Background: TPX2 is as an essential protein for mitosis, but its nuclear function is unknown.
Results: TPX2 goes to DNA double strand breaks and regulates γ-H2AX levels upon ionizing radiation.
Conclusion: We discover a novel and the first nuclear function for TPX2.
Significance: Our study provides new insights into the physiological and oncological roles of TPX2.
Keywords: Cell Cycle, Cytoskeleton, DNA Damage Response, MAPs, Mitosis, Neurons, TPX2
Abstract
The microtubule-associated protein targeting protein for Xenopus kinesin-like protein 2 (TPX2) plays a key role in spindle assembly and is required for mitosis in human cells. In interphase, TPX2 is actively imported into the nucleus to prevent its premature activity in microtubule organization. To date, no function has been assigned to nuclear TPX2. We now report that TPX2 plays a role in the cellular response to DNA double strand breaks induced by ionizing radiation. Loss of TPX2 leads to inordinately strong and transient accumulation of ionizing radiation-dependent Ser-139-phosphorylated Histone 2AX (γ-H2AX) at G0 and G1 phases of the cell cycle. This is accompanied by the formation of increased numbers of high intensity γ-H2AX ionizing radiation-induced foci. Conversely, cells overexpressing TPX2 have reduced levels of γ-H2AX after ionizing radiation. Consistent with a role for TPX2 in the DNA damage response, we found that the protein accumulates at DNA double strand breaks and associates with the mediator of DNA damage checkpoint 1 (MDC1) and the ataxia telangiectasia mutated (ATM) kinase, both key regulators of γ-H2AX amplification. Pharmacologic inhibition or depletion of ATM or MDC1, but not of DNA-dependent protein kinase (DNA-PK), antagonizes the γ-H2AX phenotype caused by TPX2 depletion. Importantly, the regulation of γ-H2AX signals by TPX2 is not associated with apoptosis or the mitotic functions of TPX2. In sum, our study identifies a novel and the first nuclear function for TPX2 in the cellular responses to DNA damage.
Introduction
Human TPX2,5 the ortholog of the targeting protein for Xenopus kinesin-like protein 2, was initially named and described as restricted expression/proliferation-associated protein 100 due to its high expression levels in proliferating cells and tissues (1). The current name of TPX2 is based on its function to regulate the Xenopus kinesin-like protein 2, a plus end-directed microtubule motor required for centrosome separation and maintenance of spindle polarity during mitosis (2–8). During cell cycle progression, TPX2 levels gradually increase with highest expression in mitosis and lowest levels in G1 phase (1–6). In mitotic human cells, TPX2 mediates the binding of the human ortholog of Xenopus kinesin-like protein 2 (i.e. kinesin family member 15) to microtubules, activates the mitotic serine-threonine Aurora A kinase, and nucleates microtubules (2–6, 9). By virtue of its microtubule-associated functions, TPX2 is defined as a microtubule-associated protein and a critical factor for spindle assembly and mitosis in mammalian cells (2–6). During interphase, lasting up to 23 h of a 24-h cell cycle (e.g. HeLa cells), TPX2 is actively transported into the nucleus via the importin α/β receptors (10, 11). This nuclear import is thought to sequester TPX2 away from cytoplasmic tubulin to prevent premature spindle assembly (10, 11). However, to date, no function has been associated with nuclear TPX2 despite the fact that TPX2 resides in the nucleus during the majority of the cell cycle.
Intriguingly, elevated levels of TPX2 have been detected in numerous cancers (ovary, lung, pancreas, bone, carcinoma, cervix, etc.), and amplification of the TPX2 gene has been suggested to promote the progression of colorectal malignancies (12–19). Conversely, TPX2 haploinsufficiency, leading to decreased levels of TPX2, significantly increases the propensity for the development of tumors in mice (20). Together, these results suggest that deregulation of TPX2 levels and functions are associated with the etiology of cancers.
Many cancers arise from genomic instability caused by disturbed responses to DNA damage (21, 22). Interestingly, TPX2 has been shown to associate with the breast cancer 1 protein (BRCA1) during cytoskeletal remodeling events (23, 24) and was identified as a potential substrate of the ataxia telangiectasia mutated (ATM) kinase as suggested by a high throughput screen that displayed over 700 other hits (25). BRCA1 and ATM are key factors of the cellular DNA damage response to DNA double strand breaks (26–28). However, a function for TPX2 in the DNA damage response has not been identified.
The DNA damage response consists of a complex network of signaling and repair pathways that maintain genomic integrity (26, 29). Experimentally, the DNA damage response is frequently studied by examining the cellular response to ionizing radiation (26, 29). Ionizing radiation induces multiple forms of DNA damage, the most lethal of which is the DNA double strand break (26, 29). Upon ionizing radiation treatment, the DNA damage response mediates cell cycle arrest (through checkpoints), facilitates DNA repair, promotes cell survival, or triggers apoptosis if the damage is too severe. At the molecular level, the DNA damage sensor protein complex MRN (composed of MRE11, Rad50, and Nijmegen breakage syndrome protein 1 (NBS1)) is recruited to the DNA double strand breaks at early stages of the DNA damage response. NBS1 then recruits the ATM kinase to the DNA double strand break (30–37). Subsequently, the ATM kinase, which becomes activated by the presence of DNA double strand breaks (38), phosphorylates multiple DNA damage response proteins including the chromatin core component Histone 2AX (H2AX) and the mediator of DNA damage checkpoint 1 (MDC1) (30–37). However, phosphorylation of H2AX can also be carried out by the DNA-dependent protein kinase (DNA-PK) (36). Upon phosphorylation at serine 139, p-H2AX (also called γ-H2AX) binds to BRCA1 C-terminal (BRCT) domains in DNA damage response proteins such as MDC1, resulting in the accumulation of MDC1 at DNA double strand break-flanking chromatin regions. MDC1 then recruits more ATM kinase to the sites of DNA lesions, thereby inducing further γ-H2AX formation. Together, this creates a positive feedback loop resulting in the amplification and spreading of H2AX phosphorylation over chromatin regions containing megabases of DNA (39–43). This DNA double strand break-flanking accumulation of γ-H2AX and MDC1 is essential to attract and retain numerous other DNA damage response factors (e.g. RNF8, RNF168, BRCA1, 53BP1, etc.) to the lesion sites (26–29), thereby facilitating the DNA damage response. The accumulation of DNA damage response molecules at DNA double strand breaks manifests as so-called ionizing radiation-induced foci, which are microscopically visualized as bright spots (29).
In this study, we aimed to determine whether alteration in TPX2 levels, as found in numerous cancers exhibiting genomic instability, is associated with an abnormal DNA damage response. Because phosphorylation of H2AX lies at the core of the DNA damage response, we analyzed γ-H2AX levels in cells with altered expression of TPX2. Using four different RNAi approaches, we found that depletion of TPX2 in cells leads to a transient increase in γ-H2AX levels following treatment with ionizing radiation. Conversely, overexpression of TPX2 decreases ionizing radiation-induced MDC1 foci and γ-H2AX levels. Importantly, these phenomena are not correlated with apoptosis and occur independently of the mitotic functions of TPX2. Furthermore, TPX2 accumulates at DNA double strand breaks and associates with the machinery of DNA damage response that controls the amplification of γ-H2AX (i.e. MDC1 and ATM). Our findings identify a novel and the first nuclear function for TPX2.
EXPERIMENTAL PROCEDURES
Western Blots
Protein extracts were isolated for specific purposes as described below. The protein concentration was estimated by the Bradford procedure (Bio-Rad) or Bio-Rad DC assay to ensure equal loading. Proteins were fractionated by SDS-PAGE and blotted on a PVDF membrane for Western blot analysis. Membranes were incubated with antibodies against actin (Chemicon), ATM (Upstate), phosphorylated ATM (p-ATM) (Ser-1981, Rockland, Epitomics), DNA-PK catalytic subunit (DNA-PKcs) (18-2, Abcam), GAPDH (HRP-conjugated antibodies) (Abcam), GFP (Santa Cruz Biotechnology), FLAG (Sigma), H2A (H-124, Santa Cruz Biotechnology), H2AX (Abcam), γ-H2AX (Ser-139; JBW301, Millipore), p-H3 (Ser-10; Millipore), H3 (Abcam), cleaved PARP1 (Millipore), NBS1 (1D7, Abcam), nuclear distribution E homolog-like 1 (Ndel1) (home-made; Ref. 44), MDC1 (Abcam), and TPX2 (184, Novus Biologicals). Quantification of band signals was performed using Quantity One software from Bio-Rad, and signals were corrected with levels of actin, GAPDH, or the non-phosphorylated form of the protein of interest. For the statistical analyses, please refer to individual figure legends.
Co-immunoprecipitations
Total protein extracts of cells were obtained by lysis of cells in ice-cold radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10 mm KCl, 1 mm EDTA, 0.5% Tween 20, 0.5% Nonidet P-40, 1 mm PMSF, 5 mm sodium fluoride, 1 mm sodium orthovanadate, and 1× protease inhibitor mixture Complete Mini, EDTA-free (Roche Applied Science)). Extracts were sonicated twice for 5 s with a Sonic Dismembrator (Model 100) at Level 4, and the protein concentration was determined using the Bio-Rad Protein (Bradford) assay. Co-immunoprecipitations and nuclear fractionation were performed using standard laboratory procedures. Antibodies used for co-immunoprecipitations were MDC1 (Abcam) and TPX2 (184 from Novus Biologicals, TPX2 serum, and KiS2). Commercially available TPX2 antibody 184 is specific for the C terminus (immunogen, aa 700–749) of TPX2 (specificity was further confirmed by peptide competition (see supplemental Fig. 5) and by depletion of TPX2 by RNAi (see Figs. 1, 4, and 5)). TPX2 serum (Ref. 45; a kind gift from Dr. D. Compton) was obtained from rabbits immunized with recombinant full-length TPX2. The monoclonal TPX2 KiS2 antibody was a kind gift from Dr. Heidebrecht (1).
FIGURE 1.
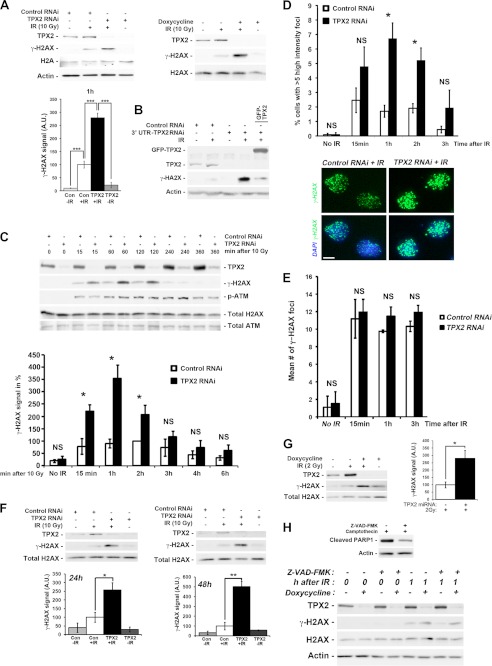
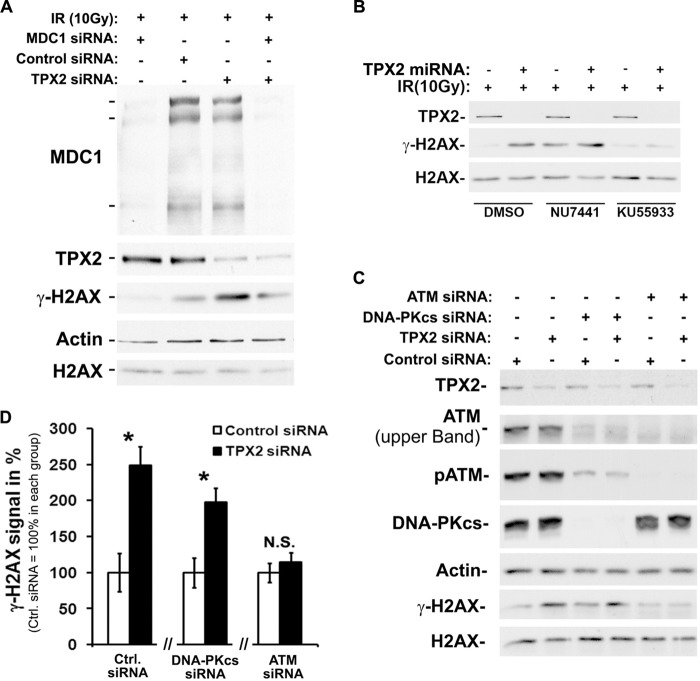
Increased ionizing radiation-dependent phosphorylation of H2AX at DNA double strand breaks upon depletion of TPX2. A, transient transfection of TPX2 siRNA or doxycycline-induced expression of an exogenous miRNA specific for TPX2 significantly increases phosphorylation of H2AX in HeLa cells 1 h after 10 Gy as indicated by Western blot analysis: control siRNA +IR (100.0 ± 13.2) versus TPX2 siRNA +IR (279.0 ± 17.6), p < 0.001, n = 4; −doxycycline +IR (100.0 ± 20.9) versus +doxycycline +IR (871.5 ± 23.4), p < 0.001, n = 4; group (mean ± S.E.), unpaired t test. B, GFP-TPX2 expression reduces the increased levels of γ-H2AX caused by depletion of TPX2 with an siRNA targeting the 3′-untranslated region of TPX2 mRNA. C, time course analysis of H2AX phosphorylation in HeLa cells depleted of TPX2 by siRNA (n = 3–5): 15 min, control siRNA (78.6 ± 31.7) versus TPX2 siRNA (221.3 ± 25.9), p < 0.05; 1 h, control siRNA (90.4 ± 18.5) versus TPX2 siRNA (354.1 ± 54.8), p < 0.05; 2 h, control siRNA (100.0 ± 0.0) versus TPX2 siRNA (206.9 ± 38.4), p < 0.05; group (mean ± S.E.), unpaired t test. Note the similar ATM activation in irradiated control and TPX2-depleted cells as indicated by the levels of p-ATM (Ser-1981). D, increase in the number of U2OS cells with more than five high intensity γ-H2AX ionizing radiation-induced foci (i.e. intensity two standard deviations above the mean focus intensity) following TPX2 depletion by siRNA, 1 and 2 h after 4 Gy: 1 h, control siRNA (1.7 ± 0.4%) versus TPX2 siRNA (6.7 ± 1.1%), p < 0.05, n = 3; 2 h, control siRNA (1.9 ± 0.3%) versus TPX2 siRNA (5.2 ± 0.9%), p < 0.05, n = 3; group (mean ± S.E.), unpaired t test. The number of cells with these high intensity ionizing radiation-induced foci declines 3 h post-IR. Representative pictures of γ-H2AX ionizing radiation-induced foci in control RNAi- and TPX2 RNAi-treated cells 2 h after 4 Gy are shown. E, TPX2 depletion in U2OS cells by siRNA does not cause a significant change in the mean number of γ-H2AX ionizing radiation-induced foci after 4 Gy (n = 3). NS, non-significant, unpaired t test, S.E. F, increased phosphorylation of H2AX 1 h after 10 Gy in caspase-3-deficient MCF-7 cells depleted of TPX2 for 24 or 48 h by siRNA (n = 3) as detected by Western blots. MCF-7 cells do not undergo ionizing radiation-induced apoptosis associated with DNA fragmentation (54, 55). G, increased phosphorylation of H2AX in TPX2-depleted HeLa cells 2 h after a non-lethal dose of 2 Gy (n = 4). H, increased γ-H2AX levels in TPX2-depleted cells 1 h after ionizing radiation are unaffected by the broad spectrum caspase inhibitor Z-VAD-fmk. Z-VAD-fmk was applied at a known effective dose (51) and inhibited PARP1 cleavage (a marker for apoptosis) in the presence of the DNA-damaging drug camptothecin (top blots). Relative quantifications of γ-H2AX signals from independent experiments are shown in bar charts (A, C, F, and G). n = number of independent experiments; NS, non-significant; *, p < 0.05, **, p < 0.01; ***, p < 0.001. Error bars represent S.E. Bar in D, 10 μm. A.U., arbitrary units; Con, control.
FIGURE 4.
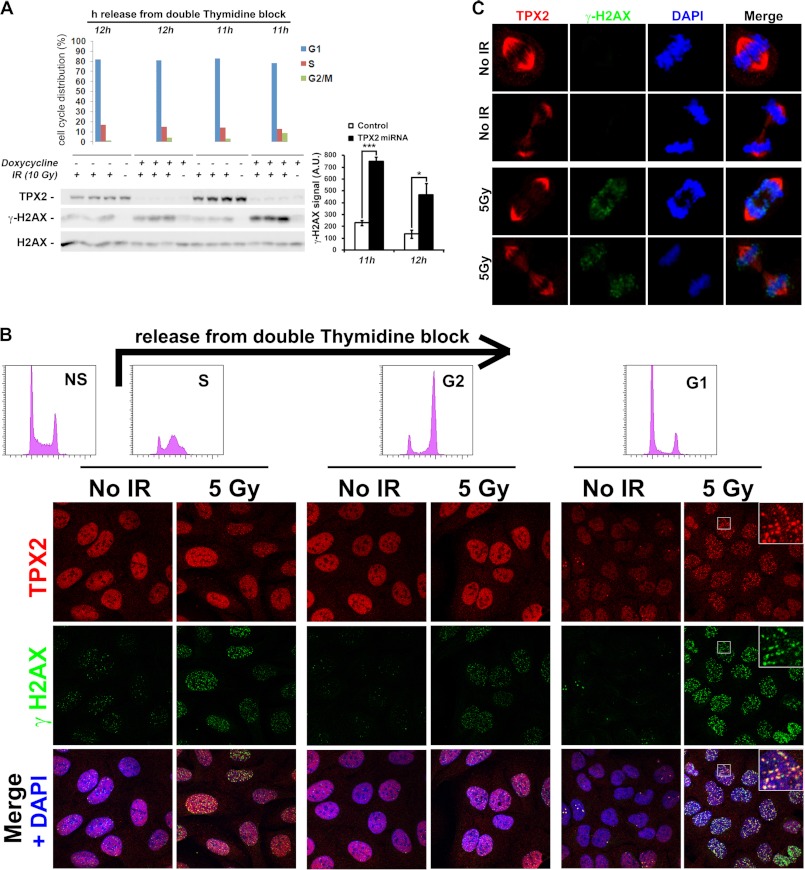
TPX2 regulates the levels of ionizing radiation-dependent γ-H2AX and forms ionizing radiation-induced foci in G1 phase cells. A, enhanced levels of γ-H2AX 1 h after 10 Gy in G1 phase HeLa cells depleted of TPX2 by doxycycline-induced TPX2 miRNA expression. Note that the γ-H2AX augmentation in these TPX2-depleted cells is ionizing radiation-dependent. Cells were synchronized using a double thymidine block, released into fresh medium, and then used at specified time points for ionizing radiation treatment as indicated. Unirradiated cells were used for flow cytometry-based cell cycle profiling (top bar charts). Relative quantifications of γ-H2AX signals from independent experiments are shown in bar charts on the right: 11 h, −doxycycline +IR (229.0 ± 21.0) versus +doxycycline +IR (748.6 ± 39.4), p < 0.001; 12 h, −doxycycline +IR (135.1 ± 34.2) versus +doxycycline +IR (466.1 ± 98.6), p < 0.05; group (mean ± S.E.), unpaired t test; n = 3 (independent experiments); *, p < 0.05; ***, p < 0.001. Error bars represent S.E. Note that 11 h after release from the thymidine block TPX2-depleted cultures contain slightly more G2/M phase cells (8.66%) than control cultures (3.25%). Thus, on the Western blot, a 12 h-11 h loading hierarchy is chosen to facilitate comparison between control 11 h versus +doxycycline 12 h (3.96% G2/M cells). Although the cell cycle profile between these two samples is highly similar, TPX2-depleted cells exhibit approximately twice the levels of γ-H2AX than control cells. B, U2OS cell cultures synchronized with a double thymidine block as in A (see flow cytometry-based cell cycle profiles in top histograms; NS, non-synchronized control) form TPX2 ionizing radiation-induced foci 1 h after irradiation that partially co-localize with γ-H2AX during G1 phase. All images were taken under identical experimental and microscopic conditions. See text for details. C, TPX2 maintains its association with the mitotic spindle in the presence of DNA double strand breaks marked by γ-H2AX. Early and late mitotic figures as identified via DAPI and TPX2 staining with and without DNA damage are shown. Commercially available TPX2 antibody 184 was used in all immunofluorescence images. See supplemental Fig. 5 and Fig. 5 for specificity of TPX2 184 antibody. A.U., arbitrary units.
FIGURE 5.
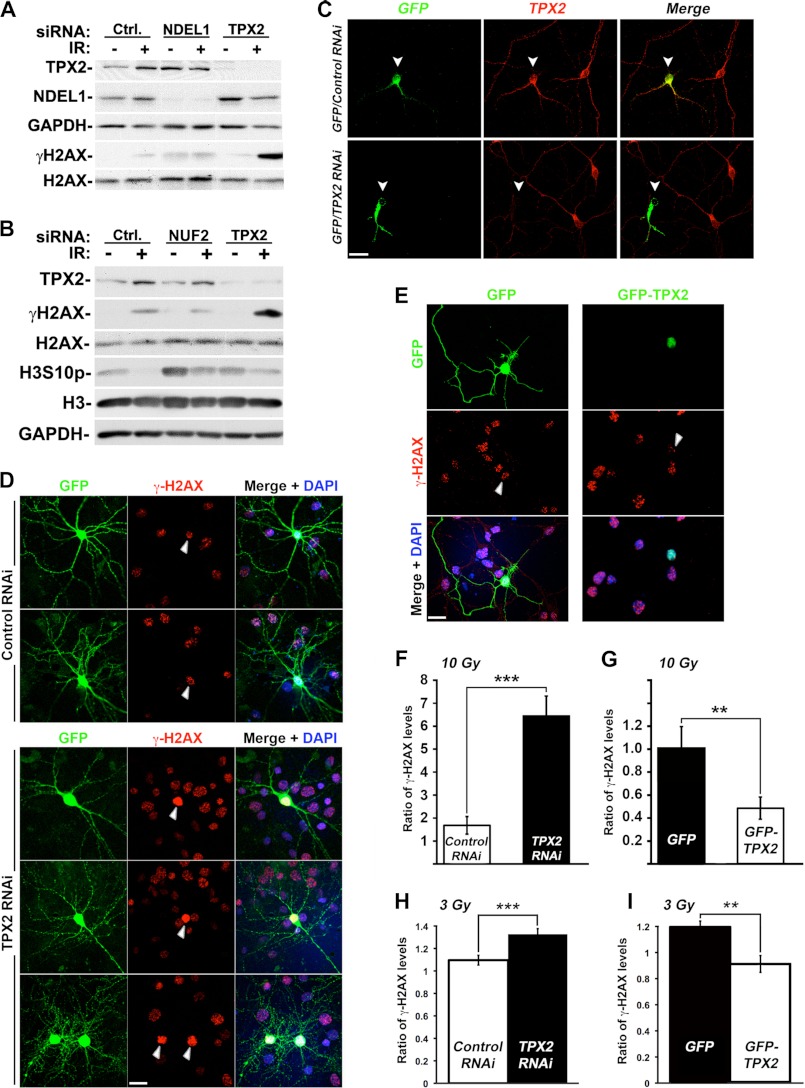
Regulation of γ-H2AX levels by TPX2 is distinct from its mitotic functions and occurs in postmitotic G0 primary neurons. A, depletion of Ndel1 does not increase γ-H2AX levels after ionizing radiation treatment. B, depletion of NUF2 does not increase γ-H2AX levels after ionizing radiation treatment. The mitotic arrest upon NUF2 depletion was confirmed with the increased levels of the mitotic marker H3S10p (see text for details). C, significant decrease in TPX2 levels in mouse primary cortical neurons co-transfected with a TPX2 shRNA-encoding construct and GFP (ratio, 5:1) as detected by immunofluorescence and confocal microscopy using commercially available TPX2 antibody 184. See supplemental Fig. 5 for the expression pattern of TPX2 in brain tissues, cellular distribution of TPX2 in primary neurons, and specificity of the TPX2 shRNA and antibody 184. D, enhanced levels of γ-H2AX in G0 postmitotic primary neurons co-transfected with vectors encoding TPX2 shRNA and GFP (ratio, 10:1) 1 h after 10 Gy compared with surrounding untransfected cells or cells co-transfected with vectors encoding a control shRNA and GFP (ratio, 10:1) as determined by immunofluorescence and confocal microscopy. E, decreased levels of γ-H2AX in G0 postmitotic primary neurons transfected with GFP-TPX2 compared with surrounding untransfected cells or cells transfected with GFP 1 h after 10 Gy as determined by immunofluorescence and confocal microscopy. Note that the γ-H2AX signals in control shRNA- and GFP-expressing neurons are of an intensity similar to that of untransfected surrounding cells. F and G, quantification of the relative changes in γ-H2AX signals in neurons in D and E expressed by the average ratio of total nuclear γ-H2AX signals of transfected neurons/average total nuclear γ-H2AX signals of non-transfected surrounding cells. F, ratio control shRNA (n = 10)/non-transfected (n = 196) = 1.7 ± 0.4 (S.E.) versus ratio TPX2 shRNA (n = 16)/non-transfected (n = 459) = 6.4 ± 0.9 (S.E.); p < 0.001, unpaired t test. G, ratio GFP (n = 9)/non-transfected (n = 37) = 1.0 ± 0.1 (S.E.) versus ratio GFP-TPX2 (n = 8)/non-transfected (n = 33) = 0.5 ± 0.1 (S.E.); p < 0.001, unpaired t test. H and I, quantification of the relative changes in γ-H2AX immunofluorescence signals in neurons co-transfected with vectors encoding a control or TPX2 shRNA and GFP (ratio, 10:1) (H) or a GFP or GFP-TPX2 construct (I) 1 h after 3 Gy. H, ratio control shRNA (n = 58)/non-transfected (n = 300) = 1.1 ± 0.1 (S.E.) versus ratio TPX2 shRNA (n = 75)/non-transfected (n = 330) = 1.3 ± 0.1 (S.E.); p < 0.001, unpaired t test. I, ratio GFP (n = 50)/non-transfected (n = 297) = 1.2 ± 0.1 (S.E.) versus ratio GFP-TPX2 (n = 47)/non-transfected (n = 420) = 0.9 ± 0.1 (S.E.); p < 0.01, unpaired t test. Values were calculated as in F and G. NS, non-significant; **, p < 0.01; ***, p < 0.001. Error bars represent S.E. Bars, 20 μm. Ctrl., control. White arrows indicate transfected cells (C–E).
Glutathione S-Transferase (GST)-TPX2 Pulldown Assays
GST-TPX2 fusion proteins were produced by cloning of TPX2 into pGEX-6.1p, expression in Escherichia coli, and purification on glutathione-Sepharose beads using standard procedures. To couple proteins to beads, 5 μg of purified GST-TPX2 or GST were incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4 °C in buffer containing 160 mm NaCl2, 16 mm Na2HPO4, 4 mm NaH2PO4, and 2 mm DTT plus 0.2 mm PMSF, 0.2 μg/ml leupeptin, and 0.2 μg/ml pepstatin. The beads were then washed with a buffer containing 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 1% (v/v) Nonidet P-40 plus 0.2 mm PMSF, 0.2 μg/ml leupeptin, 0.2 μg/ml pepstatin, 0.2 μg/ml aprotinin, and 1 μm microcystin-LR. HeLa cells were lysed in the same buffer, extracts were diluted to contain 0.25% Nonidet P-40, and 2 mg of extract were incubated with glutathione beads to which either GST-TPX2 or GST protein had been coupled. Extracts were incubated for 2 h at 4 °C and washed. Samples were analyzed by Western blot.
Chromatin Fractionation for Analysis of γ-H2AX Levels
Cells were washed twice in PBS (37 °C) and lysed in ice-cold NETN buffer (150 mm NaCl, 1 mm EDTA, 50 mm Tris-Cl, pH 7,4, 1% Nonidet P-40, and 1× protease inhibitor mixture Complete Mini, EDTA-free (Roche Applied Science)). Extracts were sonicated once for 5 s with a Sonic Dismembrator (Model 100) at Level 4, and the insoluble chromatin fraction was pelleted for 20 min at 4 °C at full speed in a table top centrifuge. The soluble NETN fraction (containing the nuclear lamins but no histones) was kept at −80 °C for further analysis, and the insoluble chromatin fraction (containing the histones but not nuclear lamins) was washed once in 1 ml of NETN buffer. The chromatin fraction was solubilized by addition of 1% SDS in PBS followed by one freeze/thaw cycle at −80 °C, incubation at 95 °C for 15 min, and sonication for 15 s. Protein concentrations were determined using the Bio-Rad DC protein assay (chromatin fraction) or Bio-Rad Protein (Bradford) assay (NETN-soluble fraction).
Ionizing Radiation
Ionizing radiation-induced DNA damage was generated using a source of 137Cs from an MDS Nordion Gammacell 1000 (for all figures except Fig. 1, D and E) or a GSR D1 from Gamma-Service Medical GmbH (for Fig. 1, D and E).
AsiSI-ER Cells
U2OS cells containing a stably integrated AsiSI-ER construct (a kind gift from Dr. Gaëlle Légube) were grown in selective medium (1 μg/ml puromycin). To induce nuclear expression of AsiSI-ER and generate site-specific DNA double strand breaks, cells were treated with 300 nm 4-hydroxytamoxifen for 4 h.
Laser Microirradiation
Laser microirradiation was carried out on a Leica SP5 confocal microscope equipped with an environmental chamber set to 37 °C and 5% CO2. U2OS cells were grown on glass coverslips. DNA double strand break-containing tracks (1.5 × n μm) were generated with a Mira mode-locked titanium-sapphire laser (λ = 800 nm; pulse length, 200 fs; repetition rate, 76 MHz; output power on the cells for DNA damage induction, 80 milliwatts). Cells were irradiated at 1-min time intervals and after 10 min immunostained with TPX2 (184, Novus Biologicals) and γH2AX (Millipore) antibodies.
Generation of Constructs and RNAi Sequences
To generate the bait-TPX2 vector (full length or aa 8–747) for the yeast two-hybrid (Y2H) assays, human TPX2 cDNA (bp 1–2241 or 22–2241) of pQE-70-TPX2 (4) was cloned into pGBKT7 (Clontech). The pGADT7-MDC1 was generated by cloning human MDC1 cDNA (bp 5049 of coding sequence to end of 3′-UTR) of pACT2-MDC1 (from Y2H screen) into pGADT7 (Clontech). Human TPX2 cDNA (bp 4–2241 or 3–2241) was cloned into pcDNA4/HISMAX (Invitrogen) or pEGFP-C1 (Clontech) to generate the His-TPX2 or GFP-TPX2 vector. RNAi sequences were selected based on the criteria of Ambion. Complementary short hairpin RNA (shRNA) sequences were commercially synthesized and cloned into pSilencer 2.0 under promoter U6 (Ambion). The sequence for the mouse TPX2 shRNA is AACACTTACCACAAAGAGACA. Sequences for the human TPX2 small interfering RNA (siRNA), TPX2 microRNA (miRNA), Ndel1 siRNA, NUF2 siRNA, and MDC1 siRNA have been described previously (Refs. 4, 17, 39, 46, and 47; see s37983 from Ambion for NUF2 siRNA). The specific ATM and DNA-PKcs siRNAs were purchased from Dharmacon. The TPX2 3′-UTR siRNA was purchased from Qiagen. A random sequence without homology to any known mRNA was used for control RNAi (44, 48–50). RNAi constructs were tested in CAD cells, primary neuronal cultures, U2OS cells, and HeLa cells by both Western blots and immunofluorescence staining.
Cell Cultures, Transfection, Induction, and Pharmacological Treatments
HeLa (ATCC), HeLa EM2-11-TPX2 (17), MCF-7 (ATCC), and U2OS cells (ATCC) were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. LAN1 cells (a kind gift from Dr. Ursula Valentiner) were cultured in RPMI 1640 medium containing l-glutamine, 10% FBS, and 1% penicillin-streptomycin. Primary cortical and hippocampal cultured postmitotic neurons were prepared from mice as described (44, 49, 50). The neurons were maintained in basal medium Eagle supplemented with 6 mg/ml glucose, 2 mm glutamine, 10 mm HEPES, 1 mm sodium pyruvate, 2% FBS, 2% penicillin-streptomycin, and 2% B-27. Transfection of cell lines with constructs or oligonucleotides was performed using LipofectamineTM 2000 reagent (Invitrogen), HiPerFect transfection reagent (Qiagen), or DharmaFECT 1 (Dharmacon) in accordance with the suppliers' recommendations. Primary neurons were either transfected with Lipofectamine 2000 or electroporated as described (44, 49). Expression of TPX2-targeting miRNA in HeLa EM2-11-TPX2 was induced by doxycycline (1 μg/ml of medium). Of note, although the doxycycline-inducible TPX2 RNAi originates from an exogenous miRNA, the mechanism of TPX2 knockdown is siRNA-based (17). Elements of miRNA precursors such as RNA polymerase II promotor, drosha processing, and Exportin 5 recognition sequences are used in this system to produce an siRNA double strand that is identical (apart from stabilizing modifications added to synthetic stealth siRNAs) to a synthetic siRNA duplex administered by transfection. However, the amount of siRNA that originates from this exogenous doxycycline-inducible miRNA is lower than what is delivered to the cell by regular transfection of synthetic siRNAs and thus maintains the cell in a more physiological context.
Target proteins were knocked down for 24 or 48 h (TPX2), 72 h (MDC1), and 96 h (ATM/DNA-PKcs) before sample processing. For the TPX2/MDC1 double knockdown, HeLa cells were transfected with MDC1 (39) or AllStars negative control (Qiagen) siRNA (10 nm) 24 h postseeding. 48 h after the initial MDC1/control knockdown, cells were transfected again with combined TPX2-control, MDC1-control, TPX2-MDC1, or control RNAi mixtures (20 nm each) and cultured for 24 h before harvesting. For the TPX2/ATM or TPX2/DNA-PKcs double knockdown, HeLa cells were transfected with ATM, DNA-PKcs, or AllStars negative control (Qiagen) siRNA (10 nm) 24 h postseeding. 72 h after the initial ATM, DNA-PKcs, or control knockdown, cells were transfected again with TPX2 or control siRNA (20 nm) and cultured for 24 h before harvesting. For pharmacological inhibition of ATM and DNA-PK, KU55933 and NU7441 (Tocris Bioscience) were dissolved in DMSO and administered to cells 45 min prior to ionizing radiation at concentrations of 8 and 5 μm, respectively. For pharmacological inhibition of caspases, Z-VAD-fmk (Santa Cruz Biotechnology; dissolved in DMSO) was administered to cells prior to ionizing radiation at a concentration of 10 μm (51).
Immunofluorescence Staining and Microscopy Analysis
Cells were fixed, permeabilized, and blocked as described (44, 48–50). Cells were stained with antibodies against γ-H2AX (Ser-139; JBW301, Millipore), TPX2 (184, Novus Biologicals), or MDC1 (Abcam). Nuclei were stained with DAPI. Images were acquired using a Nikon D-Eclipse C1 confocal microscope. Total signal intensity of γ-H2AX in primary neurons was analyzed using ImageJ software. For analysis of total number and intensity of individual γ-H2AX ionizing radiation-induced foci in U2OS cells, images were acquired using an IN Cell Analyzer 1000 (GE Healthcare) and analyzed with IN Cell Analyzer 1000 Workstation software using the Multi Target Analysis module (GE Healthcare). γ-H2AX foci with intensity above a set threshold (two standard deviations above mean focus intensity) were counted as high intensity foci.
Cell Cycle Synchronization
For G1, S, and G2 phases, HeLa EM2-11-TPX2 cells or U2OS cells were seeded 24 h before treatment with 2 mm thymidine for 20 h. Cells were then trypsinized and released in thymidine-free medium for 15 h followed by treatment with 2 mm thymidine for an additional 14 h. Early S phase-arrested cells were released into fresh medium and monitored throughout the following synchronous cell cycle via flow cytometry-based cell cycle profiling. Samples were taken at indicated cell cycle stages and used for ionizing radiation treatment and Western blot analysis or ethanol fixation, propidium iodide staining, and flow cytometry-based cell cycle profiling.
For M phase, HeLa cells were treated with 100 ng/ml nocodazole (Sigma) for 16–17 h to arrest cells at the M phase. The M phase-synchronized cells were irradiated with 10 grays (Gy) and harvested for Western blot analysis or ethanol fixation, propidium iodide staining, and flow cytometry-based cell cycle profiling.
Yeast Two-hybrid Screen and Assay
The Y2H screen in the Saccharomyces cerevisiae strain AH109 (Clontech) was carried out with the pGBKT7-TPX2 (aa 8–747) bait vector and a human fetal brain Matchmaker cDNA library cloned in pACT2 plasmids (Clontech). Sequential transformation of the bait and cDNA library was performed as described (52, 53), and Y2H experiments used combined ADE2/HIS3 high stringency reporters. The putative TPX2-interacting proteins were identified via isolation of pATC2 plasmids from colonies growing on high stringency selection medium and sequencing. For the independent Y2H assay, the AH109 yeast strain was co-transformed with pGBKT7-TPX2 (aa 1–747 or 8–747) and pGADT7-MDC1 (aa 1683 to stop codon or aa 1809 to stop codon) and processed as described above.
RESULTS
TPX2 Regulates the Levels of γ-H2AX upon Treatment with Ionizing Radiation
TPX2 associates with DNA damage response proteins such as BRCA1 and may be a target of the ATM kinase (23–25). Moreover, TPX2 levels are altered in cancers associated with aberrant cellular responses to DNA damage and genomic instability (12–19). These observations raise the question as to whether TPX2 is involved in the DNA damage response. To test this hypothesis, we analyzed the levels of γ-H2AX, a key marker of DNA damage response amplification, in HeLa cells depleted of TPX2 by siRNA and in a HeLa stable cell line expressing a doxycycline-inducible exogenous miRNA targeting specifically TPX2 mRNA (see “Experimental Procedures” for further details on this cell line and Ref. 17) following ionizing radiation. The latter system has the advantage of knocking down TPX2 in an entire population of cells. Specificities of the TPX2 siRNA and miRNA have been demonstrated previously (4, 17) and were confirmed by Western blot analysis (Fig. 1). In the absence of ionizing radiation, loss of TPX2 did not trigger an increase in γ-H2AX signal (Fig. 1, A–C, F, and H) nor did it result in higher amounts of DNA double strand breaks as detected by the neutral comet assay (supplemental Fig. 1). Interestingly, we found that a 60–95% knockdown of TPX2 significantly enhanced the levels of γ-H2AX by ∼3–9-fold, respectively, 1 h after 10 Gy when compared with irradiated control cells (Fig. 1A). Both approaches targeting different sequences of TPX2 mRNAs generated the same effect. To further support the specificity of the observed phenotype, we performed rescue experiments by expressing a GFP-TPX2 construct in HeLa cells depleted of endogenous TPX2 with a third siRNA sequence targeting the 3′-untranslated region of TPX2 mRNA. In these cells, endogenous TPX2 is depleted, but GFP-TPX2 is still expressed (Fig. 1B). Although irradiated TPX2-depleted cells exhibit increased γ-H2AX, expression of GFP-TPX2 reduces such increase (Fig. 1B) (see Fig. 5 and supplemental Fig. 5 for similar results with an additional fourth RNAi sequence). Taken together, these results provide compelling evidence that TPX2 impacts γ-H2AX levels during the DNA damage response.
To further understand the correlation between TPX2 depletion and increased γ-H2AX signals, we performed a time course analysis of ionizing radiation-triggered H2AX phosphorylation over a 6-h period. We found that both TPX2-depeleted cells and control cells exhibited similar time courses for γ-H2AX formation: both groups showed 10 Gy-triggered phosphorylation of H2AX 15 min to 2 h after ionizing radiation. Phosphorylation of H2AX started to decrease 3 h after ionizing radiation and was almost reduced to background levels 4 h post-ionizing radiation (Fig. 1C). However, knockdown of TPX2 caused dramatic increases in γ-H2AX levels at 15 min to 2 h after ionizing radiation when compared with control cells (Fig. 1C). Of note, TPX2 becomes enriched in the chromatin fraction during the course of the DNA damage response (shown in Fig. 1C; see “Experimental Procedures”).
We next analyzed γ-H2AX ionizing radiation-induced foci in TPX2-depleted U2OS cells and control cells using immunofluorescence microscopy. Ionizing radiation-induced foci were analyzed for their intensity and mean number per cell at different time points (15 min and 1, 2, and 3 h) after a non-lethal dose of 4 Gy. Note that a higher dose of 10 Gy cannot be used for this type of experiment as it interferes with the resolution of single γ-H2AX foci, thereby impeding the interpretation of the results. We found that 1 and 2 h after 4 Gy, the percentage of cells with high intensity γ-H2AX ionizing radiation-induced foci (i.e. focus intensity 2 times the standard deviation above the mean focus intensity) was markedly increased in TPX2-depleted populations compared with control cultures (Fig. 1D). 3 h post-ionizing radiation, the percentage of TPX2-depleted cells with high intensity γ-H2AX ionizing radiation-induced foci decreased to the level of control cells (Fig. 1D). These results are consistent with our Western blot time course data of γ-H2AX signals at these time points (Fig. 1C). However, we did not observe significant differences in the number of γ-H2AX ionizing radiation-induced foci between control and TPX2-depleted cells (Fig. 1E). This result suggests that the number of DNA double strand breaks is similar between the two groups and that the increased γ-H2AX signal in irradiated TPX2-depleted cells is not associated with apoptotic DNA laddering/fragmentation (which can also induce γ-H2AX but would lead to increased numbers of γ-H2AX foci). To further support the notion that the TPX2 depletion-dependent increase in ionizing radiation-triggered γ-H2AX is not linked to apoptosis, we used caspase-3-deficient MCF-7 breast cancer cells that do not undergo ionizing radiation-induced DNA fragmentation and apoptosis (54, 55). These cells still display the ionizing radiation-dependent increase in γ-H2AX following TPX2 depletion by siRNA for 24 or 48 h (Fig. 1F). Furthermore, several cell types depleted of TPX2 (i.e. HeLa and U2OS cells) that were treated with non-lethal doses of ionizing radiation (2 and 4 Gy; see Fig. 1, D, E, and G) still display up-regulated γ-H2AX signals, although they recover from these ionizing radiation doses and continue their cell cycle (supplemental Fig. 2). Finally, 1 h after ionizing radiation TPX2-depleted cells treated with the broad spectrum caspases inhibitor Z-VAD-fmk still exhibited increased levels of γ-H2AX compared to control cells expressing TPX2 (Fig.1H). Taken together, these results indicate that the absence of TPX2 causes a hyperamplification of γ-H2AX signals at DNA double strand breaks but no increase in the number of γ-H2AX ionizing radiation-induced foci (Fig. 1, A–E). This elevation of γ-H2AX signals in TPX2-depleted cells after ionizing radiation treatment is not associated with pronounced apoptosis (Fig. 1, E–H).
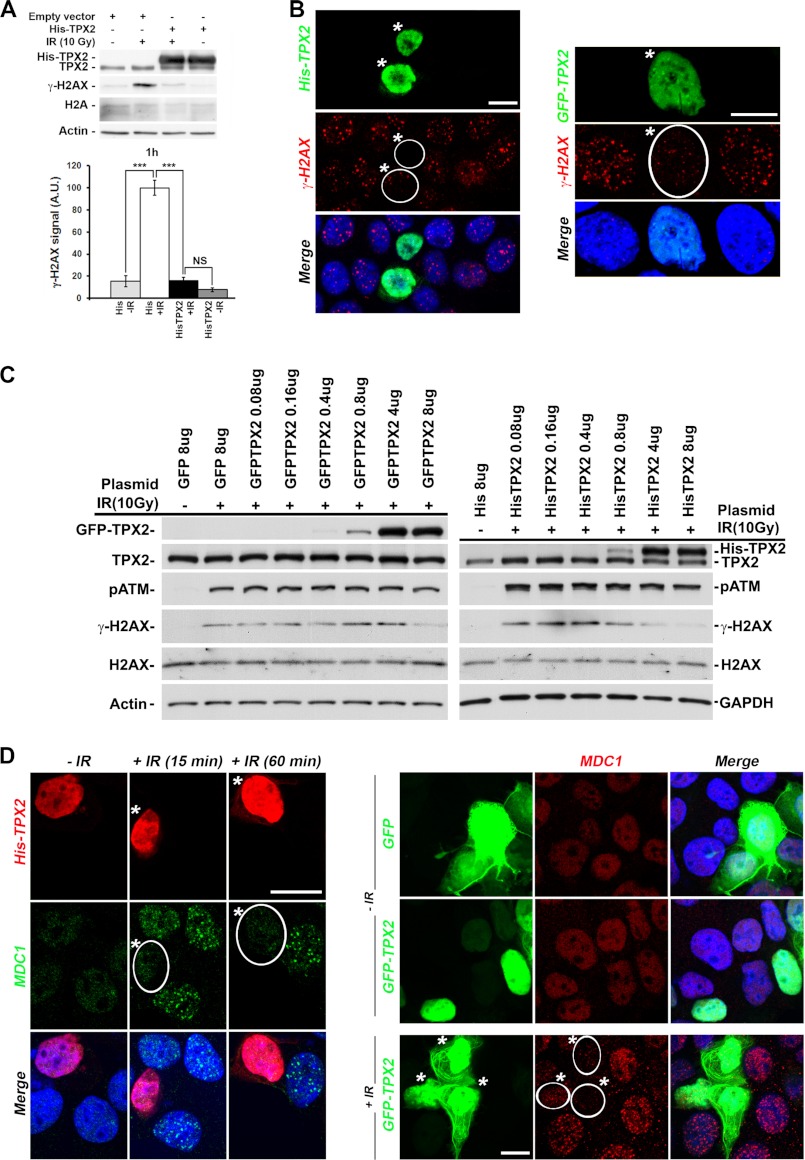
We next asked how an increase in TPX2 levels affects H2AX phosphorylation after ionizing radiation treatment. Conversely to the phenotype observed in irradiated TPX2-depleted cells, ectopic expression of a vector encoding His- or GFP-TPX2 reduced ionizing radiation-triggered phosphorylation of H2AX by ∼80% when compared with irradiated control cells as detected by Western blots (Fig. 2A) and immunofluorescence microscopy (Fig. 2B). We found that in our experimental setup a transient transfection of 4–8 μg of vector (depending on the plasmid used), corresponding grosso modo to a 1.5–2-fold overexpression of ectopic TPX2 in an asynchronous cell culture, is required to abolish γ-H2AX induction (Fig. 2C). Compatible with a model of interdependence between γ-H2AX formation and MDC1 focus formation (27, 28, 42), cells overexpressing His- or GFP-TPX2 also displayed impaired formation of MDC1 ionizing radiation-induced foci (Fig. 2D). In sum, these results indicate that changing the levels of TPX2 dramatically impacts the levels of γ-H2AX possibly through alterations in MDC1 ionizing radiation-induced focus formation.
FIGURE 2.
Effects of TPX2 overexpression on the levels of γ-H2AX and MDC1 ionizing radiation-induced foci. A and B, overexpression of His-TPX2 or GFP-TPX2 significantly decreases phosphorylation of H2AX in HeLa (A) and MCF-7 cells (B) 1 h after ionizing radiation treatment as indicated by Western blot analysis (10 Gy) (A) and immunofluorescence microscopy (B) (left panel, 2Gy; right panel, 4Gy). A, relative quantifications of γ-H2AX signals from independent experiments are shown in bar charts: control +IR (100.0 ± 6.9) versus His-TPX2 +IR (16.0 ± 2.9), p < 0.001, n = 4 (independent experiments); group (mean ± S.E.), unpaired t test. Levels of actin and H2A were used as loading controls. NS, non-significant; ***, p < 0.001. Error bars represent S.E. C, dose-dependent effects of GFP-TPX2 or His-TPX2 on the levels of ionizing radiation-induced γ-H2AX. The amount of plasmid transiently transfected per 6-cm cell culture dish is indicated. Proteins on Western blots were visualized with the indicated antibodies. TPX2 antibodies recognize endogenous and exogenous TPX2 on the same Western blot, thereby allowing comparison of absolute protein levels (see text for details). D, overexpression of His-TPX2 or GFP-TPX2 inhibits MDC1 ionizing radiation-induced focus formation in MCF-7 cells after 2 or 4 Gy (15-min recovery), respectively. Bar in B and D, 10 μm. Merged images include DAPI staining (B and D). A.U., arbitrary units. Transfected cells are indicated by white frame and asterisks (B and D).
TPX2 Localizes to DNA Double Strand Breaks
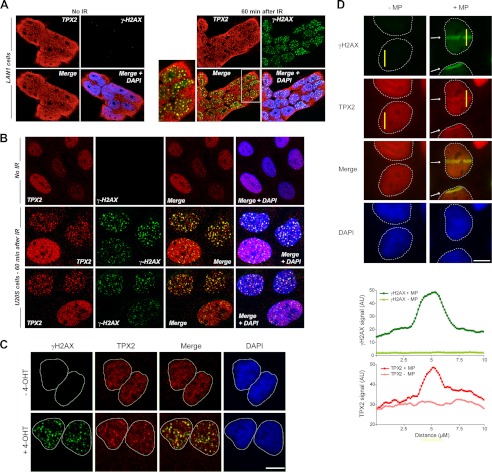
The impact of altered TPX2 levels on ionizing radiation-induced γ-H2AX formation (Figs. 1 and 2) suggests a novel function for TPX2 in the DNA damage response. Because many proteins involved in the DNA damage response accumulate at DNA double strand breaks (28, 30–37), we sought to determine the cellular distribution of TPX2 upon treatment with ionizing radiation. The hypothesis of an ionizing radiation-induced change in TPX2 localization was supported by data showing ionizing radiation-dependent enrichment of TPX2 in the chromatin fraction (Fig. 1C). Strikingly, TPX2 formed ionizing radiation-induced foci that partially co-localized with γ-H2AX ionizing radiation-induced foci in undifferentiated neuroblastoma LAN1, a subpopulation of cycling U2OS cells (Fig. 3, A and B), and primary postmitotic mouse neurons (supplemental Fig. 5E). It is noteworthy that in neuroblastoma LAN1 cells, like in neurons, TPX2 localizes to the nucleus and cytoplasm (see below and supplemental Fig. 5 for further details on the expression of TPX2 in neuronal cells). The usage of G0 primary neurons in these experiments suggests that the herein reported function of TPX2 in the DNA damage response is distinct from the mitotic functions of TPX2 because these cells do not divide (see below). Furthermore, TPX2 also formed foci positive for γ-H2AX in a system that introduces AsiSI-generated DNA double strand breaks (56) (Fig. 3C). Finally, TPX2 also accumulates at sites of DNA lesions generated via microirradiation with a multiphoton laser (57). Upon microirradiation, TPX2 was found to be enriched by ∼1.6-fold in the resulting laser tracks, which were also positive for γ-H2AX, indicating the presence of DNA double strand breaks (Fig. 3D). Taken together, these results underscore the recruitment of TPX2 to DNA double strand breaks generated by multiple stressors (ionizing radiation, a nuclease, and a laser).
FIGURE 3.
TPX2 localizes to DNA double strand breaks. A and B, TPX2 partially co-localizes with γ-H2AX-positive ionizing radiation-induced foci after 5 Gy in LAN1 (A) and U2OS cells (B), respectively (see also supplemental Fig. 5). TPX2 is found in the nucleus and cytosol in neuroblastoma LAN1 cells and neurons (see text and supplemental Fig. 5 for details). C, TPX2 co-localizes with γ-H2AX at 4-hydroxytamoxifen (4-OHT)/AsiSI-induced DNA double strand breaks. U2OS cells stably expressing AsiSI-estrogen receptor were left untreated or treated with 300 nm 4-hydroxytamoxifen for 4 h and subsequently immunostained for TPX2 and γ-H2AX. D, TPX2 accumulates in DNA double strand break-containing laser tracks (indicated by white arrows) marked by γ-H2AX. U2OS cells were either mock-treated (−MP) or microirradiated with a multiphoton laser (+MP). A representative image shows TPX2 accumulation at 10 min after irradiation. The intensity profiles of γ-H2AX and TPX2 immunofluorescence signals were measured in the yellow bars perpendicular to the laser tracks. Commercially available TPX2 antibody 184 was used in all immunofluorescence images. See supplemental Fig. 5 and Fig. 5 for specificity of TPX2 184 antibody. Bars, 10 μm. AU, arbitrary units.
TPX2 Regulates γ-H2AX Levels and Forms Ionization Radiation-induced Foci at G1 Phase
TPX2 expression levels are regulated in a cell cycle-dependent manner (4). This raises the question as to whether γ-H2AX signal amplification by TPX2 occurs at a specific cell cycle stage(s). Furthermore, not all cells in Fig. 3B exhibited TPX2 ionizing radiation-induced foci, indicating cell cycle-dependent differences. To investigate this, we synchronized HeLa cells depleted of TPX2 using a double thymidine block (see “Experimental Procedures,” Fig. 4, and supplemental Figs. 3 and 4) and analyzed γ-H2AX signal levels following 10 Gy and 1-h recovery at specific cell cycle stages. 11 and 12 h after release from the thymidine-triggered early S phase block ∼80% of control and TPX2-depleted cells were in G1 phase as determined by flow cytometry (Fig. 4 and see supplemental Fig. 3 for the profile of non-synchronized populations displaying less than 50% G1 phase cells). Significantly, TPX2-depleted G1 cells exhibited a marked increase in γ-H2AX signals following ionizing radiation treatment when compared with control populations (Fig. 4A). In populations enriched for S phase cells (∼95% 2 h after release) or G2 phase cells (∼80% 6 h after release), no significant differences in ionizing radiation-dependent γ-H2AX signals were observed between control and TPX2-depleted cultures (supplemental Fig. 4A). To analyze a potential effect of TPX2 depletion on γ-H2AX levels during mitosis, cells were treated with nocodazole to obtain populations highly enriched for M phase (∼90%). Note that the duration of mitosis in these cells is very short (less than 2 h), making it virtually impossible to capture synchronous M phase populations via release from a double thymidine block. No difference in ionizing radiation-triggered γ-H2AX was found between nocodazole-treated control or TPX2-depleted cell cultures (supplemental Fig. 4B).
In agreement with these Western blot results, we found using confocal microscopy that U2OS cell cultures synchronized at G1 phase exhibited prominent TPX2 ionizing radiation-induced foci that partially co-localized with γ-H2AX foci. However, cell cultures enriched for S or G2 phase did not form such pronounced TPX2 ionizing radiation-induced foci (Fig. 4B). It is noteworthy that a previous study suggested that the basal levels of γ-H2AX in unirradiated S and G2 phase cells are ∼3-fold higher than in G1 phase cells (58). The slope of the ionizing radiation-induced γ-H2AX response curve for G1 phase cells is in fact 2.8 times steeper than the slope for S phase cells (58). Our results in Fig. 4B are in agreement with these data (note the higher levels of γ-H2AX in unirradiated S and G2 phase cells versus G1 phase cells).
In sum, our data indicate that TPX2 regulates γ-H2AX signal amplification at G1. Of note, during mitosis, TPX2 stays associated with the mitotic spindle even in the presence of DNA double strand breaks that are marked by γ-H2AX (Fig. 4C). These results further support the idea that the function of TPX2 in regulating γ-H2AX levels is distinct from its mitotic roles (see below).
Altered Levels of γ-H2AX by TPX2 Dysfunctions Are Not Caused by Mitotic Anomalies
Cells overexpressing or lacking TPX2 display spindle abnormalities (2, 4), raising the question as to whether the alterations in levels of γ-H2AX observed in these cells were due to spindle defects and/or mitotic arrest rather than disturbances of the DNA damage response. To prove that spindle and mitotic defects are not involved in the observed γ-H2AX phenotype arising from altered TPX2 levels, we knocked down in HeLa cells two additional proteins involved in spindle biology and mitosis by siRNA and analyzed their levels of γ-H2AX upon irradiation. First, depletion of the microtubule-associated protein Ndel1, which binds TPX2 and is also involved in microtubule dynamics (59) and spindle orientation and integrity (60–62), did not increase the levels of ionizing radiation-dependent γ-H2AX (Fig. 5A). Second, depletion of the kinetochore complex member NUF2, which interferes with the attachment of the spindle to the kinetochore and triggers a strong mitotic arrest reminiscent of TPX2 deficiency (63), did not affect the levels of γ-H2AX when compared with controls (Fig. 5B). The mitotic arrest upon NUF2 or TPX2 depletion (Fig. 5B and supplemental Fig. 2) was confirmed with the increased levels of the mitotic marker Histone 3 phosphorylated at serine 10 (H3S10p). Note that although irradiated TPX2-depleted and NUF2-depleted cells exhibit similar levels of H3S10p, TPX2-depleted cells display strikingly elevated ionizing radiation-dependent γ-H2AX levels compared with NUF2-depleted cells. In sum, these data support the idea that the regulation of γ-H2AX levels by TPX2 in cycling cells is not an indirect consequence of defective mitosis but rather reflects a previously unknown function for TPX2.
To further support the notion of a novel TPX2 function distinct from its mitotic roles, we isolated and cultured primary cortical mouse neurons. These postmitotic neurons are at G0 phase, no longer cycle, and do not form a mitotic spindle. However, they still express TPX2 as evidenced by protein, RNA, and immunofluorescence analysis (supplemental Fig. 5 and Ref. 59). In neurons, TPX2 is localized in neuronal processes, cytoplasm, and nucleus (supplemental Fig. 5 and Ref. 59). The specificity of TPX2 antibody (184; Novus Biologicals) used for immunostaining was determined by peptide competition (supplemental Fig. 5) and depletion of TPX2 by RNAi (see below).
To examine the effect of TPX2 on γ-H2AX formation in mouse cortical neurons, we generated an shRNA construct targeting mouse TPX2 mRNA. The efficacy of the vector was first tested in neuroblastoma mouse CAD cells: endogenous TPX2 protein was diminished by ∼70% in cells transfected with the TPX2 shRNA vector as determined by Western blot analysis (supplemental Fig. 5). No decrease was observed in CAD cells transfected with a control plasmid encoding a random sequence that has no homology to any known messenger RNA (supplemental Fig. 5). Consistent with Western blot analysis, individual neurons co-transfected with TPX2 RNAi and GFP demonstrated a significant decrease of TPX2 as determined by confocal microscopy (Fig. 5C). In the absence of ionizing radiation, these neurons did not display γ-H2AX signals. 10 Gy-irradiated neurons transfected with the control plasmid exhibited levels of γ-H2AX similar to irradiated non-transfected surrounding cells. Importantly, 1 h after ionizing radiation neurons depleted of TPX2 by specific shRNA exhibited much higher levels of γ-H2AX than surrounding control cells and control shRNA-transfected cells (Fig. 5D). The average ratio of total nuclear γ-H2AX signals of TPX2 RNAi/untransfected cells was ∼4 times higher than the ratio found in control RNAi/untransfected cells (Fig. 5, D and F). Conversely, the ectopic expression of GFP-TPX2 abolished the 10 Gy-induced phosphorylation of H2AX in postmitotic neurons by 50% when compared with irradiated surrounding control cells. However, cells expressing GFP exhibited γ-H2AX levels similar to those of surrounding untransfected cells (Fig. 5, E and G). Using a dose of 3 Gy, we further confirmed that alterations in TPX2 levels in these neurons impact γ-H2AX amplification (Fig. 5, H and I). The average ratio of total nuclear γ-H2AX signals of 3 Gy-treated TPX2 RNAi/untransfected cells was ∼20% higher than the ratio found in irradiated control RNAi/untransfected cells (Fig. 5H). Conversely, the ectopic expression of GFP-TPX2 abolished by ∼30% the 3 Gy-induced phosphorylation of H2AX in postmitotic neurons when compared with irradiated surrounding control cells. However, cells expressing GFP exhibited γ-H2AX levels similar to those of surrounding untransfected cells (Fig. 5I). Taken together, using four different TPX2-targeting RNAi sequences and a rescue approach (Figs. 1, 4, and 5), our results confirm that TPX2 levels dictate the extent of γ-H2AX formation during the DNA damage response in a similar manner in G1 cycling cells and G0 postmitotic neurons. In sum, the observed effects of TPX2 on γ-H2AX formation during the DNA damage response are independent of the mitotic functions of TPX2.
TPX2 Associates with Proteins of the DNA Damage Response
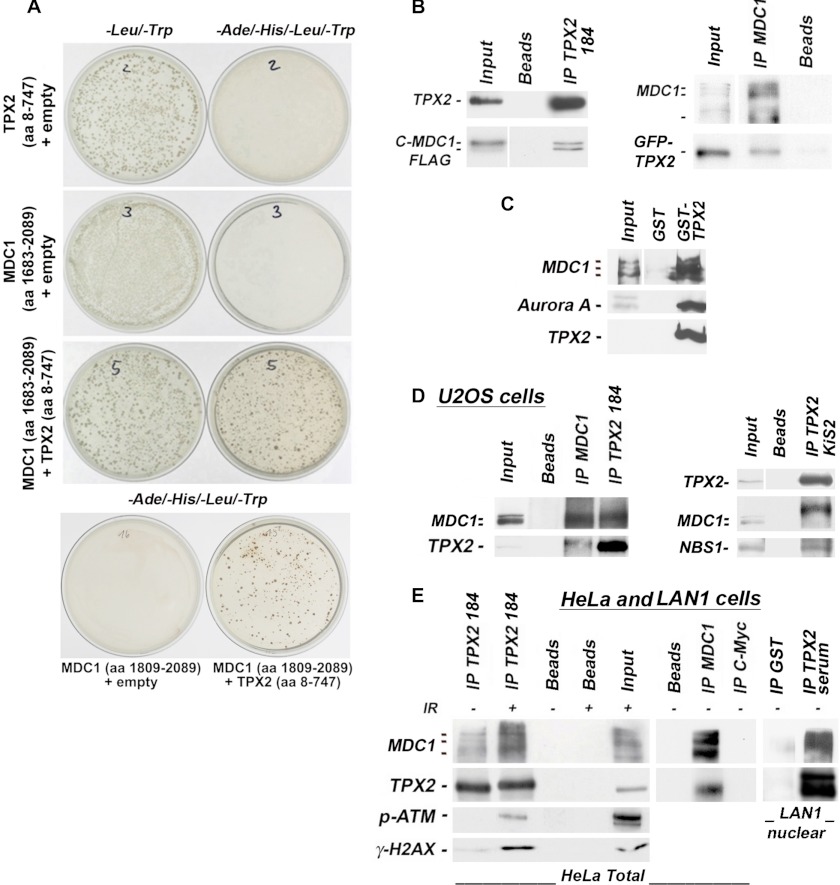
MDC1 and activated ATM mediate the amplification of γ-H2AX signals in proximity of the DNA double strand breaks (40–42). Because we establish herein that TPX2 impacts the levels of γ-H2AX (Figs. 1, 2, 4, and 5) and in light of data suggesting that TPX2 is a target of the ATM kinase (25) (ATM is also known to be in complex with MDC1), we sought to investigate a potential association between TPX2 and proteins of the DNA damage response machinery. This hypothesis was supported by an independent experimental approach in which we recovered MDC1 as a potential novel binding partner for TPX2 in a Y2H screen that used a ∼25-week-old human embryonic brain cDNA library. During that period of brain development, neuronal precursors and newly born neurons are highly susceptible to DNA damage-induced cell death and, therefore, require an efficient DNA damage response (64–67). This may explain the expression of MDC1 in the library. Using TPX2 and fragments of MDC1, we confirmed the interaction in independent Y2H assays: TPX2 (aa 8–747 and full length) binds to the C terminus of MDC1 (aa 1683–2089 and 1809–2089) (Fig. 6A), which includes the BRCT tandem domain that interacts with γ-H2AX (68). When expressed ectopically in HeLa cells, the FLAG-tagged C terminus of MDC1 (C-MDC1-FLAG; aa 1807–2089) co-immunoprecipitated with endogenous TPX2 (Fig. 6B). Moreover, ectopically expressed GFP-TPX2 was also co-immunoprecipitated with endogenous MDC1 in HeLa cells (Fig. 6B). In addition, we performed pulldown assays of MDC1 from HeLa cell lysates using purified GST and GST-TPX2 full-length (GST-TPX2) fusion proteins. MDC1 and the positive control Aurora A (69) were pulled down by GST-TPX2 but not by GST (Fig. 6C).
FIGURE 6.
TPX2 associates with MDC1, p-ATM, NBS1, and γ-H2AX. A, Y2H experiment using bait-TPX2 (aa 8–747) and prey-MDC1 (aa 1683–2089 or 1809–2089). Plasmids were co-transformed as indicated. −Leu/−Trp selection agar plates are controls for transformation efficiency. Colonies on −Ade/−His/−Leu/−Trp selection agar plates reveal an interaction between bait-TPX2 and prey-MDC1. Plates were incubated for 6 days. B, an ectopic C-terminal fragment of MDC1 (C-MDC1-FLAG; aa 1807–2089) associates with endogenous TPX2 in HeLa cells as indicated by co-immunoprecipitations with TPX2 antibody 184 from total cell lysate (left panel). Ectopic GFP-TPX2 also co-immunoprecipitates with endogenous MDC1 (right panel). The input lane for the C-MDC1-FLAG (left panel) is from a shorter exposure of the same Western blot. The input lane for the endogenous MDC1 (right panel) is from a stronger exposure of the same Western blot. C, GST-TPX2 pulls down MDC1 and the positive control Aurora A from total HeLa cell lysate. For MDC1, the input is a shorter exposure of the same blot. D, co-immunoprecipitations from U2OS cell lysates with the specified antibodies (see text for details on antibodies). TPX2 was co-immunoprecipitated with MDC1 from these cells using MDC1 antibody (left panel). MDC1 also co-immunoprecipitated with TPX2 antibody 184 (left panel). TPX2 was also found in complex with NBS1 and MDC1 species that migrate slower on SDS-PAGE gels when the co-immunoprecipitations were performed with the TPX2 KiS2 antibody (right panel; see text for further details). E, TPX2 and MDC1 associate in HeLa and LAN1 cells as detected by co-immunoprecipitations with the specified antibodies from total (left and middle panels) or nuclear (right panel) lysates. TPX2 from neuroblastoma LAN1 cells (and primary neurons) migrates as a doublet on gels (see supplemental Fig. 5). TPX2 is also found in complex with p-ATM and γ-H2AX after ionizing radiation treatment. Beads alone or antibodies against c-Myc were used as negative controls as indicated. IP, immunoprecipitation.
Next, we determined whether TPX2 forms a complex with MDC1 under physiological conditions. Co-immunoprecipitation experiments were performed on U2OS, HeLa, and neuroblastoma LAN1 cell lysates using three different antibodies specific for TPX2 (TPX2 184, TPX2 KiS2, and TPX2 serum) and antibody specific for MDC1 (for details and specificity of antibodies, see “Experimental Procedures”). As shown in Fig. 6D, TPX2 was co-immunoprecipitated with MDC1 from U2OS cell lysates using MDC1 antibody. The reverse co-immunoprecipitation using commercially available TPX2 184 antibody also recovered MDC1 in the immunoprecipitates. Furthermore, NBS1, a member of the tripartite MRN complex known to associate with MDC1 (30–37), and a slower migrating MDC1 species (suggesting post-translational modification) were also co-immunoprecipitated with TPX2 using the TPX2 KiS2 antibody (Fig. 6D). As shown in Fig. 6E, we also detected a TPX2-MDC1 complex in HeLa cell lysates and in LAN1 nuclear lysates using co-immunoprecipitations with either TPX2 184 antibody, TPX2 serum, or MDC1 antibody. To analyze the TPX2-MDC1 association in the context of the DNA damage response, HeLa cells were irradiated prior to co-immunoprecipitations with TPX2 184 antibody. Following ionizing radiation, TPX2 was also found to associate with MDC1, serine 1981 p-ATM, and its substrate γ-H2AX (Fig. 6E).
It is noteworthy that in HeLa cells the MDC1 antibody recognizes at least three bands of distinct molecular weight that may correspond to MDC1 isoforms based on entries in the UniProt database. Experimentally, treatment of HeLa cells with two different MDC1 siRNAs abolishes all these bands (Fig. 7), indicating that they are MDC1 species. However, in U2OS cells, only the lower molecular mass isoforms (∼196/198 kDa) co-immunoprecipitate with TPX2, indicating cell line-specific differences. In sum, our Y2H, pulldown, and co-immunoprecipitation data from three different cell lines and obtained with three TPX2 and MDC1 antibodies provide compelling evidence that TPX2 forms a complex with MDC1 in cells. ATM and NBS1 may be part of this novel TPX2 complex.
FIGURE 7.
The ionizing radiation-dependent increase in γ-H2AX caused by TPX2 depletion is antagonized by inhibition or knockdown of MDC1 or ATM. A, an siRNA-mediated knockdown of MDC1 antagonizes the ionizing radiation-triggered γ-H2AX hyperamplification in HeLa cells caused by TPX2 depletion (lane 3 versus lane 4). Note that during the DNA damage response TPX2 depletion and MDC1 depletion have an opposite effect on γ-H2AX levels (lane 1 versus lane 3). B, inhibition of ATM with KU55933 antagonizes the ionizing radiation-dependent increase in γ-H2AX caused by depletion of TPX2. Inhibition of DNA-PK with NU7441 does not rescue this γ-H2AX hyperamplification phenotype. C, siRNA-mediated loss of ATM abrogates the ionizing radiation-dependent increase in γ-H2AX caused by depletion of TPX2. siRNA-mediated loss of DNA-PKcs partially decreases ATM levels (as previously described in Ref. 71) but does not rescue the γ-H2AX hyperamplification caused by TPX2 depletion. D, quantification of γ-H2AX signals in C. γ-H2AX signals of control siRNA-treated cells were considered as 100% and compared with the respective TPX2 siRNA-treated cells (n = 3 (independent experiments); N.S., non-significant; *, p < 0.05, unpaired t test, S.E.). Error bars represent S.E. All cells were treated with 10 Gy and harvested after 1-h recovery. Ctrl., control.
The Effects of TPX2 on γ-H2AX Levels Can Be Antagonized by MDC1 and ATM
It is known that MDC1 is an essential factor for the amplification of γ-H2AX during the DNA damage response. Loss of MDC1 decreases ionizing radiation-dependent γ-H2AX levels (Refs. 40–42 and Fig. 7A). Herein, we establish that TPX2 associates with the H2AX phosphorylation machinery (i.e. MDC1 and ATM) at sites of lesions. Furthermore, the levels of TPX2 are inversely correlated with the levels of γ-H2AX and formation of MDC1 ionizing radiation-induced foci. In light of all these results, we hypothesized that the TPX2 RNAi phenotype (i.e. increased ionizing radiation-dependent γ-H2AX levels) should be ameliorated if MDC1 function is lost. To test this, we knocked down both MDC1 and TPX2 with RNAi and determined the levels of ionizing radiation-dependent γ-H2AX. To minimize the effects of TPX2 knockdown on mitosis, cells were treated with TPX2 RNAi for a maximum of 24 h, whereas efficient knockdown of MDC1 was achieved 72 h post-transfection (see “Experimental Procedures” for the double knockdown protocol). We found that depletion of MDC1 in irradiated TPX2-depleted cells antagonized the increased levels of γ-H2AX (triggered by the loss of TPX2) (Fig. 7A).
During the DNA damage response, H2AX can be phosphorylated by ATM and DNA-PK. To determine which kinase(s) account(s) for the increased γ-H2AX levels in irradiated TPX2-depleted cells, we treated control and TPX2 RNAi-treated cells with DMSO (control), the ATM inhibitor KU55933, or the DNA-PK inhibitor NU7441 prior to ionizing radiation. Inhibition of ATM but not DNA-PK efficiently reduced the increased γ-H2AX levels triggered by depletion of TPX2 to background levels (Fig. 7B). Of note, treatment with NU7441 similarly increased the γ-H2AX signals in TPX2-depleted and control cells compared with cells treated with DMSO, which is in agreement with a recent study (70). To support these results, we performed loss of ATM or DNA-PK function experiments using specific siRNA. To minimize the effects of TPX2 knockdown on mitosis, cells were treated with TPX2 RNAi for a maximum of 24 h, whereas efficient knockdown of DNA-PKcs and ATM was achieved 96 h post-transfection (see “Experimental Procedures” for the double knockdown protocol). Depletion of ATM but not DNA-PKcs abrogated the elevated γ-H2AX levels in irradiated TPX2-depleted cells (Fig. 7, C and D). It is noteworthy that depletion of DNA-PKcs down-regulated ATM levels as reported previously (71). However, the remaining p-ATM levels were still sufficient to trigger the TPX2 depletion-dependent γ-H2AX phenotype. Taken together, these results indicate that the up-regulation of γ-H2AX in irradiated cells depleted of TPX2 depends on ATM and MDC1. Interestingly, we did not observe a difference in ionizing radiation-dependent activation of ATM between control and TPX2-depleted cells as indicated by the unchanged levels of p-ATM (Figs. 1C and 7C). These data suggest that ATM is implicated in the γ-H2AX increase of irradiated TPX2-depleted cells, although the mode of ATM activation unlikely contributes to the phenotype (see “Discussion”). In light of our findings, we conclude that MDC1 and ATM can antagonize the effects of TPX2 on the amplification of γ-H2AX signals. It remains unclear whether TPX2 impacts the levels of γ-H2AX directly through its association with MDC1 and/or ATM (see “Discussion”).
DISCUSSION
In the present study, we found that the levels of TPX2 inversely correlate with the levels of γ-H2AX during the DNA damage response (Figs. 1, 2, 4, 5, and 7). Using four different RNAi sequences, we found that cells lacking TPX2 exhibit a significant increase in ionizing radiation-dependent phosphorylation of H2AX (1–2 h after ionizing radiation) that is accompanied by increased numbers of cells with high intensity γ-H2AX ionizing radiation-induced foci (Fig. 1). This phenotype was rescued by expression of an siRNA-insensitive TPX2 construct (Fig. 1). Conversely, cells overexpressing TPX2 display decreased ionizing radiation-dependent γ-H2AX levels and defective MDC1 ionizing radiation-induced focus formation (Fig. 2). TPX2 appears to impact γ-H2AX formation in G0 and G1 phases of the cell cycle as indicated by the use of synchronized cell cultures and postmitotic primary neurons (Figs. 4 and 5). Furthermore, consistent with a role in the DNA damage response, TPX2 localizes to DNA double strand breaks and associates with MDC1 and p-ATM (Figs. 3, 4, and 6), known key factors for γ-H2AX amplification. Finally, the TPX2 depletion and ionizing radiation-dependent increase in γ-H2AX can be antagonized by inhibition or knockdown of MDC1 or ATM (Fig. 7).
The early up-regulation of γ-H2AX observed in TPX2-depleted cells 15 min to 2 h after ionizing radiation is not linked to increased apoptosis as indicated by unchanged amounts of DNA double strand breaks before and after ionizing radiation treatment (detected by comet assay and the number of γ-H2AX ionizing radiation-induced foci; supplemental Fig. 1 and Fig. 1E), the use of non-lethal doses of ionizing radiation, the use of caspase-3-deficient MCF-7 cells that do not undergo ionizing radiation-triggered apoptosis associated with DNA fragmentation, and the use of a broad spectrum caspase inhibitor (Fig. 1). Interestingly, knockdown of TPX2 sensitizes cells to ionizing radiation, leading ultimately to increased ionizing radiation-induced apoptosis at later time points (9–12 h after ionizing radiation; data not shown). Such perspective could be thoroughly examined by therapeutic strategies combining radiotherapy and modulation of TPX2 levels to eradicate different types of cancer cells.
Previous studies have shown that mitotic cells accumulate more γ-H2AX signals than interphase cells independently of ionizing radiation (72, 73). A recent study also showed that mitotic cells exhibit an atypical DNA damage response compared with interphase cells (70). Because TPX2 is essential for mitosis, it might be argued that the increase in γ-H2AX signals in irradiated TPX2-depleted cells is caused primarily by mitotic arrest. However, depletion of Ndel1 or NUF2, which, like depletion of TPX2, leads to mitotic arrest and spindle abnormalities (60–63), did not increase ionizing radiation-dependent γ-H2AX levels (Fig. 5, A and B). Furthermore, the molecular signatures of mitotic cells and TPX2-depleted cultures with DNA lesions differ. For instance, damaged cells synchronized at M phase exhibit 53BP1 that migrates slower on SDS-PAGE gels and is hypophosphorylated at Ser-25 when compared with unsynchronized cells (70). In contrast, in irradiated TPX2-depleted cultures, 53BP1 migrates normally and does not exhibit decreased phosphorylation at Ser-25 compared with control populations (supplemental Fig. 6). Our data also clearly show that during G1 phase there is a pronounced formation of TPX2 ionizing radiation-induced foci, whereas during M phase, TPX2 stays associated with the mitotic spindle apparatus in the presence of DNA damage (Fig. 4). Finally, we found that TPX2 controls the ionizing radiation-dependent levels of γ-H2AX similarly in both cycling cells (Figs. 1–4) and postmitotic neurons that no longer enter mitosis (Fig. 5). Taken together, our results highlight a novel and unexpected role for TPX2 in the DNA damage response that is distinct from its mitotic function. This is the first nuclear function assigned to TPX2.
Based on our data showing an association between TPX2 and MDC1/ATM (Figs. 6 and 7), TPX2 could regulate γ-H2AX amplification via MDC1 and ATM. However, the accumulation of γ-H2AX signals during the DNA damage response is also influenced by the chromatin structure (74, 75). In light of the accumulation of TPX2 at ionizing radiation-induced foci (Figs. 3 and 4) and the association of TPX2 with BRCA1 (76), which has recently been shown to function as chromatin remodeling factor (76), it is possible that TPX2 regulates γ-H2AX formation via DNA double strand break-flanking chromatin remodeling. In addition, it is also possible that TPX2 impacts global chromatin architecture independently of DNA damage, thereby having an indirect effect on MDC1 ionizing radiation-induced focus formation and γ-H2AX levels once DNA double strand breaks are induced.
The time course of H2AX phosphorylation in TPX2-depleted cells differs from those observed when specific γ-H2AX protein phosphatases are depleted in which cases prolonged γ-H2AX signals are observed (77–82). In contrast, TPX2-depleted cells silence their elevated γ-H2AX signals with the same time course as control cells (Fig. 1C). An unidentified phosphatase modulating γ-H2AX amplification early after ionizing radiation could be misregulated in the absence of TPX2. In this particular context, TPX2 may impact the dephosphorylation of γ-H2AX and, thus, would regulate its steady-state levels. Alternatively, TPX2 may control H2AX phosphorylation by competing with γ-H2AX for binding to the BRCT domain of MDC1, thereby antagonizing the amplificatory function of MDC1. In support of this hypothesis, TPX2 was found to associate with the C terminus of MDC1 (aa 1809–2089) containing its BRCT domains (Fig. 6, A and B). The exact molecular mechanisms by which TPX2 impacts the ionizing radiation-dependent γ-H2AX levels remain to be determined.
TPX2 has been proposed as a biomarker and effector for cancer progression based on its elevated levels in numerous malignancies that are correlated with disease progression (12–17, 19, 83). So far, the suspected involvement of TPX2 in these diseases has been mostly linked to its functions in mitosis and activation of Aurora A (84). With the novel function of TPX2 described herein, we propose that TPX2 also contributes to cancer pathology by impacting the DNA damage response. Because phosphorylation of H2AX is essential for the amplification of the DNA damage response, it is conceivable that aberrant levels of γ-H2AX caused by abnormal expression of TPX2 (as detected in cancers) deregulate the DNA damage response. This mechanism could contribute to the genomic instability found in many cancers.
Acknowledgments
We are grateful to the members of the Lees-Miller laboratory; Drs. Oender Kamil, James Wang, Laurie Kennedy, and Hong Tran for technical help; Aaron Sheldon for help with the statistical analysis; and Felix Baerenz for regeneration of the TPX2 miRNA-expressing cell line.
This work was supported in part by a Canadian Institute of Health Research (CIHR) operating grant, an Alberta Innovates - Health Solutions (AI-HS) cancer grant (to M. D. N.), and the Medical Research Council (to T. H.).

This article contains supplemental Figs. 1–6.
- TPX2
- targeting protein for Xenopus kinesin-like protein 2
- H2A
- Histone 2A
- H2AX
- Histone 2AX
- γ-H2AX
- Ser-139-phosphorylated Histone 2AX
- MDC1
- mediator of DNA damage checkpoint 1
- ATM
- ataxia telangiectasia mutated
- DNA-PK
- DNA-dependent protein kinase
- BRCA1
- breast cancer 1 protein
- MRN
- MRE11, Rad50, and NBS1
- NBS1
- Nijmegen breakage syndrome protein 1
- BRCT
- BRCA1 C-terminal
- DNA-PKcs
- DNA-PK catalytic subunit
- H3
- Histone 3
- H3S10p
- Histone 3 phosphorylated at serine 10
- aa
- amino acid(s)
- Y2H
- yeast two-hybrid
- miRNA
- microRNA
- Z-VAD-fmk
- benzyloxycarbonyl - Val-Ala-Asp - fluoromethyl ketone
- Gy
- grays
- Ndel1
- nuclear distribution E homolog-like 1
- p-ATM
- Ser-1981-phosphorylated ATM
- IR
- ionizing radiation
- ER
- estrogen receptor.
REFERENCES
- 1. Heidebrecht H. J., Buck F., Steinmann J., Sprenger R., Wacker H. H., Parwaresch R. (1997) p100: a novel proliferation-associated nuclear protein specifically restricted to cell cycle phases S, G2, and M. Blood 90, 226–233 [PubMed] [Google Scholar]
- 2. Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83–93 [DOI] [PubMed] [Google Scholar]
- 3. Gruss O. J., Vernos I. (2004) The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol. 166, 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gruss O. J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I. W., Vernos I. (2002) Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871–879 [DOI] [PubMed] [Google Scholar]
- 5. Karsenti E. (2005) TPX or not TPX? Mol. Cell 19, 431–432 [DOI] [PubMed] [Google Scholar]
- 6. Wittmann T., Wilm M., Karsenti E., Vernos I. (2000) TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149, 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boleti H., Karsenti E., Vernos I. (1996) Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84, 49–59 [DOI] [PubMed] [Google Scholar]
- 8. Wittmann T., Boleti H., Antony C., Karsenti E., Vernos I. (1998) Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vanneste D., Takagi M., Imamoto N., Vernos I. (2009) The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 19, 1712–1717 [DOI] [PubMed] [Google Scholar]
- 10. Kahana J. A., Cleveland D. W. (2001) Cell cycle. Some importin news about spindle assembly. Science 291, 1718–1719 [DOI] [PubMed] [Google Scholar]
- 11. Schatz C. A., Santarella R., Hoenger A., Karsenti E., Mattaj I. W., Gruss O. J., Carazo-Salas R. E. (2003) Importin α-regulated nucleation of microtubules by TPX2. EMBO J. 22, 2060–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tonon G., Wong K. K., Maulik G., Brennan C., Feng B., Zhang Y., Khatry D. B., Protopopov A., You M. J., Aguirre A. J., Martin E. S., Yang Z., Ji H., Chin L., Depinho R. A. (2005) High-resolution genomic profiles of human lung cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Manda R., Kohno T., Matsuno Y., Takenoshita S., Kuwano H., Yokota J. (1999) Identification of genes (SPON2 and C20orf2) differentially expressed between cancerous and noncancerous lung cells by mRNA differential display. Genomics 61, 5–14 [DOI] [PubMed] [Google Scholar]
- 14. Warner S. L., Stephens B. J., Nwokenkwo S., Hostetter G., Sugeng A., Hidalgo M., Trent J. M., Han H., Von Hoff D. D. (2009) Validation of TPX2 as a potential therapeutic target in pancreatic cancer cells. Clin. Cancer Res. 15, 6519–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramakrishna M., Williams L. H., Boyle S. E., Bearfoot J. L., Sridhar A., Speed T. P., Gorringe K. L., Campbell I. G. (2010) Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One 5, e9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Satow R., Shitashige M., Kanai Y., Takeshita F., Ojima H., Jigami T., Honda K., Kosuge T., Ochiya T., Hirohashi S., Yamada T. (2010) Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin. Cancer Res. 16, 2518–2528 [DOI] [PubMed] [Google Scholar]
- 17. Berger S. M., Pesold B., Reber S., Schönig K., Berger A. J., Weidenfeld I., Miao J., Berger M. R., Gruss O. J., Bartsch D. (2010) Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs. Nucleic Acids Res. 38, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang H., Wang J., Tian Y., Xu J., Gou X., Cheng J. (2012) The TPX2 gene is a promising diagnostic and therapeutic target for cervical cancer. Oncol. Rep. 27, 1353–1359 [DOI] [PubMed] [Google Scholar]
- 19. Sillars-Hardebol A. H., Carvalho B., Tijssen M., Beliën J. A., de Wit M., Delis-van Diemen P. M., Pontén F., van de Wiel M. A., Fijneman R. J., Meijer G. A. (2012) TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut 61, 1568–1575 [DOI] [PubMed] [Google Scholar]
- 20. Aguirre-Portolés C., Bird A. W., Hyman A., Cañamero M., Pérez de Castro I., Malumbres M. (2012) Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 72, 1518–1528 [DOI] [PubMed] [Google Scholar]
- 21. Lengauer C., Kinzler K. W., Vogelstein B. (1998) Genetic instabilities in human cancers. Nature 396, 643–649 [DOI] [PubMed] [Google Scholar]
- 22. Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010) Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228 [DOI] [PubMed] [Google Scholar]
- 23. Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., Livingston D. M. (2006) The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell 127, 539–552 [DOI] [PubMed] [Google Scholar]
- 24. Maxwell C. A., Benítez J., Gómez-Baldó L., Osorio A., Bonifaci N., Fernández-Ramires R., Costes S. V., Guinó E., Chen H., Evans G. J., Mohan P., Català I., Petit A., Aguilar H., Villanueva A., Aytes A., Serra-Musach J., Rennert G., Lejbkowicz F., Peterlongo P., Manoukian S., Peissel B., Ripamonti C. B., Bonanni B., Viel A., Allavena A., Bernard L., Radice P., Friedman E., Kaufman B., Laitman Y., Dubrovsky M., Milgrom R., Jakubowska A., Cybulski C., Gorski B., Jaworska K., Durda K., Sukiennicki G., Lubiński J., Shugart Y. Y., Domchek S. M., Letrero R., Weber B. L., Hogervorst F. B., Rookus M. A., Collee J. M., Devilee P., Ligtenberg M. J., Luijt R. B., Aalfs C. M., Waisfisz Q., Wijnen J., Roozendaal C. E.; HEBON; EMBRACE, Easton D. F., Peock S., Cook M., Oliver C., Frost D., Harrington P., Evans D. G., Lalloo F., Eeles R., Izatt L., Chu C., Eccles D., Douglas F., Brewer C., Nevanlinna H., Heikkinen T., Couch F. J., Lindor N. M., Wang X., Godwin A. K., Caligo M. A., Lombardi G., Loman N., Karlsson P., Ehrencrona H., Wachenfeldt A.; SWE-BRCA, Barkardottir R. B., Hamann U., Rashid M. U., Lasa A., Caldés T., Andrés R., Schmitt M., Assmann V., Stevens K., Offit K., Curado J., Tilgner H., Guigó R., Aiza G., Brunet J., Castellsagué J., Martrat G., Urruticoechea A., Blanco I., Tihomirova L., Goldgar D. E., Buys S., John E. M., Miron A., Southey M., Daly M. B., Schmutzler R. K., Wappenschmidt B., Meindl A., Arnold N., Deissler H., Varon-Mateeva R., Sutter C., Niederacher D., Imyamitov E., Sinilnikova O. M., Stoppa-Lyonne D., Mazoyer S., Verny-Pierre C., Castera L., de Pauw A., Bignon Y. J., Uhrhammer N., Peyrat J. P., Vennin P., Fert Ferrer S., Collonge-Rame M. A., Mortemousque I.; GEMO Study Collaborators, Spurdle A. B., Beesley J., Chen X., Healey S.; kConFab, Barcellos-Hoff M. H., Vidal M., Gruber S. B., Lázaro C., Capellá G., McGuffog L., Nathanson K. L., Antoniou A. C., Chenevix-Trench G., Fleisch M. C., Moreno V., Pujana M. A. (2011) Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 9, e1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 26. Kurz E. U., Lees-Miller S. P. (2004) DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair 3, 889–900 [DOI] [PubMed] [Google Scholar]
- 27. van Attikum H., Gasser S. M. (2005) The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6, 757–765 [DOI] [PubMed] [Google Scholar]
- 28. van Attikum H., Gasser S. M. (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19, 207–217 [DOI] [PubMed] [Google Scholar]
- 29. Stewart G. S. (2009) Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle 8, 1532–1538 [DOI] [PubMed] [Google Scholar]
- 30. Stucki M., Jackson S. P. (2004) MDC1/NFBD1: a key regulator of the DNA damage response in higher eukaryotes. DNA Repair 3, 953–957 [DOI] [PubMed] [Google Scholar]
- 31. Stucki M., Jackson S. P. (2006) γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 5, 534–543 [DOI] [PubMed] [Google Scholar]
- 32. Khanna K. K., Lavin M. F., Jackson S. P., Mulhern T. D. (2001) ATM, a central controller of cellular responses to DNA damage. Cell Death Differ. 8, 1052–1065 [DOI] [PubMed] [Google Scholar]
- 33. Khanna K. K., Jackson S. P. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 [DOI] [PubMed] [Google Scholar]
- 34. Kim J. E., Minter-Dykhouse K., Chen J. (2006) Signaling networks controlled by the MRN complex and MDC1 during early DNA damage responses. Mol. Carcinog. 45, 403–408 [DOI] [PubMed] [Google Scholar]
- 35. Durocher D., Smerdon S. J., Yaffe M. B., Jackson S. P. (2000) The FHA domain in DNA repair and checkpoint signaling. Cold Spring Harb. Symp. Quant. Biol. 65, 423–431 [DOI] [PubMed] [Google Scholar]
- 36. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 37. Harrison J. C., Haber J. E. (2006) Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235 [DOI] [PubMed] [Google Scholar]
- 38. Bakkenist C. J., Kastan M. B. (2003) DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 39. Goldberg M., Stucki M., Falck J., D'Amours D., Rahman D., Pappin D., Bartek J., Jackson S. P. (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421, 952–956 [DOI] [PubMed] [Google Scholar]
- 40. Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 41. Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 [DOI] [PubMed] [Google Scholar]
- 42. Lou Z., Minter-Dykhouse K., Franco S., Gostissa M., Rivera M. A., Celeste A., Manis J. P., van Deursen J., Nussenzweig A., Paull T. T., Alt F. W., Chen J. (2006) MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 [DOI] [PubMed] [Google Scholar]
- 43. Lou Z., Minter-Dykhouse K., Wu X., Chen J. (2003) MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421, 957–961 [DOI] [PubMed] [Google Scholar]
- 44. Nguyen M. D., Shu T., Sanada K., Larivière R. C., Tseng H. C., Park S. K., Julien J. P., Tsai L. H. (2004) A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat. Cell Biol. 6, 595–608 [DOI] [PubMed] [Google Scholar]
- 45. Garrett S., Auer K., Compton D. A., Kapoor T. M. (2002) hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12, 2055–2059 [DOI] [PubMed] [Google Scholar]
- 46. Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. (2006) Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell 17, 680–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neumann B., Walter T., Hériché J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., Cetin C., Sieckmann F., Pau G., Kabbe R., Wünsche A., Satagopam V., Schmitz M. H., Chapuis C., Gerlich D. W., Schneider R., Eils R., Huber W., Peters J. M., Hyman A. A., Durbin R., Pepperkok R., Ellenberg J. (2010) Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shim S. Y., Samuels B. A., Wang J., Neumayer G., Belzil C., Ayala R., Shi Y., Shi Y., Tsai L. H., Nguyen M. D. (2008) Ndel1 controls the dynein-mediated transport of vimentin during neurite outgrowth. J. Biol. Chem. 283, 12232–12240 [DOI] [PubMed] [Google Scholar]
- 49. Shim S. Y., Wang J., Asada N., Neumayer G., Tran H. C., Ishiguro K., Sanada K., Nakatani Y., Nguyen M. D. (2008) Protein 600 is a microtubule/endoplasmic reticulum-associated protein in CNS neurons. J. Neurosci. 28, 3604–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., Tsai L. H. (2004) Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 [DOI] [PubMed] [Google Scholar]
- 51. Slee E. A., Zhu H., Chow S. C., MacFarlane M., Nicholson D. W., Cohen G. M. (1996) Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem. J. 315, 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gietz R. D., Woods R. A. (2002) Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol. Biol. 185, 471–486 [DOI] [PubMed] [Google Scholar]
- 53. Gietz R. D., Schiestl R. H. (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41 [DOI] [PubMed] [Google Scholar]
- 54. Kagawa S., Gu J., Honda T., McDonnell T. J., Swisher S. G., Roth J. A., Fang B. (2001) Deficiency of caspase-3 in MCF7 cells blocks Bax-mediated nuclear fragmentation but not cell death. Clin. Cancer Res. 7, 1474–1480 [PubMed] [Google Scholar]
- 55. Jänicke R. U., Sprengart M. L., Wati M. R., Porter A. G. (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 273, 9357–9360 [DOI] [PubMed] [Google Scholar]
- 56. Massip L., Caron P., Iacovoni J. S., Trouche D., Legube G. (2010) Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle 9, 2963–2972 [DOI] [PubMed] [Google Scholar]
- 57. Smeenk G., Wiegant W. W., Vrolijk H., Solari A. P., Pastink A., van Attikum H. (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 190, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. MacPhail S. H., Banáth J. P., Yu Y., Chu E., Olive P. L. (2003) Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat. Res. 159, 759–767 [DOI] [PubMed] [Google Scholar]
- 59. Mori D., Yamada M., Mimori-Kiyosue Y., Shirai Y., Suzuki A., Ohno S., Saya H., Wynshaw-Boris A., Hirotsune S. (2009) An essential role of the aPKC-Aurora A-NDEL1 pathway in neurite elongation by modulation of microtubule dynamics. Nat. Cell Biol. 11, 1057–1068 [DOI] [PubMed] [Google Scholar]
- 60. Liang Y., Yu W., Li Y., Yu L., Zhang Q., Wang F., Yang Z., Du J., Huang Q., Yao X., Zhu X. (2007) Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol. Biol. Cell 18, 2656–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pramparo T., Libiger O., Jain S., Li H., Youn Y. H., Hirotsune S., Schork N. J., Wynshaw-Boris A. (2011) Global developmental gene expression and pathway analysis of normal brain development and mouse models of human neuronal migration defects. PLoS Genet. 7, e1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vergnolle M. A., Taylor S. S. (2007) Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17, 1173–1179 [DOI] [PubMed] [Google Scholar]
- 63. DeLuca J. G., Moree B., Hickey J. M., Kilmartin J. V., Salmon E. D. (2002) hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Arbour N., Vanderluit J. L., Le Grand J. N., Jahani-Asl A., Ruzhynsky V. A., Cheung E. C., Kelly M. A., MacKenzie A. E., Park D. S., Opferman J. T., Slack R. S. (2008) Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J. Neurosci. 28, 6068–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brooks P. J. (2002) DNA repair in neural cells: basic science and clinical implications. Mutat. Res. 509, 93–108 [DOI] [PubMed] [Google Scholar]
- 66. Rass U., Ahel I., West S. C. (2007) Defective DNA repair and neurodegenerative disease. Cell 130, 991–1004 [DOI] [PubMed] [Google Scholar]
- 67. Sii-Felice K., Barroca V., Etienne O., Riou L., Hoffschir F., Fouchet P., Boussin F. D., Mouthon M. A. (2008) Role of Fanconi DNA repair pathway in neural stem cell homeostasis. Cell Cycle 7, 1911–1915 [DOI] [PubMed] [Google Scholar]
- 68. Lee M. S., Edwards R. A., Thede G. L., Glover J. N. (2005) Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the γ-H2AX histone tail. J. Biol. Chem. 280, 32053–32056 [DOI] [PubMed] [Google Scholar]
- 69. Eyers P. A., Erikson E., Chen L. G., Maller J. L. (2003) A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
- 70. Giunta S., Belotserkovskaya R., Jackson S. P. (2010) DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Peng Y., Woods R. G., Beamish H., Ye R., Lees-Miller S. P., Lavin M. F., Bedford J. S. (2005) Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 65, 1670–1677 [DOI] [PubMed] [Google Scholar]
- 72. Townsend K., Mason H., Blackford A. N., Miller E. S., Chapman J. R., Sedgwick G. G., Barone G., Turnell A. S., Stewart G. S. (2009) Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J. Biol. Chem. 284, 33939–33948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McManus K. J., Hendzel M. J. (2005) ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol. Biol. Cell 16, 5013–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cowell I. G., Sunter N. J., Singh P. B., Austin C. A., Durkacz B. W., Tilby M. J. (2007) γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One 2, e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim J. A., Kruhlak M., Dotiwala F., Nussenzweig A., Haber J. E. (2007) Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhu Q., Pao G. M., Huynh A. M., Suh H., Tonnu N., Nederlof P. M., Gage F. H., Verma I. M. (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nakada S., Chen G. I., Gingras A. C., Durocher D. (2008) PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9, 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chowdhury D., Xu X., Zhong X., Ahmed F., Zhong J., Liao J., Dykxhoorn D. M., Weinstock D. M., Pfeifer G. P., Lieberman J. (2008) A PP4-phosphatase complex dephosphorylates γ-H2AX generated during DNA replication. Mol. Cell 31, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Macůrek L., Lindqvist A., Voets O., Kool J., Vos H. R., Medema R. H. (2010) Wip1 phosphatase is associated with chromatin and dephosphorylates γH2AX to promote checkpoint inhibition. Oncogene 29, 2281–2291 [DOI] [PubMed] [Google Scholar]
- 80. Douglas P., Zhong J., Ye R., Moorhead G. B., Xu X., Lees-Miller S. P. (2010) Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates γ-H2AX. Mol. Cell. Biol. 30, 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moon S. H., Lin L., Zhang X., Nguyen T. A., Darlington Y., Waldman A. S., Lu X., Donehower L. A. (2010) Wild-type p53-induced phosphatase 1 dephosphorylates histone variant γ-H2AX and suppresses DNA double strand break repair. J. Biol. Chem. 285, 12935–12947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cha H., Lowe J. M., Li H., Lee J. S., Belova G. I., Bulavin D. V., Fornace A. J., Jr. (2010) Wip1 directly dephosphorylates γ-H2AX and attenuates the DNA damage response. Cancer Res. 70, 4112–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li B., Qi X. Q., Chen X., Huang X., Liu G. Y., Chen H. R., Huang C. G., Luo C., Lu Y. C. (2010) Expression of targeting protein for Xenopus kinesin-like protein 2 is associated with progression of human malignant astrocytoma. Brain Res. 1352, 200–207 [DOI] [PubMed] [Google Scholar]
- 84. Asteriti I. A., Rensen W. M., Lindon C., Lavia P., Guarguaglini G. (2010) The Aurora-A/TPX2 complex: a novel oncogenic holoenzyme? Biochim. Biophys. Acta 1806, 230–239 [DOI] [PubMed] [Google Scholar]