Background: The function of the p53-induced gene Ei24 in the autophagy pathway remains largely unknown.
Results: Ei24 deficiency impairs autophagic flux. Mice deficient in Ei24 show massive neuron degeneration and severe liver injury.
Conclusion: Ei24 functions in basal autophagy in clearance of aggregate-prone proteins in neurons and hepatocytes.
Significance: We revealed that EI24 is an essential component of the basal autophagy pathway.
Keywords: Autophagy, Hepatocyte, Neurodegeneration, Oligodendrocytes, p53, C. elegans, autophagosome
Abstract
Ei24 is a DNA damage response gene involved in growth suppression and apoptosis. The physiological function of Ei24, however, is poorly understood. Here we generated conditional knock-out mice of Ei24 and demonstrated that EI24 is an essential component of the basal autophagy pathway. Mice with neural-specific Ei24 deficiency develop age-dependent neurological abnormalities caused by massive axon degeneration and extensive neuron loss in brain and spinal cord. Notably, ablation of Ei24 leads to vacuolated oligodendroglial cells and demyelination of axons. Liver-specific depletion of Ei24 causes severe hepatomegaly with hepatocyte hypertrophy. Ei24 deficiency impairs autophagic flux, leading to accumulation of LC3, p62 aggregates, and ubiquitin-positive inclusions. Our study indicates that Ei24 is an essential autophagy gene and plays an important role in clearance of aggregate-prone proteins in neurons and hepatocytes.
Introduction
Ei24 (etoposide-induced 2.4 kb transcript),2 also known as PIG8 (p53-induced gene 8), encodes an ER-localized six transmembrane protein, expression of which is highly induced by the tumor suppressor protein p53 (1–3). Overexpression of Ei24 suppresses cell growth and induces apoptosis, while depletion of Ei24 results in suppression of apoptosis in response to pro-apoptotic treatment (2–4). The human EI24 gene is located on chromosome 11q23, a region frequently displaying loss of heterozygosity in several malignancies, including invasive cervical cancers, breast carcinomas and malignant melanoma (5). The tumor suppression function of Ei24 is further substantiated by reduced expression in invasive breast cancers (3, 5). However, the physiological function of Ei24 is still poorly understood.
Autophagy is an evolutionarily conserved intracellular catabolic system, involving the formation of a closed double-membrane autophagosome and its subsequent delivery to the vacuole/lysosome for degradation (6, 7). Under normal physiological conditions, autophagy occurs at a basal constitutive level, removing misfolded, or aggregate-prone proteins and damaged organelles (8). The homeostatic function of basal autophagy plays an important role in neuronal protection and tumor suppression in mammals (8, 9). Neural-specific knock-out of Atg7 or Atg5 (genes essential for autophagosome formation) causes dramatic accumulation of autophagy substrates such as p62 (also known as sequestosome 1, SQSTM1) and ubiquitin-positive aggregates in neurons, accompanied by massive neuron degeneration in various brain regions (10–12). The autophagy gene beclin 1 is a haploinsufficient tumor suppressor (8). Atg7 and Atg5 deficiency in liver cause hepatomegaly and subsequently multiple liver adenomas (13, 14). The tumor suppression function of autophagy is at least partially attributed to its elimination of p62, accumulation of which leads to persistent activation of the Nrf2 stress response and dysregulation of NF-κB signaling (14–16).
A group of evolutionarily conserved Atg proteins has been identified from yeast genetic studies that act at distinct steps of autophagosome formation (6, 7). In mammalian cells, however, the autophagy process involves more complex membrane dynamics (17). The endoplasmic reticulum (ER), Golgi apparatus, recycling endosomes, and plasma membrane all contribute to autophagosomal membranes in mammalian cells (18, 19). Among them, PI(3)P-enriched subdomains of the ER, called omegasomes, provide a platform for recruiting Atg proteins and subsequent autophagosome formation (18). The more elaborate autophagic machinery in higher eukaryotes requires Atg proteins and also metazoan-specific autophagy components. Genetic screens in Caenorhabditis elegans identified epg-4, the Ei24 homolog, as an essential autophagy gene, loss of function of which causes defective autophagic degradation of a variety of protein aggregates during embryogenesis (20). epg-4 regulates progression of omegasomes to autophagosomes (20). Ei24 is also essential for starvation-induced autophagy (20). Distinct from the role of epg-4 at the early step of autophagosome formation in C. elegans, knockdown of Ei24 by siRNA results in accumulation of degradation-defective autolysosomes (20).
Here we generated mice with tissue-specific deficiency of Ei24. Neural-specific Ei24-deficient mice exhibit massive axonal degeneration and neuronal cell loss in various brain and spinal cord regions. Ei24 deficiency causes vacuolation of oligodendroglial cells. Liver-specific Ei24-deficient mice have hepatocellular enlargement and hepatomegaly. Ei24 deficiency impairs autophagic flux, leading to accumulation of LC3, p62 aggregates, and ubiquitin-positive inclusions. Our study demonstrates that Ei24 is an essential component of basal autophagy in removing aggregate-prone proteins.
EXPERIMENTAL PROCEDURES
Mice
To construct the Ei24 targeting allele, exon 3 of Ei24 was flanked by two loxP sequences and a neomycin phosphotransferase expression cassette. The targeting vector was electroporated into 129 R1 embryonic stem (ES) cells. Three homologous recombinants were identified by Southern blotting with probes 5′ and 3′ of the genomic sequence present in the targeting vector. Heterozygous Ei24fl/wt ES cell clones were microinjected into C57BL/6N blastocysts. Chimeric offspring were backcrossed to C57BL/6N mice. Heterozygous mutant mice were outbred with C57BL/6 mice and interbred to obtain homozygous mutant mice. Cre-mediated deletion of exon 3 leads to a frameshift, producing a small truncated peptide. The following primers were used to detect wild type Ei24 and Ei24flox alleles: 5′-TAAAGTTCTTAGGACACCTCCTG-3′ (F) and 5′-AATGGAGAACTTTAGAATCTCC-3′ (R). The expected sizes are 273 bp and 377 bp, respectively.
To generate neural- or liver-specific Ei24-deficient mice, Ei24flox/flox mice were crossed with nestin-Cre or albumin-Cre mice (Jackson Laboratory), respectively. Ei24+/− mice were obtained from Ei24flox/wt; Zp3-Cre mice (Jackson Laboratory). All mice were raised under specific pathogen-free conditions in the animal facility at the National Institute of Biological Sciences, Beijing. All animal experiments were approved by the institutional committee of the National Institute of Biological Sciences, Beijing.
Antibodies
The following antibodies were used: rabbit anti-p62 (PM045, MBL), mouse anti-p62 (ab56416, Abcam), mouse anti-ubiquitin (3936, Cell Signaling), rabbit anti-GFAP (bs-0199R, Bioss), rabbit anti-LC3 (PM046, MBL) for Western, rabbit anti-LC3 (2775, Cell Signaling) for staining, mouse anti-NeuN (MAB377, Millipore), mouse anti-β tubulin III (ab7751, Abcam), mouse anti-calbindin (C9848, Sigma), mouse anti-CNPase (2′,3′-cyclic-nucleotide 3′-phosphodiesterase) (ab6319, Abcam), rabbit anti-MBP (myelin basic protein) (ab40390, Abcam), and rabbit anti-GFP (ab290, Abcam).
Behavioral Analysis
An accelerating rotarod (YLS-4C, Beijing Zhongshidichuang Science and Technology Development Co., Ltd) was used to measure motor coordination. After 2 consecutive days training, mice were put on the rolling rod with auto-acceleration ranging from 5–20 rpm within 60 s. The time when the mice fell from the rod was recorded, with a maximum observation time of 2 min.
Histology and Immunohistochemistry
Mice were perfused with 10% neutral buffered formalin (Sigma). Tissues were post-fixed, embedded in paraffin, and sectioned at 5 μm. Sections were stained with hematoxylin and eosin for histological examination or 0.1% cresyl violet for Nissl staining and examined by light microscopy (Imager A1, Zeiss).
For immunohistochemistry, sections were deparaffinized in xylene three times, then rehydrated in an ethanol series (100% × 3, 95%, and 75%). After heat-activated antigen retrieval (0.1 m citrate buffer), sections were blocked with normal goat serum. Sections were then incubated with primary antibodies at 4 °C overnight in a humidity chamber, washed in PBS three times, and incubated with fluorescently-labeled secondary antibodies in a humidified chamber for 1 h at room temperature. Finally, sections were counterstained with DAPI, mounted and examined under an epifluorescence microscope or a confocal microscope (Zeiss LSM 510 Meta plus Zeiss Axiovert zoom). Fluorescence intensity and dot numbers were analyzed by Image J software.
Cell Counting
To score the number of spinal cord motor neurons, fixed spinal cords were sequentially sectioned at 5 μm. Every 6th section was stained with 0.1% cresyl violet. Neurons with a distinct nucleus and a diameter of at least 25 μm located in the anterior horn ventral to the line tangential to the ventral tip of the central canal were counted as motor neurons. To examine cortical neurons, large pyramidal cells located in the fifth layer of the motor and sensory cortices were quantified. The number of Purkinje cells in lobules III, IV, and V were quantified and divided by the total length of the lobules. The thickness of the molecular layer was calculated by dividing the distance between lobules III and IV, or lobules V and VI, by 2.
Protein Extraction and Western Blotting
Mouse tissues were homogenized in 10 volumes of lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.1% SDS, 1% Nonidet P-40) supplemented with 1 mm PMSF and protease inhibitor mixture (Roche). After incubation on ice for 30 min, homogenates were centrifuged at 15,000 rpm for 15 min at 4 °C. Supernatants were transferred to new tubes and protein concentrations were determined by Bradford protein assay (Genstar).
For preparation of detergent-soluble and -insoluble fractions, mouse tissues were homogenized in five volumes of sucrose buffer (0.25 m sucrose, 50 mm Tris-HCl, pH 7.4, and 1 mm EDTA) supplemented with 1 mm PMSF and protease inhibitors (Roche). After centrifuging at 2500 rpm for 10 min at 4 °C, the supernatants were collected, and the protein concentration was adjusted to 4 μg/μl. An aliquot of 100-μl supernatant was mixed with 100 μl of sucrose buffer containing 1% Triton X-100, then centrifuged at 15,000 rpm for 15 min at 4 °C to separate supernatants (detergent-soluble fraction) and pellets. Pellets were dissolved in 200 μl of 1% SDS in PBS (detergent-insoluble fraction). Equal amounts (30∼40 μg) of proteins were subjected to SDS-PAGE electrophoresis and then transferred onto a PVDF membrane. After blocking with 5% nonfat milk for 1 h at room temperature, membranes were incubated with primary antibodies at 4 °C overnight, washed with PBST three times and incubated with HRP-labeled secondary antibodies for 1 h at room temperature. Immunoreactivity was detected using an Enhanced Chemiluminescent (ECL) kit (Pierce).
Quantitative RT-PCR
RNA was extracted using TRIzol (Invitrogen) and cDNA was synthesized by Super Script® III First-Strand Kit (Invitrogen). Quantitative PCR was performed on an Eppendorf Mastercycler® ep Realplex 4 using SYBR® Premix Ex TaqTM (TaKaRa).
The following primers were used: 5′-ACTTCCCTCTCGCTGTATTTGAT-3′ (F-Ei24) and 5′-TTGCTTCCGCTCTACACTCTG-3′ (R-Ei24); 5′-GCTGCCCTATACCCACATCT-3′ (F-p62) and 5′-CGCCTTCATCCGAGAAAC-3′ (R-p62); 5′-CTACCTTGCCCGAAAGCAC-3′ (F-Gstm1) and 5′-ATGTCTGCACGGATCCTCTC-3′ (R-Gstm1); 5′-ACCAAGGACACCAAGTTTCG-3′ (F-Cyp2a5) and 5′-AGAGCCCAGCATAGGAAACA-3′ (R-Cyp2a5); 5′-AGCGTTCGGTATTACGATCC-3′ (F-Nqo1) and 5′-AGTACAATCAGGGCTCTTCTCG-3′ (R-Nqo1); 5′-CTGGCTCCTAGCACCATGAAGAT-3′ (F-actin) and 5′-GGTGGACAGTGAGGCCAGGAT-3′ (R-actin).
Proteasome Activity Assay
Proteasome activity was determined using aminomethylcoumarin (AMC)-linked synthetic peptide substrates: Ac-Gly-Pro-Leu-Asp-AMC, Suc-Leu-Leu-Val-Tyr-AMC and Ac-Arg-Leu-Arg-AMC (and Boc-Leu-Arg-Arg-AMC) for caspase-like, chymotrypsin-like, or trypsin-like activity, respectively (Proteasome Substrate Pack, Enzo Life Sciences). Mice tissues were homogenized in lysis buffer (50 mm HEPES pH 7.5, 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, and 2 mm ATP). 250 μl of lysate containing equal amounts of protein (2–4 μg) were incubated for 30 min at 37 °C in the dark with 2.5 μl of each substrate (final concentration: 50 nm). The reaction was stopped by adding 252.5 μl pre-cooled 96% ethanol solution. Proteasome activity was measured by detecting fluorescence from AMC hydrolysis (380 nm excitation and 460 nm emission).
Transmission Electron Microscopy
Mice were transcardially perfused with 2% PFA/2% glutaraldehyde buffered with 0.1 m pH 7.4 phosphate buffer (PB). Tissues were separated, fixed in 2.5% glutaraldehyde, post-fixed with 1% OsO4 in PB buffer for 2 h, dehydrated with graded ethanol solutions, and embedded in Embed812. Ultrathin sections (80 nm) were stained with 2% uranyl acetate for 30 min and lead citrate for 10 min and examined using transmission electron microscopy (TecnaiTM Spirit, FEI).
Statistical Analysis
Data from at least three sets of samples were used for statistical analysis. Statistical significance was calculated by Student's t test. A p value less than 0.05 was considered significant.
RESULTS
Neural-specific Deletion of Ei24 Causes Behavioral and Motor Abnormalities
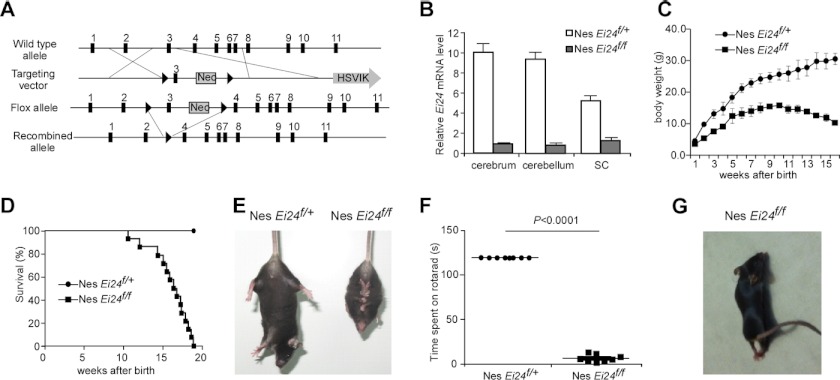
We generated Ei24 conditional knock-out mice by flanking exon 3 of Ei24 with two loxP sequences (Fig. 1A). When we crossed Ei24+/− adults (obtained from Ei24flox/wt; Zp3-Cre mice), no Ei24−/− offspring survived to the neonatal stage, suggesting an essential role of Ei24 during embryogenesis. The function of Ei24 was therefore investigated by crossing Ei24flox/flox mice with mice expressing tissue-specific Cre recombinase.
FIGURE 1.
Motor and behavioral deficits in Ei24flox/flox; nestin-Cre mice. A, scheme for generating Ei24 conditional knock-out mice. Exons are depicted by black boxes. A targeting construct was prepared by flanking exon 3 of Ei24 with the neor gene and loxP sites (triangles). The deleted allele of Ei24 was generated by Cre-mediated recombination to remove exon 3. B, Nestin Cre effectively removes exon 3 of Ei24 in brain tissues. Total RNAs were prepared from cerebrum, cerebellum, and spinal cord of Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice at 4 months of age. Transcription levels of Ei24 mRNA are normalized to Actin mRNA. Results are representative of at least three experiments. C, body weight curves of Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice over 16 weeks. Mean ± S.E. of 18 mice is shown. D, survival curves of Ei24flox/+; nestin-Cre (n = 18) and Ei24flox/flox; nestin-Cre mice (n = 18) mice over 19 weeks. E, when lifted by the tail, Ei24flox/+; nestin-Cre mice extend their limbs, while Ei24flox/flox; nestin-Cre mice have an abnormal limb-clasping reflex, adopting a bat-like posture. F, time spent on an autoaccelerating rod (rotarod). The maximum observed time was 120 s. Mean ± S.E. of 9 mice is shown. G, when Ei24flox/+; nestin-Cre mice initiate movement, their hindlimbs are held in a posteriorly hyperextended position.
Ei24flox/flox mice were crossed with nestin-Cre mice to produce animals deficient for Ei24 in the central nervous system (referred to as Ei24flox/flox; nestin-Cre) from embryonic stages (Fig. 1B). Nestin is highly expressed in neuroepithelial precursors, which give rise to neurons, astrocytes and oligodendrocytes (21). Ei24flox/flox; nestin-Cre mice appeared normal at birth, but started to show growth retardation at 2 weeks of age and had ∼50% reduction in body weight compared with Ei24flox/+; nestin-Cre littermates (referred to as control littermates in this study) by 3 months (Fig. 1C). The survival rate was also dramatically reduced; 50% of mutant mice died by ∼16 weeks after birth and all mice died by 19 weeks independent of sex (Fig. 1D). Ei24flox/flox; nestin-Cre mice exhibited motor and behavioral deficits at 6 weeks of age and became progressively worse. 8 out of 12 mutant mice at 6 weeks of age and 12 out of 12 of mutant mice at 8 weeks of age exhibited abnormal limb-clasping reflexes (Fig. 1E). All 3-month-old mutants showed poor motor coordination in a rotarod test (Fig. 1F). Mutant mice exhibited marked tremor at 8 weeks and ataxic gait at 12 weeks. At 14 weeks, the hindlimbs of resting mutant mice were flexed, like control littermates, but became hyperextended posteriorly during movement (Fig. 1G; supplemental movie). Thus, neural-specific Ei24 deficiency causes behavioral and motor defects.
Ei24flox/flox; nestin-Cre Mice Suffer Massive Axonal Degeneration and Neuronal Cell Loss
To investigate the neural defect in Ei24flox/flox; nestin-Cre mice, histological analyses were performed by hematoxylin and eosin (H&E) staining and Nissl staining of sections from cerebrum, cerebellum, and spinal cord of control and mutant mice at 4 months of age. Ei24flox/flox; nestin-Cre brains had enlarged lateral ventricles (Fig. 2A). Mutant mice showed atrophy of the cerebral cortex (Fig. 2A); the ratio of cortical to dorsoventral thickness in Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice was 0.18 ± 0.02 and 0.13 ± 0.01, respectively (p < 0.05). The white matter between the cortex and hippocampal pyramidal cells was thinner in mutants (Fig. 2A). The number of neurons in cortical layers 1–4 was greatly reduced compared with controls, as was the number of pyramidal neurons in the 5th layer of the motor and sensory cortices (Fig. 2B; supplemental Fig. S1, B and C). The hippocampal pyramidal cell layer was much thinner with reduced neuron numbers (Fig. 2C). H&E and axon-specific β-tubulin III staining revealed irregularly arranged nerve fibers in the alveus of the hippocampus (supplemental Fig. S1, D and E). Eosinophilic spheroids, representing axon swellings (10), were observed in the cortex (supplemental Fig. S1F). Vacuolated cells were observed in the cingulate cortex and to a lesser extent in other cortices, hippocampus, thalamus, and hypothalamus (Fig. 2D). Numerous vacuolated cells were found in white matter regions including the corpus callosum, the alveus of the hippocampus and the internal capsule (Fig. 2D, supplemental Fig. S1D, data not shown). The vacuolated cells were stained by the oligodendroglial marker anti-2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNPase) (Fig. 2E). Oligodendrocytes support axonal integrity and myelinate the axons of the CNS (22). Staining with anti-myelin basic protein (MBP) (which labels myelin) or anti-CNPase (which labels cell bodies and myelin) revealed fragmented myelin structures in various cerebral regions in mutant mice (Fig. 2F; supplemental Fig. S1G).
FIGURE 2.
Axonal degeneration and neuronal loss in Ei24flox/flox; nestin-Cre mice. A, Nissl staining of cortex and hippocampus of Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice at 4 months of age. The arrow indicates an enlarged lateral ventricle. Bar, 200 μm. B, Nissl staining of the fifth layer of the cortex. The graph shows the number of large pyramidal cells per mm2 in the indicated areas. Mean ± S.E. of three mice is shown. Bar, 50 μm. C, H&E staining of the hippocampal pyramidal cell layer. The number of pyramidal cells was quantified and divided by the length of the layer. Mean ± S.E. of three mice is shown. Bar, 50 μm. D, compared with control mice, Nissl staining of the cerebrum shows large numbers of vacuolated cells (arrows) in the cingulate cortex and internal capsule of Ei24flox/flox; nestin-Cre mice. Bar, 50 μm. E, anti-CNPase staining show that vacuolated cells are CNPase-positive oligodendroglial cells in the cingulate cortex and internal capsule of Ei24flox/flox; nestin-Cre mice. Bar, 10 μm. F, myelin stainning by anti-CNPase exhibits irregular arrangement in the cingulate cortex in Ei24flox/flox; nestin-Cre mice at 4 months of age. Bar, 100 μm. G, H&E staining shows that the cerebellum is less foliated (arrow) and fissured in Ei24flox/flox; nestin-Cre mice at 4 months. Bar, 500 μm. H, thickness of the molecular layer, calculated by dividing the distance between lobules III and IV, or lobules V and VI, by 2. Mean ± S.E. of five mice is shown. I, H&E staining of Purkinje cells in control and mutant mice. The number of Purkinje cells in lobules III, IV, and V was quantified and divided by the total length of the lobules. Mean ± S.E. of five mice is shown. Bar, 50 μm. J, anti-calbindin (green) and anti-MBP (red) costaining reveals that Ei24flox/flox; nestin-Cre mice exhibit dilated calbindin-positive bulbs (arrows) in the DCN region and some of the bulbs are enwrapped by MBP-labeled myelin. Bar, 10 μm. K–M, EM pictures of DCN in Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice at 4 months. K, myelinated axons of regular shape and size in the DCN region of control animals. (L–M), Degenerated axons in mutant mice. L, arrowheads indicate electron-dense amorphous structures. The arrow shows a thinned myelin sheath. M, arrowhead indicates undulated and split myelin lamellae. Note that the axons in the mutant sample are conspicuously less abundant than in the control. Bar, 1 μm (K–M). N, number of interneurons per mm2 in the lumbar spinal cord. Mean ± S.E. of five mice is shown. O, accumulation of vacuolated cells in the anterior horn of the lumbar spinal cord in mutant mice at 4 months. Bar, 100 μm. P, GFAP signal (red), in sections of cerebral cortex immunostained with anti-GFAP antibody, is stronger in mutant mice. Bar, 20 μm.
The cerebella of Ei24flox/flox; nestin-Cre mice were less foliated and fissured than controls (Fig. 2G). The molecular cell layer was thinner (Fig. 2H). Purkinje cell numbers were reduced by 40% and the remaining cells appeared shrunken (Fig. 2I). The deep cerebellar nuclei (DCN), to which Purkinje cells project axons, contained numerous large eosinophilic spheroid structures (supplemental Fig. S1H), indicative of axonal swellings (10). We further stained the DCN region with anti-calbindin, which specifically labels Purkinje axons, and found accumulation of huge bulging calbindin-positive structures in the DCN region of Ei24flox/flox; nestin-Cre mice, indicating abnormal swelling and dystrophy of Purkinje cell axon terminals (Fig. 2J). MBP staining showed that some of the dilated calbindin-positive termini were enwrapped by myelin, confirming the degeneration of Purkinje cell axons (Fig. 2J; supplemental Fig. S1, I–J). Ultrastructurally, axon numbers in the DCN region were dramatically reduced and the remaining axons showed thinned, split myelin sheaths and various signs of degeneration (Fig. 2, K--M; supplemental Fig. S1, K–N). In the anterior horn of the lumbar spinal cord, interneuron numbers were dramatically reduced in Ei24flox/flox; nestin-Cre mice but the number of large motor neurons was not significantly different (Fig. 2N; supplemental Fig. S1, O and P). Numerous vacuolated oligodendrocytes accumulated in spinal cord gray and white matter (Fig. 2O; supplemental Fig. S1Q). Expression of the glial marker GFAP (glial fibrillary acidic protein) dramatically increased in the cerebrum, cerebellum and spinal cord of mutant mice (Fig. 2P), indicating that neurodegeneration was accompanied by reactive astrogliosis.
To investigate whether the neural abnormalities in Ei24flox/flox; nestin-Cre mice increase progressively, brain and spinal cord sections from Ei24flox/flox; nestin-Cre mice and controls at 3 weeks, one month, two months, and four months were analyzed. The number of pyramidal neurons in the 5th layer of the cortex in Ei24flox/flox; nestin-Cre mice at 3 weeks of age was comparable to control littermates, but gradually decreased as the mice grew older (Fig. 3A). The number of Purkinje cells and thickness of the molecular layer progressively decreased (Fig. 3, B and C). Vacuolated oligodendrocytes in the cerebrum, cerebellum, and spinal cord were small in size and few in number in mutant mice at 3 weeks and one month of age but became larger and more numerous at 2 and 4 months of age (Fig. 3A; supplemental Fig. S2, A and B). The mutant mice also showed progressively more irregularly arranged nerve fibers in the alveus of the hippocampus, more severe axonal swelling in the DCN of the cerebellum and deterioration of axonal myelin sheaths in various regions of the brain and spinal cord (Fig. 3, D and E; supplemental Fig. 2C). Thus, neural-specific Ei24 deficiency results in progressive axonal degeneration and neuronal loss.
FIGURE 3.
Progressive deterioration in cerebrum, cerebellum and spinal cord of Ei24flox/flox; nestin-Cre mice. A, Nissl staining of the cortex in control and Ei24flox/flox; nestin-Cre mice at different ages shows progressive accumulation of vacuolated cells. Bar, 100 μm. B and C, number of Purkinje cells (B) and the thickness of the molecular layer (C) in cerebella from Ei24flox/flox; nestin-Cre mice at different ages. D, costaining of β-tubulin III (green) and p62 (red) in cerebrum shows that axonal fibers become progressively more irregularly arranged. p62 aggregates and diffuse levels of p62 in Ei24flox/flox; nestin-Cre also become dramatically elevated with age. Bar, 10 μm. E, axon terminals of Purkinje cells in the DCN region, stained by anti-calbindin, become progressively more dilated. Bar, 10 μm.
Ei24 Deficiency Impairs Autophagic Flux
We next investigated autophagic flux in mutant mice by examining levels and distribution of LC3. Brain and spinal cord extracts from Ei24flox/flox; nestin-Cre mice at 4 months showed accumulation of LC3-II, a lipidated form of LC3 that normally associates with autophagosomal membranes (Fig. 4A). Immunostaining with anti-LC3 revealed that LC3 diffusely accumulated in various regions of the brain and spinal cord of Ei24flox/flox; nestin-Cre mice at 4 months (Fig. 4B; supplemental Fig. S3, A–C). LC3 puncta were largely absent, apart from a few that accumulated in white matter regions, such as the alveus of hippocampus in the cerebrum, DCN in cerebellum and the LC (lateral column) in spinal cord (Fig. 4C; supplemental Fig. S3, A, B, D).
FIGURE 4.
Accumulation of LC3-II, p62 aggregates and ubiquitin-positive inclusions in neural-deficient Ei24 mice. A, total brain and spinal cord proteins from Ei24flox/+; nestin-Cre and Ei24flox/flox; nestin-Cre mice at 4 months of age were extracted and separated by SDS-PAGE and analyzed by immunoblotting with anti-LC3 antibodies. Results are representative of at least three experiments. B, anti-LC3 (red) staining reveals that Ei24flox/flox; nestin-Cre mice exhibit stronger diffuse LC3 signal in the DCST (dorsal corticospinal tract) of spinal cord. Bar, 10 μm. C, a few LC3 puncta colocalize with p62 aggregates in the the alveus of the hippocampus of Ei24flox/flox; nestin-Cre mice at 4 months. Bar, 10 μm. D, Ei24flox/flox; nestin-Cre mice at 4 months of age show p62 accumulation in brain and spinal cord by immunoblotting with p62 antibodies. Results are representative of at least three experiments. E, immunoblotting assay showing polyubiquitinated proteins in detergent-soluble and detergent-insoluble fractions of brain and spinal cord (SC) homogenates from control and mutant mice. Results are representative of at least three experiments. F, p62 aggregates are absent from the anterior horn of the spinal cord of Ei24flox/+; nestin-Cre mice. p62 aggregates (red) progressively accumulate in mutant mice at 3 weeks, 1 month, and 4 months of age. Levels of diffuse p62 are also elevated especially in vacuolated cells (arrow). Bar, 10 μm. G, anti-CNPase and anti-p62 costaining show that compared with controls diffuse p62 and the majority of p62 aggregates are located in CNPase-positive oligodendroglial cells in the cingulate cortex of Ei24flox/flox; nestin-Cre mice. Bar, 10 μm. H, compared with control animals, p62 (red) and ubiquitinated proteins (green) accumulate and are colocalized in cerebrum sections of Ei24flox/flox; nestin-Cre mice. p62 aggregates are more numerous than ubiquitinated protein inclusions. Bar, 10 μm.
Accumulation of p62 and ubiquitinated protein aggregates is one of the hallmarks of autophagy deficiency (23). p62 is required for formation and selective removal of ubiquitin-positive aggregates (12). Brain and spinal cord extracts from mutant mice at 4 months showed dramatic accumulation of p62 and high-molecular mass polyubiquitinated proteins (Fig. 4, D and E). p62 mRNA levels were not up-regulated in mutants (supplemental Fig. S3E). Polyubiquitinated proteins showed age-dependent progressive accumulation in Ei24flox/flox; nestin-Cre mice (supplemental Fig. S3F). Impaired proteasome function can also cause accumulation of p62 and polyubiquitinated proteins; however, proteasomal caspase-like, chymotryptic, and tryptic activities, measured using specific substrate peptides, were comparable in controls and mutants (supplemental Fig. S3G).
We next examined the distribution of p62 and ubiquitinated proteins in mutant mice. Control brain and spinal cord contained very little or no cytoplasmic p62 and ubiquitin-positive aggregates (Fig. 4F; supplemental Fig. S3, H and K). In 3-week-old Ei24flox/flox; nestin-Cre mice, the cerebrum, cerebellum and spinal cord accumulated numerous small p62 aggregates, which increased in size and number with age (Fig. 4F). p62 accumulated most heavily in the alveus of the hippocampus and the internal capsule, where vacuolated oligodendrocytes were present. p62 aggregates mainly accumulated in oligodendrocytes (identified with CNPase) and were also detected in neurons (identified with NeuN) (Fig. 4G; supplemental Fig. S3, H and I), but were hardly observed in GFAP-labeled astroglia or calbindin-stained axonal swellings (supplemental Fig. S3, J–L). The level of diffuse cytoplasmic p62 was also greatly elevated in mutant mice, especially in vacuolated oligodendrocytes (Fig. 4, C, F, G). Ubiquitin-positive aggregates colocalized with p62 aggregates in Ei24flox/flox; nestin-Cre mice, but they were fewer in number than p62 aggregates (Fig. 4H). The few LC3 puncta were colocalized with p62 aggregates (Fig. 4C; supplemental Fig. S3B). Thus, autophagy is defective in neural-specific Ei24-deficient mice.
Liver-specific Depletion of Ei24 Causes Hepatomegaly and Multiple Tumor-like Protrusions
Ei24flox/flox mice were crossed with mice expressing Cre controlled by the albumin promoter to deplete Ei24 in the liver (supplemental Fig. S4A). Ei24flox/flox; Alb-Cre mice appeared normal at birth but by 3 months their livers were greatly enlarged, with disorganized hepatic lobules and swollen appearance of hepatocytes that were mixed with normal appearance hepatocytes (Fig. 5, A–C). Unlike Ei24-deficient neuronal cells, vacuolization of hepatocytes was not obvious in Ei24flox/flox; Alb-Cre livers at 3 months. Ultrastructurally, swollen mitochondria accumulated in mutant hepatocytes (supplemental Fig. S4C). Activities of alkaline phosphatase and aspartate aminotransferase were significantly elevated in mutant mice sera, suggesting hepatic cell death (supplemental Fig. S4D). By 10 months, multiple tumor-like protrusions were detected in mutant livers (Fig. 5D). Histological analyses revealed that these protrusions were composed of enlarged hepatocytes and were demarcated from neighboring hepatocytes of normal size (Fig. 5E). However, no signs of malignancy such as abnormal nuclear morphology were detected in the protrusions.
FIGURE 5.
Liver-specific deletion of Ei24 causes hepatomegaly and autophagy defects. A, gross anatomical views of representative livers from Ei24flox/+; Alb-Cre and Ei24flox/flox; Alb-Cre mice at 3 months of age. Livers in Ei24flox/flox; Alb-Cre mice are enlarged. B, graphs showing net weight of livers and ratio of liver/body weight in control and mutant mice. Mean ± S.E. of 3 mice is shown. C, H&E-stained liver sections from control and mutant mice show disorganized hepatic lobules and swollen hepatocytes in Ei24flox/flox; Alb-Cre mice. Insets show higher magnification views. Bar, 50 μm. D, gross anatomy of livers from Ei24flox/flox; Alb-Cre mice at 10 months of age demonstrates the development of multiple tumor-like protrusions (arrows). E, H&E staining of liver sections from Ei24flox/flox; Alb-Cre mice at 10 months of age (first panel). The arrow indicates a tumor-like protrusion. Bar, 100 μm. Higher magnification images of livers from controls (second panel) and Ei24-deficient mice (third panel, protrusion region) at 10 months of age. Bar, 20 μm. F, accumulation of LC3-II, p62 and polyubiquitinated proteins in livers from Ei24flox/flox; Alb-Cre mice. Total liver and spleen proteins were extracted, separated by SDS-PAGE, and analyzed by immunoblotting with anti-LC3 and anti-p62 antibodies. Detergent-soluble and detergent-insoluble fractions from liver homogenates were immunoblotted with anti-ubiquitin and anti-p62 antibodies. G, coimmunostaining with anti-p62 (red) and anti-ubiquitin (green) antibodies reveals that p62 and ubiquitin-positive aggregates are absent from control liver sections, but accumulate heavily in liver sections of Ei24flox/flox; Alb-Cre mice. p62 aggregates colocalize with ubiquitin-positive inclusions in mutant liver. Bar, 10 μm. H, immunoblotting with anti-Nqo1 antibody shows that the Nqo1 protein level is dramatically elevated in Ei24flox/flox; Alb-Cre mice. Levels of Nrf2 remain unchanged in control and Ei24-deficient liver. I, Gstm1, Nqo1, and Cyp2a5 mRNA levels are increased in Ei24-deficient mice. The Gstm1, Nqo1, and Cyp2a5 transcription levels were normalized to actin mRNA. Results are representative of at least three experiments.
We examined autophagic flux and found that liver homogenates from Ei24flox/flox; Alb-Cre mice at 3 months contained dramatically elevated LC3-II, p62, and polyubiquitinated proteins (Fig. 5F). p62 aggregates and ubiquitin-positive inclusions accumulated and colocalized in swollen hepatocytes (Fig. 5G; supplemental Fig. S4, E and F). Diffuse p62 was much weaker than in Ei24-deficient neural cells. LC3 formed a few puncta that colocalized with p62 aggregates in mutant liver at 3 months (supplemental Fig. S4, G and H).
Ei24 Deficiency Activates the Nrf2-mediated Stress Response Pathway in Liver
Dramatic accumulation of p62 aggregates in Ei24-deficient liver prompted us to examine whether the Nrf2 stress response is activated. p62 accumulation causes sequestration of Keap1, a component of the Cullin-3-type ubiquitin ligase for Nrf2 degradation, and subsequently leads to stabilization of Nrf2 and transcriptional activation of Nrf2 target genes including those encoding various anti-oxidant and detoxifying enzymes (24). Ei24 deletion caused no changes in levels of Nrf2 (Fig. 5H). Immunoblotting showed that levels of the Nrf2 target NAD(P)H dehydrogenase quinone 1 (Nqo1) were elevated in the liver of Ei24flox/flox; Alb-Cre mice at 3 months of age (Fig. 5H). Quantitative RT-PCR showed dramatically elevated mRNA levels of Gstm1 (glutathione S-transferase), Nqo1, and Cyp2a5 (cytochrome 450), but not p62, in mutant liver (Fig. 5I, supplemental Fig. S4I). These detoxifying enzyme genes, however, were not up-regulated in the brains of Ei24flox/+; nestin-Cre mice (supplemental Fig. S4J), suggesting that persistent Nrf2 activation may be tissue specific.
DISCUSSION
Here we demonstrated that mice deficient for the metazoan-specific autophagy gene Ei24 show degeneration of neuronal populations and severe liver injury. The behavioral and motor abnormalities in mice with neural-specific Ei24 depletion are similar to, but more severe than, those in Atg5 and Atg7 conditional knock-out mice. All Ei24-deficient mice die before 19 weeks, while Atg7-deficient mice reach 28 weeks (11). Ei24 deficiency results in massive vacuolated oligodendrocytes that are not observed in Atg5 and Atg7 knock-out mice (10–12). Oligodendrocytes support axonal integrity and myelinate axons for rapid impulse propagation in the CNS, and their dysfunction, as seen in Cnp1 knock-out mice, causes axonal swelling and neurodegeneration throughout the brain (22, 25). Accumulation of p62 aggregates and ubiquitin-positive inclusions in Ei24-deficient tissues indicates that in addition to neurons, autophagy plays an important role in homeostatic maintenance of oligodendroglial cells. EI24 is an ER-localized protein and may have functions independent of autophagy, which could explain why oligodendrocytes are more severely impaired in Ei24-deficient mice than in other autophagy knockouts. Atg5 and Atg7 deficiency cause intrinsic cell-autonomous neuronal cell death (26, 27). Compared with Atg5 and Atg7 knock-out mice, Ei24-deficient mice exhibit less severe accumulation of p62 and ubiquitin-positive aggregates in neurons, which could be because the axonal degeneration and neuronal loss in Ei24 mutants are severely exacerbated by the more pronounced dysfunction of vacuolated oligodendrocytes. The pathological defects associated with Ei24 deficiency are cell-type specific. Ei24-deficient liver exhibits similar defects to Atg5- and Atg7-deficient liver, including hepatocyte hypertrophy, hepatic cell death and hepatomegaly (13, 14). However, the tumor-like protrusions in Ei24-deficient liver are composed of enlarged hepatocytes and display no signs of malignancy. These protrusions could be caused by aggregation of abnormal enlarged hepatocytes. Reduced Ei24 expression in aggressive breast cancers implicates it is a tumor suppressor (3, 5). Autophagy suppresses tumorigenesis through suppression of tumorigenic inflammation, surveillance of genome stability and elimination of p62 (14–16). The essential role of Ei24 in autophagy may contribute to its tumor suppression function in other tissues. Distinct defects in Ei24-deficient liver and brain could be caused by tissue-specific activation of Nrf2 by p62 accumulation. Anti-oxidant and detoxifying enzymes, including Nqo1, Gstm1, and Cyp2a5, are highly expressed in Ei24-deficient liver, but not in the brains of Ei24-deficient mice. Simultaneous depletion of p62 or Nrf2 significantly suppresses liver injury in Atg7-deficient mice, including increased liver weight, disorganization of lobular structures and hepatocyte hypertrophy (12, 24). However, ablation of p62 does not suppress neurodegeneration caused by autophagy deficiency (12).
The molecular role of Ei24 in the autophagy pathway has yet to be determined. EI24/EPG-4 is an ER-localized protein with six transmembrane domains. PI(3)P-enriched ER subdomains, known as omegasomes, are known to be the membrane source for autophagosome formation (18). Several ATG proteins involved in early steps of autophagy, including ULK1 and ATG14, tightly associate with the ER, and are required for formation of LC3 puncta (autophagosomes) (28). Consistent with a role in early autophagy, EPG-4 regulates the progression of omegasomes to autophagosomes (20). However, knockdown of Ei24 appears to cause a defect in the degradation step of the autophagy pathway. Enlarged nondegradative autolysosomes accumulate in Ei24 siRNA knockdown cells (20). In Ei24-deficient mice, lipidated LC3 accumulates dramatically and is diffusely localized, suggesting that Ei24 acts at an early step of autophagosome formation. The few LC3 puncta detected in Ei24-deficient mice are likely due to incorporation of LC3 into p62 aggregates via direct p62/LC3 interaction. Alternatively, p62 aggregates may accumulate in impaired autolysosomes due to a defect in the degradation step of the autophagy pathway. Ei24 is highly induced by p53 in various cell types. p53 induces autophagy activity by inhibiting mTOR activity and also by up-regulating the lysosomal protein DRAM (29, 30). Distinct from Atg genes and Ei24, DRAM is critical for induction of autophagy in the specific context of p53 activation and is not required for basal autophagy (30). The essential role of Ei24 in basal autophagy indicates that p53 may regulate autophagy activity by integrating with the core autophagic machinery.
Acknowledgments
We thank Drs. Zelda Cheung, Masaaki Komatsu, and Waguri Satoshi for helpful comments on the manuscript and Dr. Isabel Hanson for editing work.
This work was supported by the National Basic Research Program of China (2011CB910100, 2010CB835201) (to H. Z.).

This article contains supplemental Figs. S1–S4 and movie.
- Ei24
- etoposide-induced 2.4 kb transcript
- SQSTM
- sequestosome
- AMC
- aminomethylcoumarin
- MBP
- myelin basic protein
- H&E
- hematoxylin and eosin
- DCN
- deep cerebellar nuclei.
REFERENCES
- 1. Lehar S. M., Nacht M., Jacks T., Vater C. A., Chittenden T., Guild B. C. (1996) Identification and cloning of EI24, a gene induced by p53 in etoposide-treated cells. Oncogene 12, 1181–1187 [PubMed] [Google Scholar]
- 2. Gu Z., Flemington C., Chittenden T., Zambetti G. P. (2000) ei24, a p53 response gene involved in growth suppression and apoptosis. Mol. Cell. Biol. 20, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao X., Ayer R. E., Davis S. L., Ames S. J., Florence B., Torchinsky C., Liou J. S., Shen L., Spanjaard R. A. (2005) Apoptosis factor EI24/PIG8 is a novel endoplasmic reticulum-localized Bcl-2-binding protein which is associated with suppression of breast cancer invasiveness. Cancer Res. 65, 2125–2129 [DOI] [PubMed] [Google Scholar]
- 4. Mork C. N., Faller D. V., Spanjaard R. A. (2007) Loss of putative tumor suppressor EI24/PIG8 confers resistance to etoposide. FEBS Letters 2581, 5440–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gentile M., Ahnström M., Schön F., Wingren S. (2001) Candidate tumour suppressor genes at 11q23-q24 in breast cancer: evidence of alterations in PIG8, a gene involved in p53-induced apoptosis. Oncogene 20, 7753–7760 [DOI] [PubMed] [Google Scholar]
- 6. Xie Z., Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 7. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 8. Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ichimura Y., Komatsu M. (2011) Pathophysiological role of autophagy: lesson from autophagy-deficient mouse models. Exp. Anim. 60, 329–345 [DOI] [PubMed] [Google Scholar]
- 10. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 11. Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 12. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 13. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. (2011) Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inami Y., Waguri S., Sakamoto A., Kouno T., Nakada K., Hino O., Watanabe S., Ando J., Iwadate M., Yamamoto M., Lee M. S., Tanaka K., Komatsu M. (2011) Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 193, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Longatti A., Tooze S. A. (2009) Vesicular trafficking and autophagosome formation. Cell Death Differ. 16, 956–965 [DOI] [PubMed] [Google Scholar]
- 18. Tooze S. A., Yoshimori T. (2010) The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 [DOI] [PubMed] [Google Scholar]
- 19. Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D. C. (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 12, 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian Y., Li Z. P., Hu W. Q., Ren H. Y., Tian E., Zhao Y., Lu Q., Huang X. X., Yang P. G., Li X., Wang X., Kovács A. L., Yu L., Zhang H. (2010) C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 [DOI] [PubMed] [Google Scholar]
- 21. Lendahl U., Zimmerman L. B., McKay R. D. (1990) CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595 [DOI] [PubMed] [Google Scholar]
- 22. Emery B. (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782 [DOI] [PubMed] [Google Scholar]
- 23. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 25. Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P. E., Griffiths I. R., Nave K. A. (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 33, 366–374 [DOI] [PubMed] [Google Scholar]
- 26. Komatsu M., Wang Q. J., Holstein G. R., Friedrich V. L., Jr., Iwata J., Kominami E., Chait B. T., Tanaka K., Yue Z. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishiyama J., Miura E., Mizushima N., Watanabe M., Yuzaki M. (2007) Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 3, 591–596 [DOI] [PubMed] [Google Scholar]
- 28. Itakura E., Mizushima N. (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Budanov A. V., Karin M. (2008) p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M. (2006) DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134 [DOI] [PubMed] [Google Scholar]