Background: MicroRNAs control cell signaling during osteoblast differentiation.
Results: miR-218, which is highly expressed in osteoblasts and cancer cells metastatic to bone, targets three inhibitors of Wnt signaling, Sclerostin, Dickkopf2, and secreted frizzled-related protein2.
Conclusion: miR-218 promotes differentiation of normal osteoblast and the osteomimetic bone-homing properties of tumor cells.
Significance: miR-218 may be a universal stimulator of Wnt-signaling during bone development and cancer progression.
Keywords: Differentiation, Metastasis, MicroRNA, Osteoblasts, Wnt Signaling, Osteomimicry
Abstract
MicroRNAs (miRNAs) negatively and post-transcriptionally regulate expression of multiple target genes to support anabolic pathways for bone formation. Here, we show that miR-218 is induced during osteoblast differentiation and has potent osteogenic properties. miR-218 promotes commitment and differentiation of bone marrow stromal cells by activating a positive Wnt signaling loop. In a feed forward mechanism, miR-218 stimulates the Wnt pathway by down-regulating three Wnt signaling inhibitors during the process of osteogenesis: Sclerostin (SOST), Dickkopf2 (DKK2), and secreted frizzled-related protein2 (SFRP2). In turn, miR-218 expression is up-regulated in response to stimulated Wnt signaling and functionally drives Wnt-related transcription and osteoblast differentiation, thereby creating a positive feedback loop. Furthermore, in metastatic breast cancer cells but not in normal mammary epithelial cells, miR-218 enhances Wnt activity and abnormal expression of osteoblastic genes (osteomimicry) that contribute to homing and growth of cells metastatic to bone. Thus, miR-218/Wnt signaling circuit amplifies both the osteoblast phenotype and osteomimicry-related tumor activity.
Introduction
Signaling pathways activated by multiple osteogenic ligands including WNT, TGFβ/ bone morphogenetic protein (BMP),6 FGF, and Hedgehog regulate osteoblast differentiation and growth (1–5). The Wnt and BMP pathways have prominent and synergistic roles in osteoblast phenotype commitment during skeletal development. TGFβ and BMP2/7 extracellular signals result in SMAD co-receptor-mediated cytoplasmic-nuclear shuttling and activation of essential osteogenic genes including Runx2 (Cbfa1/AML3) and Osterix (1, 6). Wnt signaling transduced by LRP 5/6 and Frizzled receptor complexes, leads to nuclear translocation of β-catenin and its interaction with TCF/LEF factors to regulate transcription (2, 4). Both BMP and Wnt signaling are physiologically regulated by a number of secreted ligands and antagonists, as well as receptors and intracellular transcriptional mediators to direct bone formation (5, 7, 8).
Short non-coding microRNAs (miRNAs) have emerged as key post transcriptional repressors that support osteoblast growth and differentiation by compromising mRNA stability and/or by blocking protein translation. Conditional deletion of the miRNA processing enzyme Dicer in osteoblasts, chondrocytes, and osteoclasts has revealed an essential role for miRNAs in normal skeletal development and bone homeostasis (9–15). By binding to specific complementary sequences in the 3′-UTR of mRNAs, miRNAs control key components of osteogenic pathways (16–20).
Apart from the biological roles of BMP and Wnt signaling in bone development, these pathways are also up-regulated in breast cancer cells that grow aggressively in the bone microenvironment (21, 22). Indeed, metastatic breast cancer cells express many osteoblast related genes (osteomimicry) that facilitate homing to bone during metastasis (21). Identification of microRNAs controlling signaling pathways that support osteoblastogenesis may increase our understanding of the osteomimetic properties of bone metastatic cancer cells.
Here, we focused on miR-218 that is significantly up-regulated during osteoblast differentiation (18) and predicted to target multiple inhibitors of Wnt signaling. Because Wnt signaling is required for bone formation, we postulated that miRNA suppression of Wnt inhibitors would be pro-osteogenic. Our key finding is that miR-218 activates Wnt signaling by reducing expression of three different inhibitors, and by initiating a self-amplifying positive regulatory loop. Thus, miR-218 is a potent activator of Wnt signaling that contributes to osteoblastogenesis. Furthermore, we find that miR-218 also controls Wnt signaling to promote the osteomimicry of metastatic cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture Models
MC3T3-E1 osteoprogenitors were plated in 100-mm dishes and incubated in α-MEM with 10% FBS (Atlanta), 100 units/ml of penicillin, and 100 μg/ml of streptomycin. At confluence (day 0), these cells were treated with osteogenic differentiation media containing 10 mm β-glycerophosphate and 50 μg/ml of ascorbic acid. The differentiation media was refreshed every 48 h after the initial differentiation treatment.
Bone marrow stromal cells were isolated by flushing marrow from the femurs and tibia of 6–8-weeks-old C57/BL mice. The BMSCs were cultured in 100-mm plates in DMEM supplemented with 20% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin, 2 mm l-glutamine. After several passages to deplete hematopoietic cells, the stromal cells were transduced with a Lentivirus carrying the green fluorescent protein and pre miR-218, changing media every other day until cells reach 90% confluence. BMSCs were re-plated into 6 wells in growth media. At 80–100% confluence, differentiation media was added (day 0) (20% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin, 2 mm l-glutamine, 50 μg/ml ascorbic acid, 3 mm β-glycerophosphate). For both the miRNA analysis and quantitative real-time PCR, cells were harvested at the indicated days.
MCF10A epithelial cells were cultured in d-MEM supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml streptomycin and MDA-MB-231 metastatic breast cancer cells in α-MEM supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin as described.
Treatments
Confluent MC3T3 cells were treated with 5 ng/ml TGFβ, 100 ng/ml BMP2, and 10 μm TDZD-8 (GSK-3β inhibitor) (Alexis Biochemicals, 270–354-M005) to activate Wnt signal for 48 h in 10% FBS α-MEM with 50 μg/ml ascorbic acid and 10 mm β-glycerophosphate. To inhibit Wnt signaling MDA-MB-231 cells were treated with 30 μm small molecule inhibitor that destabilizes β-catenin, CCT036477 (Enzo Life Sciences)(23). Lentivirus clones; control (PMIRH000PA-1), miRNA-218 (PMIRH218-2PA-1) and anti-miR Lentivirus (miRZip-218 anti-miR, MZIP218-PA-1) were purchased from System Biosciences. Lentivirus plasmids were transfected by using Fugene 6 (Promega). miRNAs and anti-miRNAs (purchased from Ambion) were introduced into the cells at 50 nm by oligofectamin (Invitrogen). Lentivirus transduction was described elsewhere (16).
Real Time PCR
Total RNA was isolated from various experimental conditions using TRIzol (Invitrogen) following the manufacturer's instructions. Total RNA (0.5–1 μg) was reverse transcribed into cDNA using Oligo (dT) primers and used to analyze by real time qPCR. SYBR Green Master Mix (Applied Biosystems, Inc.) was used to detect the expression of gene markers. For miRNA detection, QuantimiR-RT kit (Systems Biosciences (SBI) was utilized according to manufacturer's instruction. The relative expressions were calculated by ΔCT method normalized to GAPDH or U6 expression to show absolute values of mRNAs or miRNA. Primers were synthesized from Invitrogen and sequences are listed in supplemental Table S1.
DNA Construction and Luciferase Assay
The 3′-UTR ( = 90–100 bps) for putative targets, Tob1, Sost, Dkk2, and Sfrp2 genes were synthesized with flanking 5′ SpeI and 3′ MluI restriction sites. Double-stranded annealed DNA fragments were phosphorylated and ligated to pMIR-Reporter Luciferase Plasmid (Applied BioSystems). Transformants were grown up and plasmid DNAs were confirmed by sequencing. The Luciferase assay was conducted by co-transfecting miR-218 (100 ng), NS miR (100 ng), and anti-miR-218 (100 ng) with WT and mutant pMIR-Reporter Luciferase DNA (200 ng) construct into MC3T3 cells. WT and mutants sequence of each 3′-UTR are shown (supplemental Table S2). Renilla Luciferase plasmid (Promega) was transfected to normalize the relative luciferase values. The transfected cells were incubated for 36 h to determine luciferase activity.
Northern Blots of miR-218
Northern blot analysis of miRNAs was done as described (17). RNA blots from MC3T3 osteoblasts at indicated time points were hybridized with radiolabeled antisense probe of miR-218. The blots were reprobed with U6 small RNA as control to determine the relative expression of miR-218.
Western Blots
Equal amounts of total protein were loaded into a 10% SDS-PAGE transferred onto a PVDF membrane probed with indicated primary antibody for SOST (R&D systems, AF1589), SFRP2 (Abcam, ab86379), TOB1 (Santa Cruz Biotechnology, sc-33192), and DKK2 (Abcam, ab38594), Lamin B1 (Invitrogen, 33-2000). After incubation with secondary antibody, antibody-bound protein complex in the membrane was detected with chemiluminescence reagent.
Top Flash Reporter Assay
Wild type and mutant top flash β-catenin LUC reporter (Addgene, Inc., Cambridge MA) containing 8 TCF/LEF binding sites (200 ng) was co-transfected with NS, miR-218, and anti-miR-218 along with 5 ng of Renilla LUC reporter plasmid using Fugene (Promega) in MC3T3 osteoblasts (60% confluency). The transfection was performed according to the manufacturer's protocol. Transfected cells were incubated at 37 °C for 36 h and assayed for relative luciferase activity normalized with Renilla values.
RESULTS
MiR-218 Is a Potent Activator of Osteoblast Differentiation
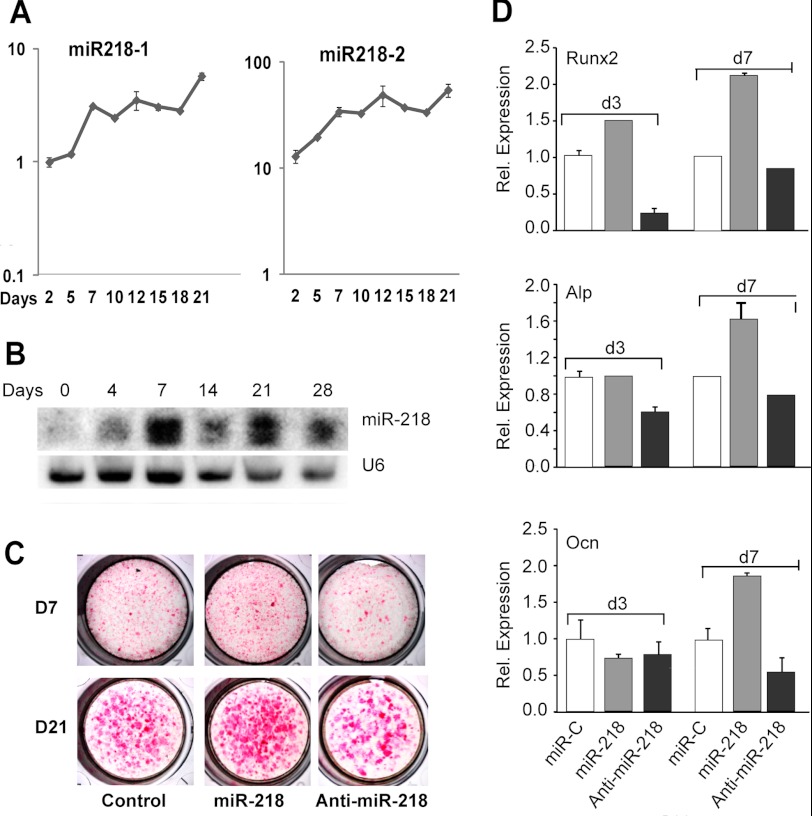
Differentiation of MC3T3 preosteoblasts is characterized by three stages of phenotypic maturation: day 7 (post-confluency, early differentiation), day 12 (mature osteoblasts), and days 15–21 (mineralization). During this maturation program, multiple microRNAs are up-regulated (18), including the pre-miRNAs miR-218–1 and 218–2 (Figs. 1, A and B and supplemental Fig. S1). The functional activity of miR-218 was assessed in MC3T3 pre-osteoblasts following forced expression of this miRNA or its corresponding anti-miRNA using lentiviral vectors. Elevation of miR-218 increased while the anti-miR diminished differentiation reflected by alkaline phosphatase (Alp) activity, a marker of osteoblast maturation (Fig. 1C). Similarly, expression of mRNAs for Alp, the osteogenic transcription factor Runx2, and the mineralization marker osteocalcin (Bglap/Ocn) were all significantly increased at day 3 (proliferation) and/or day 7 with exogenous miR-218 but not with anti-miR-218 (Fig. 1D). Thus, miR-218 promotes osteoblast differentiation.
FIGURE 1.
MicroRNA 218 expression and function during osteoblast differentiation. A, total RNA was analyzed for expression of precursor miR-218-1 and 218-2 by real time PCR at the indicated time points using MC3T3 cells. Absolute expression (y axis) was normalized to U6 small RNA. B, Northern blot analysis shows the expression of mature miR-218 during mouse calvarial preosteoblast MC3T3 differentiation with U6 RNA expression used as an equal loading control. C, effect of miR and anti-miR-218 on the histochemical staining of Alp activity at 7 and 21 days in MC3T3 differentiation is shown. D, MC3T3 cells were transduced with miR-218 and anti-miR-218 Lentivirus. Total RNA was analyzed by real time qRT-PCR for mRNA expression profile of bone marker genes at d3 and d7. The Runx2 transcription factor, Alp, and Ocn are represented as markers of early, mid, and late stages of osteoblast lineage cells.
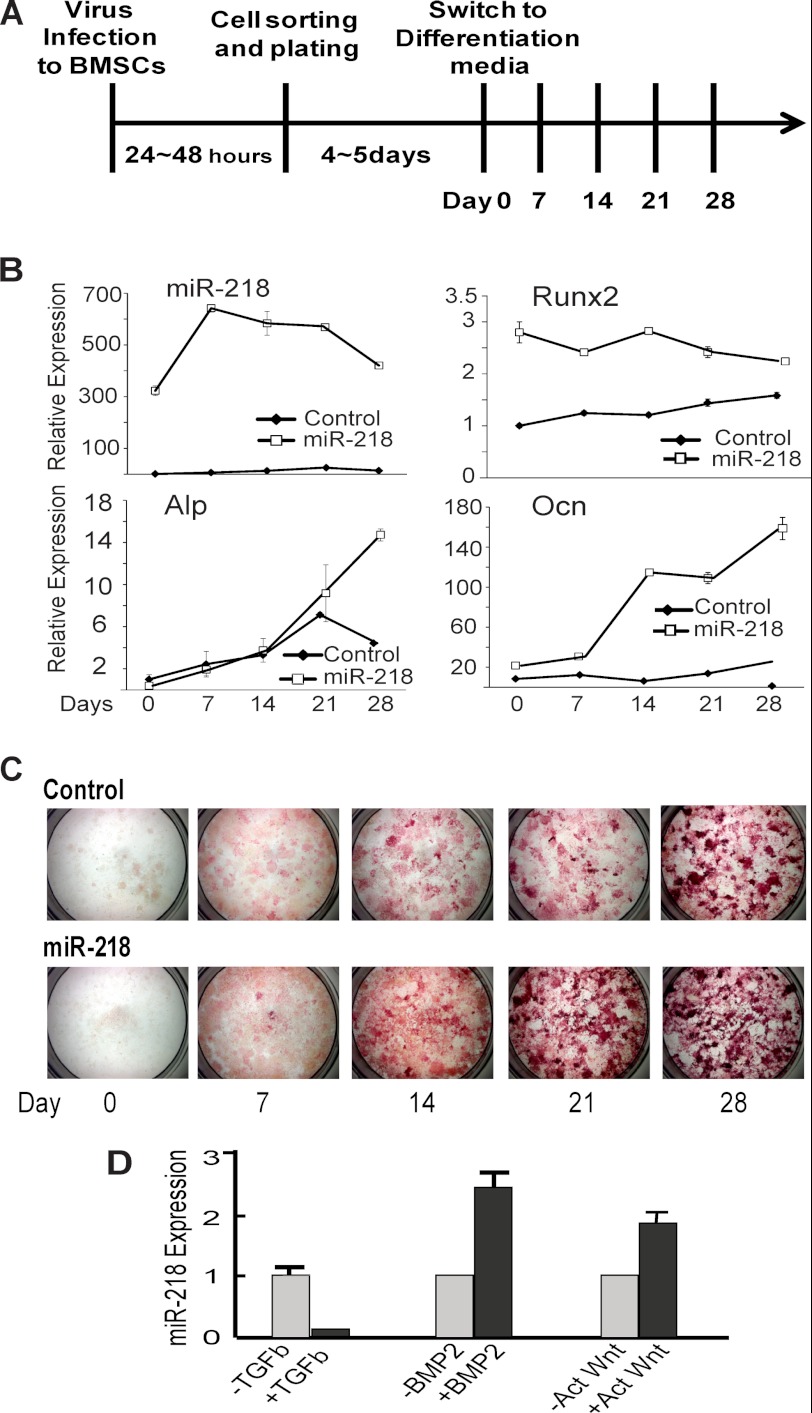
We addressed the role of miR-218 as an inducer of osteogenic lineage commitment and progression using multipotent bone marrow-derived mesenchymal stem cells (BMSCs). Introduction of miR-218 into BMSCs resulted in increased Runx2 levels, indicating that miR-218 is sufficient to commit BMSCs to the osteoblast lineage (Fig. 2A,B). Upon induction of differentiation, exogenous miR-218 and elevated Runx2 expression were each sustained throughout the time course (Fig. 2B). miR-218 dramatically increased both Alp and Bglap (Ocn) mRNAs at the late stage of phenotype maturation. In addition, miR-218 strongly enhanced ALP enzyme activity at both early and late stages of the differentiation time course (Fig. 2C). None the less, the data demonstrate that expression of miR-218 is a positive effect on BMSCs to support both commitment and progression of osteogenesis. Of note, we performed anti-miR-218 treatment of BMSCs. This resulted in apoptosis of the cells beginning 24 h after addition, as well as 2 lower concentrations of anti-miR-218. We used both lentivirus expressing anti-miR-218 and the anti-miR oligonucleotide with the same result (data not shown). It appears that committed osteoprogenitor MC3T3 cells (Fig. 1D) may be protected from this anti-miR effect that occurs with undifferentiated BMSCs.
FIGURE 2.
miR-218 positively regulates osteoblastic differentiation in BMSCs. A, timeline of BMSCs transduced with control and miR-218 Lentivirus and experimental treatments. B, qRT-PCR analysis for miR-218 expression and mRNA profile of bone marker genes in control and cells infected with miR-218 harvested at 0, 7, 14, 21, and 28 days. C, histochemical staining for Alp activity, the osteoblast differentiation marker essential for matrix mineralization, collected at the indicated days. D, MC3T3 cells were treated with 5 ng/ml TGFβ, 100 ng/ml BMP2, and 10 mm TDZD-8, a Wnt activator for 48 h. Total miR-218 RNA expression was analyzed using U6 RNA to normalize expression levels.
We examined the responsiveness of endogenous miR-218 expression to signaling by TGFβ, BMP, and Wnt, which are potent paracrine regulators of bone formation. BMP and Wnt, both inducers of osteoblast differentiation, up-regulated miR-218 expression, while TGFβ, a negative regulator of osteogenesis (1), suppressed miR-218 expression (Fig. 2D). These findings indicate that Wnt and BMP2 are upstream regulators of miR-218 and suggest that miR-218 may be mediator of these bone anabolic pathways to control both commitment and progression of osteogenesis.
miR-218 Is a Positive Regulator of Wnt Signaling
To address the mechanism by which miR-218 regulates osteoblast differentiation, we examined predicted targets of miR-218 relevant to bone formation. We focused on targets that are inhibitory to bone formation. Four key regulators of BMP and Wnt signaling that met this criterion appeared in the top 12% of a cohort of ∼800 putative targets: a negative regulator of BMP2 signaling, Tob1, and three Wnt inhibitors, Dkk2, Sfrp2, Sost. Each of these potential targets for miR-218 is expressed at significant levels in osteoblasts except for Sost in BMSCs (supplemental Fig. S2). This may be due to differentiation of pluripotent primary BMSCs compared with a committed calvarial cell line. Furthermore, Wnt proteins were enriched and supplemental Table S3 are compared with other GO categories targeted by miR-218. The binding site of miR-218 through the seed sequences within the 3′-UTR for each target mRNA is illustrated in supplemental Table S2.
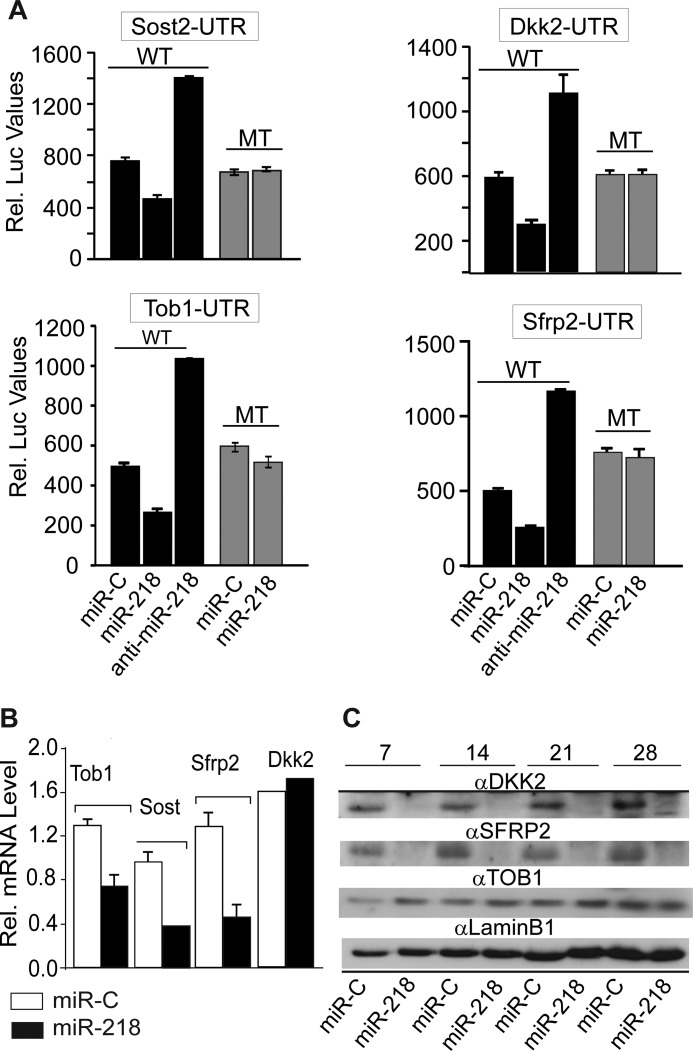
Because the default activity of miRNAs is to repress protein translation by binding to the 3′-UTR of mRNAs, functional testing was carried out using the microRNA-target mRNA UTR reporter assay. This study revealed significant down-regulation of Luc activity (40–60%) for Dkk2, Sost, Sfrp2, and Tob1 UTRs in MC3T3 cells in the presence of ectopic miR-218 (Fig. 3A). No down-regulation occurred when the miR binding sites in each of the potential target UTRs were mutated. Contrary to the decrease in UTR-LUC activity caused by miR-218 overexpression, anti-miR-218 increased LUC activity by at least 2-fold. These results indicated that these inhibitors are direct targets of miR-218 in osteoblasts. We therefore examined expression and protein levels for each target in response to miR-218 and found reduced mRNA levels of 3 targets, Tob, Sfrp2, and Sost, but not Dkk2 (Fig. 3B). Western blot studies revealed that DKK2 and SFRP2 protein levels were significantly decreased by miR-218 overexpression (Fig. 3C) during the differentiation of BMSCs. miR-218 down-regulated DKK2 protein while the mRNA levels did not change, DKK2 repression occurred strictly through inhibition of protein translation. Sost mRNA, but not protein, is expressed in most culture models, which we confirmed in these MC3T3 studies with a validated antibody (24). However, miR-218 reduced Sost mRNA levels by 66% (Fig. 3B), which together with the Luc reporter assay indicates Sost is a direct target. Although Tob1 mRNA was reduced by nearly 50% (Fig. 3B) by miR-218 expression, TOB1 protein was not significantly changed when normalized by LAMIN B1 protein level (Fig. 3C). Taken together, these results indicate miR-218 stimulates osteoblast differentiation by targeting multiple Wnt pathway inhibitors.
FIGURE 3.
Dkk2, Sfrp2, Sost, and Tob1 are direct targets of miR-218. A, activity of the 3′-UTR Luc reporter for each target sequence transfected with wild type (WT) and mutated (MT) UTR constructs along with nonspecific miR control (miR-C), miR-218, and anti-miR-218. Relative Luciferase activity was normalized with Renilla luciferase activity and expressed in relative luminescence units. We note that an expected increase in reporter activity is not observed with mutant constructs due to the presence of 3–4 other miRNA recognition sites in the cloned sequences containing the miR-218 binding site, thereby contributing to inhibition of stimulated activity. B, regulation on the endogenous expression of Tob1, Sost, Sfrp2, and Dkk2 by miR-218. Analysis of total RNA for the relative expression miR-218 targets by real time qRT-PCR in MC3T3 preosteoblast cells 72 h post transfection using miR-C and miR-218. Relative expressions were normalized to Gapdh levels. C, Western blot analysis of proteins from bone marrow stromal cells transduced with control and miR-218 Lentivirus, then cultured in differentiation media for 28 days. The protein profiles were normalized with anti-Lamin B1 antibody.
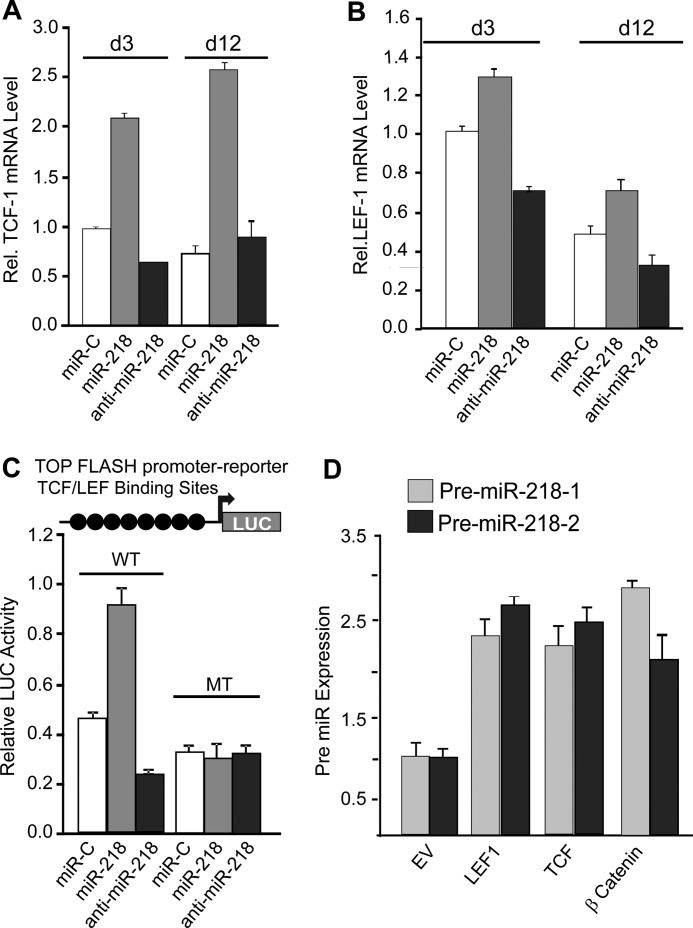
To identify a mechanism for miR-218 and Wnt signaling in osteoblasts, we first examined whether transcriptional mediators of activated Wnt signaling, Tcf-1 and Lef-1, respond to miR-218 (Fig. 4, A and B). Exogenous expression of miR-218 robustly increased endogenous Tcf-1 and Lef-1 expression, while treatment of anti-miR-218 markedly decreased Lef-1 expression. Tcf-1 reduction by the anti-miR did not occur in post proliferative cells (Fig. 4A). Direct regulation of Wnt signaling was further supported by miR-218 induced activation of Wnt-Luc reporter by 2-fold, whereas the anti-miR-218 decreased Wnt activity also by 2-fold (Fig. 4C). These effects were abolished when the Wnt responsive elements were mutated. Together these data support a feed forward mechanism for stimulation of Wnt signaling by miR-218 through down-regulation of inhibitors of Wnt signaling that is reflected by elevated Tcf1 and Lef1. Protein levels of β-catenin and TCF1 were also induced by miR-218, concomitant with elevated RUNX2 as an indicator of osteoblast differentiation (supplemental Fig. S3). In addition to these direct targets of Wnt signaling (25, 26), we examined two other Wnt signaling targets, Cyclin D1 and Axin2, a feedback inhibitor of Wnt signaling (supplemental Fig. S4). Cyclin D1 is increased by miR-218 and decreased by the anti-miR. Axin2, an inhibitor and well documented feedback regulator of Wnt signaling, becomes decreased by miR-218, rather than be stimulated as reported when Wnt signaling is activated (27). This result suggests that miR-218 may target Axin2, but according to current databases, the 3′-UTR of Axin2 does not contain a miR-218 recognition sequence. Thus miR-218 which has hundreds of targets, may regulate Axin2 mRNA indirectly by other mechanisms, such as targeting a transcription factor which induces Axin2. In conclusion, the decrease in Axin2 is consistent with the anabolic effect of miR-218 on osteoblast differentiation.
FIGURE 4.
Wnt signaling and miR-218 feed forward and feedback regulation. A, expression of Tcf-1 mRNA and B, Lef-1 mRNA in MC3T3 cells transduced with miR-218 and anti-miR-218 Lentivirus. Cells were analyzed at the indicated days. C, miR and anti-miR-218 affect Wnt reporter activity. WT and Mutant Top flush Wnt reporter were transfected with miR-C, miR-218, and anti-miR-218 in MC3T3 for 24 h. Relative luciferase activity was measured and plotted. D, relative expression of pre-miRNAs, miR-218-1, and miR-218-2. Both are up-regulated in response to exogenous expression of LEF, TCF, and β-catenin, mediators of canonical Wnt signaling. Expression was normalized by U6.
Because stimulated Wnt signaling increases endogenous miR-218 expression (see Fig. 2D), we investigated whether transcriptional mediators of Wnt signaling induced miR-218 levels (Fig. 4D). Indeed, overexpression of LEF1, TCF1, or β-catenin induced both primary miR-218–1 and -218–2 by 2–2.5-fold (Fig. 4D). These studies demonstrate that Wnt signaling induces miR-218 and in turn miR-218 increases Wnt activity. Thus, miR-218 and Wnt signaling are coupled by a feed forward-positive feedback loop to form a biological regulatory circuit.
miR-218 Promotes Breast Cancer Osteomimicry through Wnt Signaling
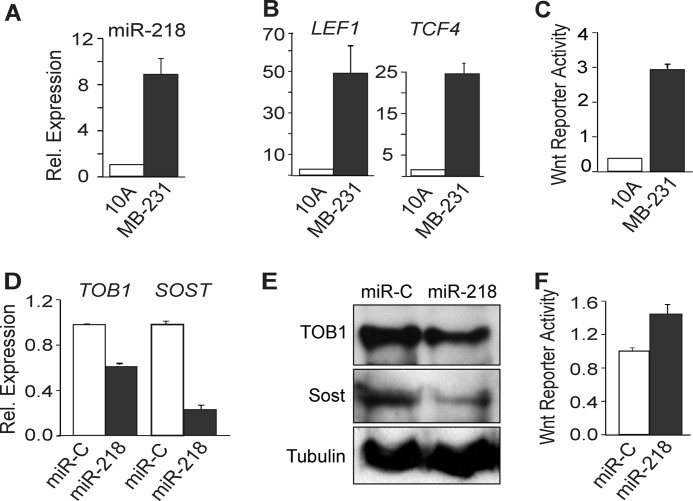
The Wnt signaling pathway is involved in cancer pathogenesis (21, 22). Because genes expressed in osteoblasts are also expressed in breast cancer cells (osteomimicry), we interrogated the potential contribution of the miR-218/Wnt regulatory axis in metastatic cancer cells that home to bone. While miR-218 expression was low in normal MCF-10A mammary epithelial cells, levels were increased 10 fold in highly metastatic MDA-MB-231 cells (Fig. 5A). Significantly, the Wnt transcriptional mediators LEF1 and TCF4 were elevated by more than 20-fold in the MDA-MB-231 cells (Fig. 5B). The Wnt responsive Top Flash reporter assay established that endogenous Wnt signaling was highly activated in MDA-MB-231 cells (Fig. 5C). Hence, high miR-218 positively correlated with robust stimulation of the Wnt pathway in bone-homing breast cancer cells.
FIGURE 5.
miR-218 expression positively correlates with Wnt signaling. A–C, endogenous expression profile in normal breast epithelial cells (MCF10A) and metastatic breast cancer cells (MDA-MB-231), which were cultured under standard conditions to confluency and compared for: (A) miR-218 expression which was detected by qRT-PCR and normalized to U6 small RNA. B, expression of Wnt target genes LEF1 and TCF4 qRT-PCR analysis normalized to GAPDH as internal control. C, Wnt reporter activity in MCF10A and MDA-MB-231 cells. D and E, miR-218 functional activity: D, MDA-MB-231 cells were transfected with control miRNA (miR-C) or miR-218 and analyzed by qRT-PCR of the targets TOB1 and SOST. E, TOB1 and SOST protein levels were analyzed by Western blot in miR-C- and miR-218-transfected MDA-MB-231 cells. F, miR-218 stimulates Wnt signaling activity in MDA-231 cells transfected with TOP-flash Wnt responsive reporter. Relative firefly luciferase activity was measured and plotted using Renilla luciferase as control.
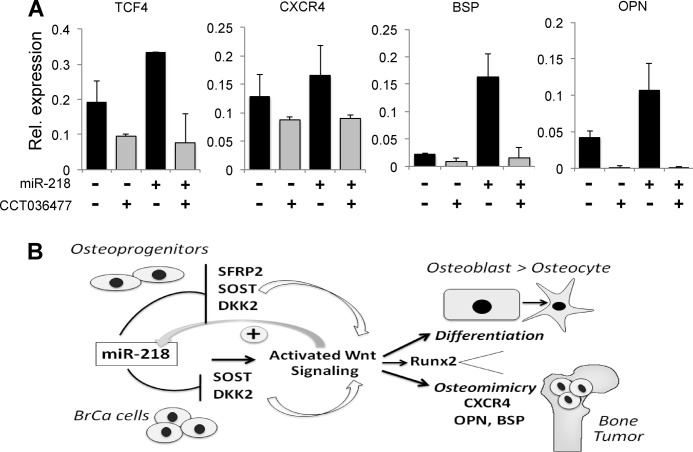
To address miR-218 function in attenuating Wnt signaling inhibitors, we ectopically expressed miR-218 in MDA-MB-231 cells and examined mRNA levels of miR-218 target genes. Elevation of miR-218 markedly decreased the mRNA and protein levels of TOB1 and SOST, while SFRP2 and DKK2 were not detected in these cells (Fig. 5, D and E). Consequently, Wnt Top Flash reporter activity was increased (Fig. 5F) beyond high basal levels of Wnt signaling in MDA-MB-231 cells (Fig. 5C). We also examined the effect of miR-218 on expression of several genes that are elevated in serum of breast cancer patients and correlated with bone metastasis, including bone sialoprotein (BSP), osteopontin (OPN), and a chemokine receptor CXCR4 (22, 28, 29). All three genes were markedly increased by ectopic expression of miR-218 (Fig. 6A). Inhibition of Wnt signaling in MDA-MB-231 cells by a small molecule inhibitor abolished this miR-218-induced elevation, further validating that the activation of metastatic genes is mediated by Wnt signaling (Fig. 6A). We then tested whether miR-218 had the potential to change the properties of a MCF-10A epithelial cells or MCF-7 non metastatic cells. We treated both cell lines with miR-218 and did not find a significant increase in either cell line for either metastasis or bone related genes (data not shown). These findings are possibly due to very low basal Wnt signaling activity. These observations suggest miR-218 expression is functional only in cells that have bone metastatic potential. We conclude that the miR-218/Wnt loop fuels Wnt signaling to enhance expression of metastatic and osteomimetic genes in aggressive breast cancer cells that home to bone. Our data suggest that miR-218 directs a Wnt signaling loop to promote differentiation in osteoblast lineage cells and expression of osteomimetic genes in breast cancer cells (Fig. 6B).
FIGURE 6.
miR-218 promotes osteomimetic properties in metastatic breast cancer cells. A, MDA-MB-231 cells were transfected with control miRNA (miR-C) or miR-218 and treated with small molecule Wnt signaling inhibitor (CCT036477). Expression of Wnt target gene TCF4 and metastatic and osteomimetic markers CXCR4, OPN, and BSP was analyzed by qRT-PCR. GAPDH was used as internal control. B, schematic illustration of the feed-forward-/positive feedbackward loop between miR-218 and Wnt signaling that constantly renews Wnt signaling through regulation of multiple Wnt inhibitors. In turn, Wnt signaling increases miR-218 expression. This miR-218/Wnt circuit operates in normal osteoblast lineage cells to promote differentiation from an osteoprogenitor to the final osteocyte stage (top panel), as well as in highly bone seeking MDA-MB-231 breast cancer cells to induce expression of bone related genes (osteomimicry) that facilitate tumor growth in bone (41). CXCR4, chemokine receptor 4; Runx2, a target of miR-218 (19) is also highly expressed in osteoblasts and MDA-MB-231 in response to activated Wnt signaling (25).
DISCUSSION
Here, we establish that miR-218 is a central regulatory node within a positive feedback loop that optimizes Wnt signaling to promote osteoblast differentiation. miR-218 induces Wnt signaling by downregulating inhibitors of the canonical Wnt pathway, while stimulation of Wnt signaling increases expression of miR-218. Wnt signaling is also pathologically activated during breast tumorigenesis and metastasis. Significantly, we find that the miR-218/Wnt regulatory circuit also operates in breast cancer cells resulting in increased expression of osteoblastic genes (osteomimicry). Therefore, the miR-218/Wnt signaling axis defines a molecular network that promotes osteogenesis and contributes to tumor properties in the bone microenvironment.
miR-218 and Osteogenic Differentiation
Bone formation is a dynamic developmental process that is stimulated by the interplay of several bone anabolic ligands (e.g. BMPs and Wnts) and attenuated by corresponding antagonists to generate interlocked regulatory mechanisms (30–32). miR-218 is a potent osteo-miR expressed in mesenchymal derived BMSCs and MC3T3 cells and continuously up-regulated during osteoblast differentiation, thus functioning throughout the course of osteogenesis. Furthermore, miR-218 is induced by Wnt and BMP2 signaling and suppressed by the osteogenic inhibitor TGFβ, consistent with miR-218 mediating the bone anabolic effects of Wnt and BMP2. We propose that miR-218 coordinates the complex biological interactions among osteogenic signaling pathways to control the progression of osteoblastogenesis.
Constitutively active Wnt signaling increases bone mass, increases expression of bone marker genes and stimulates fracture healing (33, 34). Our studies provide evidence that miR-218 directly activates Wnt signaling demonstrated by a striking increase in TCF-1, β-catenin and Runx2, as well as by an increase in Wnt signaling reporter by miR-218 expression. Furthermore, inhibition of Wnt signaling decreases the Wnt promoting effects of miR-218. The potent osteogenic properties of miR-218 that result from directly targeting and down-regulating three inhibitors of Wnt signaling, are consistent with loss of function phenotypes of these targets in mouse models. Mutation of the miR-218 targets SFRP2 and SOST increases bone mass in vivo (31, 35, 36), while down-regulation of DKK2 is required for osteoblast maturation in vivo and ex vivo (37). These skeletal phenotypes are entirely consistent with the repression of these Wnt inhibitors by miR-218. Furthermore, we found that Axin2, an inhibitor of Wnt receptor signaling, is decreased by miR-218 up-regulation of the Wnt pathway. Of significance, the Axin null mouse has increased bone mass due to up-regulated osteoblast activity (38). We note that other miRNAs expressed in osteoblasts also target different Wnt inhibitors, e.g. DKK1 by miR-29 and miR-335–5p (39, 40). Thus, multiple miRNAs including miR-218 form a network that regulates Wnt signaling at multiple levels for bone formation.
The osteogenic properties of miR-218 are conclusive, yet the molecular regulation of two bone-related targets is rather complex. For example, apart from controlling Wnt inhibitors, miR-218 can target the 3′-UTRs of the BMP/Smad inhibitor TOB1 (this work) and the BMP/Smad responsive transcription factor RUNX2 (19), thus potentially mediating the known cross-coupling between BMP signaling and Wnt signaling (7). However, Tob1 mRNA is clearly reduced by miR-218, but its protein levels are not. In addition, Runx2 mRNA is not sensitive to miR-218 and its protein levels are in fact increased. Thus, TOB1 protein is stabilized and the miR-218 stimulation of Wnt signaling overrides the repression of Runx2 activity to enhance osteoblast differentiation. This mechanism, presumably occurs by up-regulated Wnt signaling which directly increases Runx2 transcription (25). We conclude that while miR-218 is predicted to control both BMP and Wnt signaling pathways, its activity in osteoblasts is selectively directed to activating Wnt signaling.
miR-218 and Osteomimetic Gene Expression
Our results indicate that the Wnt regulatory osteogenic properties of miR-218 may contribute to cancer cells that metastasize to bone by stimulating cytokine receptors that facilitate homing to the bone micro-environment (osteomimicry) (21, 41, 42) and expression of bone-related ECM proteins. miR-218 is highly expressed in metastatic breast cancer cells, but not in normal mammary epithelial cells. The enhanced levels of miR-218 increase Wnt signaling and expression of both bone sialoprotein (BSP) and osteopontin (OPN), which are Wnt signaling responsive proteins and serve as clinical markers for bone metastasis (28, 42). In addition miR-218 in MDA-MB-231 cells may facilitate metastasis of aggressive breast cancer cells by up-regulating CXCR4, a crucial cytokine supporting breast cancer homing to bone and mediating tumor growth in bone (29). Interestingly, SOST is highly expressed in untreated cancer cells and miR-218 not only mediates the oncogenic deregulation of Wnt signaling by downregulating SOST, but also by decreasing levels of the Smad inhibitor TOB. Unlike normal osteoblasts where miR-218 selectively controls Wnt but not BMP/Smad signaling, this molecular selectivity appears to be compromised in breast cancer. Taken together, miR-218 may contribute to bone tumor growth and the accompanying metastatic bone disease in patients (21).
The multifunctional role of miR-218 as an osteo-miR in controlling bone formation defines it as a member of a growing master class of specialized miRNAs that control differentiation and tissue development, including muscle, skin, and fat (43–45). In addition, a growing number of miRs have been revealed as signatures for different diseases including cancers (46–48). Although miR-218 functions as a tumor-suppressive “oncomiRNA” to suppress certain cancers (49, 50), elevated miR-218 expression has also previously been associated with estrogen receptor positive breast tumors indicating a possible role in certain breast cancer phenotypes (51, 52). Consistent with latter findings, miR-218 promotes metastasis-related molecular properties in MDA-MB-231 aggressive breast cancer cells. Hence, the biological activity of miR-218 clearly depends on the cellular micro-environment
The biological activity of miR-218 is controlled in part by its expression from two separate copies, miR-218–1 and miR-218–2, that are respectively located in the Slit2 and Slit3 genes (49, 53). These findings raise the possibility that the functions of these two host genes and their corresponding passenger mi-RNAs are functionally coupled. While this relationship has been observed for regulation of neuronal guidance and vascular formation (54, 55), our findings suggest that this relationship between host gene and passenger miRNA is complex. Slit2 is expressed in osteoblasts (56); however we observed here that the levels of miR-218–1 and miR-218–2 are not coupled to their respective host genes Slit2 or Slit3 in bone.
Further miR-218 complexity is found within cancer cells. miR-218 was reported to function as a tumor suppressor through regulation of the Slit-Robo pathway in some cancers, but not others (49). The Slit2 gene harboring miR-218 is expressed in metastatic breast cancer cells and is associated with metastasis to brain, but not to bone (50), whereas both Wnt signaling and estrogen are linked to bone metastasis (22, 52). These results suggest that the expression of the two forms of miR-218 arising from its host genes serve different biological functions than their host genes in osteoblasts and breast cancer cells.
In summary, the linkage of miR-218 to the Wnt, BMP2, and Runx2 regulators of bone biology and cancer cells metastatic to bone supports the emerging concept that miRNAs control interlocking regulatory pathways that are requisite for maintaining a normal tissue environment. The enrichment of tissue differentiation pathways and various cancers that involve miR-218 (supplemental Table S3) suggests diverse functions for miR-218 that include influencing bone tissue homeostasis and tumor biology.
Acknowledgments
We thank members of our laboratories for stimulating discussions on procedures and Judy Rask for manuscript preparation.
This work was supported, in whole or in part by National Institutes of Health Grants R01AR039588 (to G. S. S. and J. B. L.), R37DE012528 (to J. B. L.), R01AR049069 (to A. V. W.), and P01 CA 082834 (to G. S. S.).

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
- BMP
- bone morphogenetic protein
- miRNA
- microRNA
- UTR
- untranslated region
- Alp
- alkaline phosphatase
- BMSC
- bone marrow-derived mesenchymal stem cell
- Ocn
- osteocalcin
- OPN
- osteopontin
- BSP
- bone sialoprotein.
REFERENCES
- 1. Komori T. (2011) Signaling networks in RUNX2-dependent bone development. J. Cell. Biochem. 112, 750–755 [DOI] [PubMed] [Google Scholar]
- 2. Monroe D. G., McGee-Lawrence M. E., Oursler M. J., Westendorf J. J. (2012) Update on Wnt signaling in bone cell biology and bone disease. Gene 492, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ornitz D. M. (2005) FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 16, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Long F. (2008) Targeting intercellular signals for bone regeneration from bone marrow mesenchymal progenitors. Cell Cycle 7, 2106–2111 [DOI] [PubMed] [Google Scholar]
- 5. Canalis E. (2009) Growth factor control of bone mass. J. Cell Biochem. 108, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen G., Deng C., Li Y. P. (2012) TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 8, 272–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshida Y., Tanaka S., Umemori H., Minowa O., Usui M., Ikematsu N., Hosoda E., Imamura T., Kuno J., Yamashita T., Miyazono K., Noda M., Noda T., Yamamoto T. (2000) Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103, 1085–1097 [DOI] [PubMed] [Google Scholar]
- 8. Long F. (2008) When the gut talks to bone. Cell 135, 795–796 [DOI] [PubMed] [Google Scholar]
- 9. Gaur T., Hussain S., Mudhasani R., Parulkar I., Colby J. L., Frederick D., Kream B. E., van Wijnen A. J., Stein J. L., Stein G. S., Jones S. N., Lian J. B. (2010) Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 340, 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi T., Lu J., Cobb B. S., Rodda S. J., McMahon A. P., Schipani E., Merkenschlager M., Kronenberg H. M. (2008) Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizoguchi F., Izu Y., Hayata T., Hemmi H., Nakashima K., Nakamura T., Kato S., Miyasaka N., Ezura Y., Noda M. (2010) Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell Biochem. 109, 866–875 [DOI] [PubMed] [Google Scholar]
- 12. Sugatani T., Hruska K. A. (2009) Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 284, 4667–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taipaleenmaki H., Bjerre L., Chen L., Kauppinen S., Kassem M. (2011) REVIEW TOPIC ON MECHANISMS IN ENDOCRINOLOGY MicroRNAs: Targets for enhancing osteoblast differentiation and bone formation. Eur. J. Endocrinol. [DOI] [PubMed] [Google Scholar]
- 14. Kapinas K., Delany A. M. (2011) MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res. Ther. 13, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lian J. B., Stein G. S., van Wijnen A. J., Stein J. L., Hassan M. Q., Gaur T., Zhang Y. (2012) miRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 8, 212–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hassan M. Q., Gordon J. A., Beloti M. M., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2010) A network connecting Runx2, SATB2, and the miR-23a∼27a∼24–2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. U.S.A. 107, 19879–19884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Z., Hassan M. Q., Volinia S., van Wijnen A. J., Stein J. L., Croce C. M., Lian J. B., Stein G. S. (2008) A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. U.S.A. 105, 13906–13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z., Hassan M. Q., Jafferji M., Aqeilan R. I., Garzon R., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 284, 15676–15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Xie R. L., Croce C. M., Stein J. L., Lian J. B., van Wijnen A. J., Stein G. S. (2011) A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc. Natl. Acad. Sci. U.S.A. 108, 9863–9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr., Pang K., Brownell D. R., Harding S., Mitani S., Ruvkun G., Yates J. R., 3rd, Mello C. C. (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354 [DOI] [PubMed] [Google Scholar]
- 21. Weilbaecher K. N., Guise T. A., McCauley L. K. (2011) Cancer to bone: a fatal attraction. Nat. Rev. Cancer 11, 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sethi N., Kang Y. (2011) Dysregulation of developmental pathways in bone metastasis. Bone 48, 16–22 [DOI] [PubMed] [Google Scholar]
- 23. Ewan K., Pajak B., Stubbs M., Todd H., Barbeau O., Quevedo C., Botfield H., Young R., Ruddle R., Samuel L., Battersby A., Raynaud F., Allen N., Wilson S., Latinkic B., Workman P., McDonald E., Blagg J., Aherne W., Dale T. (2010) A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res. 70, 5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonewald L. F. (2011) The amazing osteocyte. J. Bone Miner. Res. 26, 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gaur T., Lengner C. J., Hovhannisyan H., Bhat R. A., Bodine P. V., Komm B. S., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2005) Canonical WNT signaling promotes osteogenesis by directly stimulating RUNX2 gene expression. J. Biol. Chem. 280, 33132–33140 [DOI] [PubMed] [Google Scholar]
- 26. Wu C. I., Hoffman J. A., Shy B. R., Ford E. M., Fuchs E., Nguyen H., Merrill B. J. (2012) Function of Wnt/β-catenin in counteracting Tcf3 repression through the Tcf3-β-catenin interaction. Development 139, 2118–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacDonald B. T., Tamai K., He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ibrahim T., Leong I., Sanchez-Sweatman O., Khokha R., Sodek J., Tenenbaum H. C., Ganss B., Cheifetz S. (2000) Expression of bone sialoprotein and osteopontin in breast cancer bone metastases. Clin. Exp. Metastasis 18, 253–260 [DOI] [PubMed] [Google Scholar]
- 29. Wang J., Loberg R., Taichman R. S. (2006) The pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis. Cancer Metastasis Rev. 25, 573–587 [DOI] [PubMed] [Google Scholar]
- 30. Kamiya N., Kobayashi T., Mochida Y., Yu P. B., Yamauchi M., Kronenberg H. M., Mishina Y. (2010) Wnt inhibitors Dkk1 and Sost are downstream targets of BMP signaling through the type IA receptor (BMPRIA) in osteoblasts. J. Bone Miner. Res. 25, 200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamiya N., Ye L., Kobayashi T., Mochida Y., Yamauchi M., Kronenberg H. M., Feng J. Q., Mishina Y. (2008) BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development 135, 3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krause C., Korchynskyi O., de Rooij K., Weidauer S. E., de Gorter D. J., van Bezooijen R. L., Hatsell S., Economides A. N., Mueller T. D., Löwik C. W., ten Dijke P. (2010) Distinct modes of inhibition by sclerostin on bone morphogenetic protein and Wnt signaling pathways. J. Biol. Chem. 285, 41614–41626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoeppner L. H., Secreto F. J., Razidlo D. F., Whitney T. J., Westendorf J. J. (2011) Lef1ΔN binds β-catenin and increases osteoblast activity and trabecular bone mass. J. Biol. Chem. 286, 10950–10959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niziolek P. J., Farmer T. L., Cui Y., Turner C. H., Warman M. L., Robling A. G. (2011) High-bone-mass-producing mutations in the Wnt signaling pathway result in distinct skeletal phenotypes. Bone 49, 1010–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sathi G. A., Inoue M., Harada H., Rodriguez A. P., Tamamura R., Tsujigiwa H., Borkosky S. S., Gunduz M., Nagatsuka H. (2009) Secreted frizzled related protein (sFRP)-2 inhibits bone formation and promotes cell proliferation in ameloblastoma. Oral Oncol. 45, 856–860 [DOI] [PubMed] [Google Scholar]
- 36. Li X., Ominsky M. S., Niu Q. T., Sun N., Daugherty B., D'Agostin D., Kurahara C., Gao Y., Cao J., Gong J., Asuncion F., Barrero M., Warmington K., Dwyer D., Stolina M., Morony S., Sarosi I., Kostenuik P. J., Lacey D. L., Simonet W. S., Ke H. Z., Paszty C. (2008) Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J. Bone Miner. Res. 23, 860–869 [DOI] [PubMed] [Google Scholar]
- 37. Li X., Liu P., Liu W., Maye P., Zhang J., Zhang Y., Hurley M., Guo C., Boskey A., Sun L., Harris S. E., Rowe D. W., Ke H. Z., Wu D. (2005) Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat. Genet. 37, 945–952 [DOI] [PubMed] [Google Scholar]
- 38. Yan Y., Tang D., Chen M., Huang J., Xie R., Jonason J. H., Tan X., Hou W., Reynolds D., Hsu W., Harris S. E., Puzas J. E., Awad H., O'Keefe R. J., Boyce B. F., Chen D. (2009) Axin2 controls bone remodeling through the β-catenin-BMP signaling pathway in adult mice. J. Cell Sci. 122, 3566–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kapinas K., Kessler C., Ricks T., Gronowicz G., Delany A. M. (2010) miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 285, 25221–25231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang J., Tu Q., Bonewald L. F., He X., Stein G., Lian J., Chen J. (2011) Effects of miR-335–5p in modulating osteogenic differentiation by specifically down-regulating Wnt antagonist DKK1. J. Bone Miner. Res. 26, 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rucci N., Teti A. (2010) Osteomimcry: how tumor cells try to deceive the bone. Front. Biosci. 2, 907–915 [DOI] [PubMed] [Google Scholar]
- 42. Fedarko N. S., Jain A., Karadag A., Van Eman M. R., Fisher L. W. (2001) Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clin. Cancer Res. 7, 4060–4066 [PubMed] [Google Scholar]
- 43. Botchkareva N. (2012) MicroRNA/mRNA regulatory networks in the control of skin development and regeneration. Cell Cycle 11, [DOI] [PubMed] [Google Scholar]
- 44. Liu N., Olson E. N. (2010) MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bengestrate L., Virtue S., Campbell M., Vidal-Puig A., Hadaschik D., Hahn P., Bielke W. (2011) Genome-wide profiling of microRNAs in adipose mesenchymal stem cell differentiation and mouse models of obesity. PLoS ONE 6, e21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sayed D., Abdellatif M. (2011) MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 [DOI] [PubMed] [Google Scholar]
- 47. Nana-Sinkam S. P., Croce C. M. (2011) Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol. Oncol. 5, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. White N. M., Fatoohi E., Metias M., Jung K., Stephan C., Yousef G. M. (2011) Metastamirs: a stepping stone towards improved cancer management. Nat. Rev. Clin. Oncol. 8, 75–84 [DOI] [PubMed] [Google Scholar]
- 49. Tie J., Pan Y., Zhao L., Wu K., Liu J., Sun S., Guo X., Wang B., Gang Y., Zhang Y., Li Q., Qiao T., Zhao Q., Nie Y., Fan D. (2010) MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS. Genet. 6, e1000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schmid B. C., Rezniczek G. A., Fabjani G., Yoneda T., Leodolter S., Zeillinger R. (2007) The neuronal guidance cue Slit2 induces targeted migration and may play a role in brain metastasis of breast cancer cells. Breast Cancer Res. Treat. 106, 333–342 [DOI] [PubMed] [Google Scholar]
- 51. Lowery A. J., Miller N., Devaney A., McNeill R. E., Davoren P. A., Lemetre C., Benes V., Schmidt S., Blake J., Ball G., Kerin M. J. (2009) MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 11, R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chimge N. O., Baniwal S. K., Little G. H., Chen Y. B., Kahn M., Tripathy D., Borok Z., Frenkel B. (2011) Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Res. 13, R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alajez N. M., Lenarduzzi M., Ito E., Hui A. B., Shi W., Bruce J., Yue S., Huang S. H., Xu W., Waldron J., O'Sullivan B., Liu F. F. (2011) MiR-218 suppresses nasopharyngeal cancer progression through down-regulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 71, 2381–2391 [DOI] [PubMed] [Google Scholar]
- 54. Small E. M., Sutherland L. B., Rajagopalan K. N., Wang S., Olson E. N. (2010) MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ. Res. 107, 1336–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Devine C. A., Key B. (2008) Robo-Slit interactions regulate longitudinal axon pathfinding in the embryonic vertebrate brain. Dev. Biol. 313, 371–383 [DOI] [PubMed] [Google Scholar]
- 56. Sun H., Dai K., Tang T., Zhang X. (2009) Regulation of osteoblast differentiation by slit2 in osteoblastic cells. Cells Tissues Organs 190, 69–80 [DOI] [PubMed] [Google Scholar]