Background: The guanine nucleotide exchange factor that activates Rab6A GTPase is not known.
Results: Ric1-Rgp1 binds Rab33B-GTP and catalyzes nucleotide exchange on Rab6A GTPase.
Conclusion: Ric1 and Rgp1 are needed to generate and maintain active Rab6A on the Golgi.
Significance: This evidence supports the presence of a Rab cascade on the Golgi complex of human cells.
Keywords: Golgi, GTPase, Guanine nucleotide exchange factor (GEF), Membrane trafficking, Rab proteins, Rab GTPase cascade
Abstract
Rab GTPases are master regulators of membrane trafficking events and template the directionality of protein transport through the secretory and endocytic pathways. Certain Rabs recruit the guanine nucleotide exchange factor (GEF) that activates a subsequent acting Rab protein in a given pathway; this process has been termed a Rab cascade. We show here that the medial Golgi-localized Rab33B GTPase has the potential to link functionally to the late Golgi, Rab6 GTPase, by its capacity for association with Ric1 and Rgp1 proteins. In yeast, Ric1p and Rgp1p form a complex that catalyzes guanine nucleotide exchange by Ypt6p, the Rab6 homolog. Human Ric1 and Rgp1 both bind Rab6A with preference for the GDP-bound conformation, characteristic of a GEF. Nevertheless, both Ric1 and Rgp1 proteins are needed to catalyze nucleotide exchange on Rab6A protein. Ric1 and Rgp1 form a complex, but unlike their yeast counterparts, most of the subunits are not associated, and most of the proteins are cytosolic. Loss of Ric1 or Rgp1 leads to destabilization of Rab6, concomitant with a block in Rab6-dependent retrograde transport of mannose 6-phosphate receptors to the Golgi. The C terminus of Ric1 protein contains a distinct binding site for Rab33B-GTP, supporting the existence of a Rab cascade between the medial and trans Golgi. This study thus identifies a GEF for Rab6A in human cells.
Introduction
Rab GTPases are master regulators of the secretory and endocytic pathways (1–3). They recruit effector proteins to the surfaces of specific membrane compartments to catalyze transport vesicle formation, motility, docking, and fusion. Rab GTPases are active with GTP bound, and guanine nucleotide exchange factors (GEFs)3 are key to facilitating the release of bound GDP to convert Rab proteins into their active conformations. Identification of the cognate GEFs for the more than 60 human Rab GTPases is important for our understanding of membrane traffic (4).
The ordered steps of membrane traffic are likely templated by a process referred to as a “Rab cascade” (5–7). Several examples of Rab GEF cascades have been described in which one Rab, in its GTP-bound state, recruits the GEF that can activate the next Rab GTPase along the pathway. For example, the yeast Golgi Rab, Ypt32, recruits the Sec2p GEF to activate the subsequent acting, Sec4p Rab GTPase (5). We have proposed that Rab cascades order the compartments of the polarized Golgi complex and help create this organelle (8). Delineation of the relationships between GEFs and upstream Rab proteins will enable us to define the way in which cells establish membrane trafficking pathways.
Rab6 is a Golgi-associated Rab GTPase that is important for normal Golgi structure and function (9, 10). It is present on the surface of transport vesicles en route to the cell surface where it links transport vesicles to motor proteins (11). It is also needed to anchor tethering factors such as GCC185 to the Golgi surface to facilitate the receipt of incoming transport vesicles to that compartment (12, 13). Studies of the yeast Rab6 homolog, Ypt6p, have led to the identification of Ric1p and Rgp1p as a complex that catalyzes Ypt6p nucleotide exchange (14). These authors showed that RIC1 is linked to YPT6 genetically; Ric1p binds Rgp1p, and this complex binds Ypt6p with preference for the GDP-bound form. The Ric1p-Rgp1p complex, but not the individual polypeptides, catalyzes nucleotide exchange by Ypt6p in vitro and is needed for Ypt6p localization to the Golgi in yeast cells (14).
Human cells express Ric1p and Rgp1p orthologs that have not been studied to date. We show here that human Ric1 and Rgp1 proteins also interact with one another, via the central domain of Ric1 protein. Ric1 and Rgp1 proteins can each bind independently to Rab6 with preference for the GDP-bound forms; nevertheless, Rab6-specific GEF activity is only seen when both polypeptides are present. Loss of Ric1 or Rgp1 from cells destabilizes Rab6 protein, decreases its abundance on the Golgi complex, and at least in the case of Rgp1, interferes with the retrograde transport of mannose 6-phosphate receptors from late endosomes to the Golgi. Consistent with Ric1-Rgp1 participating in a cascade of Rab proteins, we find that Ric1 interacts with Rab33B GTPase, a protein localized to the medial Golgi complex, in a location where it can template the formation of a Rab6 membrane compartment.
EXPERIMENTAL PROCEDURES
Plasmids
GFP-Rgp1 was obtained by PCR amplification from an IMAGE clone (ID-3533857, Open Biosystems) and cloned into the EcoRI and BamHI sites of pEGFP-C1 (Clontech) containing a GFP tag at the N terminus. Myc-Ric1 was obtained by PCR amplification from pFlag-CMV-2-CIP150, a gift of Dr. Tatsuo Takeya (Nara Institute of Science and Technology, Nara, Japan) and cloned into the AscI and PacI sites of modified pCS2+ vector (Invitrogen) containing a 6×Myc tag at the N terminus. FLAG-Rab6A was generated from His-Rab6 (13) and cloned into the AscI and PacI sites of modified pCS2+ vector containing a 3×FLAG tag at the N terminus. The GDP-locked (T27N) and GTP-locked (Q72L) mutants of Rab6A were generated by site-directed mutagenesis (Stratagene). GFP-Rab33B and GFP-Rab4A were described (15).
For bacterial expression, full-length His-Rgp1 and GST-Rgp1 were subcloned into the AscI and PacI sites of modified pET-14b vector (Novagen) containing a His6 tag or into pGEX-6P1 (GE Healthcare) containing a GST tag at the N terminus, respectively. GST-Ric1-1–364, GST-Ric1-365–719, GST-Ric1-720–1344, and GST-Ric1-365–1344 constructs were PCR-amplified and ligated into pGEX-6P1. His-Ric1-365–1344 and His-Ric1-1032–1366 was PCR-amplified and ligated into pET-14b. His-Rab6A T27N was generated by site-directed mutagenesis. GST-Rab33B and GST-Rab9A were described previously (15, 16); GST-Rab6A was generated from FLAG-Rab6A. For insect cell expression, FLAG-Rgp1 and Myc-Ric1 were PCR-amplified and ligated under the p10 promoter (Pp10) or polyhedrin promoter (PPH) of the pFastBac Dual expression vector (Invitrogen), respectively.
Protein Expression and Purification
All constructs were purified from Rosetta2 (DE3) cells (Novagen). Bacteria transformed with His-Rgp1, His-Ric1-1032–1344, or His-Rab6A T27N were grown at 37 °C to an A600 = 0.5. The cells were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside and grown for an additional 4 h at 22 °C. Harvested cells were resuspended in cold lysis buffer (50 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5 mm MgCl2, 0.1 mm dithiothreitol (DTT)) plus 20 mm imidazole, and 10 μm GDP (for Rab constructs) supplemented with 1 mm phenylmethanesulfonyl fluoride (PMSF) and lysed by a single pass at 20,000 p.s.i. through an EmulsiFlex-C5 (Avestin). Cleared lysates (20,000 rpm, 45 min at 4 °C in a JA-20 rotor; Beckman Coulter) were incubated with nickel-nitrilotriacetic acid agarose (Qiagen) for 1 h at 4 °C. The resin was then washed with the same buffer and eluted with lysis buffer containing 200 mm imidazole. Fractions containing desired proteins were pooled and desalted using PD-10 columns (GE Healthcare). The samples were then concentrated using an Amicon Ultra spin (Millipore), brought to 10% (v/v) glycerol, aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C.
Bacteria transformed with GST-Rgp1, GST-Ric1-1–364, GST-Ric1-365–719, GST-Ric1-720–1344, and GST-Ric1-365–1344 were grown at 37 °C to an A600 = 0.5. The cells were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside and grown for an additional 4 h at 22 °C. Harvested cells were resuspended in cold buffer (50 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 1 mm MgCl2, 1 mm DTT) supplemented with 1 mm PMSF and lysed as above. Cleared lysates were incubated with glutathione-Sepharose (GE Healthcare) for 2 h at 4 °C. The resin was then washed with the same buffer and eluted with 50 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 1 mm MgCl2, 1 mm DTT, and 20 mm glutathione. Proteins were processed as above and stored at −80 °C. GST-Rab6A, GST-Rab33B, and GST-Rab9A expression and purification were as described previously (15).
Baculovirus expression of Ric1-Rgp1 complex was according to the manufacturer (Invitrogen). Sf9 cells were infected with recombinant bacmid, and the passage 3 viral supernatant was harvested, spun to remove the cell debris, brought to 5% (v/v) fetal bovine serum, and stored at 4 °C for subsequent infections. Sf9 cells infected with passage 3 viral supernatant were harvested after 48 h and lysed in buffer C (Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 1% Triton X-100, and 10% glycerol) supplemented with 1 mm PMSF, 2.3 μm leupeptin, 1.5 μm pepstatin, and 150 nm aprotinin. Clarified lysates were incubated with FLAG antibody-agarose (Sigma-Aldrich) for 2 h at 4 °C. The resin was then washed with buffer C and eluted with PBS containing 0.1 mg/ml 3×FLAG peptide (Sigma-Aldrich). The eluted sample was used immediately for GEF assays or stored on ice for up to 48 h.
Cell Culture, Transfections, and Sucrose Gradient Flotation
HEK293T and HeLa cells from the American Type Culture Collection were cultured at 37 °C, 5% CO2 in DMEM (Gibco) supplemented with 7.5% fetal bovine serum, 100 units penicillin, and 100 μg/ml streptomycin. HEK293T cells were transfected with polyethyleneimine (Polysciences); HeLa cells were transfected using FuGENE 6 (Promega). Sucrose gradient flotation was as described (17).
Antibodies
Rabbit anti-GFP (Molecular Probes); mouse anti-FLAG and mouse anti-α-tubulin (Sigma-Aldrich); mouse anti-His (Qiagen); rabbit anti-Rab6 (C-19; Santa Cruz Biotechnology); mouse anti-Rab33 (D5; Frontier Institute); and mouse anti-GM130 (BD Biosciences) were purchased. Mouse anti-Myc (9E10) and mouse (2G11) anti-cation-independent mannose 6-phosphate receptor (MPR) were described previously (18). Rabbit anti-GDP dissociation inhibitor-β antibody was described previously (19). HRP-goat anti-mouse and goat anti-rabbit antibodies were from Bio-Rad. Alexa Fluor 488-anti-rabbit and Alexa Fluor 555- and 647-anti-mouse antibodies were from Molecular Probes.
Co-immunoprecipitation and Binding Assays
HEK293T cells expressing Rgp1, Ric1, and/or Rab6A constructs were harvested after 24 h and lysed in Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 1 mm DTT, 1% Triton X-100, and 10% glycerol containing 1 mm PMSF, 2.3 μm leupeptin, 1.5 μm pepstatin, and 150 nm aprotinin. After centrifugation at 12,000 × g for 10 min, 2 mg of extracts were precleared with protein A-Sepharose (GE Healthcare) at 4 °C for 30 min. The precleared extracts were incubated with either GFP-binding protein-conjugated agarose (for GFP-Rgp1) or FLAG antibody-conjugated agarose (for FLAG-Rab6A) for 2 h at 4 °C. Immobilized proteins were washed with the same buffer, eluted with 2× Laemmli buffer, and resolved by SDS-PAGE. Bound proteins were detected by immunoblotting.
Glutathione-Sepharose-bound GST-Ric1 (100 nm) was incubated with His-Rgp1 (1 μm) for 1 h at 20 °C in binding buffer (50 mm HEPES, pH 7.4, 150 mm KCl, 5 mm MgCl2, and 0.1 mg/ml BSA). The complexes were washed in binding buffer, eluted with 2× Laemmli, and analyzed by immunoblotting. Binding of GST-Rgp1 and GST-Ric1-720–1344 with His-Rab6A T27N was the same except that the buffer was supplemented with 50 μm GDP and the concentrations of GST-Rgp1 and GST-Ric1 were 2 μm; His-Rab6A T27N was 1 μm. GST-Rab6A, GST-Rab33B, and GST-Rab9A were loaded with GTPγS or GDP (16), and 1 μm of active Rabs were incubated with His-Ric1-1032–1344 (0.5 μm); binding was performed as above. In Figs. 4B and 8E, bound Rab GTPase was determined by quantitative immunoblot using anti-His and Alexa Fluor 647 anti-mouse antibody; bands were quantified using a VersaDoc system (Bio-Rad) and Adobe Photoshop CS3 (Adobe Systems) software.
FIGURE 4.
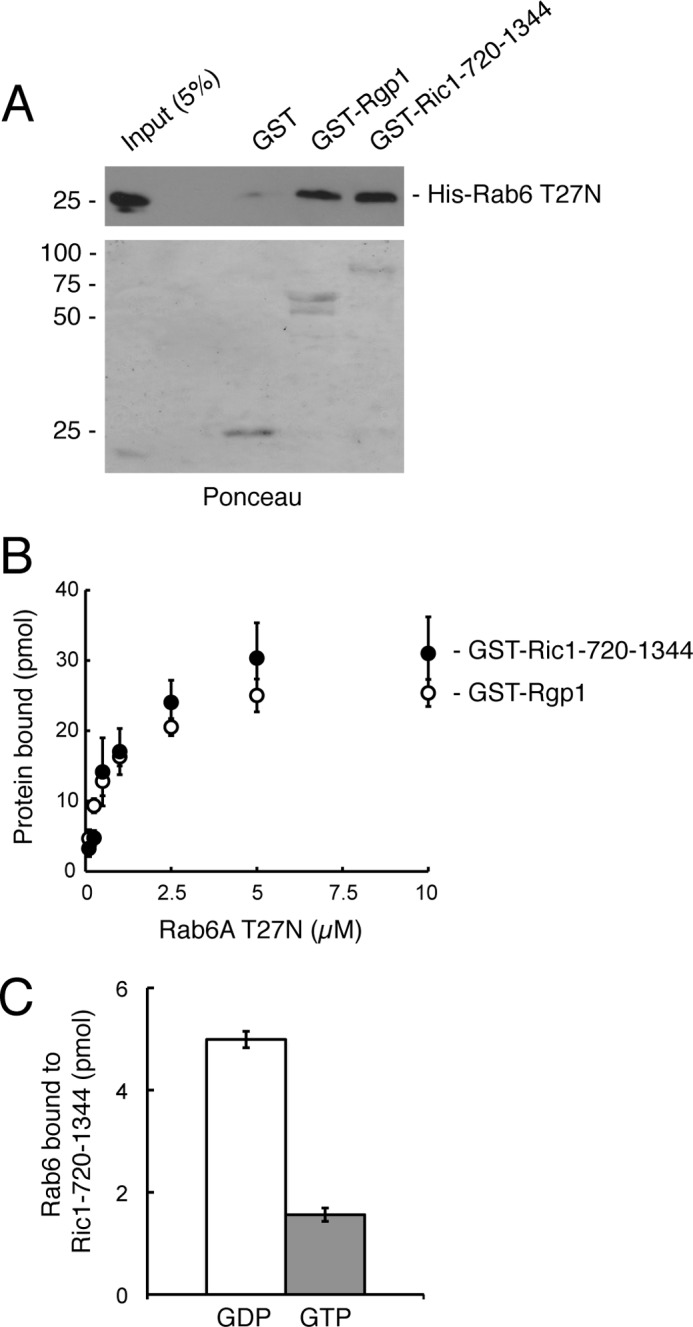
Rgp1 and Ric1-720–1344 interact directly with Rab6A-GDP in vitro. A, GST, GST-Rgp1, or GST-Ric1-720–1344 proteins were immobilized on glutathione-Sepharose followed by incubation with His-Rab6A T27N protein. Bound material was analyzed by immunoblotting with anti-His antibody (upper panel). Eluted GST, GST-Rgp1, and GST-Ric1-720–1344 proteins were detected by Ponceau S staining (lower panel). B, binding of Rab6 T27N (1 μm) to GST-Rgp1 or GST-Ric1-720–1344 determined as in A. Error bars represent S.E. from two independent experiments. C, GST-Ric1-720–1344 immobilized on glutathione-Sepharose was incubated with His-Rab6A loaded with either [3H]GDP or [35S]GTPγS. Ric1-bound Rab6A was detected by scintillation counting. Error bars represent S.E. from two independent experiments.
FIGURE 8.
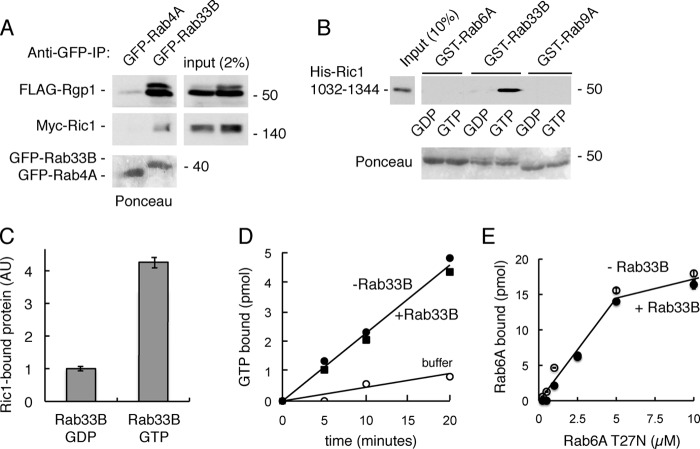
Ric1 is an effector of Rab33B. A, HEK293T cells were co-transfected with either GFP-Rab4A or GFP-Rab33B and FLAG-Rgp1 and Myc-Ric1 for 24 h. GFP-Rab4A and GFP-Rab33B were immunoprecipitated (IP) with GFP-binding protein-agarose followed by immunoblotting with anti-FLAG and anti-Myc antibodies to detect co-immunoprecipitated FLAG-Rgp1 and Myc-Ric1. B, GST-Rab6A, GST-Rab33B, and GST-Rab9A were preloaded with either GDP or GTPγS and immobilized on glutathione-Sepharose followed by incubation with His-Ric1-1032–1344 protein. Bound material was analyzed by immunoblotting with anti-His antibody. Eluted GST-Rab6A, GST-Rab33B, and GST-Rab9A proteins were detected by Ponceau S staining. C, quantification of Ric1 binding from immunoblot analysis. Error bars represent S.E. of the three independent experiments. AU, arbitrary units. D, Rab6A (50 pmol) was incubated with 20 units of Ric1-Rgp1 complex (corresponding to 10 pmol of Rgp1) that had been preincubated with or without 5 μm Rab33B-GTPγS at 30 °C. Protein-bound nucleotide was determined. Shown are the means from duplicate experiments, and S.E. was <5%. E, GST-Ric1-720–1344 (1 μm) was immobilized on glutathione-Sepharose followed by incubation with His-Rab6A T27N protein ± Rab33B-GTPγS. Bound material was analyzed by immunoblotting with anti-His antibody and quantified. Error bars represent S.E. from two independent experiments. Numbers at right indicate the mobility of prestained marker proteins of the mass indicated in kDa.
Cell Fractionation and Gel Filtration Chromatography
HeLa cells co-expressing GFP-Rgp1 and Myc-Ric1 were washed three times with PBS and one time with 10 mm HEPES, pH 7.4, followed by 15 min in 10 mm HEPES, pH 7.4. Cells were harvested by scraping in 10 mm HEPES, pH 7.4, 150 mm KCl, 5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol plus protease inhibitors and homogenized with 10 passes through a 27-gauge needle. A postnuclear supernatant was obtained by centrifugation at 3000 rpm at 4 °C for 5 min. The postnuclear supernatant was further centrifuged at 95,000 rpm for 20 min at 4 °C, and the supernatant (cytosol) was subjected to gel filtration chromatography on a Superdex 200 FPLC column (GE Healthcare) controlled by ÄKTApurifier (GE Healthcare). Gel filtration of the immunopurified Ric1-Rgp1 complex from Sf9 insect cells was on the same column. Fractions were subjected to immunoblot analysis, and band intensities were quantified using Adobe Photoshop.
Nucleotide Binding and GEF Assays
Purified Rab GTPases were loaded with [γ-35S]GTP (16) to determine their nucleotide binding capacity. Active Rabs were incubated with Rgp1, Ric1, or both proteins in 50 μl containing 20 mm HEPES, pH 7.4, 150 mm KCl, 2.5 mm MgCl2, 0.1 mg/ml BSA, 20 μm GTPγS, and 2 μCi of [γ-35S]GTP at 30 °C, and aliquots were removed at various times and quenched by the addition of ice-cold 20 mm HEPES, pH 7.4, 100 mm NaCl, and 25 mm MgCl2. The quenched samples were assayed by filter binding followed by liquid scintillation counting in BioSafe-II scintillation fluid (Research Products International) using an LS-6500 liquid scintillation counter (Beckman Coulter). Nucleotide exchange is reported in pmol of GTP bound.
RNA Interference
siGENOME nontargeting small interfering RNA (siRNA) and siRNA oligonucleotides targeting the ORF regions of Rgp1 (5′-CAGTGATGGCCGAGGGAAA-3′) and Ric1 (5′-GCACCTATCTAGAGAGCAA-3′) were from Dharmacon. Transfections were performed using 50% confluent cells with 40 nm siRNA using DharmaFECT (Dharmacon). 72 h after transfection, cells were processed for immunofluorescence microscopy.
Immunofluorescence
HeLa cells seeded on coverslips and depleted for endogenous Rgp1 and Ric1 were washed twice in PBS and fixed for 15 min in 3.7% formaldehyde in 200 mm HEPES-NaOH, pH 7.4. After fixation, cells were washed twice in PBS and permeabilized with 0.1% Triton X-100 in PBS for 3 min. Coverslips were washed twice in PBS, blocked with 1% BSA in PBS (blocking buffer) for 30 min, and incubated with anti-Rab6, anti-GM130, and anti-cation-independent MPR antibodies diluted in blocking buffer for 1 h at room temperature. Coverslips were washed, incubated with Alexa Fluor 488 or Alexa Fluor 647 secondary antibodies diluted in blocking buffer for 1 h, and then washed and mounted using Mowiol (Polysciences). Imaging was performed with a Leica TCS SP2 SE confocal scanner in conjunction with a Leica DM6000 B upright scope (with attached Leica HCX PL apochromatic 63×/NA 1.4 objective) and a Leica CTR 6000 confocal control box. This setup was controlled by Leica Control software (Leica Microsystems). Images were processed using ImageJ.
RESULTS
To explore the activity of human Ric1 and Rgp1, we expressed the proteins in human cells and first tested their association. N-terminally Myc-tagged Ric1 was co-expressed in HEK293T cells with either GFP or GFP-tagged Rgp1, and GFP-tagged proteins were collected using immobilized single chain anti-GFP llama antibody. As shown in Fig. 1A, this procedure precipitated about 20–25% of GFP and GFP-Rgp1 proteins; Myc-Ric1 (∼5%) co-precipitated with GFP-Rgp1 but not GFP. This experiment shows that at least a portion of Ric1 associates with Rgp1 in HEK293T cells.
FIGURE 1.
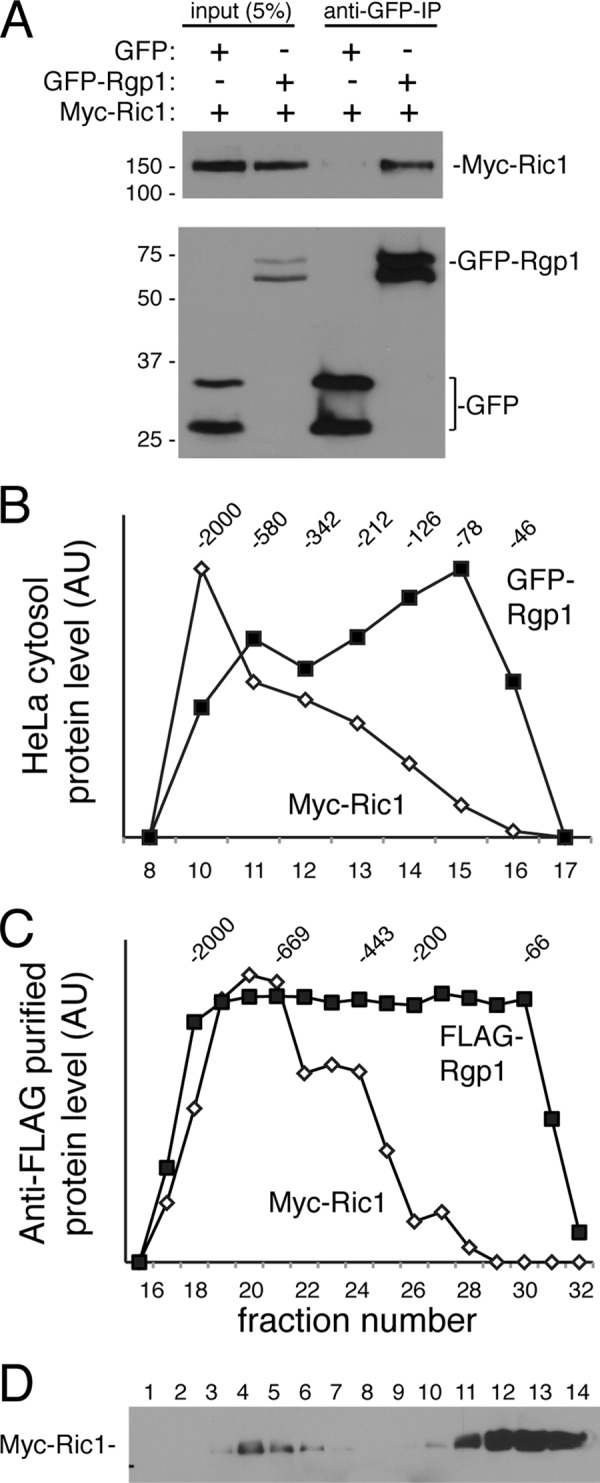
Rgp1 and Ric1 interact in vivo. A, HEK293T cells were transfected with either GFP or GFP-Rgp1 with Myc-Ric1 for 24 h, and Rgp1 was immunoprecipitated (IP) with anti-GFP-binding protein-conjugated agarose followed by immunoblotting with anti-Myc antibody to detect Myc-Ric1 (top panel) or anti-GFP antibody to detect GFP-Rgp1 (bottom panel). Double bands are seen in the lower panel because the samples are only heated to 37 °C to decrease aggregate formation seen at higher temperatures. Numbers at left indicate the mobility of prestained marker proteins of the mass indicated in kDa. B, Superdex 200 gel filtration of co-transfected HeLa cell cytosol; migration of GFP-Rgp1 or Myc-Ric1 was determined by immunoblot. Numbers at the top indicate migration of marker proteins in kDa. AU, arbitrary units. C, Superdex 200 chromatography of anti-FLAG immunopurified FLAG-Rgp1/Myc-Ric1 complexes obtained after co-expression in Sf9 cells. D, immunoblot after sucrose gradient flotation of an extract of HeLa cells transfected with Myc-Ric1. The top of the gradient is fraction 1.
We noted that Ric1 and Rgp1 proteins migrated anomalously if samples were boiled prior to SDS-PAGE; samples were instead warmed to 37 °C, which yielded double bands for GFP and GFP-Rgp1 (Fig. 1A). The tendency of the proteins to aggregate upon boiling suggested that Ric1 and Rgp1 may form larger complexes in cells. Gel filtration of cytosol from HeLa cells expressing Myc-Ric1 or GFP-Rgp1 revealed that both proteins chromatographed as larger, heterodisperse assemblies (Fig. 1B). Much of the ∼150-kDa Myc-Ric1 gene product chromatographed with an apparent mass of >700 kDa; the ∼68-kDa GFP-Rgp1 chromatographed as a monomeric species as well as larger forms that may be associated with Ric1 protein. This pattern was also seen when the proteins were co-expressed in Sf9 insect cells and isolated by anti-FLAG antibody adsorption and elution (Fig. 1C). Consistent with the co-immunoprecipitation results, a significant fraction of FLAG-Rgp1 protein did not co-chromatograph with Ric1 protein. Thus, ∼20% of exogenous Myc-Ric1 may associate with GFP-Rgp1 in cytosol. Moreover, sucrose gradient flotation of whole cell extracts indicated that less than ∼15% of Myc-Ric1 or FLAG-Rgp1 (not shown) proteins were membrane-associated (Fig. 1D).
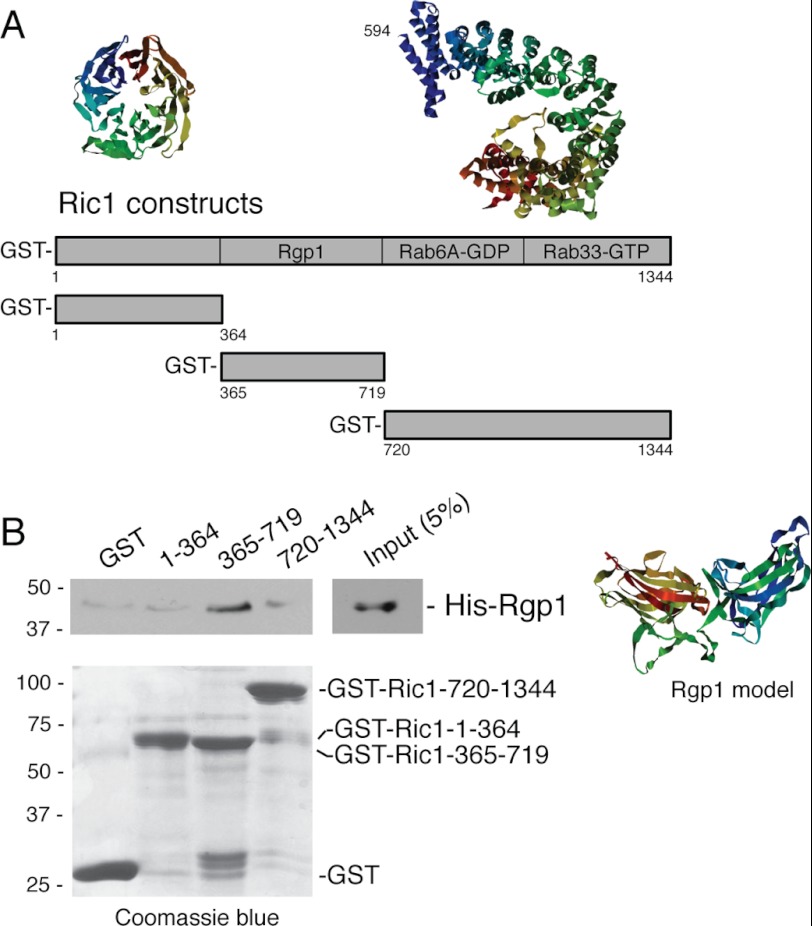
The Protein Model Portal was used to explore the predicted structure of Ric1 protein, which was found to contain a possible WD40-like, β propeller at its N terminus and a helical solenoid at its C terminus. This is reminiscent of the structure of clathrin and coatomer coat proteins as well as certain transport vesicle-tethering proteins such as Dsl1 (20). To identify the region in Ric1 needed for Rgp1 association, we relied on the predicted structures to generate GST fusion constructs representing the N-terminal propeller (residues 1–364), a central domain composed of residues 365–719, and the C-terminal α solenoid (residues 720–1344). The constructs were expressed in bacteria (Fig. 2B, lower panel); only the middle construct showed significant binding to His-tagged Rgp1 protein also purified after expression in bacteria. Thus, the central domain of Ric1 interacts with Rgp1 protein.
FIGURE 2.
Ric1 interacts directly with Rgp1 via the Ric1 central domain (365–719). A, schematic representation of Ric1 constructs used. Multiple Ric1 isoforms of 1423, 1386, 1344, and 1165 can be found in PubMed. Structure prediction of the N-terminal (1–364) and C-terminal (594–1415) domains of Ric1 using the modeling tool, Protein Model Portal, is shown. Rgp1, Rab6A-GDP and Rab33B-GTP binding sites discovered in this study are also indicated (see “Results”). B, GST and GST-Ric1 constructs were immobilized on glutathione-Sepharose followed by incubation with His-Rgp1 as described under “Experimental Procedures.” Bound material was analyzed by immunoblotting with anti-His antibody; GST and GST-Ric1 fragments were detected by Coomassie Blue staining. Numbers at left indicate the mobility of prestained marker proteins of the mass indicated in kDa. Structure prediction for Rgp1 residues 2–346 (out of 391) is shown.
Previous work on yeast Ric1-Rgp1 showed that the complex bound Ypt6 with preference for the GDP-bound form, but those experiments did not determine which of the two polypeptides bound Ypt6 (14). GFP-Rgp1 and Myc-Ric1, in extracts obtained after expression in HEK293T cells, displayed binding to Rab6 with preference for Rab6A T27N, which binds GDP more than GTP (Fig. 3). Binding was seen when the Ric1 and Rgp1 proteins were expressed alone or in combination. In these experiments, we cannot rule out the presence of the endogenous partner protein assembling with either Ric1 or Rgp1.
FIGURE 3.
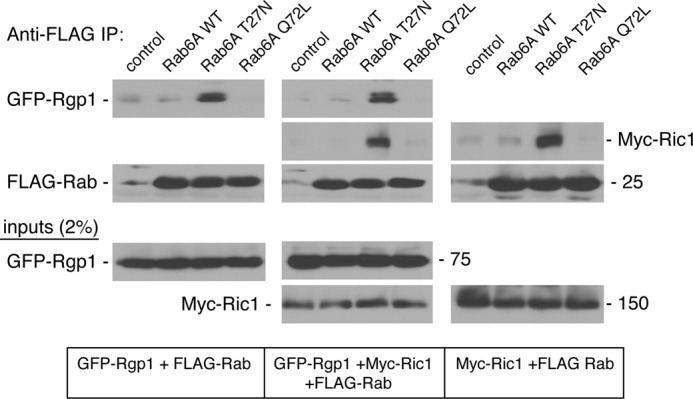
Rgp1 and Ric1 interact with GDP-bound Rab6A in vivo. HEK293T cells were transfected with FLAG vector, FLAG-Rab6A wild type, GDP-preferring FLAG-Rab6A T27N, or GTP-locked FLAG-Rab6A Q72L and GFP-Rgp1 alone (left panels), Myc-Ric1 alone (right panels), or both (middle panels) for 24 h. FLAG-Rab6A was immunoprecipitated (IP) with anti-FLAG antibody-agarose followed by immunoblotting with anti-GFP and anti-Myc antibodies to detect co-immunoprecipitated GFP-Rgp1 and Myc-Ric1, respectively. Immunoprecipitated FLAG-Rab6A levels were assessed by immunoblotting with anti-FLAG antibody. Inputs (2%) are shown below.
Direct binding of Rab6A to each of the subunits was shown using purified proteins (Fig. 4). In these experiments, bacterially expressed and purified GST-Rgp1 or GST-Ric1 solenoid (720–1344), but not GST control, retained purified His-Rab6 T27N protein, as determined by immunoblot analysis (Fig. 4A). These experiments show that both Ric1 and Rgp1 bind Rab6A-GDP and can do so even in the absence of the corresponding partner protein.
The binding of Rab6A-GDP to Rgp1 and Ric1-720–1344 was saturable with Kd values of ∼1 μm (Fig. 4B). The preference of Ric1-720–1344 for Rab6A-GDP over Rab6A-GTP was further confirmed using purified GST-Ric1-720–1344 and Rab6A preloaded with either GDP or GTPγS. Ric1-720–1344 bound with 3-fold preference to Rab6-GDP when compared with Rab6-GTP (Fig. 4C).
To test whether Ric1-Rgp1 showed Rab6A GEF activity, we attempted to generate recombinant Ric1 and Rgp1 proteins (Table 1). The smaller, full-length Rgp1 protein could be prepared in GST- and His-tagged forms, but we were not successful in preparing either His-tagged or GST-tagged versions of full-length Ric1 protein from bacteria. For Ric1 protein, the longest form produced in bacteria was GST-Ric1-365–1344. None of these proteins, tested alone or mixed as indicated, showed GEF activity when assayed at 500 nm with 1 μm active Rab6A protein (Table 1). Full-length Ric1 and Rgp1 were produced successfully upon expression in HEK293T cells, but the proteins were not obtained in significant yield and also failed to display GEF activity when tested in vitro. To circumvent these issues, we generated a baculovirus that would permit co-expression of Ric1 and Rgp1 in insect cells.
TABLE 1.
Summary of protein expression and purification
ND, not detected.
| Proteins purified and tested by GEF assay | Source | Rab6A GEF activity |
|---|---|---|
| Fusion protein | ||
| GFP-Rgp1 and Myc-Ric1 | Mammalian cells | ND |
| FLAG-Rgp1 and Myc-Ric1 | Mammalian cells | ND |
| FLAG-Rgp1 and GFP-Ric1 | Mammalian cells | ND |
| FLAG-Rgp1 and Ric1-GFP | Mammalian cells | ND |
| GFP-Rgp1 and Ric1-Myc | Mammalian cells | ND |
| Rgp1-GFP and Ric1-Myc | Mammalian cells | ND |
| Rgp1-GFP and Ric1-Myc | In vitro translation | ND |
| GST-Rgp1 full length | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| His-Rgp1 full length | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| GST-Ric1–720–1344 | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| His-Ric1–720–1344 | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| GST-Ric1–365–1344 | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| GST-Ric1–365–1344 and His-Rgp1 | Bacteria (500 nm enzyme, 1 μm Rab6) | ND |
| FLAG-Rgp1 and Myc-Ric1 | Insect cells (100 nm Rgp1, 1 μm Rab6) | 0.2 pmol of GTP exchanged/mina |
| Additional purified proteins | ||
| GST-Ric1–1–364 | Bacteria | |
| GST-Ric1–365–719 | Bacteria | |
| His-Ric1–1032–1344 | Bacteria | |
| His-Rgp1-N | Bacteria | |
| His-Rgp1-C | Bacteria | |
| Proteins that could not be purified | ||
| His-Ric1 full length | Bacteria | |
| His-Ric1–1–364 | Bacteria | |
| His-Ric1–365–719 | Bacteria | |
| His-Ric1–720–1031 | Bacteria | |
| GST-Ric1 full length | Bacteria | |
| GST-Ric1–720–1031 | Bacteria | |
| GST-Ric1–1032–1344 | Bacteria | |
a Assayed at 30 °C; see Fig. 5.
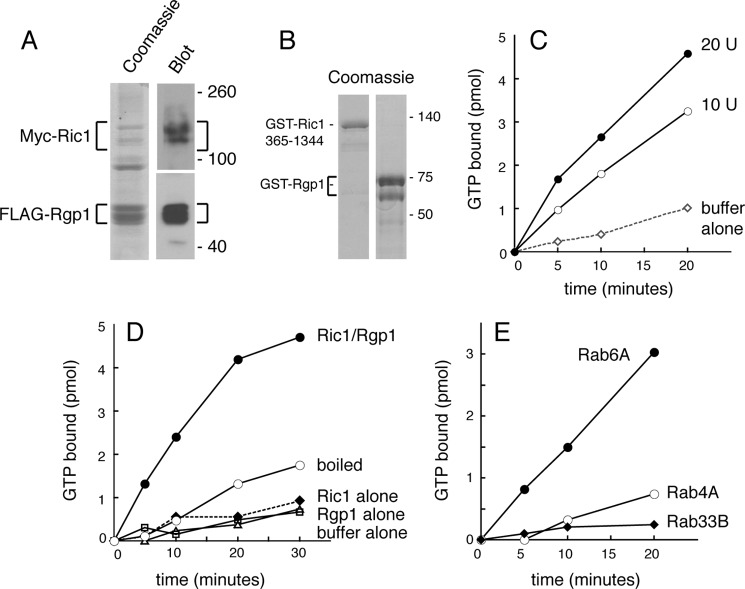
Fig. 5A (left lane) shows a Coomassie Blue-stained gel of the protein complex isolated from insect cells. Purification was accomplished by anti-FLAG affinity chromatography; although both polypeptides could be detected in the purified fractions, significantly more FLAG-Rgp1 was present in the purified material (Fig. 5A; see also Fig. 1C). Nevertheless, the addition of the purified Ric1-Rgp1 complex enhanced the rate of GTP binding to Rab6A protein in a concentration-dependent manner (Fig. 5C). Activity was due to protein as it was lost upon boiling of the enzyme sample (Fig. 5D). GEF activity was specific for Rab6A and not seen for the early endosome Rab4A protein or for the medial Golgi Rab33B protein (Fig. 5E). Activity required the presence of both polypeptides and was not seen if proteins purified separately were simply mixed (Fig. 5, B and D). Thus, proper assembly and/or post-translational modification may be necessary for recovery of the active Ric1-Rgp1 GEF. We cannot exclude the possibility that the co-expressed proteins bring with them an additional component during purification as a few other bands were seen on the Coomassie Blue-stained gel (Fig. 5A) that may or may not represent degradation products of Ric1 and Rgp1.
FIGURE 5.
Ric1-Rgp1 complex has guanine nucleotide exchange activity toward Rab6A in vitro. A, Ric1-Rgp1 complex was purified from insect cells with anti-FLAG antibody-agarose followed by immunoblotting with anti-FLAG and anti-Myc antibodies to detect Rgp1 and Ric1, respectively (right panel). Coomassie Blue staining of the purified complex is shown at the left. Double bands are in part due to not boiling samples to avoid aggregation. B, Coomassie Blue-stained gel of purified GST-Ric1-365–1344 (left lane) and GST-Rgp1 (right lane). C, Rab6A (50 pmol) was incubated with 10 (10 U) or 20 units (20 U) of Ric1-Rgp1 complex (corresponding to 5 or 10 pmol of Rgp1) at 30 °C. Protein-bound nucleotide was determined. D, Rab6A (50 pmol) was incubated with Rgp1 (40 pmol), Ric1-365–1344 (40 pmol), Ric1-Rgp1 complex (20 units), or boiled Ric1-Rgp1 complex (20 units) at 30 °C. Nucleotide exchange was determined as above. E, Rab6A or Rab4A or Rab33B (50 pmol) was incubated with Ric1-Rgp1 complex (20 units) at 30 °C. Nucleotide exchange was determined as above. Shown are the means from at least two independent experiments, and S.E. was <5%.
Cellular Role of Ric1-Rgp1
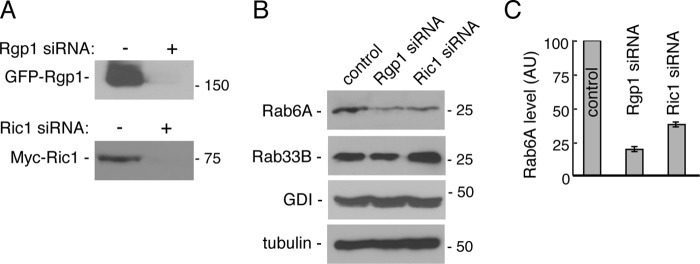
If Ric1-Rgp1 functions as a Rab6A GEF in cells, its absence should influence Rab6A protein function. Antibodies were not available to monitor siRNA depletion of the endogenous proteins; however, under conditions where exogenously expressed Ric1 or Rgp1 were completely depleted (Fig. 6A), Rab6A protein levels were decreased 60–75% (Fig. 6, B and C). This effect was specific for Rab6A as Rab33B, another Golgi Rab, was not decreased and even increased slightly upon Ric1 siRNA treatment (Fig. 6B). It is known that the ability of a Rab to bind to key effectors stabilizes Rab proteins (16); thus, the destabilization of Rab6A seen upon Rab6A GEF loss is not entirely unexpected. This finding supports the proposal that Ric1-Rgp1 is a Rab6A GEF in cells.
FIGURE 6.
Rgp1 and Ric1 regulate cellular levels of Rab6A. A, HEK293T cells were transfected with either Rgp1 or Ric1 siRNA. 12 h after transfection, cells were transfected with either GFP-Rgp1 or Myc-Ric1 for 24 h. Levels of Rgp1 and Ric1 were then assessed by immunoblotting with anti-GFP antibody to detect GFP-Rgp1 (top panel) or anti-Myc antibody to detect Myc-Ric1 (bottom panel). B, HeLa cells were transfected with control, Rgp1, or Ric1 siRNAs for 72 h, and the levels of endogenous Rab6A and Rab33B were assessed by immunoblotting with anti-Rab6A and anti-Rab33B antibodies. Equal protein loading was verified by immunoblotting for GDP dissociation inhibitor-β (GDI) (for Rab6A) or α-tubulin (for Rab33B). C, quantification of Rab6A. Data represent the average of three independent experiments. Error bars represent S.E. of the three independent experiments. AU, arbitrary units.
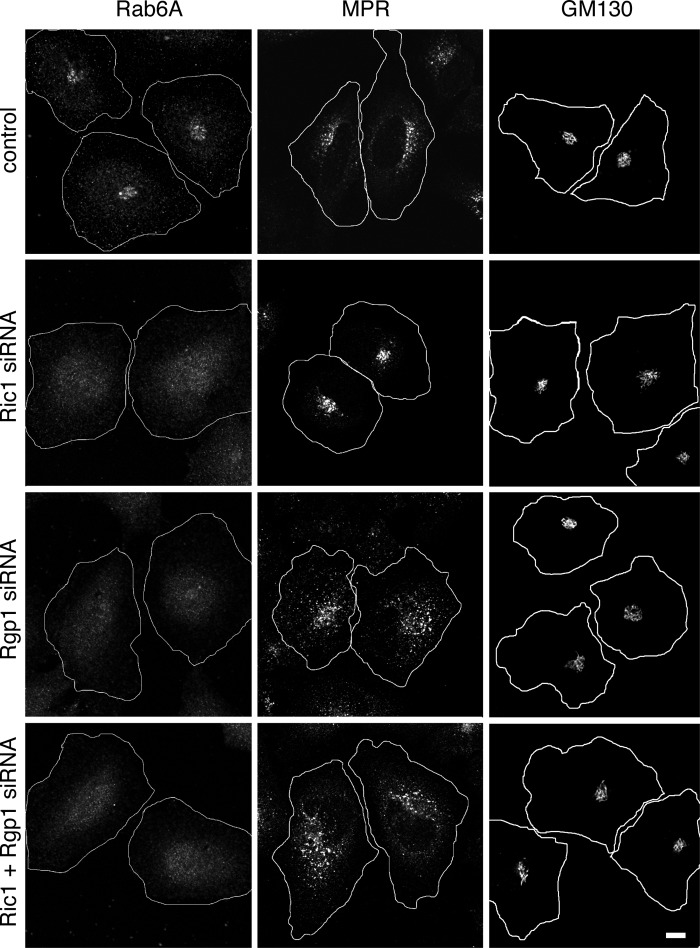
The Rab6A protein level decrease seen by immunoblotting of siRNA-treated cell extracts was also detected by immunofluorescence microscopy; perinuclear Golgi staining of Rab6A was lost upon depletion of Ric1, Rgp1, or Ric1 and Rgp1 proteins (Fig. 7, left column). Under these conditions, the structure of the Golgi, as monitored by the localization of the protein GM130, was essentially normal (Fig. 7, right column). Transport of mannose 6-phosphate receptors from late endosomes back to the Golgi complex relies upon Rab6 to localize the tethering factor, GCC185, to the trans Golgi network (13). For this reason, loss of Rab6 would be predicted to interfere with mannose 6-phosphate receptor trafficking. Indeed, depletion of Rgp1 or both Rgp1 and Ric1 led to the accumulation of mannose 6-phosphate receptors in peripheral structures (Fig. 7, middle column), some of which represent transport intermediates (21). Depletion of Ric1 alone did not show the dispersal of mannose 6-phosphate receptors seen upon depletion of Rgp1; this is most likely due to the decreased ability of the Ric1 siRNA to decrease Rab6A levels in cells relative to Rgp1 siRNA (Fig. 6). Nevertheless, these data confirm the importance of Ric1-Rgp1 on Rab6A stability and localization to the Golgi complex, where it can recruit factors needed for retrograde trafficking steps.
FIGURE 7.
Effect of Rgp1 and Ric1 depletion on Rab6A at the Golgi, cation-independent MPR transport, and Golgi structure (GM130). HeLa cells were transfected with control or Rgp1 or Ric1 or both siRNAs for 72 h and processed for immunofluorescence microscopy. Shown are confocal micrographs of a single Z-stack of endogenous Rab6A (left column), CI-MPR (middle column), and GM130 (right column). Bar, 10 μm.
Rab6A Connects to Rab33B in a Rab Cascade
Rab GTPase cascades (3, 5–7) can template the formation of an ordered series of compartments in the secretory and endocytic pathways and have been proposed to underlie the creation of the Golgi complex (8). Thus, it is important to explore which Rab GTPases may be key to establishment of the Golgi and how they are ordered in a possible cascade. Rab33B is the only Rab that has been localized specifically to the medial Golgi, and this protein is a candidate to direct the localization of the subsequent acting Rab6A GTPase. We were delighted to discover that Rab33B binds to Ric1 and Rgp1 proteins after expression in mammalian cells (Fig. 8). For these experiments, GFP-Rab33B was co-expressed in cells with Ric1 and Rgp1 proteins; binding was monitored by collecting the GFP-Rab33B using immobilized GFP-binding protein. As shown in Fig. 8A, co-expressed FLAG-Rgp1 and Myc-Ric1 could be retained by immobilized GFP-Rab33B but not GFP-Rab4A after expression in mammalian cells.
To ensure that binding was direct, we tested whether purified, recombinant His-Ric1 residues 1032–1344 could also bind Rab33B. As shown in Fig. 8B, this Ric1 C-terminal fragment showed significant binding to GST-Rab33B but not GST-Rab9A. Moreover, binding was stronger to Rab33B-GTP than to Rab33B-GDP (Fig. 8, B and C), demonstrating that Ric1 is an effector of Rab33B protein. Importantly, the fragment that bound to Rab33-GTP failed to bind to Rab6A-GDP or GTP, demonstrating that Rab33B interaction occurs via a distinct site on Ric1 protein (Fig. 2A).
Rab33B preloaded with GTPγS did not influence the ability of Ric1-Rgp1 to catalyze nucleotide exchange on Rab6A protein (Fig. 8D). In addition, Rab33B-GTP did not alter the binding of Ric1-720–1344 to Rab6A-GDP (Fig. 8E). These experiments suggest that Rab33B binding may be used for localization rather than activation of the Ric1-Rgp1 GEF complex.
DISCUSSION
We have shown here that human Ric1 and Rgp1 proteins can associate with one another and together, comprise a GEF for Rab6A. The yeast orthologs of these proteins comprise a GEF for the Rab6A homolog, Ypt6p (14). In support of Ric1-Rgp1 functioning as a Rab6A GEF in cells, depletion of the proteins destabilized Rab6A and interfered with the retrograde transport of mannose 6-phosphate receptors. In addition, we have shown that Ric1 and Rgp1 can each bind Rab6A-GDP independently; at least Ric1 can also bind Rab33B-GTP directly, to form a possible Rab cascade at the Golgi. Binding of Rab33B did not compete with binding for Rab6A, consistent with the proteins occupying a distinct binding site on Ric1 protein. Thus, we have identified four distinct domains in Ric1: an N-terminal, putative β propeller followed by independent binding sites for Rgp1, Rab6A-GDP, and Rab33B-GTP (Fig. 2A, top).
We were surprised to find that unlike the yeast orthologs, most Ric1 and Rgp1 were not Golgi-associated, and both were found predominantly in the cytosol. Moreover, the majority of the polypeptides did not form a stoichiometric complex, and much of Ric1 chromatographed with an apparent Mr of greater than 700,000. That the proteins might carry out diverse and possibly independent functions is implied from their gene expression in various human tissues. According to the BioGSP expression profile, Ric1 is uniformly expressed across all tissues and cell types tested, whereas Rgp1 is much more highly expressed in CD56+ natural killer cells, dendritic cells, and CD19+ B cells, and to a lesser extent in the thyroid, colon, and prostate. This suggests that Ric1 and Rgp1 likely play distinct roles in these cell and tissue types (see also Ref. 22).
A connection between Rab33B and Rab6 has been seen previously. Storrie and colleagues (23) reported that Rab33B overexpression leads to loss of Rab6 from the Golgi. It is possible that overexpressed Rab33B sequesters Ric1-Rgp1 complex so that it is unable to activate Rab6 appropriately. In an attempt to confirm interaction of Ric1-Rgp1 with Rab33B in cells, we co-expressed the proteins and tested whether expression of Rab33B increases the association of Ric1 or Rgp1 with the Golgi complex. Upon co-expression of Rab33B with either Ric1 or Rgp1, we were not able to detect a significant increase in Ric1 or Rgp1 Golgi association. It is possible that binding is sensitive to the liquid nitrogen procedure used to wash out cytosolic proteins prior to fixation for light microscopy. Nevertheless, interaction of the proteins with Rab33B-GTP was confirmed in immunoprecipitation experiments (Fig. 8), and a specific Rab33B binding site was identified in Ric1 protein.
Although the GEFs for Sec4p (Sec2p), Rab5-Rab21 (Rabex5), and Rab35 (DENND1B) are monomeric, several other Rab GTPase guanine nucleotide exchange factors are multisubunit enzymes (4). For example, the mammalian Rab1 GEF, TRAPP (transport protein particle) complex, contains nine polypeptide subunits, and four of these contribute to GEF activity (24). The Ypt7 GEF is composed of Ccz1p and Mon1p subunits that are both required for activity, and one of the subunits associates with the homotypic fusion and protein sorting (HOPS) tethering complex (25). In the case of the monomeric GEFs, the proteins interact with other polypeptides that catalyze vesicle docking and also contribute to GEF localization. By encoding Rab GEF activity in a multisubunit complex, Ric1 and Rgp1 proteins can harbor additional binding sites for upstream Rab proteins (in this case, Rab33B), and perhaps also vesicle-tethering factors. How Ric1 and Rgp1 engage Rab6 on the Golgi and help Rab6 mediate anterograde and retrograde transport, as well as Golgi structure maintenance, represent important questions for future investigation.
This work was supported, in whole or in part, by National Institutes of Health Grant DK37332 (to S. R. P.).
- GEF
- guanine nucleotide exchange factor
- MPR
- mannose 6-phosphate receptor
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Hutagalung A. H., Novick P. J. (2011) Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 3. Mizuno-Yamasaki E., Rivera-Molina F., Novick P. (2012) GTPase networks in membrane traffic. Annu. Rev. Biochem. 81, 637–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ortiz D., Medkova M., Walch-Solimena C., Novick P. (2002) Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 157, 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rivera-Molina F. E., Novick P. J. (2009) A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 14408–14413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 8. Pfeffer S. R. (2010) How the Golgi works: a cisternal progenitor model. Proc. Natl. Acad. Sci. U.S.A. 107, 19614–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barr F. A. (2009) Rab GTPase function in Golgi trafficking. Semin. Cell Dev. Biol. 20, 780–783 [DOI] [PubMed] [Google Scholar]
- 10. Storrie B., Micaroni M., Morgan G. P., Jones N., Kamykowski J. A., Wilkins N., Pan T. H., Marsh B. J. (2012) Electron tomography reveals Rab6 is essential to the trafficking of trans-Golgi clathrin and COPI-coated vesicles and the maintenance of Golgi cisternal number. Traffic 13, 727–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grigoriev I., Splinter D., Keijzer N., Wulf P. S., Demmers J., Ohtsuka T., Modesti M., Maly I. V., Grosveld F., Hoogenraad C. C., Akhmanova A. (2007) Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 13, 305–314 [DOI] [PubMed] [Google Scholar]
- 12. Bensen E. S., Yeung B. G., Payne G. S. (2001) Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol. Biol. Cell 12, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burguete A. S., Fenn T. D., Brunger A. T., Pfeffer S. R. (2008) Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 132, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siniossoglou S., Peak-Chew S. Y., Pelham H. R. (2000) Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19, 4885–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nottingham R. M., Ganley I. G., Barr F. A., Lambright D. G., Pfeffer S. R. (2011) RUTBC1: A Rab9 effector that activates GTP hydrolysis by Rab32 and Rab33b proteins. J. Biol. Chem. 286, 33213–33222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aivazian D., Serrano R. L., Pfeffer S. (2006) TIP47 is a key effector for Rab9 Localization. J. Cell Biol. 173, 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Díaz E., Schimmöller F., Pfeffer S. R. (1997) A novel Rab9 effector required for endosome-to-TGN transport. J. Cell Biol. 138, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ganley I. G., Espinosa E., Pfeffer S. R. (2008) A Syntaxin 10-containing SNARE complex distinguishes two distinct transport routes from endosomes to the trans Golgi network. J. Cell Biol. 180, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dirac-Svejstrup A. B., Soldati T., Shapiro A. D., Pfeffer S. R. (1994) Rab-GDI presents functional rab9 to the intracellular transport machinery and contributes selectivity to rab9 membrane recruitment. J. Biol. Chem. 269, 15427–15430 [PubMed] [Google Scholar]
- 20. Field M. C., Sali A., Rout M. P. (2011) Evolution: On a bender–BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 193, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown F. C., Schindelhaim C. H., Pfeffer S. R. (2011) GCC185 plays independent roles in Golgi structure maintenance and AP-1-mediated vesicle tethering. J. Cell Biol. 194, 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akiyama M., Ishida N., Ogawa T., Yogo K., Takeya T. (2005) Molecular cloning and functional analysis of a novel Cx43 partner protein CIP150. Biochem. Biophys. Res. Commun. 335, 1264–1271 [DOI] [PubMed] [Google Scholar]
- 23. Starr T., Sun Y., Wilkins N., Storrie B. (2010) Rab33B and Rab6 are functionally overlapping regulators of Golgi homeostasis and trafficking. Traffic 11, 626–636 [DOI] [PubMed] [Google Scholar]
- 24. Barrowman J., Bhandari D., Reinisch K., Ferro-Novick S. (2010) TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11, 759–763 [DOI] [PubMed] [Google Scholar]
- 25. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]