Background: Hyaluronidase-4 is a chondroitin sulfate-specific endo-β-N-acetylgalactosaminidase.
Results: The amino acid residues responsible for the substrate specificity of hyaluronidase-4 were identified.
Conclusion: The combination of the amino acid residues at 261–265 and glutamine 305 was essential.
Significance: These results could help to generate artificial chondroitin sulfate hydrolases, which recognize specific structures.
Keywords: Chondroitin Sulfate, Glycosaminoglycan, Hyaluronate, Hydrolases, Proteoglycan, Hyaluronidase
Abstract
Human hyaluronidase-4 (hHYAL4), a member of the hyaluronidase family, has no hyaluronidase activity, but is a chondroitin sulfate (CS)-specific endo-β-N-acetylgalactosaminidase. The expression of hHYAL4 is not ubiquitous but restricted to placenta, skeletal muscle, and testis, suggesting that hHYAL4 is not involved in the systemic catabolism of CS, but rather has specific functions in particular organs or tissues. To elucidate the function of hyaluronidase-4 in vivo, mouse hyaluronidase-4 (mHyal4) was characterized. mHyal4 was also demonstrated to be a CS-specific endo-β-N-acetylgalactosaminidase. However, mHyal4 and hHYAL4 differed in the sulfate groups they recognized. Although hHYAL4 strongly preferred GlcUA(2-O-sulfate)-GalNAc(6-O-sulfate)-containing sequences typical in CS-D, where GlcUA represents d-glucuronic acid, mHyal4 depolymerized various CS isoforms to a similar extent, suggesting broad substrate specificity. To identify the amino acid residues responsible for this difference, a series of human/mouse HYAL4 chimeric proteins and HYAL4 point mutants were generated, and their preference for substrates was investigated. A combination of the amino acid residues at 261–265 and glutamine at 305 was demonstrated to be essential for the enzymatic activity as well as substrate specificity of mHyal4.
Introduction
Chondroitin sulfate (CS)2 chains are linear polymers composed of the repeating disaccharide unit 4GlcUAβ1–3GalNAcβ1, which are sulfated at different positions in various combinations (1, 2). CS chains are covalently linked to a wide range of core proteins, forming proteoglycans, which are ubiquitous components of the extracellular matrix of connective tissues and are also found at the surface of a variety of cell types (3, 4). They are involved in the regulation of various biological processes such as cell proliferation, differentiation, and migration, cell-cell recognition, extracellular matrix deposition, and tissue morphogenesis (1, 5, 6). The biological functions involved in such events have attracted much attention in connection with the mechanism of CS biosynthesis (7). However, not only biosynthesis but also catabolism is important for the regulation of the biological functions of CS.
Recently, we have demonstrated that human hyaluronidase-4 (hHYAL4), a member of the hyaluronidase family, is a CS-specific endo-β-N-acetylgalactosaminidase (8). hHYAL4 exhibited hydrolytic activity toward CS chains and degraded them into oligosaccharides, but did not degrade hyaluronan (HA), chondroitin (Chn), dermatan sulfate (DS), or heparan sulfate. The expression of hHYAL4 mRNA is not ubiquitous but restricted to placenta, skeletal muscle (9), and testis (see “Results”), suggesting that hHYAL4 is not involved in the systemic catabolism of CS, but rather has specific functions in particular organs or tissues. The biological functions of hHYAL4 have not been clarified.
In contrast to hHYAL4, hHYAL1 prefers HA to CS and digests CS to a limited extent (10). Although hHYAL1 and hHYAL4 have significant homology in amino acid sequence (39%) (10), the residues responsible for the difference in substrate specificity have not been identified. However, the amino acid residues required for the enzymatic activity of hHYAL1 have been determined from structural information obtained by x-ray crystallography and site-directed mutagenesis (11). Because hHYAL4 is the only CS-specific endo-type hydrolase identified to date, it is important to elucidate the amino acid residues critical to the enzyme recognition of substrates as well as catalytic activity.
In the present study, we characterized mouse hyaluronidase-4 (mHyal4) to elucidate the function of hyaluronidase-4 in vivo. As expected, mHyal4 was also a CS-specific endo-β-N-acetylgalactosaminidase. However, the sulfated positions in CS recognized by mHyal4 and hHYAL4 were different. hHYAL4 strongly preferred CS-D as a substrate, which is rich in the disulfated disaccharide unit GlcUA(2-O-sulfate)-GalNAc(6-O-sulfate), to other CS variants, whereas mHyal4 depolymerized CS-A, CS-C, and CS-D to similar extents, suggesting a broad substrate specificity. To identify the amino acid residues responsible for this difference in specificity, a series of human/mouse HYAL4 chimeras and site-directed mutants were prepared and characterized.
EXPERIMENTAL PROCEDURES
Materials
The following sugars and enzymes were purchased from Seikagaku Corp. (Tokyo, Japan): CS-A from whale cartilage, DS from pig skin, CS-C from shark cartilage, Chn, a chemically desulfated derivative of CS-A, CS-Dfrom shark cartilage, CS-E from squid cartilage, heparan sulfate from bovine kidney, seven unsaturated standard disaccharides derived from CS, chindroitinase (CSase) ABC from Proteus vulgaris (EC 4.2.2.20), and CSase AC-II from Arthrobacter aurescens (EC 4.2.2.5). HA of human umbilical cord was obtained from Sigma. COS-7 cells were from Health Science Research Resources Bank (Osaka, Japan). SuperdexTM peptide and 200 10/300 GL columns were obtained from GE Healthcare. Fluorescein 5(6)-isothiocyanate (FITC)-labeled GAG isoforms, including Chn, CS-A, DS, CS-C, CS-D, CS-E, and HA, were prepared as described (12). All of the FITC-labeled GAGs were sensitive to at least CSase ABC and can be degraded into small oligosaccharides as examined by gel filtration on a column of Superdex peptide (data not shown).
Analysis of Expression Pattern of hHYAL4 and mHyal4
The hHYAL4 or mHyal4 transcript in multiple tissues was amplified from the MTC Multiple Tissue Panels (Clontech) by PCR using as primers 5′-CGACCACAGTGGGCCCGGAACTGGAACTC-3′ and 5′-CCTGCAGCTCCCAAGGCAGCACTTTCTCC-3′, or 5′-ACACAAGCTTACAAGTACACCTG-3′ and 5′-GGTTCACTTTCGTACAGTTCTCC-3′, respectively. Each PCR was carried out with the Takara ExTaq® DNA polymerase (Takara Bio Inc., Otsu, Japan) in the presence of 5% (v/v) dimethyl sulfoxide for 40 cycles at 96 °C for 30 s, 53 °C for 30 s, and 72 °C for 60 s. PCR products were analyzed by 3% agarose gel electrophoresis.
Cloning of mHyal4 cDNA
Total RNA was extracted from mouse testis with an illustra RNAspin Mini RNA Isolation Kit (GE Healthcare), and cDNA was synthesized from 1 μg of total RNA using Moloney Murine Leukemia Virus reverse transcriptase (Promega) and an oligo(dT) primer (13). The putative full-length open reading frame encoding mouse Hyal4 was amplified from the mouse testicular cDNA by two rounds of PCR using specific primers corresponding to the sequences in the 5′- and 3′-noncoding regions. The first PCR was performed with the primers, 5′-CTAACTCCAGTCTATATGTGGC-3′ and 5′-CAGTCCTTAAACTGCTACCTAG-3′. The second PCR was performed with the nested primers, 5′-ACCCAAGGAATAGCTATTCACC-3′ and 5′-TTATAAGGCCTCTCAGAGGAA-3′. Each PCR was carried out with the KOD-Plus DNA polymerase (Toyobo, Tokyo, Japan) in the presence of 5% (v/v) dimethyl sulfoxide for 30 cycles at 96 °C for 30 s, 51 °C for 30 s, and 68 °C for 2.5 min. The amplified cDNA fragment of expected size (∼1.4 kbp) was subcloned into a pGEM®-T Easy vector (Promega) and sequenced by Hokkaido System Science (Sapporo, Japan).
Analysis of Expression of mHyal4 in Embryos
cDNA was synthesized from 1 μg of total RNA extracted from C57BL/6 mouse embryo at 18.5 days as described above. The mHyal4 transcript was amplified from the embryo cDNA by two rounds of PCR using the specific primers described above. The first PCR was performed with the primers 5′-ACACAAGCTTACAAGTACACCTG-3′ and 5′-TCTTGCAAAGGTGACGACTGCACAC-3′. The second PCR was performed with the nested primers, 5′-ACACAAGCTTACAAGTACACCTG-3′ and 5′-GGTTCACTTTCGTACAGTTCTCC-3′. Each PCR was carried out with the KOD-Plus DNA polymerase in the presence of 5% (v/v) dimethyl sulfoxide for 30 cycles at 96 °C for 30 s, 52 °C for 30 s, and 68 °C for 1 min. The chromosomal sex of individual embryos was confirmed by PCR of the male-specific Sry gene (14), using the primers 5′-CAGCCTCATCGGAGGGCTAAAGTG-3′ and 5′-TGCAGGTGCCCAGTGGGGATATCA-3′. The PCR was carried out with the Takara ExTaq polymerase in the presence of 5% (v/v) dimethyl sulfoxide for 30 cycles at 95 °C for 30 s, 67 °C for 42 s, and 74 °C for 30 s.
Construction of an Expression Vector Containing a cDNA Fragment Encoding a Soluble Form of mHyal4
The DNA fragment, which encodes a putative mHyal4 protein lacking both the first N-terminal 33 amino acids (a hydrophobic region) and the last C-terminal 19 amino acids (the putative glycosylphosphatidylinositol (GPI)-anchored region) was amplified by PCR using a 5′-primer containing an in-frame BamHI site (5′-CGGGATCCTCCCTAAAACCTGCCGA-3′) and a 3′-primer containing a BamHI site (5′-CGGGATCCTCAAGAGGAAAGGGAAAGCC-3′), and subcloned into the BamHI site of the expression vector p3×FLAG-CMV-8 (Sigma), resulting in the fusion of mHyal4 to the preprotrypsin leader sequence and the FLAG tag sequence present in the vector. The p3×FLAG-CMV-8/hHYAL4 construct was prepared as described previously (8).
Plasmid Construction Expressing Chimeric Proteins of hHYAL4/mHyal4
The chimeric hHYAL4/mHyal4 plasmids were obtained by overlapping extension PCR (15). Briefly, fragments from the genes to be recombined were generated separately by PCR using primers designed (supplemental Table S1) so that the ends of the products contain complementary sequences. When these PCR products are mixed, denatured, and reannealed, the strands having the matching sequences at their 3′-ends overlap and act as primers for each other. Extension of this overlap by DNA polymerase produces a molecule in which the original sequences are fused together. Finally, the fusion molecule is amplified by conventional primers matching sequences at its end.
Generation of hHYAL4 or mHyal4 Point Mutants
Point mutants of hHYAL4 or mHyal4 were generated from the wild-type p3×FLAG-CMV-8/hHYAL4 or p3×FLAG-CMV-8/mHyal4 construct, respectively, using the QuikChange Site-directed Mutagenesis kit (Stratagene) according to the manufacturer's protocol. Primer sequences used to create the site-directed mutants are given in supplemental Table S1.
Expression of Recombinant Proteins
The expression plasmid was introduced into COS-7 cells (2.2 × 106) using FuGENE 6 (Roche Diagnostics) according to the manufacturer's instructions. After 3 days of culture at 37 °C, 1 ml of the culture medium was collected and incubated with 10 μl of anti-FLAG® M2 affinity gel (Sigma) overnight at 4 °C. Recombinant proteins were eluted from anti-FLAG® M2 affinity resin with 0.1 m acetate buffer (pH 3.5), and the eluates were neutralized with Tris-HCl buffer (pH 9.5). The protein was subjected to SDS-PAGE and analyzed by Western blotting (supplemental Fig. S1) (12).
Measurement of the Enzymatic Activity
The cells transfected with each plasmid were cultured for 3 days, and the recombinant enzyme protein was purified from the spent medium with anti-FLAG® M2 affinity resin as described (8). Recombinant proteins were eluted from anti-FLAG® M2 affinity resin with 0.1 m acetate buffer (pH 3.5), and the eluates were neutralized with Tris-HCl buffer (pH 9.5). The amounts of the protein in the eluates were measured using the Micro BCA Protein Assay kit (Thermo Scientific).
The purified resin was incubated with ∼10 μg of FITC-labeled glycosaminoglycan (GAG) isoforms. The mixture was incubated at 37 °C for 12 h and analyzed by gel filtration chromatography on a Superdex 200 column equilibrated with 0.2 m NH4HCO3 (8). Nonlabeled Chn, CS-A, CS-C, CS-D CS-E, heparan sulfate, and HA were incubated individually with the purified enzyme-bound resin or the eluted protein (125 ng) at 37 °C in 50 mm acetate buffer (pH 4.5) for 30 min. The resin was then removed by filtration using an Ultrafree-MC filter, each sample was labeled with 2-aminobenzamide (2AB) (16), and excess 2AB-derivatizing reagents were removed by extraction with chloroform (16). The 2AB derivatives were digested by CSase AC-II (5 mIU) in 50 mm sodium acetate buffer (pH 6.0), and the digests were analyzed by anion-exchange HPLC on an amine-bound silica PA03 column (4.6 × 250 mm; YMC Co., Kyoto, Japan) using a linear gradient of NaH2PO4 from 16 to 800 mm over 60 min at a flow rate of 1 ml/min. Eluates were monitored by measuring fluorescence at excitation and emission wavelengths of 330 and 420 nm, respectively. The CS-degrading activity was assessed based on the proportion (percentage) of 2AB-labeled disaccharide formed as described (8). The reaction rate was measured as moles of the products formed/min, and apparent Michaelis-Menten constants were determined by fitting the data to the Michaelis-Menten equation (V = Vmax[CS disaccharide]/Km + [CS disaccharide]).
Immunostaining
COS-7 cells transiently transfected with mHyal4 were cultured on coverslips and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. Sperm suspension isolated from epididymis of adult C57BL/6 mouse was spread onto glass slides, air-dried at room temperature, and then fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. The coverslips or glass slides were blocked with 3% bovine serum albumin in PBS for 45 min at room temperature, incubated with rabbit anti-mHyal4 polyclonal antibody (dilution 1:100; Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C, and then with Alexa Fluor 568® goat anti-rabbit IgG (dilution 1:200; Molecular Probes). Nucleus was stained with 4′,6-diamino-2-phenylindole (DAPI) (Invitrogen) for 5 min at room temperature. Images were captured using a fluorescence microscope, BZ-9000 (Keyence, Osaka, Japan).
RESULTS
Tissue Distribution and Cellular Localization of HYAL4
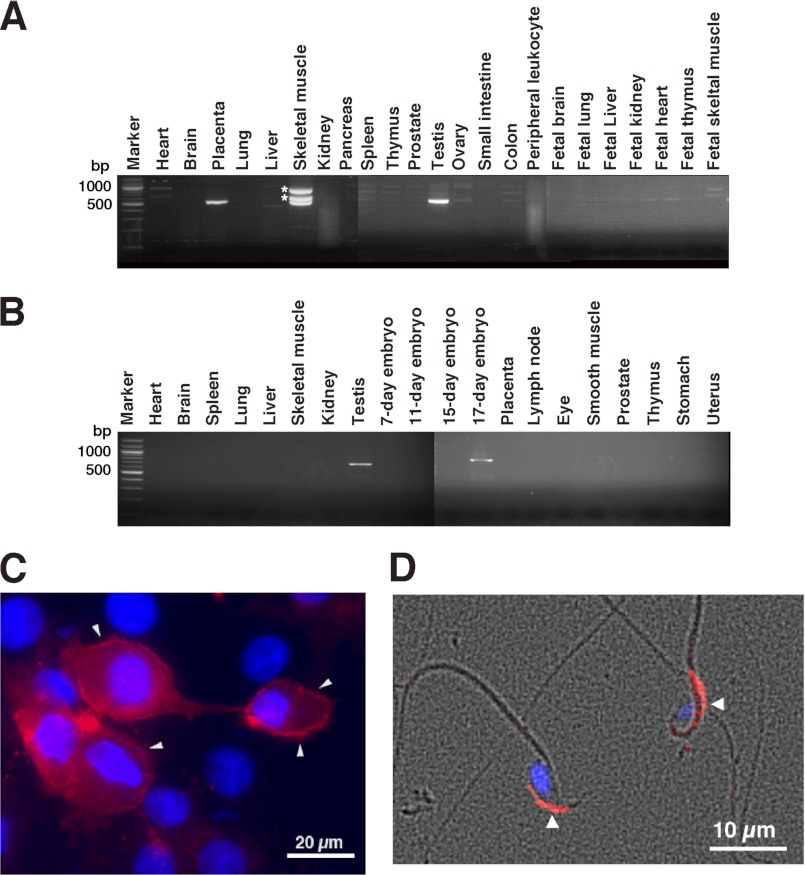
Although CS exists ubiquitously in mammalian tissues, hHYAL4 was reported to be differentially expressed in placenta and skeletal muscle. The expression patterns of hHYAL4 and mHyal4 were studied using multiple tissue cDNA panels. In addition to placenta and skeletal muscle, hHYAL4 mRNA was detected in testis (Fig. 1A). The expression of mHyal4 was not ubiquitous either, being restricted to testis and 17-day-old embryos (Fig. 1B). These observations indicate specific temporal functions of HYAL4 in particular organs or tissues.
FIGURE 1.
Analysis of the expression pattern of hHYAL4 or mHyal4 mRNA and the cellular localization of mHyal4. The expression pattern of hHYAL4 (A) or mHyal4 (B) mRNA was examined by PCR using cDNA from various tissues. Because three bands were detected in the PCR product using cDNA from human skeletal muscle, they were purified separately from the gel and subjected to the DNA sequence analysis. The upper two bands indicated by asterisks were not derived from the hHYAL4 mRNA but rather nonspecifically amplified PCR bands (data not shown). To examine the cellular localization of mHyal4, COS-7 cells transiently expressing mHyal4 (C) as well as mouse sperm (D) were stained with anti-mHyal4 antibody (red) and DAPI (blue). A merge of the phase contrast and fluorescence images of sperms is depicted in D. The strong staining at the periphery of the COS-7 cells (C, arrowheads) indicates the presence of mHyal4 at the cell surface. The mHyal4 protein was observed in the anterior head of sperm (D, arrowheads).
To examine whether only male but not female embryos express mHyal4, mouse embryos (embryonic day 18) were collected, the chromosomal sex of individual embryos was determined by PCR using specific primers for the male-specific Sry gene, and cDNAs prepared from individual embryos were subjected to reverse transcription-PCR to analyze the expression of mHyal4. The mHyal4 mRNA was detected in not only male but also female embryos (data not shown), indicating that mHyal4 appears to function in other tissue(s)/organ(s) besides testis, which remains to be identified.
Because both hHYAL4 and mHyal4 possess a putative GPI-anchored domain in the C-terminal region and appear to be GPI-anchored proteins, they may function on the cell surface, although the optimum pH of hHYAL4 is 4.5–5.0. To examine the cellular localization of mHyal4, COS-7 cells were transfected with a vector containing mHyal4 and stained using anti-mHyal4 polyclonal antibodies. The periphery of the cells was labeled under nonpermeabilized conditions, suggesting the cell surface expression of mHyal4 (Fig. 1C). The cells were also stained intracellularly after permeabilization, although the intracellular staining did not match that of the late endosome or lysosome marker, LAMP-2 (data not shown).
The localization of mHyal4 on the cell surface might be an artifact because it was forcibly overexpressed in COS-7 cells. To investigate the natural localization of mHyal4 protein in sperm, immunofluorescent staining was performed. It was observed at the surface of the sperm heads, presumably on the acrosomal membrane (Fig. 1D). mHyal4 appears to be tethered on acrosomal membranes of mouse sperm through a GPI anchor as PH-20 does (18).
Demonstration of the CS-degrading Activity
To investigate whether mHyal4 is also a CS-specific hydrolase as demonstrated for hHYAL4, a soluble form of the protein was generated with a FLAG tag by replacing the putative signal sequence with a cleavable leader sequence of preprotrypsin and by deleting the putative GPI-anchored domain as described under “Experimental Procedures.” The soluble recombinant protein was expressed in COS-7 cells at 37 °C. The protein secreted in the medium was adsorbed onto an anti-FLAG® M2 affinity gel for eliminating endogenous glycosidases, and then the protein-bound resin was used as an enzyme source.
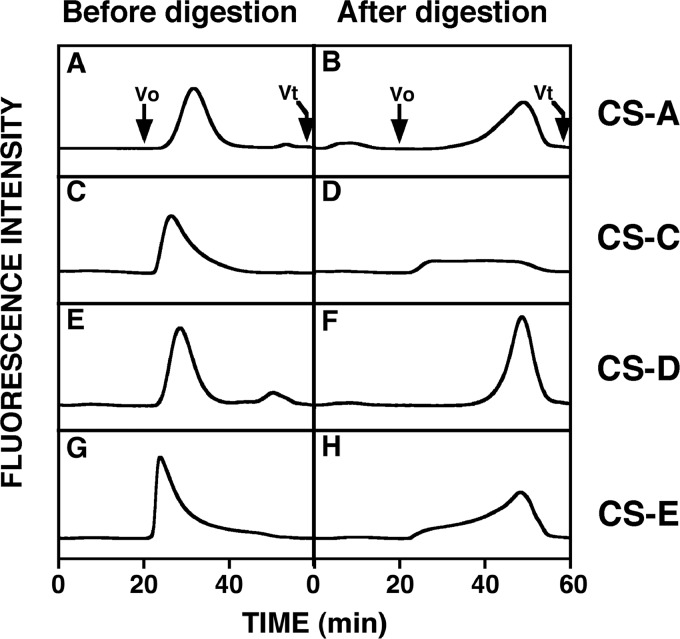
The bound fusion protein was assayed for its CS-degrading activity at 37 °C for 12 h using FITC-GAG isoforms as substrates, and each digest was analyzed by gel filtration HPLC on a Superdex 200 column (Fig. 2). When FITC-CS-A, -CS-D, and -CS-E were used as substrates, the eluted positions of the fluorescent peaks were shifted to approximately 50 min upon digestion with mHyal4. The peak of FITC-CS-C became broader after digestion with mHyal4 (Fig. 2D), although the total area of the peak did not change significantly. However, FITC-Chn, -DS, and -HA were hardly depolymerized by mHyal4 (data not shown). Thus, CS-specific degrading activity was detected for mHyal4 as demonstrated previously for hHYAL4 (8). Although hHYAL4 showed only weak activity toward FITC-CS-A as described previously (8), it was completely depolymerized by mHyal4, indicating the substrate specificity of hHYAL4 and mHyal4 to be different.
FIGURE 2.
Gel filtration HPLC of the FITC-labeled GAG isoforms digested with mHyal4. FITC-labeled CS-A (A and B), CS-C (C and D), CS-D (E and F), and CS-E (G and H) were analyzed by gel filtration HPLC on a column of Superdex 200 before (A, C, E, and G) and after (B, D, F, and H) incubation with the purified mHyal4 protein and monitored by measuring the fluorescent intensity of FITC with excitation and emission wavelengths of 490 and 520 nm, respectively. V0, void volume; Vt, total volume.
The effects of pH on the mHyal4 activity were examined by incubating mHyal4 with CS-A (5 μg) over a range of pH values from 3.5 to 6.5 (supplemental Fig. S2). The results indicated the optimum pH of mHyal4 to be 4.5, similar to that of hHYAL4 (pH 4.5–5.0) (8).
Kinetic Analysis of CS-degrading Activity of mHyal4
To compare the difference in substrate specificity between hHYAL4 and mHyal4, a kinetic analysis of the CS-degrading activity of mHyal4 was performed. The initial reaction rates and substrate concentrations (as disaccharide) were used for the analysis with Lineweaver-Burk plots (supplemental Fig. S3). The apparent Michaelis-Menten constants as well as Vmax values for CS-A, CS-C, and CS-D were determined and are shown in Table 1. The apparent Km values toward CS-A and CS-D were similar, indicating that mHyal4 equally recognizes CS-A and CS-D as its substrates. In contrast, the apparent Km value of hHYAL4 toward CS-A was ∼3 times that toward CS-D as reported previously (8), suggesting that the structural preference of the substrates by mHyal4 is different from that by hHYAL4 and that the former has a broader specificity than the latter.
TABLE 1.
Kinetic parameters of the recombinant mHyal4
| Substrate | Apparent Km | Apparent Vmax |
|---|---|---|
| mm as disaccharides | pmol/min | |
| CS-A | 0.66 | 47.4 |
| CS-C | 2.16 | 43.9 |
| CS-D | 0.50 | 34.4 |
Analysis of the Reducing Terminal Structure in the Degradation Products
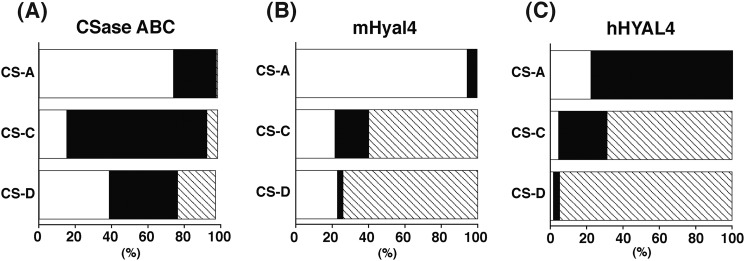
To further characterize the substrate specificity of hHYAL4 and mHyal4, the reducing terminal structure of oligosaccharide products generated by digestion with mHyal4 was analyzed. Nonlabeled CS-A, CS-C, or CS-D (20 μg) was incubated with hHYAL4 or mHyal4 (125 ng of protein) for 30 min, and then each digest was derivatized with 2AB and analyzed by anion-exchange HPLC after digestion with CSase AC-II (Fig. 3). Reactions with hHYAL4 or mHyal4 were terminated by the time when <5% of the substrate was consumed to measure the initial burst kinetics of the enzymatic activity. Although the major disaccharide unit in the intact polysaccharide of CS-A, CS-C, and CS-D is an A (74%), C (77%), and C (38%) unit, respectively (Fig. 3A), the major reducing terminal disaccharide structure of the hHYAL4 digest was a C (78%), D (69%), and D (95%) unit, respectively (Fig. 3C). This observation was consistent with our previous results, which demonstrated that hHYAL4 is fond of D units and to some extent C units as the disaccharide located on the nonreducing side of the cleavage site (8). The major reducing terminal disaccharide structure of the mHyal4 digest of CS-C or CS-D was also a D unit (60 and 74%, respectively) (Fig. 3). However, that of the digest of CS-A with mHyal4 was not C but A (95%), which was completely different from the results obtained using hHYAL4, indicating that mHyal4 prefers A units to C units on the nonreducing side of the cleavage site. Furthermore, when the incubation of the mHyal4 with the substrate was prolonged, the proportion of the reducing terminal disaccharide structure of mHyal4 digests became similar to the disaccharide composition of the intact CS polysaccharide used as a substrate, although that of the hHYAL4 digest did not change even after a long incubation (data not shown), suggesting that mHyal4 has broader substrate specificity than hHYAL4, consistent with the results of the kinetic analysis (Table 1).
FIGURE 3.
The reducing terminal disaccharide structure generated by digestion of CS variants with mHyal4. A, the disaccharide composition of the intact CS variants was determined by anion-exchange HPLC after digestion with CSase ABC followed by 2AB-labeling. B and C, CS-A, CS-C, and CS-D were digested individually with mHyal4 (B) or hHYAL4 (C). Each digest was derivatized with 2AB and analyzed by anion-exchange HPLC after digestion with CSase AC-II. Disaccharides derived from the reducing termini were quantified based on the fluorescent intensity of the peaks. The width of each box corresponds to the proportion of each disaccharide unit. Open, closed, and hatched boxes indicate A, C, and D units, respectively. The data were obtained in the linear range because reactions were terminated by the time when <5% of the substrate was consumed.
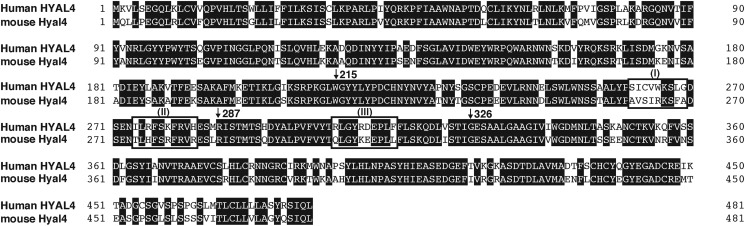
Examination of the Domains in hHYAL4/mHyal4 Responsible for Recognizing Substrates
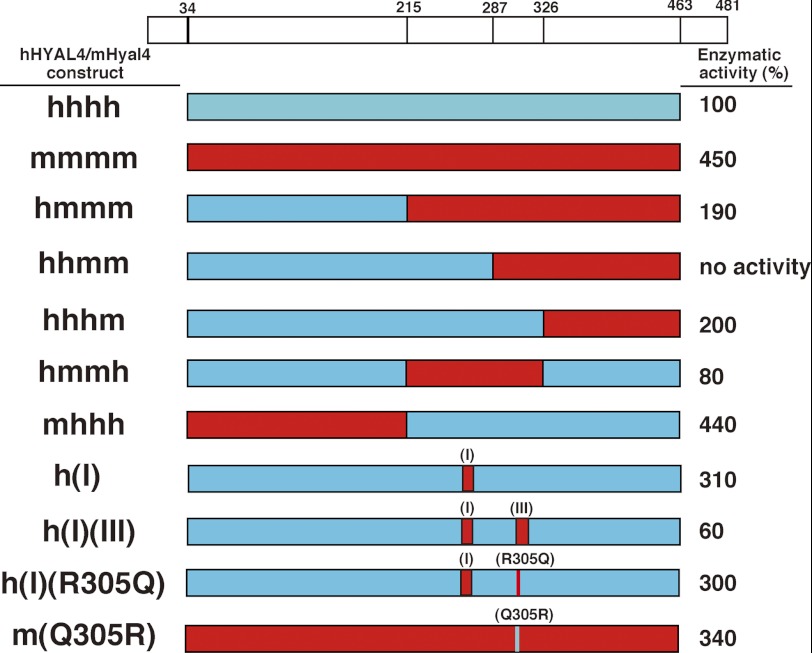
Although hHYAL4 and mHyal4 differ in substrate specificity, their amino acid sequences show as much as 80% identity (Fig. 4). To identify the residues responsible for recognizing substrates, a series of hHYAL4/mHyal4 chimeras were constructed, and their enzymatic activities toward CS-A were assayed (Fig. 5). Based on the major reducing terminal structure of oligosaccharides generated by the digestion of CS-A with the hHYAL4/mHyal4 chimeras, the structural recognition of the chimeras was classified into a mouse type or a human type. If A or C units were detected as the major structure, the specificity was defined as the mouse type or human type, respectively. As shown in Fig. 4, HYAL4 can be divided into four domains, which were separated by amino acid residues 215, 287, and 326. Constructs were designated based on the combination and order of the above human and mouse domain sequences. The constructs hmmm/hmmh and hhhm showed a similar substrate specificity to mHyal4 and hHYAL4, respectively (Fig. 6), indicating that the central regions of hHYAL4 and mHyal4 are responsible for their substrate specificity. The construct hhmm showed no enzymatic activity, suggesting the combination of the second and third domains to be critical. The steric conformation of the active site might be impaired by the irregular combination of the second and third domains.
FIGURE 4.
Comparison of mHyal4 with hHYAL4. A multiple sequence alignment of hHYAL4 with mHyal4 was conducted using the ClustalW program (version 1.83). Boxes (I), (II), and (III) indicate the amino acid residues that were different between mHyal4 and hHYAL4. Downward arrows indicate the positions of junctions in the hHYAL4/mHyal4 sequences.
FIGURE 5.
Enzymatic activity of hHYAL4/mHyal4 chimeras toward CS-A. Scale drawings of hHYAL4 (red boxes), mHyal4 (blue boxes), and hHYAL4/mHyal4 chimeras are shown. Nonlabeled CS-A was incubated individually with the purified protein (125 ng each) at 37 °C in 50 mm acetate buffer, pH 4.5, for 30 min. Enzymatic activities of mHyal4 and hHYAL4/mHyal4 chimeras toward CS-A are indicated at the right and expressed as a percentage of that of hHYAL4. The top diagram shows the positions of the signal peptide (amino acids 1–33), the putative GPI-anchored region (amino acids 463–481), and the amino acid positions where hHYAL4 cDNA was recombined with mHyal4 cDNA. Boxes indicated by (I) and (III) represent the regions where amino acid residues were replaced.
FIGURE 6.
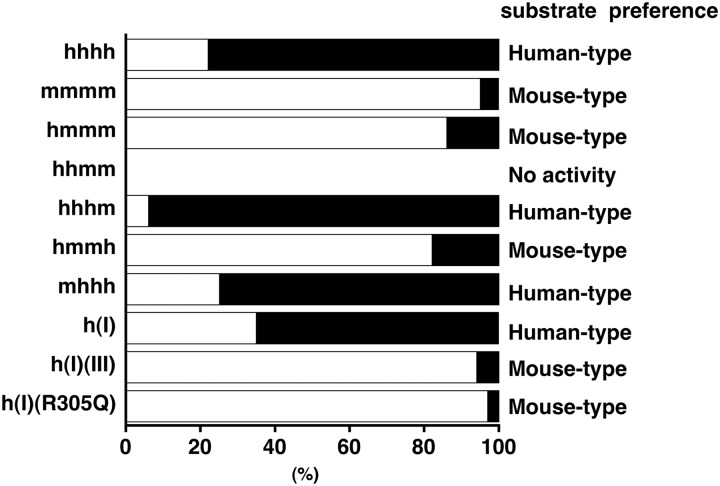
Proportion of the reducing terminal disaccharide structure of oligosaccharide products generated by digestion of CS-A with hHYAL4/mHyal4 chimeras or point mutants. CS-A was digested individually with hHYAL4/mHyal4 chimeras or point mutants. Each digest was derivatized with 2AB and analyzed by anion-exchange HPLC after digestion with CSase AC-II. Disaccharides derived from the reducing termini were quantified based on the fluorescent intensity of the peaks. The width of open and filled boxes corresponds to the proportion of A and C units, respectively. The data obtained were in the linear range because reactions were terminated by the time when <5% of the substrate was consumed. Because hHYAL4 and mHyal4 predominantly recognize C and A units, respectively, as the disaccharide unit on the nonreducing side of the cleavage site, structural preference (human type or mouse type) was assessed based on the predominant disaccharide structure detected at the reducing end of the oligosaccharide products and is shown on the right.
Analysis of the Amino Acid Residues Responsible for Recognizing Substrates
The analysis of the enzymatic activity of hHYAL4/mHyal4 chimeras indicated that the amino acid sequence between 215 and 326 is responsible for the structural recognition of the substrates by HYAL4. To identify the residues critical to substrate recognition, hHYAL4 mutants containing a substituent of a single amino acid or a single domain of the mouse sequence were constructed, and their structural specificity was investigated. Amino acid residues indicated by box (I), (II), or (III) in Fig. 4 were especially different in the corresponding regions between hHYAL4 and mHyal4. To determine which domain supports the structural recognition of HYAL4, the constructs h(I), h(II), h(III), h(I)(II), h(I)(III), h(II)(III), and h(I)(II)(III) were generated, and their substrate specificity was characterized (Fig. 6 and Table 2). Although the construct h(I)(II)(III) showed slightly weak enzymatic activity toward CS-A compared with the wild-type hHYAL4 (Fig. 5 and Table 2), the major reducing terminal disaccharide of the oligosaccharides generated by the digestion was an A unit (93%), suggesting that its substrate preference is the mouse type rather than human type (Fig. 6 and Table 2). The major reducing terminal disaccharide structure of the oligosaccharide generated by digestion with the construct h(I), h(II), or h(I)(II) was a C unit (65, 80, and 64%, respectively), indicating a preference similar to that of hHYAL4 (Fig. 6 and Table 2). In contrast, that of the oligosaccharide generated by the digestion with construct h(I)(III) was an A unit (94%), showing the mouse-type specificity. However, the constructs h(III) and h(II)(III) showed no enzymatic activity (Fig. 5 and Table 2), consistent with the results for the construct hhmm. Taken together, domain III is essential for the enzymatic activity but not sufficient for the substrate specificity of mHyal4. The combination of domains I and III appears to be responsible for the different structural preference between the human and mouse types.
TABLE 2.
Enzymatic activity of hHYAL4/mHyal4 mutants
Nonlabeled CS-A was incubated individually with the purified protein (125 ng each) at 37 °C in 50 mm acetate buffer, pH 4.5, for 30 min. Enzymatic activities of hHYAL4/mHyal4 mutants toward CS-A are expressed as a percentage of that of hHYAL4.
| Constructs | Relative activity | Proportion of A unita | Substrate preference |
|---|---|---|---|
| % | |||
| hHYAL4 (hhhh) | 100 | 22 | Human type |
| mHyal4 (mmmm) | 450 | 95 | Mouse type |
| h(I) | 310 | 35 | Human type |
| h(II) | 140 | 20 | Human type |
| h(III) | NDb | ND | No activity |
| h(I)(II) | 420 | 36 | Human type |
| h(I)(III) | 60 | 94 | Mouse type |
| h(II)(III) | ND | ND | No activity |
| h(I)(II)(III) | 90 | 93 | Mouse type |
| h(I)(III/Q305R) | 460 | 30 | Human type |
| h(I)(III/K309R,E310D) | 150 | 96 | Mouse type |
| h(I)(III/L314F) | 70 | 95 | Mouse type |
| h(I)(R305Q) | 300 | 97 | Mouse type |
| m(A261S,V262I,S263G,I264V,R265W)(Q305R) | 63 | 47 | Human type |
a The proportion of the reducing terminal disaccharide of oligosaccharides generated by the digestion of CS-A.
b ND, not detected.
To determine the amino acid residue(s) in domain III responsible for recognizing substrates, the structural specificity of other mutants, which were generated by introducing mutations in domain III of h(I)(III), was investigated. The constructs h(I)(III/Q305R), h(I)(III/K309R,E310D), and h(I)(III/L314F) were produced and examined as to whether their specificity was converted from the mouse type to the human type. As shown in Table 2, the major reducing terminal disaccharide structure of the digest of CS-A with h(I)(III/Q305R), h(I)(III/K309R,E310D), or h(I)(III/L314F) was a C (70%), A (96%), or A (95%) unit, respectively. The substrate specificity of the construct h(I)(III) was converted from the mouse type to the human type upon site-directed mutagenesis at position 305, suggesting glutamine 305 to be essential for the structural specificity characteristic of mHyal4. To confirm that the amino acid 305 is critical for the substrate specificity of mHyal4, characterization of the construct h(I)(R305Q) was also performed. The major reducing terminal disaccharide structure of the digest of CS-A with h(I)(R305Q) was an A unit (97%) (Table 2), indicating that the human-type specificity of h(I) was converted to the mouse type by the single replacement of Arg305 with Gln305.
To characterize the amino acid residue(s) in domain I responsible for recognizing substrates, the substrate specificity of mutants generated by introducing mutation(s) in domain I of h(R305Q) (Table 3), was investigated. All the mutants, which exhibited hydrolytic activity toward CS-A, showed the mouse-type specificity (data not shown), indicating that glutamine 305 is critical for the structural preference of mHyal4. Taking the hydrolytic activity of h(I)(R305Q) toward CS-A as 100%, the relative enzymatic activity of the constructs generated is shown in Table 3. Although the construct h(L268F,G269A)(R305Q) had no hydrolytic activity, the constructs h(S261A,I262V,G263S)(R305Q), h(I262V,G263S,V264I)(R305Q), and h(G263S,V264I,W265R)(R305Q) showed weak but significant activity, suggesting that residues 261–265 are important, but residues 267 and 268 are not essential for the catalytic activity. Therefore, it was concluded that the combination of the residues at 261–265 and glutamine 305 is essential for the catalytic activity as well as the structural recognition of mHyal4. To confirm the indispensable role of these residues for the substrate recognition, the mHyal4 mutant containing substituents of these amino acids of the human sequence, m(A261S,V262I,S263G,I264V,R265W)(Q305R), was constructed. This mutant showed the substrate preference of the human type (Table 2).
TABLE 3.
Enzymatic activity of the hHYAL4 mutants generated by introducing mutation(s) in domain I of h(R305Q)
Nonlabeled CS-A was incubated individually with the purified protein (125 ng each) at 37 °C in 50 mm acetate buffer, pH 4.5, for 30 min. The hydrolytic activity of the construct h(I)(R305Q) toward CS-A is taken as 100%. All constructs having the hydrolytic activity exhibited the mouse-type specificity.
| Constructs | Relative activity |
|---|---|
| h(I)(R305Q) | 100 |
| h(S261A,I262V,G263S,V264I,W265R)(R305Q) | 23 |
| h(S261A,I262V,G263S)(R305Q) | 6 |
| h(I262V,G263S,V264I)(R305Q) | 4 |
| h(G263S,V264I,W265R)(R305Q) | 16 |
| h(S261A,I262V)(R305Q) | NDa |
| h(G263S)(R305Q) | ND |
| h(V264I,W265R)(R305Q) | ND |
| h(L268F,G269A)(R305Q) | ND |
| h(R305Q) | ND |
a ND, not detected.
DISCUSSION
In this study, we demonstrated the catalytic activity of mHyal4 for the first time and proved the enzyme to be a CS-specific endo-β-N-acetylgalactosaminidase. Although its human counterpart hHYAL4 is also a CS-specific hydrolase, the substrate specificity of mHyal4 was broader than that of hHYAL4 (8). hHYAL4 recognizes CS-D and CS-C but rarely CS-A. In contrast, mHyal4 depolymerized CS-A, CS-C, and CS-D at a similar rate (Fig. 3). Hence, this enzyme will be a better experimental tool than hHYAL4 for depolymerizing CS chains in tissues and cells without degrading HA to investigate the specific functions of CS. Approaches using a bacterial CS lyase against glial scar CS/DS proteoglycans have received much attention in the treatment of acute spinal cord injuries (19, 20). However, the bacterial CS lyase is not a hydrolase but an eliminase and depolymerizes not only CS but also HA. mHyal4, a CS-specific hydrolase, may also be more useful for treating acute spinal cord injury than the bacterial lyase (19).
The optimal pH of mHyal4 was 4.5 (supplemental Fig. S2), similar to that of hHYAL4 (pH 5.0), suggesting the enzyme to be active mostly in lysosomes. However, HYAL4 does not seem to be involved in the systemic catabolism of CS in lysosomes because the expression of mHyal4 and hHYAL4 is not ubiquitous but restricted to placenta, skeletal muscle, and testis (9) (Fig. 1), and to testis and 17-day-old embryos (Fig. 1), respectively. Because CS exists ubiquitously in mammals, HYAL4 may have specific temporal functions in particular organs or tissues. Disaccharide composition of CS from human skeletal muscle has never been reported. Although CS from human placenta has been characterized by Achur et al. (21), D disaccharide units preferred by hHYAL4 were not detected and might be a minor component (<1%). Because hHYAL4 also recognizes C unit-containing structures to some extents, C units which account for 20% of placenta CS may be the major substrate for HYAL4 in human placenta.
mHyal4 appears to play a special role in the testes of adult mice. It may be involved in the maturation of sperm or during fertilization by degrading CS chains in the cumulus cell-oocyte complex (COC). For successful fertilization, a HYAL, PH-20, tethered to the plasma membrane on the head of the sperm by the GPI anchor must degrade the matrix of the expanded COC (22). mHyal4 may contribute to the penetration of the COC matrix by degrading CS. A significant amount of CS is present in the matrix of COC.3 Kim et al. (23) reported that mouse hyaluronidase-5 (Hyal5) was also exclusively expressed in the testis and enzymatically active in the pH range 5–7 based on HA zymography analysis, suggesting to function as a HA hydrolase in the matrix of COC and compensate in part for the function of PH-20. Because the human genome lacks the Hyal5 gene, hHYAL4 and PH-20 may cooperate to disperse COC by degrading CS and HA, respectively.
hHYAL4 and mHyal4 possess a putative GPI-anchored domain in the C-terminal region. They may function at the cell surface, although the optimum pH of mHyal4 is 4.5 (supplemental Fig. S2). Notably, HYAL2, which is a GPI-anchored molecule but has an optimal pH of 3.8 (24), has been assigned to the plasma membrane with CD44 (25, 26). HA-degrading activity of HYAL2 was detected in the membrane fraction of the cells co-expressing HYAL2 and CD44 at pH 6.0–7.0 (27). Therefore, we investigated the cellular distribution of this enzyme. COS-7 cells were transfected with a vector containing the mHyal4 gene and stained with a (polyclonal) anti-mHyal4 antibody. The periphery of the cells was labeled under nonpermeabilized conditions, suggesting the cell surface expression of mHyal4 (Fig. 1C). To detect the enzymatic activity of Hyal4 in cells, we have generated Chinese hamster ovary cell lines that stably express Hyal4. CS and HS chains were purified from the Hyal4-expressing cells as well as control cells, and their total amounts were compared. Although the amount of HS chains was not significantly reduced, that of CS from the Hyal4-expressing Chinese hamster ovary cells decreased dramatically compared with the control cells (supplemental Table S2), suggesting that Hyal4 most likely hydrolyzes CS chains even in the cells. Therefore, Hyal4 appears to be enzymatically active in vivo, and hydrolyzes CS chains in testis or some other organs, or on the surface of sperm under physiological conditions.
In the present study, we identified the amino acid residues essential for the structural recognition of substrates by HYAL4. The amino acid sequences of hHYAL4 and mHyal4 showed 80% identity with only 98 of 481 residues differing. Although hHYAL4 predominantly recognizes D and C units as the disaccharides located on the nonreducing side of the cleavage site (8), mHyal4 has a broader substrate specificity and recognizes A units as well as D and C units. The 4-O-sulfate group on the GalNAc residue appears to be tolerated by mHyal4 but not recognized by hHYAL4. Based on an analysis of the enzymatic activity of hHYAL4/mHyal4 chimeras and site-directed mutants, glutamine 305 in mHyal4 was identified as essential for the recognition of the GalNAc(4-O-sulfate) structure in the CS polysaccharides. These results should help to generate artificial CS hydrolases, which recognize specific CS structures and could be useful for elucidating novel roles of CS.
Whereas HYAL4 exclusively depolymerizes CS, HYAL1 has been reported to degrade both HA and CS (18). Identification of the amino acid residues responsible for distinguishing GalNAc from GlcNAc is the next objective. Studies of sequence homology between HYAL4 and HYAL1 may reveal the amino acid residues. Based on the three-dimensional modeling of human HYALs, Jedrzejas and Stern (10) have claimed that Cys263 in hHYAL4 replaced by Tyr247 in hHYAL1 may cause the distinct substrate specificity of hHYAL4. The Cys263 in HYAL4 reported by Jedrzejas and Stern (10) (accession number AF009010) was replaced by Gly263 in the sequence of the HYAL4 gene cloned by us (accession number AB470346) and Mammalian Gene Collection Program Team (accession numbers: BC104788 and BC104790) as well as in human genomic DNA (accession number: NT_007933). It, however, should be noted that the Tyr247 residue in hHYAL1 is conserved among all HYAL family members except for HYAL4. In the sequence of the mHyal4 gene, it is also replaced by Ser263. To investigate the involvement of the Tyr residue in the recognition of HA as a substrate, point mutants h(G263Y) as well as m(S263Y) were generated, and their substrate specificities were characterized. Based on unpublished observations,4 m(S263Y) hydrolyzed not only CS but also HA, indicating that replacement of Ser by Tyr conferred the HA-degrading activity to mHyal4. However, h(G263Y) still depolymerized only CS but not HA. The Tyr residue appears to be partially involved in the recognition of GlcNAc residue in HA, but it is not sufficient to govern the substrate specificity. Investigation of the amino acid residues contributing to the recognition of the difference between CS and HA is in progress.
The estimated location of the amino acid residues of mHyal4 was three-dimensionally investigated in reference to a bee venom hyaluronidase using PyMOL (28). Based on the crystal structure of the bee venom hyaluronidase with a HA tetrasaccharide (Protein Data Bank ID code 1fcv), the amino acid residues corresponding to positions 261–265 and the glutamine at 305 of mHyal4 are located close to and appear to interact with the HA tetrasaccharide substrate (supplemental Fig. S4). Therefore, three-dimensional study also supports that those amino acids are essential to the substrate specificity of mHyal4. However, the crystal structure of mHyal4 should be analyzed to investigate the precise interaction of mHyal4 with CS oligosaccharides.
Recently, Zhang et al. (11) identified the active site residues of hHYAL1 based on the crystal structure of the enzyme in conjunction with an assessment of the effects of site-directed mutagenesis. The results showed that Glu131 and Tyr202 are likely proton donors for the hydroxyl leaving group and a substrate binding determinant, respectively, and that glycosylation on Asn350 is necessary for the enzymatic activity. Accordingly, point mutations in hHYAL4 were generated at Glu147 (E147Q), Tyr218 (Y218A), and Asn368 (N368A), which correspond to Glu131, Tyr202, and Asn350 in HYAL1, respectively, to investigate their individual contributions to the mechanism of enzymatic action. Mutagenesis of these residues completely eliminated the enzymatic activity (supplemental Table S3), indicating that the catalytic mechanism of hHYAL4 is similar to that of hHYAL1.
Some endo-type GAG hydrolases are overexpressed in pathological situations. Up-regulation of heparanase, endo-β-glucuronidase specific for heparan sulfate, enhances tumor growth, angiogenesis, and metastasis (29). In some cancer specimens, HYAL1 is expressed exclusively in tumor cells (30). HYAL1 has been implicated in the promotion of angiogenesis through the dissipation of accumulated HA within tumors, which may facilitate its recognition as an angiogenic signal (30, 31). In addition, it has been shown that HA oligosaccharides generated by digestion with HYAL1 are involved in inflammation, tumor migration, tumor apoptosis, and so forth (30). Some reports showed that CS oligosaccharides also have biological functions. Oligosaccharides derived from CS-E but not intact CS-E, enhance CD44 cleavage and tumor cell motility (32). Thus, HYAL4 may be highly expressed under pathological conditions, and HYAL4 itself or CS oligosaccharides generated by HYAL4 may contribute to such biological events. Accumulating evidence has demonstrated various biological functions of GAG oligosaccharides (33, 34). Investigation of the expression pattern of HYAL4 in some diseases may provide a key for elucidating biological functions of HYAL4.
This work was supported in part by Grant-in-aid for Scientific Research C-24590071 (to S. Y.) and the Matching Program for Innovations in Future Drug Discovery and Medical Care (to K. S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), and the Mizutani Foundation for Glycoscience (to S. Y.), Tokyo, Japan.

This article contains supplemental Figs. S1–S4 and Tables S1–S3.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) AB524072.
A. Miyazaki, T. Kaneiwa, S. Mizumoto, K. Sugahara, and S. Yamada, unpublished data.
T. Kaneiwa, K. Sugahara, and S. Yamada, unpublished data.
- CS
- chondroitin sulfate
- 2AB
- 2-aminobenzamide
- Chn
- chondroitin
- COC
- cumulus cell-oocyte complex
- CSase
- chondroitinase
- DS
- dermatan sulfate
- GAG
- glycosaminoglycan
- GPI
- glycosylphosphatidylinositol
- HA
- hyarulonan
- hHYAL
- human hyaluronidase
- mHyal4
- mouse hyaluronidase-4.
REFERENCES
- 1. Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. (2003) Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 13, 612–620 [DOI] [PubMed] [Google Scholar]
- 2. Sugahara K., Yamada S. (2000) Structure and function of oversulfated chondroitin sulfate variants: unique sulfation patterns and neuroregulatory activities. Trends Glycosci. Glycotechnol. 12, 321–349 [Google Scholar]
- 3. Couchman J. R. (2010) Transmembrane signaling proteoglycans. Annu. Rev. Cell Dev. Biol. 26, 89–114 [DOI] [PubMed] [Google Scholar]
- 4. Iozzo R. V. (1998) Matrix proteoglycans: from molecular design to cellular function. Annu. Rev. Biochem. 67, 609–652 [DOI] [PubMed] [Google Scholar]
- 5. Esko J. D., Selleck S. B. (2002) Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 6. Rauch U., Kappler J. (2006) Chondroitin/dermatan sulfates in the central nervous system: their structures and functions in health and disease. Adv. Pharmacol. 53, 337–356 [DOI] [PubMed] [Google Scholar]
- 7. Uyama T., Kitagawa H., Sugahara K. (2007) in Comprehensive Glycoscience (Kamerling J. P., ed) Vol. 3, pp. 79–104, Elsevier, Amsterdam [Google Scholar]
- 8. Kaneiwa T., Mizumoto S., Sugahara K., Yamada S. (2010) Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology 20, 300–309 [DOI] [PubMed] [Google Scholar]
- 9. Csóka A. B., Scherer S. W., Stern R. (1999) Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics 60, 356–361 [DOI] [PubMed] [Google Scholar]
- 10. Stern R., Jedrzejas M. J. (2006) Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L., Bharadwaj A. G., Casper A., Barkley J., Barycki J. J., Simpson M. A. (2009) Hyaluronidase activity of human Hyal1 requires active site acidic and tyrosine residues. J. Biol. Chem. 284, 9433–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaneiwa T., Yamada S., Mizumoto S., Montaño A. M., Mitani S., Sugahara K. (2008) Identification of a novel chondroitin hydrolase in Caenorhabditis elegans. J. Biol. Chem. 283, 14971–14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akatsu C., Mizumoto S., Kaneiwa T., Maccarana M., Malmström A., Yamada S., Sugahara K. (2011) Dermatan sulfate epimerase 2 is the predominant isozyme in the formation of the chondroitin sulfate/dermatan sulfate hybrid structure in postnatal developing mouse brain. Glycobiology 21, 565–574 [DOI] [PubMed] [Google Scholar]
- 14. Koopman P., Münsterberg A., Capel B., Vivian N., Lovell-Badge R. (1990) Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348, 450–452 [DOI] [PubMed] [Google Scholar]
- 15. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 16. Kinoshita A., Sugahara K. (1999) Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal. Biochem. 269, 367–378 [DOI] [PubMed] [Google Scholar]
- 17. Kawashima H., Atarashi K., Hirose J., Yamada S., Sugahara K., Miyasaka M. (2002) Oversulfated chondroitin/dermatan sulfates containing GlcAβ1/IdoAα1–3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J. Biol. Chem. 277, 12921–12930 [DOI] [PubMed] [Google Scholar]
- 18. Csoka A. B., Frost G. I., Stern R. (2001) The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 20, 499–508 [DOI] [PubMed] [Google Scholar]
- 19. Bradbury E. J., Moon L. D, Popat R. J., King V. R., Bennett G. S., Patel P. N., Fawcett J. W., McMahon S. B. (2002) Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 416, 636–640 [DOI] [PubMed] [Google Scholar]
- 20. Alilain W. J., Horn K. P., Hu H., Dick T. E., Silver J. (2011) Functional regeneration of respiratory pathways after spinal cord injury. Nature 475, 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Achur R. N., Valiyaveettil M., Alkhalil A., Ockenhouse C. F., Gowda D. C. (2000) Characterization of proteoglycans of human placenta and identification of unique chondroitin sulfate proteoglycans of the intervillous spaces that mediate the adherence of Plasmodium falciparum-infected erythrocytes to the placenta. J. Biol. Chem. 275, 40344–40356 [DOI] [PubMed] [Google Scholar]
- 22. Lin Y., Mahan K., Lathrop W. F., Myles D. G., Primakoff P. (1994) A hyaluronidase activity of the sperm plasma membrane protein PH-20 enables sperm to penetrate the cumulus cell layer surrounding the egg. J. Cell Biol. 125, 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim E., Baba D., Kimura M., Yamashita M., Kashiwabara S., Baba T. (2005) Identification of a hyaluronidase, Hyal5, involved in penetration of mouse sperm through cumulus mass. Proc. Natl. Acad. Sci. U.S.A. 102, 18028–18033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lepperdinger G., Strobl B., Kreil G. (1998) HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 273, 22466–22470 [DOI] [PubMed] [Google Scholar]
- 25. Rai S. K., Duh F. M., Vigdorovich V., Danilkovitch-Miagkova A., Lerman M. I., Miller A. D. (2001) Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. U.S.A. 98, 4443–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duterme C., Mertens-Strijthagen J., Tammi M., Flamion B. (2009) Two novel functions of hyaluronidase-2 (Hyal2) are formation of the glycocalyx and control of CD44-ERM interactions. J. Biol. Chem. 284, 33495–33508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harada H, Takahashi M. (2007) CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J. Biol. Chem. 282, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 28. DeLano W. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 29. Vlodavsky I., Korner G., Ishai-Michaeli R., Bashkin P., Bar-Shavit R., Fuks Z. (1990) Extracellular matrix-resident growth factors and enzymes: possible involvement in tumor metastasis and angiogenesis. Cancer Metastasis Rev. 9, 203–226 [DOI] [PubMed] [Google Scholar]
- 30. Stern R. (2008) Hyaluronidases in cancer biology. Semin. Cancer Biol. 18, 275–280 [DOI] [PubMed] [Google Scholar]
- 31. Lokeshwar V. B., Rubinowicz D., Schroeder G. L., Forgacs E., Minna J. D., Block N. L., Nadji M., Lokeshwar B. L. (2001) Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J. Biol. Chem. 276, 11922–11932 [DOI] [PubMed] [Google Scholar]
- 32. Sugahara K. N., Hirata T., Tanaka T., Ogino S., Takeda M., Terasawa H., Shimada I., Tamura J., ten Dam G. B., van Kuppevelt T. H., Miyasaka M. (2008) Chondroitin sulfate E fragments enhance CD44 cleavage and CD44-dependent motility in tumor cells. Cancer Res. 68, 7191–7199 [DOI] [PubMed] [Google Scholar]
- 33. Yamada S., Sugahara K. (2008) Potential therapeutic application of chondroitin sulfate/dermatan sulfate. Curr. Drug Discov. Technol. 5, 289–301 [DOI] [PubMed] [Google Scholar]
- 34. Stern R., Asari A. A., Sugahara K. N. (2006) Hyaluronan fragments: an information-rich system. Eur. J. Cell Biol. 85, 699–715 [DOI] [PubMed] [Google Scholar]