Background: Protein-tyrosine pseudokinase 7 (PTK7) is an essential component of the Wnt pathway.
Results: PTK7 is regulated by an intricate post-translational mechanism that, in addition to MT1-MMP and ADAM proteolysis, involves γ-secretase and proteasomes, and these events contribute to cell invasion.
Conclusion: Proteolytic processing of PTK7 involves several distinct membrane proteinases.
Significance: Therapeutic inhibition of PTK7 shedding could slow cancer progression.
Keywords: ADAM ADAMTS, Invasion, Membrane Enzymes, Metalloprotease, Wnt Pathway, MT1-MMP, PCP, PTK7
Abstract
The membrane PTK7 pseudokinase, a component of both the canonical and noncanonical/planar cell polarity Wnt pathways, modulates cell polarity and motility in biological processes as diverse as embryo development and cancer cell invasion. To determine the individual proteolytic events and biological significance of the ectodomain shedding in the PTK7 function, we used highly invasive fibrosarcoma HT1080 cells as a model system. Current evidence suggested a likely link between PTK7 shedding and cell invasion in our HT1080 cell model system. We also demonstrated that in HT1080 cells the cleavage of the PTK7 ectodomain by an ADAM proteinase was coupled with the membrane type-1 matrix metalloproteinase (MT1-MMP) cleavage of the PKP621↓LI site in the seventh Ig-like domain of PTK7. Proteolytic cleavages led to the generation of two soluble, N-terminal and two matching C-terminal, cell-associated fragments of PTK7. This proteolysis was a prerequisite for the intramembrane cleavage of the C-terminal fragments of PTK7 by γ-secretase. γ-Secretase cleavage was predominantly followed by the efficient decay of the resulting C-terminal PTK7 fragment via the proteasome. In contrast, in HT1080 cells, which overexpressed the C-terminal PTK7 fragment, the latter readily entered the nucleus. Our data imply that therapeutic inhibition of PTK7 shedding may be used to slow cancer progression.
Introduction
Cells exhibit a well coordinated arsenal of migration and invasion mechanisms. Understanding of these mechanisms is a step toward prevention and therapy of a multitude of diseases. Regardless of the extensive earlier research, our understanding of certain central molecules of the Wnt pathway, a vital pathway in both normal development and disease, is limited. The catalytically inert protein-tyrosine kinase 7 pseudokinase (PTK7)3 (also known as colon carcinoma kinase-4/CCK-4 in humans), is an intrinsic component of the Wnt pathway and a member of a receptor protein-tyrosine pseudokinase (RPTK) family. Mutant mice with a 1–114 PTK7 truncation die perinatally with severe defects (1, 2). It is believed as of now that PTK7 regulates both the canonical Wnt and the noncanonical Wnt/planar cell polarity (Wnt/PCP) pathways (3–5). In different biological systems, PTK7 interacts, either directly or indirectly, with plexins, semaphorins, the receptor for activated-C kinase 1 (RACK1) and protein kinase C δ1 (PKCδ1), Wnt3a, Wnt8, and β-catenin (3, 5–10). The full-length membrane 1070-residue long PTK7 (Swiss-Prot number Q13308.2) consists of seven extracellular immunoglobulin-like (Ig) domains (residues 31–684), a 685–704 stalk region, a 705–725 transmembrane region, a 726–795 juxtamembrane region, and a 796–1070 catalytically inactive cytoplasmic tyrosine kinase (PTK) domain that shares over 40% identity with muscle-specific kinase (MuSK) (11).
PTK7 expression is elevated in the fetal colon and cancer tissues, including colon, gastric, and lung carcinomas (12–14) and also in acute myeloid leukemia blast cells (15). In contrast, the expression of PTK7 is decreased or lost in metastastic melanomas (16) and breast carcinomas (17). Deletions of chromosome 6p, where the human PTK7 gene is located (6p21.1), were also found in a number of cancers (18, 19). The exact function of PTK7 in cancer remains unclear.
Matrix metalloproteinases (MMPs) and ADAMs (A Disintegrin And Metalloproteinase) belong to zinc endopeptidases of the metzincin superfamily (20, 21). Membrane type-1 matrix metalloproteinase (MT1-MMP) is a prototypic member of a membrane-type MMP subfamily. MT1-MMP is distinguished from soluble MMPs by a C-terminal transmembrane domain and a cytoplasmic tail (22, 23). Because of its accumulation at the leading and trailing cell edges, MT1-MMP functions as the main mediator of the proinvasive pericellular proteolysis in migrating, polarized cells. MT1-MMP proteolysis degrades extracellular matrix (ECM) proteins, activates soluble MMPs, and controls the functionality of cell adhesion and signaling receptors, including CD44, integrins, low density lipoprotein receptor-related protein 1 (LRP1), transglutaminase, and PTK7 (24–33). As a result, proinvasive, protumorigenic MT1-MMP usurps tumor growth control and stimulates cancer cell invasion and metastasis (27, 34–36). MT1-MMP also plays a critical role in normal development: in contrast with other MMP knockouts with minor developmental defects, MT1-MMP null mice are dwarfs and die at adulthood (37, 38).
In humans, the ADAM family includes 19 catalytically active members. ADAMs exhibit a disintegrin domain, a metalloproteinase domain, an EGF-like domain, a transmembrane domain, and a cytoplasmic domain. The disintegrin-like domain can bind integrins or other receptors. The metalloproteinase-like domain contains a consensus active site sequence (39, 40).
MMPs and ADAMs are synthesized as inactive zymogens (39, 41). Once activated, MMPs/ADAMs can be inhibited by tissue inhibitors of metalloproteinases (TIMPs). Four individual species of TIMPs are known in humans (TIMP-1, -2, -3, and -4) (42). The protease/TIMP balance is a major factor regulating the net metalloproteinase activity in vivo.
Our studies revealed that direct interactions between cellular MT1-MMP and PTK7 constitute an evolutionary conserved master switch in directional cell motility. Thus, the PTK7 ectodomain was efficiently shed in a variety of cell lines, in which MT1-MMP was overexpressed, and also in human fibrosarcoma HT1080 cells with high endogenous levels of MT1-MMP (43, 44). MT1-MMP cleaved the exposed PKP621↓LI sequence of the seventh Ig-like domain of the full-length membrane PTK7 and generated, as a result, an N-terminal, soluble, 65-kDa ectodomain fragment (N-PTK7–65) and the C-terminal membrane-attached, 50-kDa fragment (C-PTK7–50) that included the pseudokinase domain. MT1-MMP proteolysis of PTK7 reversed the anti-invasive function of the full-length PTK7 (43, 44).
It is expected that MT1-MMP, however, is not a singular PTK7 sheddase and that additional proteinases also contribute to the proteolysis of cellular PTK7. To determine whether the endogenous proteinases, which are distinct and additional to MT1-MMP, contribute significantly to the PTK7 shedding, we used highly migratory fibrosarcoma HT1080 cells. Here, we provide evidence for the previously uncharacterized proteolytic mechanisms, which also contribute to the regulation of PTK7 functionality and cancer cell invasion.
MATERIALS AND METHODS
Cells, Antibodies, and Reagents
Original human fibrosarcoma HT1080 cells (HT1080 cells) and normal mammary epithelial 184B5 cells (184B5 cells) were from ATCC (Manassas, VA). A goat polyclonal antibody (catalog number AF4499) against the N-terminal 31–199 portion of PTK7 was from R&D Systems. A murine monoclonal 3G4 antibody (catalog number MAB1767) against the catalytic domain of MT1-MMP was from Millipore. The function blocking, active site-targeting human recombinant MT1-MMP antibody (DX2400) was kindly provided by Dyax. A murine monoclonal antibody and a goat FITC-conjugated antibody to the V5 tag were from Invitrogen. A murine monoclonal FLAG M2 antibody, the FLAG M2 antibody-agarose beads, a monoclonal actin antibody, and phorbol 12-myristate 13-acetate (PMA) were from Sigma. Rabbit ADAM17/TACE (tumor necrosis factor-α converting enzyme) polyclonal antibodies were from Abcam and Cell Signaling. The ADAM17 inhibitor INCB3619 was kindly provided by Incyte. Recombinant TIMP-1 was from Invitrogen. Recombinant TIMP-3 and TIMP-4 were from R&D Systems. Recombinant TIMP-2 was isolated earlier (45). γ-Secretase inhibitor IX (N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester) was from Calbiochem. The irreversible proteasome inhibitor lactacystin was from Enzo Life Sciences.
Cloning and Mutagenesis
HT1080 cells transfected with the full-length MT1-MMP (MT1-MMP cells), the full-length PTK7-V5-HIS and PTK7-FLAG constructs (PTK7-V5 and PTK7-FLAG cells, respectively), and the L622D-FLAG PTK7 mutant with the inactivated MT1-MMP cleavage site (L622D-FLAG cells) were characterized earlier (43, 44). The shRNA constructs for the MT1-MMP silencing and the required scrambled shRNA controls were obtained and characterized earlier (33, 45, 46). Briefly, the MT1-MMP shRNA target sequence 5′-GAAGCCUGGCUACAGCAAUAU-3′ was cloned into the psiLentGene Puromycin vector (Promega) and the resulting construct was then used to transfect HT1080 cells. Stably transfected cells (shMT1-MMP cells) were selected using 2 μg/ml of puromycin. MT1-MMP levels were determined in the selected cells using Western blotting with the 3G4 antibody. To obtain the construct coding for the 726–1070 cytoplasmic sequence of PTK7 (PTK7726–1070), we truncated the PTK7-V5-HIS template sequence using 5′-TGCAAGAAGCGCTGC-3′ and 5′-CGGCTTGCTGTCCAC-3′ as the forward and reverse primers, respectively, in the PCR. The resulting constructs were subcloned into the pcDNA3.1D/V5-His-TOPO directional TOPO expression vector (Invitrogen). The full-length PTK7-FLAG and PTK7-V5 constructs were also transiently expressed in HT1080, MT1-MMP, and shMT1-MMP cells. For this purpose, cells (1 × 105/well) were seeded in wells of a 12-well tissue culture plate. Cells were then transfected with the PTK7 constructs using Lipofectamine 2000 (Invitrogen). In 24 h, cells were incubated in serum-free DMEM for an additional 16 h, with or without the stimulation by PMA (50 ng/ml). The levels of the soluble PTK7 fragments were measured in the medium samples using Western blotting with the PTK7 antibody. In our studies, we also used 184B5 cells infected with the MT1-MMP-V5 lentiviral construct (184B5-MT1 cells), which were isolated earlier (47).
Detection of the Soluble PTK7 Ectodomain in the Medium Samples
Cells were grown to reach a 60–90% confluence in wells of a 12-well tissue culture plate (BD Biosciences). Cells were washed with and then transferred to fresh serum-free DMEM (0.5 ml/well). PMA (50 ng/ml) and the inhibitors were added to the cells at the indicated concentrations and incubation was continued for an additional 16 h. The medium aliquots were then withdrawn and centrifuged to remove cell debris. The supernatant was concentrated until dryness using a SpeedVac. To remove salt, the pellet was washed using acetone, dissolved in 1% SDS, and centrifuged to remove insoluble material (14,000 × g, 10 min). The supernatant aliquots were separated by SDS-gel electrophoresis in 4–12% NuPAGE gels (Invitrogen). The soluble species of PTK7 were detected by Western blotting with a goat polyclonal PTK7 antibody. The blots were scanned and digitized. The band density was measured using ImageJ.
Total Cell Lysates
Cells were lysed using 0.1 ml of 20 mm Tris-HCl, pH 7.4, containing 1% deoxycholate, 1% IGEPAL, 150 mm NaCl, a protease inhibitor mixture III (Sigma), 1 mm phenylmethylsulfonyl fluoride, and 10 mm EDTA. The samples were centrifuged at 4 °C (14,000 × g, 15 min). The protein concentration in the supernatant samples was adjusted to 1 mg/ml. The sample aliquots (30 μg of total protein each) were separated by SDS-gel electrophoresis in 4–12% NuPAGE gels and analyzed by Western blotting with the specific primary antibodies followed by horseradish peroxidase-conjugated species-specific secondary antibody (Fitzgerald) and the 3,3′,5,5″-tetramethylbenzidine-M substrate (BioFX).
Cell Surface Biotinylation
Cell surface proteins were biotinylated by incubating cells for 1 h on ice in PBS containing 0.1 mg/ml of EZ-Link sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate. Cells were lysed in 20 mm Tris-HCl, 150 mm NaCl, 1% deoxycholate, and 1% octylphenoxypolyethoxyethanol (IGEPAL), pH 7.4, supplemented with a protease inhibitor mixture set III (Sigma), 1 mm phenylmethylsulfonyl fluoride, and 10 mm EDTA. Biotinylated proteins were precipitated from cell lysates using streptavidin-agarose beads (Sigma). Biotinylated proteins were eluted from the beads using 2× SDS sample loading buffer (125 mm Tris-HCl, pH 6.8, 4% SDS, 0.005% bromphenol blue, 20% glycerol, and 20 mm DTT).
Gelatin Zymography of MMP-2
To determine the status of endogenous MMP-2, cells were incubated in serum-free medium for 16 h. Medium samples were analyzed by gelatin zymography in 0.1% gelatin, 10% acrylamide gels as described earlier (46).
Immunofluorescence
Cells grown on a microscope coverglass (Fisher) were fixed using 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked in 1% casein. Cells were stained using the indicated primary antibodies (1:1,000 dilution) for 16 h at 4 °C followed by Alexa Fluor 488-or Alexa Fluor 594-conjugated species-specific secondary antibodies (Molecular Probes; 1:500 dilution). The Alexa Fluor 594-conjugated phalloidin (Molecular Probes; 1:500 dilution) was used to visualize the actin cytoskeleton. The specimens were mounted in the Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Images were acquired on an Olympus BX51 fluorescence microscope equipped with a MagnaFire digital camera and MagnaFire 2.1C software (Olympus).
Cell Invasion Assay
To assess cell locomotion, we employed a modified invasion assay. For this purpose, rat tail type I collagen (BD Biosciences, 0.3 mg/ml) was used to coat the 8-μm pore size membranes of the wells of a 24-well plate (0.1 ml/well). The solution was allowed to evaporate to dryness in the tissue culture hood. The collagen layer was then re-hydrated for 1 h using 0.1 ml of DMEM. Cells (1 × 105/80 μl) in ice-cold serum-free DMEM were mixed with 20 μl of ice-cold collagen (3 mg/ml) and the sample was placed into the upper chamber. The ice-cold 10% FBS-containing DMEM (0.6 ml, used as a chemoattractant) was placed in the lower chamber. Serum-free DMEM (0.6 ml) was used as a control. Cells were allowed to invade for 16 h at 37 °C in a CO2 incubator. Under our experimental conditions, over 50% HT1080 cells migrated to the membrane undersurface. The cells were then stained for 10 min using 0.5 ml of 0.2% crystal violet in 20% methanol. The cells on the upper membrane surface were removed with a cotton swab. The dye from the cells that migrated onto the lower surface of the membrane was extracted with 1% SDS (0.25 ml). The resulting OD570 nm was measured using a plate reader. To acquire the statistically significant data, the assays were performed in triplicate in three independent experiments.
RESULTS
PTK7 Ectodomain Is Processed at Two Distinct Cleavage Sites
To evaluate in more detail the processing of membrane PTK7 by the endogenous proteinases, we specifically selected to use highly invasive HT1080 cells. In the course of our study, we used cells expressing full-length PTK7 tagged with either the C-terminal FLAG or V5 tags (PTK7-V5 or PTK7-FLAG cells). We also used the cells expressing the L622D-FLAG PTK7 mutant resistant to MT1-MMP processing at the PKP621↓LI sequence (L622D-FLAG cells). To observe the effects of overexpression of MT1-MMP on PTK7, we employed cells stably transfected with the wild-type full-length MT1-MMP construct (MT1-MMP cells), whereas MT1-MMP transcriptional silencing was achieved in cells that stably expressed the shMT1-MMP construct (shMT1-MMP cells).
The shed PTK7 fragments were detected in the medium samples using Western blotting with goat polyclonal antibody against the N-terminal sequence of PTK7. Two, 65 and 70 kDa, shed PTK7 forms were detected in PTK7-V5 cells (N-PTK7–65 and N-PTK7–70, respectively, Fig. 1A, lane 1).
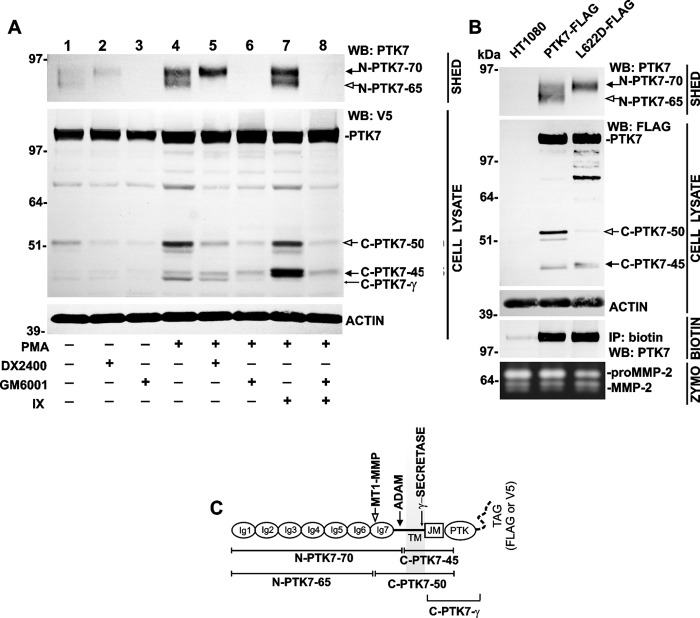
FIGURE 1.
The PTK7 ectodomain shedding at the two distinct cleavage sites is followed by γ-secretase cleavage. A, the individual and combined effects of GM6001 (a broad spectrum metalloproteinase inhibitor), DX2400 (a function blocking MT1-MMP antibody), and inhibitor IX (a γ-secretase inhibitor) on PTK7 shedding (SHED) and residual cellular PTK7 fragments (CELL LYSATE) in intact and PMA-stimulated PTK7-V5 cells. Actin, loading control. B, the generation of the N-PTK7–65 ectodomain fragment and the matching C-PTK7–50 fragment was blocked in the MT1-MMP-resistant PTK7 L622D mutant. Top panel, PTK7 fragments in medium (SHED) and total cell lysate (CELL LYSATE) in HT1080, PTK7-FLAG, and L622D-FLAG cells. Actin, loading control. Middle panel, the levels of biotinylated membrane PTK7 in HT1080, PTK7-FLAG, and L622D-FLAG cells (BIOTIN). Bottom panel, gelatin zymography of the medium samples (ZYMO). The N-PTK7–65 fragment and the matching C-PTK7–50 fragment are shown by open arrows. The N-PTK7–70 fragment and the matching C-PTK7–456 fragment are shown by solid arrows. The C-PTK7-γ γ-secretase fragment is shown by a thin arrow. WB, Western blotting. C, a model of the full-length membrane PTK7 with the MT1-MMP and ADAM cleavage sites. Cleavage of the PKP621↓LI site in the seventh Ig-like domain (MT1-MMP, open arrow) generates the N-terminal N-PTK7–65 ectodomain fragment and the matching C-terminal C-PTK7–50 fragment. Cleavage of the peptide sequence that is proximal to the plasma membrane (ADAM, solid arrow) generates the N-terminal N-PTK7–70 ectodomain fragment and the matching C-terminal C-PTK7–45 fragment. Intramembrane cleavage by γ-secretase (thin arrow) generates the cytoplasmic C-PTK7-γ fragment. Ig, immunoglobulin-like domains 1–7. TM, transmembrane domain. JM, juxtamembrane region. PTK, catalytically inactive pseudokinase domain.
PMA (50 ng/ml), a well studied inducer of receptor shedding (48–51), significantly increased the levels of both soluble PTK7 fragments (Fig. 1A, lane 4). The levels of N-PTK7–65 and N-PTK7–70 in the medium directly correlated with the levels of the C-terminal, C-PTK7–50 and C-PTK7–45, PTK7 fragments in the total cell lysate. The generation of N-PTK7–65 and the corresponding C-PTK7–50 were blocked by DX2400, the MT1-MMP-specific function-blocking antibody (Fig. 1A, lanes 2 and 5). On the contrary, the production of N-PTK7–70 was resistant to DX2400. Shedding of both N-PTK7–65 and N-PTK7–70 was completely blocked by GM6001 (Ilomastat), a broad spectrum MMP inhibitor (Fig. 1A, lanes 3 and 6).
In PTK7-FLAG cells, PMA stimulated the release of N-PTK7–65 and N-PTK7–70 fragments in the medium and accumulation of the matching C-PTK7–50 and C-PTK7–45 fragments in cell lysates (Fig. 1B). In contrast, in L622D-FLAG cells, which exhibited the inactivated PKP621↓LI MT1-MMP cleavage site, PMA stimulated the shedding of N-PTK7–70 alone (Fig. 1B). The presence of soluble N-PTK7–70 in the medium was directly correlated with the matching C-PTK7–45 fragment alone in the cell lysate. Cell surface biotinylation followed by the capture of the biotin-labeled proteins on streptavidin beads and Western blotting of the captured proteins demonstrated an exceedingly low level of endogenous PTK7 in the original HT1080 cells. In turn, transfection of the latter with the PTK7-FLAG and L622D-FLAG constructs resulted in a high level of PTK7 in PTK7-FLAG and L622D-FLAG cells, respectively (Fig. 1B). Overexpression of the PTK7-FLAG and L622D-FLAG constructs did not affect MT1-MMP-dependent conversion of the 68-kDa proenzyme into the 62-kDa enzyme of MMP-2. Accordingly, we suggested that PTK7 did not directly interfere with MT1-MMP catalytic activity (Fig. 1B).
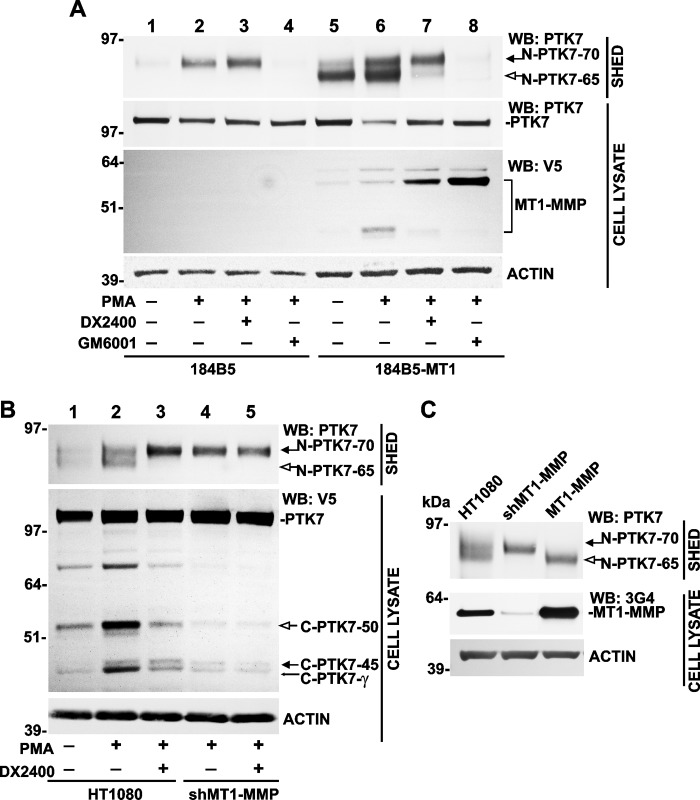
To corroborate the data we obtained using HT1080 cells, we next employed normal mammary epithelial 184B5 cells. We specifically selected 184B5 cells for our experiments because these cells endogenously express significant levels of PTK7. In our experiments, we used both noninvasive 184B5 and invasive 184B5-MT1 cells. The latter acquired invasive capabilities because of the stable overexpression of MT1-MMP (47). We determined that the levels of the released PTK7 ectodomain was exceedingly low in nonstimulated 184B5 cells (Fig. 2A, lane 1). PMA (50 ng/ml) increased the release of the soluble N-PTK7–70 fragment in 184B5 cells (Fig. 2A, lane 2). The PMA-induced release of the N-PTK7–70 fragment was not affected by DX2400 (Fig. 2A, lane 3) but, in contrast, the release of this fragment was completely repressed by GM6001 (Fig. 2A, lane 4) in 184B5 cells. In turn, in 184B5-MT1 cells the soluble N-PTK7–65 fragment was predominantly detected in medium samples (Fig. 2A, lane 5). Consistent with our observations in HT1080 cells, PMA stimulated the release of both soluble N-PTK7–65 and N-PTK7–70 fragments in 184B5-MT1 cells (Fig. 2A, lane 6). In agreement with its anti-MT1-MMP specificity, DX2400 inhibited shedding of the N-PTK7–65 fragment alone (Fig. 2A, lane 7), whereas GM6001 completely blocked the release of both N-PTK7–65 and N-PTK7–70 fragments in 184B5-MT1 cells (Fig. 2A, lane 8). In agreement with the role of MT1-MMP in PTK7 ectodomain shedding, DX2400 and GM6001 efficiently inhibited the catalytic activity and, consequently, the autocatalytic degradation of MT1-MMP in 184B5-MT1 cells. As a result of MT1-MMP inhibition by GM6001 and DX2400, the levels of the MT1-MMP enzyme increased but the levels of the MT1-MMP 45-kDa degradation fragment decreased in 184B5-MT1 cells (Fig. 2A, lanes 7 and 8). We concluded that MT1-MMP contributed to release of the N-PTK7–65 fragment, whereas a metalloproteinase, which was additional and distinct from MT1-MMP, contributed to release of the N-PTK7–70 ectodomain fragment in both HT1080 and 184B5 cells.
FIGURE 2.
MT1-MMP specifically generates the N-PTK7–65 and matching C-PTK7–50 fragments. A, effect of the specific MT1-MMP inhibitor (DX2400) and the wide-range hydroxamate metalloproteinase inhibitor (GM6001) on PTK7 shedding in 184B5 and 184B5-MT1 cells. Intact and PMA-stimulated cells were coincubated with DX2400 and GM6001. The levels of the soluble N-PTK7–65 and N-PTK7–70 ectodomain fragments were measured in the medium samples (SHED) using Western blotting with the PTK7 antibody. The status of the overexpressed MT1-MMP-V5 construct was assessed in cell lysates (CELL LYSATE) using Western blotting with the V5 antibody. Actin, loading control. B, silencing and activity inhibition of MT1-MMP using shRNA and DX2400 in HT1080 and shMT1-MMP cells transiently transfected with the PTK7-V5 construct. C, MT1-MMP silencing specifically inhibits the N-PTK7–65 shedding. PTK7-FLAG construct was transiently expressed in HT1080, shMT1-MMP, and MT1-MMP cells. MT1-MMP was measured in the cell lysates (CELL LYSATE) using Western blotting (WB) with the 3G4 antibody. The N-PTK7–65 fragment and the matching C-PTK7–50 fragment are shown by open arrows. The N-PTK7–70 fragment and the matching C-PTK7–45 fragment are shown by solid arrows. The C-PTK7-γ γ-secretase fragment is shown by a thin arrow.
Shedding of the PTK7 Ectodomain Is followed by γ-Secretase Cleavage of the PTK7 C-terminal Portion
Because γ-secretase cleavage follows the PMA-induced shedding of multiple receptors (52–57), we investigated if γ-secretase cleaved the cell-associated, C-terminal fragments of PTK7. To inhibit the putative γ-secretase cellular activity, we used a specific γ-secretase inhibitor (Inhibitor IX; IC50 = 115 nm against γ-secretase). When compared with the PMA-stimulated PTK7-V5 cells (Fig. 1A, lane 4), the inhibition of γ-secretase caused an accumulation of the C-PTK7–45 fragment with a concomitant gross decline in the levels of the low molecular weight fragment labeled as C-PTK7-γ (Fig. 1A, lane 7). The MMP inhibitor (GM6001) prevented generation of the C-PTK7-γ species (Fig. 1A, lanes 6 and 8). These results imply that metalloproteinase proteolysis of the PTK7 ectodomain is required for γ-secretase cleavage of the C-terminal portion of PTK7.
Consistent with the role of MT1-MMP in PTK7 ectodomain shedding, MT1-MMP silencing blocked the generation of N-PTK7–65 and C-PTK7–50 in shMT1-MMP cells (Fig. 2, B, lanes 4 and 5, and C). In a similar way, in HT1080 cells transiently transfected with the PTK7-V5 construct, inhibition of MT1-MMP activity using DX2400 abrogated the release of the soluble N-PTK7–65 fragment into the medium and the accumulation of the C-PTK7–50 fragment in the cell lysate (Fig. 2B, lane 3). In agreement, both the inhibition of MT1-MMP activity by the DX2400 antibody (Figs. 1A and 2B) and transcriptional silencing of MT1-MMP by the shMT1-MMP construct (Fig. 2B) blocked the accumulation of C-PTK7-γ, suggesting that proteolysis of the PTK7 ectodomain by MT1-MMP was required for the follow-on cleavage of the cell-associated C-terminal PTK7 portion by γ-secretase (Figs. 1A and 2B).
While our manuscript was under review, ADAM17 and γ-secretase were reported to contribute to proteolysis of PTK7 in colon cancer SW480 cells (58). According to this paper, ADAM17 cleavage of the Glu689-Ser690 scissile bond in membrane PTK7 generated the 100-kDa soluble ectodomain fragment. This fragment roughly corresponds to our N-PTK7–70. According to the data of Na et al. (58), γ-secretase cleaved the Gly721-Leu722 PTK7 sequence and, as a result, generated the fragment that corresponded to the C-PTK7-γ fragment we observed in HT1080 cells. The characteristics of the PTK7 proteolytic fragments are summarized in Table 1.
TABLE 1.
Proteolytic fragments of PTK7
The fragments we identified in our work were named according to their theoretical molecular weight. These fragments were compared to those identified by Na et al. (58).
| PTK7 fragment | Sequence position | Calculated mass | Corresponding fragment in the study by Na et al. |
|---|---|---|---|
| Da | |||
| N-PTK7–70 | 31–689 | 73,050.65 | sPTK7, 100 kDa |
| N-PTK7–65 | 31–621 | 65,455.93 | Not identified |
| C-PTK7–50 | 622–1070 | 49,746.09 | Not identified |
| C-PTK7–45 | 690–1070 | 42,264.53 | CTF1 |
| C-PTK7-γ | 722–1070 | 39,124.82 | CTF2 |
Proteasomal Degradation follows the Cleavage of the C-terminal PTK7 Fragment by γ-Secretase
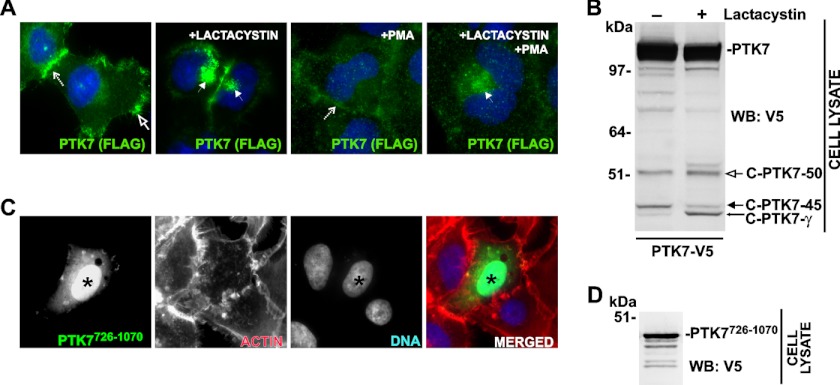
We next examined if the C-terminal PTK7 proteolytic fragments are subjected to the proteasomal degradation. The intact and PMA-stimulated PTK7-FLAG cells were coincubated with lactacystin, a selective inhibitor that irreversibly alkylates subunit X of the 20 S proteasome (59). To specifically visualize the C-terminal portion of the PTK7-FLAG construct, cells were stained with the FLAG antibody. In both intact and PMA-stimulated cells, lactacystin caused accumulation of PTK7-FLAG immunoreactivity in the perinuclear region that corresponds to typical localization of the proteasomes (Fig. 3A). Under our experimental conditions, we did not observe any significant FLAG immunoreactivity in the nucleus. Similar observations were obtained in PTK7-V5 cells in which the PTK7-V5 construct was stained with V5 antibody (not shown). Inhibition of the proteasomal degradation of the C-terminal PTK7 portion by lactacystin correlated with accumulation of the C-PTK7-γ fragment in PTK7-V5 cells (Fig. 3B).
FIGURE 3.
Proteasomal degradation of the C-terminal PTK7 fragments. A, proteasomal inhibitor lactacystin induced an accumulation of PTK7-FLAG in the perinuclear region. PMA stimulation did not induce the PTK7 translocation to the nuclei. FLAG immunoreactivity (green) of the PTK7-FLAG construct is shown in PMA-induced and lactacystin-treated PTK7-FLAG cells. The dashed, open, and solid arrows point to cell-cell junctions, membrane ruffles, and the perinuclear region, respectively. B, lactacystin induces an accumulation of C-PTK7-γ in lactacystin-treated PTK7-V5 cells. The total cell lysate samples were analyzed by Western blotting (WB) with a V5 antibody. C, the recombinant PTK7726–1070 construct (green) accumulates in the nucleus in the transiently transfected HT1080 cells. The recombinant PTK7726–1070 construct was tagged with a V5 tag. Actin (phalloidin) and the nuclei (DAPI) are red and blue, respectively. An asterisk indicates the nucleus in the cell that expressed the PTK7726–1070 construct. D, expression of the recombinant PTK7726–1070 construct in HT1080 cells. Cell lysate samples were analyzed by Western blotting with a V5 antibody.
The Recombinant Cytoplasmic PTK7726–1070 Translocated into the Cell Nucleus
To determine the fate, cellular localization, and potential individual function of the C-terminal portion of PTK7, we designed the PTK7726–1070 construct. The construct was then expressed in HT1080 cells. The PTK7726–1070 construct comprised a 726–795 juxtamembrane region and a 796–1070 kinase domain and C terminally tagged with a V5 tag. Although the C-PTK7-γ fragment predominantly underwent proteasomal degradation in PTK7-FLAG and PTK7-V5 cells, the recombinant, transiently expressed PTK7726–1070 construct predominantly accumulated in the nuclei of HT1080 cells (Fig. 3C). A high expression level of the construct was readily confirmed using Western blotting with a V5 antibody (Fig. 3D). It is likely that low levels of the C-PTK7-γ fragment in PTK7-FLAG and PTK7-V5 cells that were generated as a result of γ-secretase cleavage limited our ability of detecting this fragment in the nucleus.
Shedding of the PTK7 Ectodomain Might Contribute to Cell Invasion
To get an additional level of mechanistic understanding of the PTK7 ectodomain shedding, we then determined the effect of TIMPs, natural protein inhibitors of MMPs and ADAMs (39, 42). TIMP-1 is an inefficient inhibitor of MT1-MMP and many ADAMs excluding ADAM10 (ki = 0.1 nm) (40, 60). TIMP-2 and, especially TIMP-3 are highly potent inhibitors of MT1-MMP and ADAM17 (40, 42, 61–65). Certain ADAM family members, including ADAM8 and ADAM9, are not inhibited by the TIMPs, including TIMP-4 (66). TIMP-4, however, displays a very high inhibitory potency against MT1-MMP (ki ≤ 100 pm) (67) but a modest inhibitory activity against ADAM17 (ki = 120–180 nm) (68).
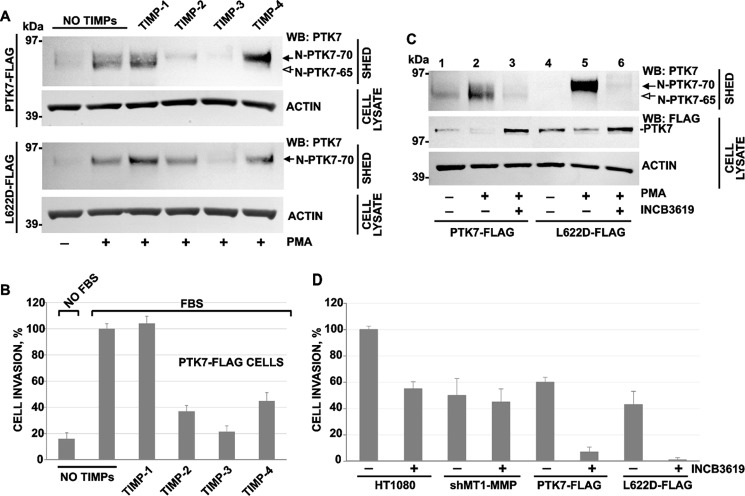
TIMPs were tested using both PTK7-FLAG and L622D-FLAG cells. The cells were stimulated with PMA (50 ng/ml) in the presence of TIMPs-1, -2, -3, and -4 (50 ng/ml each). In 16 h, levels of the N-PTK7–65 and N-PTK7–70 fragments were measured in medium using Western blotting with the PTK7 antibody (Fig. 4A). TIMP-1 did not affect PTK7 shedding. TIMP-2 significantly reduced the release of N-PTK7–65 by PTK7-FLAG cells. TIMP-3 reduced shedding of both the N-PTK7–65 and N-PTK7–70 fragments in PTK7-FLAG cells and the N-PTK7–70 fragment in L622D-FLAG cells. TIMP-4 repressed the release of N-PTK7–65 in PTK7-FLAG cells (Fig. 4A). To specifically determine the effect of TIMPs on PTK7-FLAG cell invasion, we employed a modified invasion assay using Transwells (Costar) coated with type I collagen gel. The individual TIMPs (50 ng/ml each) were added to both the upper and bottom chambers. There was a direct correlation of the shed PTK7 levels with cell invasion (Fig. 4, A and B). TIMP-2 and TIMP-4, which inhibited N-PTK7–65 shedding alone, reduced cell invasion by ≈60% as compared with intact PTK7-FLAG cells (=100%). TIMP-3 that inhibited shedding of both the N-PTK7–65 and N-PTK7–70 fragments reduced cell invasion by ≈80%. TIMP-1 did not inhibit PTK7 shedding and had no effect on cell invasion (Fig. 4B).
FIGURE 4.
Metalloproteinase inhibitors differentially regulate PTK7 shedding and cell invasion. A, effect of TIMP-1, -2, -3, and -4 on PTK7 shedding. Cells were stimulated with PMA (50 ng/ml) in the presence of the TIMPs (50 ng/ml each). Medium samples (SHED) and total cell lysates (CELL LYSATE) were analyzed using Western blotting (WB) with a PTK7 antibody. The N-PTK7–65 and N-PTK7–70 fragments are shown by open and solid arrows, respectively. Actin, loading control. B, effect of TIMP-1, -2, -3, and -4 on invasion of PTK7-FLAG cells in collagen. The levels of cell invasion are shown in percent (±S.D., error bars) relative to the untreated PTK7-FLAG cell control (=100%). Where indicated, TIMP-1, -2, -3, and -4 (50 ng/ml each) were added to the upper and lower chambers. FBS (10%) in the lower chamber was used as an attractant. The assays were performed in triplicate. C, INCB3619 (5 μm) repressed the release of the soluble N-PTK7–65 and N-PTK7–70 fragments (open and solid arrows, respectively) in PTK7-FLAG and L622D-FLAG cells. Where indicated, cells were stimulated with PMA (50 ng/ml). Medium samples (SHED) and total cell lysates (CELL LYSATE) were analyzed using Western blotting (WB) with the PTK7 and FLAG M2 antibodies, respectively. D, INCB3619 inhibited cell invasion in Transwells coated with type I collagen. HT1080, shMT1-MMP, PTK7-FLAG, and L622D-FLAG cells migrated toward 10% FBS in the lower chamber. Where indicated, INCB3619 (5 μm) was added to the upper and lower chambers. The levels of cell invasion are shown in percent (±S.D., error bars) compared with the untreated HT1080 cell control (= 100%). The assays were performed in triplicate.
To support these observations, we also employed INCB3619, a potent inhibitor of ADAMs (ADAM17; IC50 = 14 nm and ADAM10, IC50 = 22 nm (69, 70)) and MMPs (MT1-MMP; IC50 = 772 nm MT1-MMP). INCB3619 (5 μm) efficiently blocked the release of both the soluble N-PTK7–65 and N-PTK7–70 fragments in PMA-stimulated PTK7-FLAG and L622D-FLAG cells and, reciprocally, rescued the presentation of the full-length membrane PTK7 in these cells (Fig. 4C).
We also studied the effect of INCB3619 (5 μm) on cell invasion in HT1080, shMT1-MMP, PTK7-FLAG, and L622D-FLAG cells (Fig. 4D). INCB3619 reduced the invasion rate of the original HT1080 cells by ≈50%. In agreement with our previous data (43, 44), silencing of MT1-MMP (shMT1-MMP cells) caused a roughly 50% reduction in cell invasion compared with HT1080 cells. INCB3619 did not decrease the cell invasion rate of MT1-MMP-deficient shMT1-MMP cells. Overexpression of the full-length PTK7 alone (PTK7-FLAG cells) reduced cell invasion by 40% compared with HT1080 cells. INCB3619 reduced the invasion rate of PTK7-FLAG cells much further. Expression of the MT1-MMP-uncleavable L622D PTK7 mutant alone (L622D-FLAG cells) reduced the invasion by 60%, whereas INCB3619 caused a near complete inhibition of invasion in these cells (Fig. 4D).
Specifically, a direct comparison of shMT1-MMP cells (in which MT1-MMP expression was silenced and the endogenous PTK7 expression was low) with PTK7-FLAG cells (in which MT1-MMP expression was high and PTK7 was overexpressed) suggested a role of PTK7 in cell invasion (Fig. 4D). In general, the results of our cell invasion tests correlated with the status of PTK7 in these cells (Fig. 4, C and D).
DISCUSSION
Our earlier studies provided evidence that the full-length membrane PTK7 and, especially the MT1-MMP uncleavable L622D mutant inhibited cell invasion and altered the actin cytoskeleton in HT1080 cells (44). Specifically, our earlier data suggested that there was a link between efficiency of the proteolysis of the PTK7 ectodomain and cell invasion. Thus, the Chz PTK7 mutant exhibited an additional MMP cleavage site in the ectodomain. As compared with the wild-type and especially L622D mutant PTK7, the Chz mutant was efficiently shed from the cell surface and promoted cell invasion (43). We determined that the PTK7 ectodomain was a major cleavage target of invasion-promoting MT1-MMP at the cell surface.
In our present study we focused on achieving a deeper understanding of the proteolytic regulation of the membrane PTK7 and on the effect of PTK7 processing in cell invasion. For these purposes, we specifically employed highly invasive HT1080 cells in our study. The original HT1080 cells express low levels of endogenous PTK7 but significant endogenous levels of active MT1-MMP and ADAMs. These parameters allowed us to readily manipulate the levels of wild-type and mutant PTK7 in transfected cells.
To understand the regulatory proteolysis of membrane PTK7 in more detail, we performed our studies in intact and PMA-stimulated HT1080 cells that co-expressed MT1-MMP, the wild-type membrane 1–1070 PTK7, and PTK7 mutants with the modified putative proteinase cleavage sites. In addition, we used the cells with silenced MT1-MMP and the individual ADAM family members. To discriminate between the individual steps of PTK7 processing, TIMPs-1, -2, -3, and -4, the function-blocking MT1-MMP monoclonal antibody (DX2400), small-molecule ADAM, and MMP inhibitor (INCB3619), inhibitors of the proteasome and γ-secretase (lactacystin and inhibitor IX, respectively), were widely used in our study.
We established that the PTK7 ectodomain was processed at two distinct cleavage sites. The first cleavage caused the release of the soluble N-PTK7–65 ectodomain fragment and the generation of the matching cell-associated C-PTK7–50 species was the result of MT1-MMP proteolysis. The second cleavage in the C-terminal portion of the PTK7 ectodomain was performed by ADAMs. This cleavage led to the release of the soluble N-PTK7–70 fragment and to generation of the matching cell-associated C-PTK7–45 form. Based on the size of the PTK7 fragment we predicted that ADAM-dependent cleavage took place in the 678–704 C-terminal region of the PTK7 ectodomain that was adjacent to the cell membrane, an appropriate location for the ADAM-dependent excisions. In agreement, the data on inhibition of PTK7 shedding by TIMPs and INCB3619, an MMP and ADAM inhibitor, support this prediction. Accordingly, while our article was under review, ADAM17 was reported to contribute to PTK7 ectodomain shedding in colon cancer SW480 cells (58).
In our experiments, however, TIMP-4, which displays an inhibitory activity against ADAM17 (ki = 120–180 nm) (68), did not repress ADAM-mediated generation of the N-PTK7–70 fragment. Therefore, we hypothesize that because of the functional redundancy in the ADAM family, multiple ADAMs, rather than a singular principal ADAM member alone, contributed to PTK7 ectodomain shedding in HT1080 cells. Conversely, a high sensitivity of the C-terminal portion of the PTK7 ectodomain to cleavages suggests an importance of ADAM proteolysis in the regulation of the PTK7 functionality. It is likely that in different cell systems the distinct ADAM family members would function as PTK7 sheddases.
Our experiments also suggested that MT1-MMP and ADAM proteolysis of PTK7 was a prerequisite for the follow-on intramembrane γ-secretase cleavage of the C-terminal membrane portion of PTK7. This portion was then decayed via proteasomal degradation in our cell system. On the contrary, the recombinant PTK7726–1070 construct that is similar to the γ-secretase fragment of PTK7 readily entered the nuclei in HT1080 cells. However, in contrast with the observation by others in colon cancer SW480 cells (57), this fragment did not stimulate invasion of HT1080 cells. We suspect that the pre-existing high invasive potential of HT1080 cells did not allow us to readily identify the proinvasive effect of PTK7726–1070 in our cell system.
It is generally accepted that cell invasion is regulated by the MT1-MMP-dependent ECM proteolysis. It is also established that actin cytoskeleton remodeling and contractility are involved in the regulation of cell motility. It is likely that both ECM proteolysis and cytoskeleton dynamics contribute to cell invasiveness. Based on our previous and current observations, it is now tempting to suggest that ectodomain shedding and processing of PTK7 contributes to cell invasion via affecting the cytoskeleton and that this event is additional to the ECM proteolysis by MT1-MMP and ADAMs (44). Thus, full-length PTK7 inhibits actin cytoskeleton contractility, whereas by shedding the PTK7 ectodomain the metalloproteinase activity reverses this inhibitory effect. In agreement, MT1-MMP transcriptional silencing reduced invasion of PTK7-deficient HT1080 shMT1-MMP cells by 50%. We suspect that this effect is a result of the repressed ECM degradation. On the other hand, overexpression of the PTK7-FLAG that reduced cytoskeleton contractility (43, 44) inhibited cell invasion also by 50%, although the MT1-MMP activity was not affected. Accordingly, we now suspect that this effect we observed in the HT1080 PTK7-FLAG cells is linked to the PTK7 effect on the cytoskeleton rather than related to ECM degradation. When both overexpression of the PTK7-FLAG (and L622D-FLAG) and the pharmacological inhibition of the metalloproteinase activity were combined, the invasion of cells was blocked completely. We believe that the stabilization of membrane PTK7 at the cell surface is an important factor that controls cell invasion by targeting actin cytoskeleton contractility (43, 44).
Based on our results, it is likely that metalloproteinase inhibitors would suppress cancer cell invasion most efficiently in malignancies, which express PTK7. Using HT1080 and 184B5 cells as a model, our studies recognized novel steps in the stepwise proteolytic regulation of PTK7 functionality and generated testable hypotheses. The latter can be experimentally pursued to fully understand the precise molecular events the aberrations of which result in cell motility defects in pathological conditions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA77470, CA83017, and CA157328 (to A. Y. S.).
- PTK7
- protein-tyrosine kinase 7
- ADAM
- A Disintegrin And Metalloproteinase
- C-PTK7–45 and C-PTK7–50
- C-terminal 45- and 50-kDa PTK7 proteolytic fragments, respectively
- Inhibitor IX
- N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane type-1 matrix metalloproteinase
- N-PTK7–65 and N-PTK7–70
- N-terminal 65- and 70-kDa PTK7 proteolytic fragments, respectively
- PMA
- phorbol 12-myristate 13-acetate
- PTK
- protein-tyrosine kinase
- TIMP-1
- -2, -3 and -4, tissue inhibitor of metalloproteinases-1, -2, -3 and -4
- ECM
- extracellular matrix.
REFERENCES
- 1. Lu X., Borchers A. G., Jolicoeur C., Rayburn H., Baker J. C., Tessier-Lavigne M. (2004) PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature 430, 93–98 [DOI] [PubMed] [Google Scholar]
- 2. Yen W. W., Williams M., Periasamy A., Conaway M., Burdsal C., Keller R., Lu X., Sutherland A. (2009) PTK7 is essential for polarized cell motility and convergent extension during mouse gastrulation. Development 136, 2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Puppo F., Thomé V., Lhoumeau A. C., Cibois M., Gangar A., Lembo F., Belotti E., Marchetto S., Lécine P., Prébet T., Sebbagh M., Shin W. S., Lee S. T., Kodjabachian L., Borg J. P. (2011) Protein-tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 12, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winberg M. L., Tamagnone L., Bai J., Comoglio P. M., Montell D., Goodman C. S. (2001) The transmembrane protein off-track associates with Plexins and functions downstream of Semaphorin signaling during axon guidance. Neuron 32, 53–62 [DOI] [PubMed] [Google Scholar]
- 5. Peradziryi H., Kaplan N. A., Podleschny M., Liu X., Wehner P., Borchers A., Tolwinski N. S. (2011) PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling. EMBO J. 30, 3729–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whitford K. L., Ghosh A. (2001) Plexin signaling via off-track and rho family GTPases. Neuron 32, 1–3 [DOI] [PubMed] [Google Scholar]
- 7. Toyofuku T., Zhang H., Kumanogoh A., Takegahara N., Yabuki M., Harada K., Hori M., Kikutani H. (2004) Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat. Cell Biol. 6, 1204–1211 [DOI] [PubMed] [Google Scholar]
- 8. Shnitsar I., Borchers A. (2008) PTK7 recruits dsh to regulate neural crest migration. Development 135, 4015–4024 [DOI] [PubMed] [Google Scholar]
- 9. Wehner P., Shnitsar I., Urlaub H., Borchers A. (2011) RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327 [DOI] [PubMed] [Google Scholar]
- 10. Lhoumeau A. C., Puppo F., Prébet T., Kodjabachian L., Borg J. P. (2011) PTK7. A cell polarity receptor with multiple facets. Cell Cycle 10, 1233–1236 [DOI] [PubMed] [Google Scholar]
- 11. Jung J. W., Ji A. R., Lee J., Kim U. J., Lee S. T. (2002) Organization of the human PTK7 gene encoding a receptor protein-tyrosine kinase-like molecule and alternative splicing of its mRNA. Biochim. Biophys. Acta 1579, 153–163 [DOI] [PubMed] [Google Scholar]
- 12. Mossie K., Jallal B., Alves F., Sures I., Plowman G. D., Ullrich A. (1995) Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene 11, 2179–2184 [PubMed] [Google Scholar]
- 13. Gorringe K. L., Boussioutas A., Bowtell D. D. (2005) Novel regions of chromosomal amplification at 6p21, 5p13, and 12q14 in gastric cancer identified by array comparative genomic hybridization. Genes Chromosomes Cancer 42, 247–259 [DOI] [PubMed] [Google Scholar]
- 14. Endoh H., Tomida S., Yatabe Y., Konishi H., Osada H., Tajima K., Kuwano H., Takahashi T., Mitsudomi T. (2004) Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J. Clin. Oncol. 22, 811–819 [DOI] [PubMed] [Google Scholar]
- 15. Müller-Tidow C., Schwäble J., Steffen B., Tidow N., Brandt B., Becker K., Schulze-Bahr E., Halfter H., Vogt U., Metzger R., Schneider P. M., Büchner T., Brandts C., Berdel W. E., Serve H. (2004) High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets. Clin. Cancer Res. 10, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 16. Easty D. J., Mitchell P. J., Patel K., Flørenes V. A., Spritz R. A., Bennett D. C. (1997) Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int. J. Cancer 71, 1061–1065 [DOI] [PubMed] [Google Scholar]
- 17. Su Y. A., Yang J., Tao L., Nguyen H., He P. (2010) Undetectable and decreased expression of KIAA1949 (Phostensin) encoded on chromosome 6p21.33 in human breast cancers revealed by transcriptome analysis. J. Cancer 1, 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Piao Z., Lee K. S., Kim H., Perucho M., Malkhosyan S. (2001) Identification of novel deletion regions on chromosome arms 2q and 6p in breast carcinomas by amplotype analysis. Genes Chromosomes Cancer 30, 113–122 [PubMed] [Google Scholar]
- 19. Baudrier-Régnier A., Bodenant C., Proust F., Delangre T., Hemet J., Laquerrière A. (2000) An isochromosome 6p in a primary meningeal malignant melanoma. Cancer Genet Cytogenet 119, 80–82 [DOI] [PubMed] [Google Scholar]
- 20. Duffy M. J., McKiernan E., O'Donovan N., McGowan P. M. (2009) Role of ADAMs in cancer formation and progression. Clin. Cancer Res. 15, 1140–1144 [DOI] [PubMed] [Google Scholar]
- 21. Stocker W., Grams F., Baumann U., Reinemer P., Gomis-Ruth F. X., McKay D. B., Bode W. (1995) The metzincins–topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc peptidases. Protein Sci. 4, 823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 23. Itoh Y., Seiki M. (2006) MT1-MMP. A potent modifier of pericellular microenvironment. J. Cell Physiol. 206, 1–8 [DOI] [PubMed] [Google Scholar]
- 24. Morrison C. J., Butler G. S., Rodríguez D., Overall C. M. (2009) Matrix metalloproteinase proteomics. Substrates, targets, and therapy. Curr. Opin. Cell Biol. 21, 645–653 [DOI] [PubMed] [Google Scholar]
- 25. Rodríguez D., Morrison C. J., Overall C. M. (2010) Matrix metalloproteinases. What do they not do? New substrates and biological roles identified by murine models and proteomics. Biochim. Biophys. Acta 1803, 39–54 [DOI] [PubMed] [Google Scholar]
- 26. Hotary K., Li X. Y., Allen E., Stevens S. L., Weiss S. J. (2006) A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 20, 2673–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hotary K. B., Allen E. D., Brooks P. C., Datta N. S., Long M. W., Weiss S. J. (2003) Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell 114, 33–45 [DOI] [PubMed] [Google Scholar]
- 28. Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sabeh F., Ota I., Holmbeck K., Birkedal-Hansen H., Soloway P., Balbin M., Lopez-Otin C., Shapiro S., Inada M., Krane S., Allen E., Chung D., Weiss S. J. (2004) Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J. Cell Biol. 167, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Belkin A. M., Akimov S. S., Zaritskaya L. S., Ratnikov B. I., Deryugina E. I., Strongin A. Y. (2001) Matrix-dependent proteolysis of surface transglutaminase by membrane-type metalloproteinase regulates cancer cell adhesion and locomotion. J. Biol. Chem. 276, 18415–18422 [DOI] [PubMed] [Google Scholar]
- 31. Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deryugina E. I., Ratnikov B. I., Postnova T. I., Rozanov D. V., Strongin A. Y. (2002) Processing of integrin αv subunit by membrane type 1 matrix metalloproteinase stimulates migration of breast carcinoma cells on vitronectin and enhances tyrosine phosphorylation of focal adhesion kinase. J. Biol. Chem. 277, 9749–9756 [DOI] [PubMed] [Google Scholar]
- 33. Rozanov D. V., Savinov A. Y., Williams R., Liu K., Golubkov V. S., Krajewski S., Strongin A. Y. (2008) Molecular signature of MT1-MMP. Transactivation of the downstream universal gene network in cancer. Cancer Res. 68, 4086–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seiki M., Koshikawa N., Yana I. (2003) Role of pericellular proteolysis by membrane-type 1 matrix metalloproteinase in cancer invasion and angiogenesis. Cancer Metastasis Rev. 22, 129–143 [DOI] [PubMed] [Google Scholar]
- 35. Rowe R. G., Li X. Y., Hu Y., Saunders T. L., Virtanen I., Garcia de Herreros A., Becker K. F., Ingvarsen S., Engelholm L. H., Bommer G. T., Fearon E. R., Weiss S. J. (2009) Mesenchymal cells reactivate Snail1 expression to drive three-dimensional invasion programs. J. Cell Biol. 184, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szabova L., Chrysovergis K., Yamada S. S., Holmbeck K. (2008) MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene 27, 3274–3281 [DOI] [PubMed] [Google Scholar]
- 37. Holmbeck K., Bianco P., Yamada S., Birkedal-Hansen H. (2004) MT1-MMP. A tethered collagenase. J. Cell Physiol. 200, 11–19 [DOI] [PubMed] [Google Scholar]
- 38. Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99, 81–92 [DOI] [PubMed] [Google Scholar]
- 39. Mochizuki S., Okada Y. (2007) ADAMs in cancer cell proliferation and progression. Cancer Sci. 98, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murphy G. (2009) Regulation of the proteolytic disintegrin metalloproteinases, the ”Sheddases.“ Semin. Cell Dev. Biol. 20, 138–145 [DOI] [PubMed] [Google Scholar]
- 41. Duffy M. J., Mullooly M., O'Donovan N., Sukor S., Crown J., Pierce A., McGowan P. M. (2011) The ADAMs family of proteases. New biomarkers and therapeutic targets for cancer? Clin. Proteomics 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brew K., Nagase H. (2010) The tissue inhibitors of metalloproteinases (TIMPs). An ancient family with structural and functional diversity. Biochim. Biophys. Acta 1803, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Golubkov V. S., Aleshin A. E., Strongin A. Y. (2011) Potential relation of aberrant proteolysis of human protein-tyrosine kinase 7 (PTK7) chuzhoi by membrane type 1 matrix metalloproteinase (MT1-MMP) to congenital defects. J. Biol. Chem. 286, 20970–20976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Golubkov V. S., Chekanov A. V., Cieplak P., Aleshin A. E., Chernov A. V., Zhu W., Radichev I. A., Zhang D., Dong P. D., Strongin A. Y. (2010) The Wnt/planar cell polarity protein-tyrosine kinase-7 (PTK7) is a highly efficient proteolytic target of membrane type-1 matrix metalloproteinase. Implications in cancer and embryogenesis. J. Biol. Chem. 285, 35740–35749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sounni N. E., Rozanov D. V., Remacle A. G., Golubkov V. S., Noel A., Strongin A. Y. (2010) Timp-2 binding with cellular MT1-MMP stimulates invasion-promoting MEK/ERK signaling in cancer cells. Int. J. Cancer 126, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Golubkov V. S., Boyd S., Savinov A. Y., Chekanov A. V., Osterman A. L., Remacle A., Rozanov D. V., Doxsey S. J., Strongin A. Y. (2005) Membrane type-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J. Biol. Chem. 280, 25079–25086 [DOI] [PubMed] [Google Scholar]
- 47. Golubkov V. S., Chekanov A. V., Savinov A. Y., Rozanov D. V., Golubkova N. V., Strongin A. Y. (2006) Membrane type-1 matrix metalloproteinase confers aneuploidy and tumorigenicity on mammary epithelial cells. Cancer Res. 66, 10460–10465 [DOI] [PubMed] [Google Scholar]
- 48. Kveiborg M., Instrell R., Rowlands C., Howell M., Parker P. J. (2011) PKCα and PKCδ regulate ADAM17-mediated ectodomain shedding of heparin binding-EGF through separate pathways. PLoS One 6, e17168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kohutek Z. A., diPierro C. G., Redpath G. T., Hussaini I. M. (2009) ADAM-10-mediated N-cadherin cleavage is protein kinase Cα dependent and promotes glioblastoma cell migration. J. Neurosci. 29, 4605–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sundberg C., Thodeti C. K., Kveiborg M., Larsson C., Parker P., Albrechtsen R., Wewer U. M. (2004) Regulation of ADAM12 cell-surface expression by protein kinase Cϵ. J. Biol. Chem. 279, 51601–51611 [DOI] [PubMed] [Google Scholar]
- 51. Le Gall S. M., Auger R., Dreux C., Mauduit P. (2003) Regulated cell surface pro-EGF ectodomain shedding is a zinc metalloprotease-dependent process. J. Biol. Chem. 278, 45255–45268 [DOI] [PubMed] [Google Scholar]
- 52. Guo S., Liu M., Gonzalez-Perez R. R. (2011) Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim. Biophys. Acta 1815, 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Killar L., White J., Black R., Peschon J. (1999) Adamalysins. A family of metzincins including TNF-α converting enzyme (TACE). Ann. N.Y. Acad. Sci. 878, 442–452 [DOI] [PubMed] [Google Scholar]
- 54. Yañez-Mó M., Sánchez-Madrid F., Cabañas C. (2011) Membrane proteases and tetraspanins. Biochem. Soc. Trans. 39, 541–546 [DOI] [PubMed] [Google Scholar]
- 55. Lichtenthaler S. F., Haass C., Steiner H. (2011) Regulated intramembrane proteolysis. Lessons from amyloid precursor protein processing. J. Neurochem. 117, 779–796 [DOI] [PubMed] [Google Scholar]
- 56. Montes de Oca-B. P. (2010) Ectdomain shedding and regulated intracellular proteolysis in the central nervous system. Cent. Nerv. Syst. Agents Med. Chem. 10, 337–359 [DOI] [PubMed] [Google Scholar]
- 57. Zolkiewska A. (2008) ADAM proteases. Ligand processing and modulation of the Notch pathway. Cell Mol. Life Sci. 65, 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Na H. W., Shin W. S., Ludwig A., Lee S. T. (2012) The cytosolic domain of protein-tyrosine kinase 7 (PTK7), generated from sequential cleavage by a disintegrin and metalloprotease 17 (ADAM17) and γ-secretase, enhances cell proliferation and migration in colon cancer cells. J. Biol. Chem. 287, 25001–25009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fenteany G., Schreiber S. L. (1998) Lactacystin, proteasome function, and cell fate. J. Biol. Chem. 273, 8545–8548 [DOI] [PubMed] [Google Scholar]
- 60. Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. (2000) The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 473, 275–279 [DOI] [PubMed] [Google Scholar]
- 61. Zhao H., Bernardo M. M., Osenkowski P., Sohail A., Pei D., Nagase H., Kashiwagi M., Soloway P. D., DeClerck Y. A., Fridman R. (2004) Differential inhibition of membrane type 3 (MT3)-matrix metalloproteinase (MMP) and MT1-MMP by tissue inhibitor of metalloproteinase (TIMP)-2 and TIMP-3 rgulates pro-MMP-2 activation. J. Biol. Chem. 279, 8592–8601 [DOI] [PubMed] [Google Scholar]
- 62. Lin E. A., Liu C. J. (2010) The role of ADAMTSs in arthritis. Protein Cell 1, 33–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lim N. H., Kashiwagi M., Visse R., Jones J., Enghild J. J., Brew K., Nagase H. (2010) Reactive-site mutants of N-TIMP-3 that selectively inhibit ADAMTS-4 and ADAMTS-5. Biological and structural implications. Biochem. J. 431, 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kwak H. I., Mendoza E. A., Bayless K. J. (2009) ADAM17 co-purifies with TIMP-3 and modulates endothelial invasion responses in three-dimensional collagen matrices. Matrix Biol. 28, 470–479 [DOI] [PubMed] [Google Scholar]
- 65. Jones G. C., Riley G. P. (2005) ADAMTS proteinases. A multi-domain, multi-functional family with roles in extracellular matrix turnover and arthritis. Arthritis Res. Ther. 7, 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Amour A., Knight C. G., English W. R., Webster A., Slocombe P. M., Knäuper V., Docherty A. J., Becherer J. D., Blobel C. P., Murphy G. (2002) The enzymatic activity of ADAM8 and ADAM9 is not regulated by TIMPs. FEBS Lett. 524, 154–158 [DOI] [PubMed] [Google Scholar]
- 67. Bigg H. F., Morrison C. J., Butler G. S., Bogoyevitch M. A., Wang Z., Soloway P. D., Overall C. M. (2001) Tissue inhibitor of metalloproteinases-4 inhibits but does not support the activation of gelatinase A via efficient inhibition of membrane type 1-matrix metalloproteinase. Cancer Res. 61, 3610–3618 [PubMed] [Google Scholar]
- 68. Lee M. H., Rapti M., Murphy G. (2005) Total conversion of tissue inhibitor of metalloproteinase (TIMP) for specific metalloproteinase targeting. Fine-tuning TIMP-4 for optimal inhibition of tumor necrosis factor-α-converting enzyme. J. Biol. Chem. 280, 15967–15975 [DOI] [PubMed] [Google Scholar]
- 69. Zhou B. B., Peyton M., He B., Liu C., Girard L., Caudler E., Lo Y., Baribaud F., Mikami I., Reguart N., Yang G., Li Y., Yao W., Vaddi K., Gazdar A. F., Friedman S. M., Jablons D. M., Newton R. C., Fridman J. S., Minna J. D., Scherle P. A. (2006) Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 10, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fridman J. S., Caulder E., Hansbury M., Liu X., Yang G., Wang Q., Lo Y., Zhou B. B., Pan M., Thomas S. M., Grandis J. R., Zhuo J., Yao W., Newton R. C., Friedman S. M., Scherle P. A., Vaddi K. (2007) Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin. Cancer Res. 13, 1892–1902 [DOI] [PubMed] [Google Scholar]