data suggest that the residues on H7 of hLysRS that are important for interaction with CA-CTD are also important in maintaining the dimer interface.
Our results extend the current model of a tRNALys packaging complex (30) where HIV-1 assembly at the plasma membrane involves complex formation between genomic RNA, Gag, Gag-Pol, tRNALys, and hLysRS, with LysRS specifically interacting with Gag. It has been shown that Gag alone is sufficient for the incorporation of LysRS into Gag viral-like particles, whereas GagPol is additionally required for the incorporation of primer tRNALys. The details of this complex network of interactions are still unknown, and the presence of multiple pools of Gag actively undertaking several functions to aid in the viral life cycle has been suggested (31). This is supported by the packaging of several host factors by Gag in addition to LysRS including ABCE 1, cyclophilin A, Tsg 101, and Alix, which are also critical for viral infectivity (5, 32–36). Previously, it has been suggested that newly synthesized LysRS is packaged into HIV-1 (18), but how LysRS is diverted from its normal role in translation is still unclear.
Aminoacyl-tRNA synthetases have been shown to function in a wide array of cellular processes that are distinct from aminoacylation and changes in oligomeric state, as well as post-translational modifications have been shown to regulate these alternate functions (37–39). An intriguing observation that highlights the dynamic nature of LysRS within the cell, is illustrated by its mobilization to the nucleus (37). In the MSC, LysRS interacts with the dimeric p38 protein in a unique α2β1:β1α2 geometry; which is designed to control both retention and mobilization of LysRS from the MSC (16). In response to an immunological challenge, specific phosphorylation of Ser-207 of hLysRS has been shown to result in release of LysRS from the MSC and enhanced diadenosine tetraphosphate synthesis. Nuclear translocation of hLysRS is followed by transcriptional activation of genes via interaction with microphthalmia-associated transcription factor (37). HIV-1 CA (40–42) has also been shown to readily adopt different dimerization modes (43) and to interact with a variety of cellular proteins (44).
In summary, we have identified residues in H7 that, when altered, promote formation of monomeric hLysRS that is less efficient in tRNA aminoacylation and displays decreased GagΔp6 binding affinity and reduced packaging into HIV-1. Thus, the H7 residues identified in the present study appear to have a dual function in both the canonical and noncanonical roles of hLysRS. Previous studies identified alternative monomeric LysRS variants that retained interaction with Gag (21, 23). To reconcile the new findings with these previous studies suggesting that monomeric LysRS is the functional form packaged into virus, we hypothesize that the dimerization state of hLysRS is perturbed in the pool of synthetase molecules that are packaged in a manner that facilitates binding to Gag. Whether post-translational modifications of LysRS are altered during HIV-1 infection, thereby modulating the oligomerization state of LysRS and facilitating interaction with components of the tRNALys packaging complex, remains to be tested.
Acknowledgments
Plasmids encoding HIV-1 CA-CTD were a gift from Dr. Wesley I. Sundquist (University of Utah). We thank Dr Jingjing Li for preparing the HIV-1 biotin-WM CA-CTD mutant and development of the ELISA assay. We also thank Dr. Christopher P. Jones and Joseph Weinstein-Webb for purifying GagΔp6.
This work was supported, in whole or in part, by National Institutes of Health Grant AI077387 (to L. K. and K. M.-F.).

This article contains supplemental text and Table S1.
- CA
- capsid
- hLysRS
- human lysyl-tRNA synthetase
- CTD
- C-terminal domain
- FA
- fluorescence anisotropy
- MSC
- multi-synthetase complex
- DLS
- dynamic light scattering.
REFERENCES
- 1. Mak J., Kleiman L. (1997) Primer tRNAs for reverse transcription. J. Virol. 71, 8087–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swanstrom R., Wills J. W. (1997) Synthesis, assembly, and processing of viral proteins, pp. 263–334, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 3. Kleiman L., Halwani R., Javanbakht H. (2004) The selective packaging and annealing of primer tRNALys3 in HIV-1. Curr HIV Res. 2, 163–175 [DOI] [PubMed] [Google Scholar]
- 4. Mak J., Jiang M., Wainberg M. A., Hammarskjöld M. L., Rekosh D., Kleiman L. (1994) Role of Pr160Gag-Pol in mediating the selective incorporation of tRNA(Lys) into human immunodeficiency virus type 1 particles. J. Virol. 68, 2065–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cen S., Khorchid A., Javanbakht H., Gabor J., Stello T., Shiba K., Musier-Forsyth K., Kleiman L. (2001) Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75, 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gabor J., Cen S., Javanbakht H., Niu M., Kleiman L. (2002) Effect of altering the tRNALys,3 concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 76, 9096–9102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo F., Cen S., Niu M., Javanbakht H., Kleiman L. (2003) Specific inhibition of the synthesis of human lysyl-tRNA synthetase results in decreases in tRNALys incorporation, tRNA3,Lys annealing to viral RNA, and viral infectivity in human immunodeficiency virus type 1. J. Virol. 77, 9817–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Javanbakht H., Cen S., Musier-Forsyth K., Kleiman L. (2002) Correlation between tRNALys3 aminoacylation and its incorporation into HIV-1. J. Biol. Chem. 277, 17389–17396 [DOI] [PubMed] [Google Scholar]
- 9. Cen S., Javanbakht H., Niu M., Kleiman L. (2004) Ability of wild-type and mutant lysyl-tRNA synthetase to facilitate tRNALys incorporation into human immunodeficiency virus type 1. J. Virol. 78, 1595–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 11. Eriani G., Dirheimer G., Gangloff J. (1990) Aspartyl-tRNA synthetase from Escherichia coli. Cloning and characterisation of the gene, homologies of its translated amino acid sequence with asparaginyl- and lysyl-tRNA synthetases. Nucleic Acids Res. 18, 7109–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cusack S., Yaremchuk A., Tukalo M. (1996) The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript. Anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 15, 6321–6334 [PMC free article] [PubMed] [Google Scholar]
- 13. Onesti S., Miller A. D., Brick P. (1995) The crystal structure of the lysyl-tRNA synthetase (LysU) from Escherichia coli. Structure. 3, 163–176 [DOI] [PubMed] [Google Scholar]
- 14. Guo M., Ignatov M., Musier-Forsyth K., Schimmel P., Yang X. L. (2008) Crystal structure of tetrameric form of human lysyl-tRNA synthetase. Implications for multisynthetase complex formation. Proc. Natl. Acad. Sci. U.S.A. 105, 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park S. G., Ewalt K. L., Kim S. (2005) Functional expansion of aminoacyl-tRNA synthetases and their interacting factors. New perspectives on housekeepers. Trends Biochem. Sci. 30, 569–574 [DOI] [PubMed] [Google Scholar]
- 16. Fang P., Zhang H. M., Shapiro R., Marshall A. G., Schimmel P., Yang X. L., Guo M. (2011) Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc. Natl. Acad. Sci. U.S.A. 108, 8239–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaminska M., Shalak V., Francin M., Mirande M. (2007) Viral hijacking of mitochondrial lysyl-tRNA synthetase. J. Virol. 81, 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halwani R., Cen S., Javanbakht H., Saadatmand J., Kim S., Shiba K., Kleiman L. (2004) Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 78, 7553–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobbi L., Octobre G., Dias J., Comisso M., Mirande M. (2011) Association of mitochondrial Lysyl-tRNA synthetase with HIV-1 GagPol involves catalytic domain of the synthetase and transframe and integrase domains of Pol. J. Mol. Biol. 410, 875–886 [DOI] [PubMed] [Google Scholar]
- 20. Javanbakht H., Halwani R., Cen S., Saadatmand J., Musier-Forsyth K., Gottlinger H., Kleiman L. (2003) The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 278, 27644–27651 [DOI] [PubMed] [Google Scholar]
- 21. Kovaleski B. J., Kennedy R., Hong M. K., Datta S. A., Kleiman L., Rein A., Musier-Forsyth K. (2006) In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J. Biol. Chem. 281, 19449–19456 [DOI] [PubMed] [Google Scholar]
- 22. Kovaleski B. J., Kennedy R., Khorchid A., Kleiman L., Matsuo H., Musier-Forsyth K. (2007) Critical role of helix 4 of HIV-1 capsid C-terminal domain in interactions with human lysyl-tRNA synthetase. J. Biol. Chem. 282, 32274–32279 [DOI] [PubMed] [Google Scholar]
- 23. Guo M., Shapiro R., Morris G. M., Yang X. L., Schimmel P. (2010) Packaging HIV virion components through dynamic equilibria of a human tRNA synthetase. J. Phys. Chem. B. 114, 16273–16279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Na Nakorn P., Treesuwan W., Choowongkomon K., Hannongbua S., Boonyalai N. (2011) In vitro and in silico binding study of the peptide derived from HIV-1 CA-CTD and LysRS as a potential HIV-1 blocking site. J. Theor. Biol. 270, 88–97 [DOI] [PubMed] [Google Scholar]
- 25. Wei M., Yang Y., Niu M., Desfosse L., Kennedy R., Musier-Forsyth K., Kleiman L. (2008) Inability of human immunodeficiency virus type 1 produced in murine cells to selectively incorporate primer formula. J. Virol. 82, 12049–12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Datta S. A., Rein A. (2009) Preparation of recombinant HIV-1 Gag protein and assembly of virus-like particles in vitro. Methods Mol. Biol. 485, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dewan V., Liu T., Chen K. M., Qian Z., Xiao Y., Kleiman L., Mahasenan K. V., Li C., Matsuo H., Pei D., Musier-Forsyth K. (2012) Cyclic peptide inhibitors of HIV-1 capsid-human lysyl-tRNA synthetase interaction. ACS Chem. Biol. 7, 761–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stello T., Hong M., Musier-Forsyth K. (1999) Efficient aminoacylation of tRNALys,3 by human lysyl-tRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res. 27, 4823–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rein A., Datta S. A., Jones C. P., Musier-Forsyth K. (2011) Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 36, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kleiman L., Jones C. P., Musier-Forsyth K. (2010) Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 584, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chu H., Wang J. J., Spearman P. (2009) Human immunodeficiency virus type-1 gag and host vesicular trafficking pathways. Curr. Top. Microbiol. Immunol. 339, 67–84 [DOI] [PubMed] [Google Scholar]
- 32. Zimmerman C., Klein K. C., Kiser P. K., Singh A. R., Firestein B. L., Riba S. C., Lingappa J. R. (2002) Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415, 88–92 [DOI] [PubMed] [Google Scholar]
- 33. Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C. T., Sodroski J., Göttlinger H. G. (1994) Functional association of cyclophilin A with HIV-1 virions. Nature 372, 363–365 [DOI] [PubMed] [Google Scholar]
- 34. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., Sundquist W. I. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 35. Strack B., Calistri A., Craig S., Popova E., Göttlinger H. G. (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 [DOI] [PubMed] [Google Scholar]
- 36. Franke E. K., Yuan H. E., Luban J. (1994) Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372, 359–362 [DOI] [PubMed] [Google Scholar]
- 37. Yannay-Cohen N., Carmi-Levy I., Kay G., Yang C. M., Han J. M., Kemeny D. M., Kim S., Nechushtan H., Razin E. (2009) LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34, 603–611 [DOI] [PubMed] [Google Scholar]
- 38. Park S. G., Kim H. J., Min Y. H., Choi E. C., Shin Y. K., Park B. J., Lee S. W., Kim S. (2005) Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl. Acad. Sci. U.S.A. 102, 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park S. G., Schimmel P., Kim S. (2008) Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. U.S.A. 105, 11043–11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ganser-Pornillos B. K., von Schwedler U. K., Stray K. M., Aiken C., Sundquist W. I. (2004) Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 78, 2545–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ganser-Pornillos B. K., Yeager M., Sundquist W. I. (2008) The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pornillos O., Ganser-Pornillos B. K., Kelly B. N., Hua Y., Whitby F. G., Stout C. D., Sundquist W. I., Hill C. P., Yeager M. (2009) X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivanov D., Tsodikov O. V., Kasanov J., Ellenberger T., Wagner G., Collins T. (2007) Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 104, 4353–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mascarenhas A. P., Musier-Forsyth K. (2009) The capsid protein of human immunodeficiency virus. Interactions of HIV-1 capsid with host protein factors. FEBS J. 276, 6118–6127 [DOI] [PubMed] [Google Scholar]