Background: Iron-storage Dps proteins are expressed in prokaryotes to confer resistance to specific stressful conditions.
Results: Dps from Porphyromonas gingivalis (PgDps) is able to bind heme through a conserved cysteine and protects DNA against H2O2-mediated degradation.
Conclusion: The heme sequestration property of PgDps confers resistance to heme toxicity.
Significance: This Dps protection strategy may be common among members of the order Bacteroidales.
Keywords: DNA Damage, Ferritin, Heme, Ligand-binding Protein, Oxidative Stress, Protein Structure, X-ray Scattering, DNA Protection, Dps
Abstract
The widely expressed DNA-protective protein from starved-cells (Dps) family proteins are considered major contributors to prokaryotic resistance to stress. We show here that Porphyromonas gingivalis Dps (PgDps), previously described as an iron-storage and DNA-binding protein, also mediates heme sequestration. We determined that heme binds strongly to PgDps with an apparent Kd of 3.7 × 10−8 m and is coordinated by a single surface-located cysteine at the fifth axial ligand position. Heme and iron sequestered in separate sites by PgDps provide protection of DNA from H2O2-mediated free radical damage and were found to be important for growth of P. gingivalis under excess heme as the only iron source. Conservation of the heme-coordinating cysteine among Dps isoforms from the Bacteroidales order suggests that this function may be a common feature within these anaerobic bacteria.
Introduction
Porphyromonas gingivalis is a Gram-negative anaerobic bacterium implicated in the pathogenesis of chronic periodontal disease, an inflammatory condition involving the supporting structures of the teeth (1). Recently, the organism has been associated with systemic diseases, including cardiovascular disease, rheumatoid arthritis, and also pre-term delivery of low birth weight infants (2–4). Like most microbial pathogens, P. gingivalis relies upon iron for a wide range of metabolic and signaling functions. Because of defects in synthesis of the tetrapyrrole ring, P. gingivalis is also a porphyrin auxotroph (5). Heme, being the most abundant source of iron and porphyrin in the mammalian host, is an essential nutrient for survival of this organism in vivo. Thus, P. gingivalis possesses several heme uptake systems to scavenge heme from host hemoproteins, to store heme on the surface of the organism, and for translocation into the protoplasm (6–10).
Paradoxically, internalized heme can mediate damage to cellular structures. Because of the high redox potential of free heme, high concentrations can cause protein inactivation, fatty acid oxidization, and DNA damage through peroxidase-like and monooxygenase-like activities (11). Furthermore, the release of iron during heme degradation can cause oxidative damage by the production of hydroxyl radicals via the Fenton reaction (Fe2+ + H2O2 → Fe3+ + OH•). Indeed, a number of organisms, including Staphylococcus aureus, have been reported to be sensitive to heme toxicity (12, 13).
Tolerance to heme in bacteria has been attributed to a number of the following mechanisms: (i) regulation of heme uptake; (ii) heme export; (iii) heme sequestration, and (iv) heme degradation (11). Heme uptake in P. gingivalis is inextricably linked with iron metabolism and is up-regulated in growth under iron limitation (14). The resulting influx of heme requires other mechanisms to neutralize heme toxicity, including sequestration and degradation. In Gram-negative bacteria, protoplasmic heme-binding proteins associated with heme uptake systems, such as the HemS family, have been proposed to act as a heme sequestration or degradation system (11). A BLAST search of the P. gingivalis genome failed to identify orthologs of HemS. Furthermore, the fate of intracellular heme in P. gingivalis is currently not known, and heme degradation pathways described in other bacteria such as the heme oxygenase family, including pigA (15), isdG (16), and hemO (17), are not present in P. gingivalis.
Through our study of heme-binding proteins in P. gingivalis (6), a number of heme-binding proteins were isolated from P. gingivalis lysate using heme-agarose purification and identification by peptide mass fingerprinting. One of the major bands with an apparent molecular mass at 18 kDa was identified as a Dps3 protein homolog. This protein has been reported previously in P. gingivalis as a DNA-binding protein, protecting cells from hydrogen peroxide attack (18). The widely expressed protoplasmic Dps proteins belong to the ferritin superfamily and are considered to be major contributors to prokaryotic resistance to general and specific stress conditions, especially oxidative stress (19). However, as an iron-storage protein, the capacity of P. gingivalis Dps (PgDps) to protect against the oxidative stress mediated by heme is unknown.
In this study, we describe a previously unknown heme binding property of PgDps. Spectroscopic analysis and structural modeling indicate that binding of heme is coordinated via a conserved surface cysteine. This was verified by site-directed mutagenesis. PgDps mediates tolerance to heme toxicity during growth of P. gingivalis using heme as the only iron source. At low heme concentrations, PgDps improves the efficiency of heme utilization, and at high heme concentrations, it prevents heme toxicity. Unlike most known Dps family proteins, DNA protection by PgDps is contributed by free heme chelation and ferroxidase activity rather than assembly of a protein shell via DNA binding as for other described Dps family proteins.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
P. gingivalis wild-type strain W83 and mutant derivatives were grown in enriched Tryptic Soy Broth (eTSB; per liter (w/v): 30 g of trypticase soy broth, 5 g of yeast extract, 0.5 g of l-cysteine, 2 mg of menadione, pH 7.5, and supplemented with hemin at various concentrations) or eTSB blood agar (eTSB medium plus 15 g/liter agar and 3% defibrinated sheep blood) at 37 °C in an anaerobic chamber (Don Whitley Scientific, Shipley, UK) with an atmosphere of 80% N2, 10% CO2, 10% H2. Escherichia coli strain DH5α was used for all plasmid construction work or BL21(DE3) as the expression host. All E. coli were grown in Luria-Bertani (LB) broth or agar. For antibiotic selection in E. coli, ampicillin was used at 100 μg/ml, together with 50 μg/ml kanamycin or 300 μg/ml erythromycin.
Cloning, Overexpression, and Purification of P. gingivalis Dps Protein
The P. gingivalis dps gene (PG0090) was amplified by PCR from strain W83 genomic DNA and cloned into pET24d(+) using XhoI/NcoI restriction sites. Primers used for the construction are listed in the Table 1. The stop codon of the Dps gene (dps) was replaced by a PreScission protease cleavage site (LEVLFQ↓GP) followed by a C-terminal Strep-tag II (WSHPQFEK). The resulting pETdps plasmid was checked by DNA sequencing, and the correct construct was transformed into E. coli BL21(DE3) expression host. Expression cultures were grown at 37 °C in LB broth with 50 μg/ml kanamycin. Cells were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside at A600 of 0.7 and collected 3 h after induction. Cells were harvested by centrifugation at 6500 × g for 15 min and resuspended in cold 50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, pH 8.0, at 4 °C and lysed by pulse sonication in an ice bath. The soluble fraction of the lysate was collected by ultracentrifugation at 150,000 × g, 30 min, 4 °C, and purified over a Strep-Tactin-Sepharose (IBA, Goettingen, Germany). Bound PgDps was eluted using 2.5 mm desthiobiotin, and the Strep-tag II was removed by HRV 3C protease (Novagen, Melbourne, Australia). De-tagged PgDps was passed through a glutathione-Sepharose (GE Healthcare) followed by passage over a Strep-Tactin-Sepharose to remove the protease and cleaved tags, respectively. Purified protein was buffer exchanged into 50 mm Tris-HCl, pH 8.0, 150 mm NaCl using a PD-10 desalting column (GE Healthcare).
TABLE 1.
Primers and probes
| Primers or probes | Nucleotide sequence (5′–3′)a | Product or target |
|---|---|---|
| Primers | ||
| ermKpnIF | aaGGTACCCCCGATAGCTTC | ermFermAM cassette |
| ermSphIR | attGCATGCCGAAGCTGTCAGTAGTATACC | ermFermAM cassette |
| tetQKpnIF | tcGGTACCAACCCCGTTATATACATTCATGT | tetQ cassette |
| tetQSphIR | actGCATGCCAAGTTCTAATGCTTCTATCTC | tetQ cassette |
| dpsfrASacIF | aaGAGCTCATAGAACTGGAATCTGCCAAG | pWDdps construction |
| dpsfrAKpnIR | caGGTACCAATCTTCTACGTTTTTAATTGTGTTCC | pWDdps construction |
| dpsfrBSphIF | atGCATGCTACGCTGCCAAGTAAAAAC | pWDdps and pWCdps construction |
| dpsfrBHindIIIR | ccAAGCTTCTCCGAACATCTCAATACC | pWDdps and pWCdps construction |
| dpsfrCEcoRIF | gtGAATTCGGACTTCTTCTGGAAAGCA | pWCdps construction |
| dpsfrCSacIR | ttGAGCTCTCACTCTTGGGTTTCTGC | pWCdps construction |
| dpsFrCF | CGATACGGGAGTCTTCAATATC | Crossover confirmation PCR |
| dpsFrBR | ATACCTGCGAAGGGTAGAT | Crossover confirmation PCR |
| dpsF | AAAAGATTCTTGAAGTAACGGGTT | Sequencing primer |
| dpsR | TCCAAACGAGCTTCTCTTG | Sequencing primer |
| DpsCteminus XhoIR | aCTCGAGTTATTTTTCAAACTGCGGATGGCTCCACGGGCCCTGAAACAGCACTTCCAGCTTGGCAGCGTAGGCAGA | pETdps construction |
| DpsNcoIF | agaCCATGGgtATGAAAAAGATTCTTGAAGTAACG | pETdps construction |
| DpsC101AFA | GAACTCGTTGCGGCTGCAAGTACGCTGAAG | pDpsC101A mutagenesis |
| DpsC101AFB | AGTACGCTGAAGAATGTGAC | pDpsC101A mutagenesis |
| DpsC101ARA | TGCAGCCGCAACGAGTTCGTGCTCTTC | pDpsC101A mutagenesis |
| DpsC101ARB | GTGCTCTTCCTTCACTTCTG | pDpsC101A mutagenesis |
| RTdpsF | GTGAAGGAAGAGCACGAACTCG | qPCR of dps |
| RTdpsR | CTGCCTCACCGGCAACTTC | qPCR of dps |
| RT16sF | TCGGTAAGTCAGCGGTGAAAC | qPCR of 16S RNA |
| RT16sR | ATTGCGAAGGCAGCTTGC | qPCR of 16S RNA |
| Fluorescent-quencher probes | ||
| FAMRTdpsPb | CGCTGCAGATCATCATGGCGAAGGAGCGTGC | qPCR probe for dps |
| CY5RT16sPc | AGCGCTCAACGTTCAGCCTGCCGTTGA | qPCR probe for 16S RNA |
a Restriction sites are in bold; PrescissionTM cleavage site is in capital italics; Strep-tag II is underlined, and irrelevant bases are in lowercase.
b 5′-Fluorescent dye is 6-carboxyfluorescein (FAM), and 3′-quencher dye is IowaBlackTM FQ (Integrated DNA Technologies Inc., Coralville, IA).
c 5′-Fluorescent dye is Cy5TM, and 3′-quencher dye is IowaBlackTM RQ (Integrated DNA Technologies Inc.).
dps-deletion Mutant Construction
A 1.25-kb upstream fragment and a 1.1-kb downstream fragment to the dps gene were amplified from chromosomal DNA of P. gingivalis W83 strain by PCR and inserted into SacI/KpnI and SphI/HindIII sites in pUC19, respectively. A 2.2-kb ermF/ermAM cassette was amplified from the plasmid pVA2198 (20) and inserted into KpnI/SphI sites in the modified pUC19 to create plasmid pWDdps. Constructed plasmids were verified by DNA sequencing prior to being electroporated into P. gingivalis W83 for homologous recombination into the genome to create a deletion of the dps gene. Transformed cells were selected on 5 μg/ml erythromycin eTSB blood agar plates. Resistant clones were further confirmed for deletion of the dps gene by PCR and DNA sequencing and are designated as Δdps mutants. The dps-complemented mutant was constructed in the same way except that the upstream fragment included the dps gene. A 2.6-kb tetQ cassette amplified from plasmid pNFD13-2 (21) was used to replace the ermF/ermAM cassette to create plasmid pWCdps. The complementation plasmid was electroporated into the Δdps mutant for homologous recombination and reinsertion of the dps gene. Tetracycline-resistant clones (designated dps+ mutants) were selected on 1 μg/ml tetracycline eTSB blood agar, and genotype was confirmed by PCR and DNA sequencing. All primers used in the genetic manipulations are listed in Table 1.
Mutation of Cys-101 to Alanine in PgDps
The single Cys-101 residue in PgDps was mutated to alanine using the pETdps plasmid and SLIM mutagenesis (22) to generate plasmid pDpsC101A. DNA sequencing was used to confirm the desired mutation. Primers used are listed in Table 1. Protein expression and purification of the PgDpsC101A mutant follow the same procedures as described for the wild-type PgDps.
Growth Experiments
The cultures of P. gingivalis wild-type W83, dps deletion mutant (Δdps), and complementation mutant (dps+) were prepared and inoculated for growth curve analysis as described previously (6). Briefly, the start cultures were adjusted to A600 of 1.0 prior to inoculation at 1:25 into eTSB medium supplemented with 25, 5, or 0.5 μm heme in individual Teflon screw-cap test tubes. All media used in the growth studies contained 125 μm dipyridyl to chelate free iron ions. Tubes were capped tightly and incubated at 37 °C in a water bath. At predetermined time points, tubes were vortexed, and absorbance at 600 nm was recorded using a Beckman DU 640 spectrophotometer. Three independent experiments were performed in triplicate. Cells were collected at A600 of 1.0 for Western blot analysis, and mRNA was stabilized with RNAprotectTM (Qiagen, Melbourne, Australia) prior to RNA extraction using the RNAqueous kit (Ambion, Melbourne, Australia) for quantitative RT-PCR analysis.
Quantitative RT-PCR
Total RNA from stabilized cells was extracted using the RNAqueous micro kit as per the manufacturer's instructions (Ambion, Melbourne, Australia). Reverse transcription was carried out on 2 μl of total RNA using the AffinityScript quantitative PCR cDNA synthesis kit (Stratagene, Melbourne, Australia) in a volume of 10 μl. Quantitative PCR was carried out in 25-μl singleplex reactions using 5 μl of a 1:100 dilution of the cDNA along with TaqMan probe/primers against dps or 5 μl of a 1:200 dilution of cDNA for the calibrator gene 16S ribosomal RNA (see primers and probes in Table 1) on a Mx3005P real time PCR system using the Brilliant® II quantitative PCR master mix (Stratagene). Calibrations of the primers and probes along with normalization of the data were as described previously (23).
Size-exclusion Chromatography with Multi-angle Laser Light Scattering (SEC-MALLS)
SEC-MALLS data were collected for purified PgDps, PgDps-heme (molar ratio 1:1), with and without 2 mm TCEP. Typically, 400 μl of protein sample at 100 μm was loaded onto a Superdex 200 10/300 GL gel filtration column coupled to a miniDAWN light-scattering unit (Wyatt Technology, Santa Barbara, CA) and a DSP refractometer (Optilab, Phoenix, AZ). The system was equilibrated with buffer containing 50 mm Tris-HCl, pH 8.0 and 150 mm NaCl prior to each experiment. To reduce the base line, two 0.22- and 0.1-μm filters were used for on-line filtration of the mobile phase. To determine the molecular mass of each species, the system was calibrated to an absolute scale using the intrinsic Rayleigh scattering of toluene. A uniform refractive index to concentration gradient (dn/dc) of 0.185 ml/g was assumed for all proteins. The calibration was verified using a sample of bovine serum albumin on the system prior to analysis of the samples.
Small-angle X-ray Scattering (SAXS) Data Acquisition, Analysis, and Modeling
Recombinant PgDps protein sample freshly purified from SEC-MALLS was collected, and a 0.41 mg/ml sample was analyzed immediately at 20 °C using a SAXSess instrument (Anton Paar, Graz, Austria) with a10-mm line collimation and a charge-coupled device detector. All data reduction and analysis followed the procedures described in Jeffries et al. (24). The low resolution ab initio shape reconstructions were performed using the program DAMMIF (25) as described in Jeffries et al. (26). The normalized spatial discrepancy (NSD) value of a set of 10 DAMMIF models was 0.517 ± 0.005.
Homology Modeling
A homology model of monomeric PgDps was generated using the Phyre protein structure prediction server (27). The Phyre E-value is generated internally from the Phyre profile-profile alignment algorithm, and it is different from the E-value from BLAST. The top scoring homology model from Phyre was used as the final model for the PgDps monomer. Twelve monomer models were then assembled following the same subunit arrangements and a 3-fold symmetry as in Protein Data Bank code 3IQ1 to form the PgDps dodecamer. In general, the dodecamer consists of four sets of identical trimers. The goodness-of-fit of homology model for the dodecamer to the SAXS data was evaluated using the program CRYSOL (28).
PgDps Heme Binding Assays
Tetramethylbenzidine (TMBZ) Heme Staining
Heme solutions were prepared immediately before use. Fresh 10 mm stock solution of hemin (purity >98% HPLC, Fluka, Sydney, Australia) in 0.1 m NaOH was diluted into 50 mm Tris-HCl, 150 mm NaCl, pH 8.0, at 1:1000 ratio for binding assays. PgDps was pre-incubated with heme for 15 min prior to loading onto native polyacrylamide gel or boiled in SDS-PAGE sample buffer with or without 2% β-mercaptoethanol for denaturing gels. Gels were stained with TMBZ as described previously (6). The peroxidase activity of the bound heme in PgDps promotes H2O2 oxidation of TMBZ to a visible blue precipitate on the gel.
UV-Visible Absorption Spectroscopy
All absorbance spectra were recorded in an ultra-micro quartz cuvette (100 μl) with a 10-mm path length (Jenway, Staffordshire, UK) using a Beckman DU800 spectrophotometer (Beckman Coulter, Indianapolis, IN).
Fluorescence Quenching
All spectral determinations were carried out in a quartz cuvette (1 ml) with a 10-mm path length (Starna Ltd., Sydney, Australia) using a Luminescence spectrometer LS 50B (PerkinElmer Life Sciences). Tryptophan fluorescence intensity from 500 nm PgDps or PgDpsC101A was recorded three times at each heme titration (100 nm) in 50 mm Tris-HCl, 150 mm NaCl, pH 8.0 (using fresh 10 mm heme stock dissolved in 0.1 m NaOH). Similarly, 100 nm heme was sequentially titrated into 500 nm PgDps in 50 mm PIPES-HCl, 150 mm NaCl, pH 6.5 (using fresh 10 mm heme stock dissolved in DMSO). The three spectra were averaged to produce the final spectrum. Tryptophan fluorescence was observed using an excitation wavelength of 295 nm, and fluorescence intensity was measured between 250 and 500 nm. Solvent effect and dilution effect were considered in the data analysis. Inner filter effects were corrected using sequential titration of heme stock into 200 nm N-acetyltryptophanamide control. For each set of titration data, the wavelength that gave the largest intensity changes between the apo- and bound-states was chosen for analysis (29). Thus, 344 nm was chosen for the PgDps pH 8.0 data set or 351 nm for PgDps at pH 6.5 and 358 nm for the PgDpsC101A data set. The dissociation constant (Kd) in each titration experiment was obtained by plotting the corrected fluorescence intensity at each titration step against the concentration of heme and fitted to the Equation 1, assuming 1:1 binding stoichiometry using OriginPro 8.6 software (30).
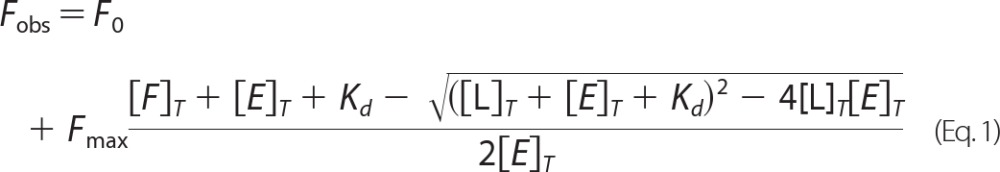 |
where Fobs is the observed fluorescence; F0 is the initial fluorescence; Fmax is the maximum amplitude of fluorescence quenching; [L]T is the total ligand concentration; [E]T is the total concentration of protein, and Kd is the apparent dissociation constant.
Solid-phase Ligand Binding Assay
Heme was used to coat polystyrene plates (Starna Ltd., Sydney, Australia) in 0.1 m sodium bicarbonate buffer, pH 9.0, overnight at room temperature. Plates were blocked with freshly prepared 0.05% Tween 20 in 20 mm Tris-HCl, 500 mm NaCl, pH 7.5, at room temperature for 30 min. Serial dilutions of apo-PgDps were applied onto the plate in 50 mm Tris-HCl, 150 mm NaCl, pH 8.0, and incubated at 37 °C for 1 h. Bound PgDps was detected by anti-Dps polyclonal antibodies (a kind gift from Drs. K. Nakayama and M. Shoji, Nagasaki University, Japan) and secondary alkaline phosphatase-conjugated antibody. Development was monitored by hydrolysis of the substrate p-nitrophenyl phosphate in 5 mm Tris-HCl, pH 9.5, at 405 nm using a Microplate reader (Bio-Rad). The apparent dissociation constant Kd was defined as the heme concentration corresponding to half the maximum response.
Ferene S Iron Staining
PgDps was preincubated with ferrous ammonium sulfate in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl prior to loading onto the native gel. The PgDps-iron complex was detected by 1 mm ferene S in 15 mm thioglycolic acid and 2% (w/w) acetic acid.
Ferroxidase Activity by PgDps
Fresh 300 μm ferrous ammonium sulfate in aqueous solution was added to 5 μm PgDps in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl. The kinetics of ferrous iron oxidation to Fe3+ by PgDps at room temperature were measured at 305 nm using a Beckman DU 640 spectrophotometer (Beckman Coulter, Indianapolis, IN). Data were analyzed using Prism 3.0 (GraphPad Software Inc., La Jolla, CA).
DNA Protection Assay
DNA protection against oxidative stress was assessed in vitro using 10 nm of 2.2-kb PCR-amplified linear DNA. Purified DNA was preincubated with (or without) 30 μm apo-PgDps and 10 μm heme or 10 μm Fe(NH4)2SO4 in PBS at 37 °C in an anaerobic chamber for 1 h in the dark. Subsequently, 10 mm hydrogen peroxide was added to the mixture and incubated anaerobically at 37 °C in the dark for 15 min before being mixed with TAE buffer prior to removal from the anaerobic environment. The mixture was separated on 1% agarose gel in TAE buffer and DNA visualized by ethidium bromide staining.
DNA Binding Assay
Agarose gel retardation assays were used to assess nonspecific DNA binding by PgDps. 300 ng of purified 2.2-kb linear DNA was preincubated with 30 μm PgDps with or without 300 μm heme or 1.5 mm Fe(NH4)2SO4 in PBS at 4 °C and 37 °C for 1 h. Subsequently, the mixtures were separated on 1% agarose in TAE buffer. As an additional control, iron-loaded PgDps was purified through a PD-10 desalting column (GE Healthcare) prior to incubation with DNA as above.
Statistical Analysis
Prism version 3.03 software (GraphPad Software Inc.) was used for all statistical analyses. Quantitative PCR data and protein expression-integrated densities from Western blots were compared using one-way analysis of variance with Bonferroni's correction and 95% confidence intervals. p values of less than 0.05 were considered significant.
RESULTS
Recombinant PgDps Protein Exists as a Stable Dodecamer
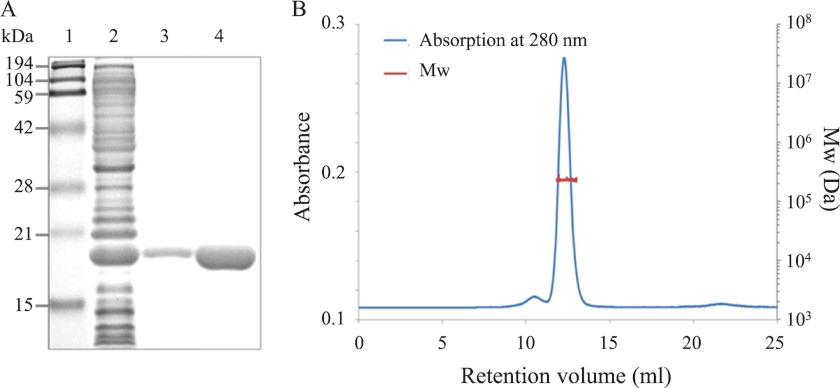
Recombinant Strep-tagged PgDps protein was expressed in E. coli, and the C-terminal affinity tag, Strep-tag II, was removed by HRV-3C protease digestion at an engineered cleavage site to eliminate potential tag interference. The de-tagged PgDps presented as a single band with molecular mass of ∼18 kDa (corresponding to the predicted mass of monomeric PgDps) under reduced denaturing conditions on SDS-PAGE (Fig. 1A). However, SEC-MALLS data indicate that native PgDps is monodisperse in solution with a molecular mass of 224 ± 3.5 kDa (Fig. 1B). Thus, similar to other Dps family proteins (31), PgDps self-assembles into a dodecamer or 12-subunit complex. Because of the presence of a single unpaired cysteine in each monomer, disulfide bridge formation was a possible contributor in the dodecamer architecture observed. Therefore, the oligomerization status of PgDps was re-evaluated by pretreating with the irreversible reducing agent TCEP before analysis by SEC-MALLS. The TCEP-treated PgDps was found to elute at a similar retention volume, with the same molecular mass by SEC-MALLS as for non-treated PgDps dodecamer (data not shown). These results suggest that dodecameric formation of PgDps is independent of a cysteine disulfide link.
FIGURE 1.
Recombinant PgDps protein exists in solution as a dodecamer. A, purified recombinant PgDps with or without affinity Strep-tag II appears as a monomer on denaturing SDS-polyacrylamide gel. Lane 1, molecular mass markers; lane 2, E. coli lysate; lane 3, purified tagged PgDps; lane 4, de-tagged PgDps. B, molecular mass determination of PgDps by SEC-MALLS. The elution profile is shown as the blue line. The red line superimposed on the main elution peak indicates the molecular weight distribution of the protein. The experimental molecular mass for PgDps is 224 kDa with a polydispersity index of 1.001 ± 0.021, consistent with a dodecameric complex.
PgDps Dodecamer Has a Spherical Shape with a Hollow Core
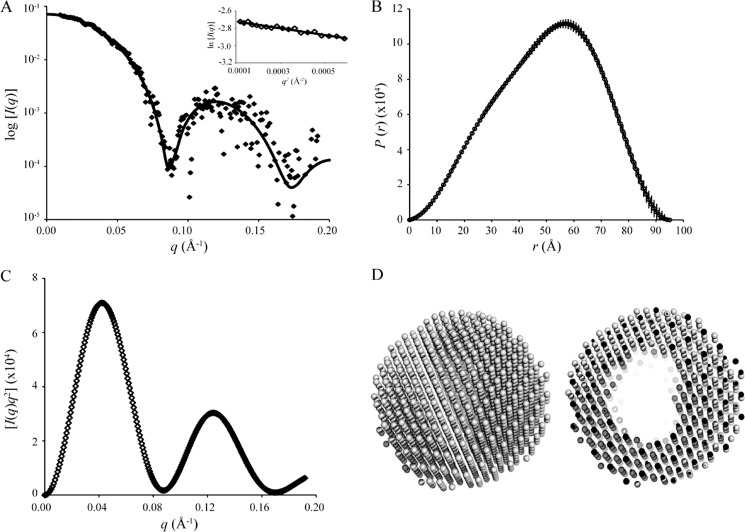
SAXS data were collected for PgDps dodecamer samples taken from SEC-MALLS-eluted fractions, and the associated structural parameters are summarized in Table 2. The scattering profile I(q) (Fig. 2A) shows a typical scattering feature of a spherical particle with I(q) decreasing rapidly from its maximum at I(0) and oscillating with increasing q. Guinier analysis of the scattering data gives the expected linear fit for a monodisperse solution of identical particles and yields a radius of gyration (Rg) value of 37.9 ± 0.3 Å (Fig. 2A, inset). The atom-pair distribution function P(r) calculated using GNOM (Fig. 2B) (32) shows a single maximum at r ∼56.5 Å, with a shoulder at smaller r ∼30 Å and a maximum linear dimension (Dmax) of 95 Å. These features are consistent with a spherical shape with a hollow core (33). The Rg value determined from the second moment of P(r) agrees with the value obtained from the Guinier analysis (Table 2).
TABLE 2.
Structural parameters determined from SAXS experiments
| Structural parameters | |
| I(0) (from P(r)) | 0.0700 ± 0.0007 cm−1 |
| Rg (from P(r)) | 37.9 ± 0.3 Å |
| I(0) (from Guinier) | 0.0697 ± 0.0007 cm−1 |
| Rg (from Guinier) | 37.7 ± 0.4 Å |
| Dmax | 95 Å |
| Molecular mass (Mr) determination | |
| Partial specific volume | 0.75 cm3 g−1 |
| Contrast (Δρ) | 2.734 × 1010 cm−2 |
| Concentration from A280 nm | 0.41 mg ml−1 |
| Mr from I(0) | 245 kDa |
| Mr from AutoPorod | 216 kDa |
| Mr for dodecamer calculated from sequence | 215 kDa |
FIGURE 2.
SAXS profiles and ab initio shape reconstruction of dodecameric PgDps. A, scattering data, I(q) versus q, of PgDps (in dots), corrected for beam geometry effects, and the corresponding fit of the homology model to the data (χ2 = 0.74). Inset shows Guinier plot and associated linear fit (solid line) to the data for the Guinier region, qRg <1.3. B, P(r) profile of PgDps, calculated for a maximum linear dimension, Dmax, of 95 Å. C, Kratky plot of the scattering data is characteristic of a spherically shaped scattering particle. D, ab initio shape reconstruction calculated from the scattering data from PgDps represented by the averaged and filtered dummy atom models (from DAMMIF). The left panel shows an external view indicating that PgDps is a sphere; the right panel shows a sliced view revealing the hollow interior.
A Kratky plot (I(q)q2 versus q) (Fig. 2C) provides an indication of the shape and folded state of a protein. Stably folded spherical proteins, or protein assemblies, typically yield multiple Gaussian peaks with decreasing intensity as q increases. The Kratky plot of PgDps displayed perfect Gaussian peaks with minimal I(q)q2 decay close to zero, confirming that the dodecamer is folded and is spherical. The molecular mass of the dodecamer determined using AutoPorod (34) or from I(0) (35) is in reasonable agreement with the SEC-MALLS results that indicate PgDps is present in solution as a stable dodecamer (Table 2). Ten ab initio reconstructed models were averaged and filtered to arrive at the final model, which also supports the conclusion from SAXS data analysis that PgDps has a spherical shape with a hollow core (Fig. 2D).
PgDps Is a Heme-binding Protein
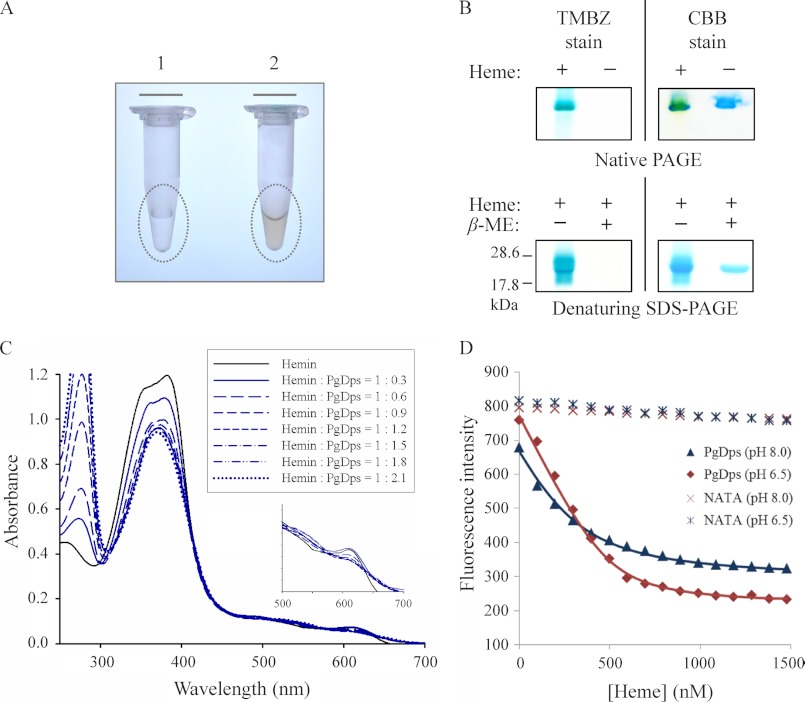
Recombinant Strep-tagged Dps protein was expressed in E. coli with and without 1 mm δ-aminolevulinic acid (δ-ALA) and 40 μm FeCl3 supplementation in LB broth. In the presence of δ-ALA, purified tagged PgDps exhibited a reddish coloration and was found to be heme-bound (holo-PgDps), in contrast to the colorless heme-free form (apo-PgDps) when expressed without δ-ALA (Fig. 3A). As a precursor of heme biosynthesis, δ-ALA supplementation along with Fe3+ has been shown to increase heme production in E. coli and holo-hemoprotein synthesis (36). Further assays were carried out to confirm and evaluate heme binding properties of PgDps. De-tagged apo-PgDps was pre-incubated with heme prior to separation on native PAGE and denaturing SDS-polyacrylamide gels under reducing or non-reducing conditions. Gels were initially stained with TMBZ to detect the presence of heme, followed by Coomassie Brilliant Blue G-250 staining for protein. As shown in Fig. 3B, heme was found to be associated with PgDps under native conditions and also under non-reducing, denaturing conditions. However, denaturation under reducing conditions containing β-mercaptoethanol resulted in the loss of heme binding, suggestive of a role for the single cysteine of PgDps in heme coordination (Fig. 3B).
FIGURE 3.
PgDps binds ferric heme with a high affinity. A, eluted fraction of purified recombinant Strep-tagged PgDps from E. coli cultures supplemented without (tube 1) or with (tube 2) δ-aminolevulinic acid. The reddish colorization of sample in tube 2 indicates the expressed PgDps is complexed with heme. B, pseudo-peroxidase activity of PgDps-heme complex as detected by TMBZ staining of native PAGE and denaturing SDS-polyacrylamide gels. PgDps was pre-incubated with heme for 15 min prior to loading onto native polyacrylamide gel or boiled in SDS-PAGE sample buffer with or without 2% β-mercaptoethanol (β-ME) in denaturing SDS-PAGE. Gels were re-stained with Coomassie Brilliant Blue G-250 (CBB) to show the apo-PgDps. C, changes in UV-visible spectra of hemin upon sequential titration of apo-PgDps at pH 8.0. D, maximal tryptophan fluorescence quenching of apo-PgDps at 344 nm, pH 8.0, or 351 nm, pH 6.5, by sequential titrations of heme as described under “Experimental Procedures.” Background quenching was obtained using N-acetyltryptophanamide (NATA) under the same conditions.
Titration of PgDps into heme solution revealed a dose-dependent and saturable change in the heme UV-visible absorbance spectrum. Hemin blank in 50 mm Tris-HCl, 150 mm NaCl, pH 8.0, displayed a typical absorption spectrum with a major Soret peak at 385 nm and a shoulder at 365 nm representing a mixture of dimeric and monomeric ferric heme at this pH (Fig. 3C). Addition of apo-PgDps to heme caused a progressive consolidation of the Soret bands into a single peak at 368 nm with a progressive decrease in the maximum absorption intensity, confirming the specific binding between PgDps and heme (Fig. 3C). The observed change in the Soret region was essentially complete at ∼1:1 ratio of hemin/PgDps, with no further change seen upon addition of excess apo-PgDps.
Shifts in the visible spectrum were also observed with holo-PgDps by the emergence of peaks at 514, 544, and 650 nm indicative of high spin ferric heme and the disappearance of the bisheme-associated band around 610 nm (Fig. 3C, inset).
The strength of heme association was further investigated by quenching of intrinsic tryptophan fluorescence of PgDps. Sequential titrations of ferric heme into apo-PgDps resulted in a dose-dependent progressive and saturable quenching of PgDps intrinsic tryptophan fluorescence. This was accompanied by a red shift of the maximum tryptophan emission wavelength from 356 to 370 nm at pH 8.0 and 360 to 394 nm at pH 6.5 (data not shown). As heme exists predominantly in the monomeric form at pH 6.5 (37), determination of the dissociation constant (Kd) was more accurate at this pH rather than at pH 8.0 where dominant bisheme species will cause underestimation of the Kd values. Consequently, the maximal quenching of tryptophan fluorescence (351 nm) at pH 6.5 was plotted against heme concentrations and could be fitted into a 1:1 binding model to allow determination of the Kd at equilibrium of (3.72 ± 0.5) × 10−8 m (Fig. 3D). A similar value was derived from a solid-phase ligand binding assay (data not shown), confirming the accuracy of the observed Kd.
Heme Coordination by PgDps Is via the Only Cysteine
The frequency and intensity of the Soret band are known to be dependent on the electronic field of the fifth and sixth axial ligands of the heme iron. The character of holo-PgDps spectral shift (Fig. 3C) is reminiscent of a fifth coordination of the ferric heme iron by a single cysteine as seen in the heme regulatory motifs present in a number of heme-sensing transcriptional regulators (38, 39).
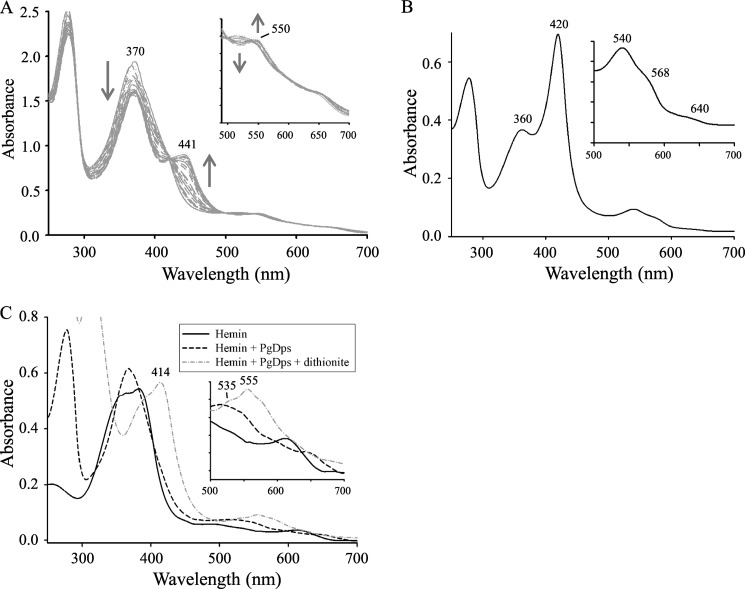
Phosphines have been used to probe the nature of cysteine-coordinated axial ligands of the heme iron in hemoproteins such as cytochrome P-450 and chloroperoxidase (40). TCEP is a hydrophilic trialkyl phosphine that is commonly used for the reduction of protein disulfide bonds to the corresponding thiols. Furthermore, TCEP is also a phosphine and acts as a potential polydentate for transition metal ions (41). Previously, it has been shown that binding of phosphines to ferric heme-thiolate complexes such as in cytochrome P-450CAM or chloroperoxidase produces a hyperporphyrin spectrum with a “split Soret” giving two maxima at 375 and 446 nm with a minor peak in the Q bands at 553 nm (42). Thus, to further validate the cysteine coordination of PgDps, TCEP was used to confirm the endogenous thiolate ligation of the proximal side of ferric heme in PgDps. Titrations of ferric PgDps-heme complex with the phosphine TCEP induced spectral changes with the appearance of a single isosbestic point in the Soret region (Fig. 4A). This result indicates that a single phosphine adduct is formed in each case without any detectable intermediate. Phosphine binding converts the single broad Soret peak of the ferric PgDps-heme at 368 nm to a “split” Soret with two peaks at 370 and 441 nm. As TCEP concentration was increased, the intensity of the Soret peak at 441 nm increased and the peak at 370 nm decreased. Concomitantly, the intensities of high spin ferric heme iron spectral feature peaks at 514, 540, and 650 nm decreased with the emergence of low spin ferric heme peak at 550 nm (Fig. 4A). The phosphine adduct UV-visible spectra of ferric PgDps resembled a phosphine-thiolate coordinated ferric heme, and they are analogous to the hyperporphyrin spectra reported earlier by Sono et al. (42) for phosphine binding trans to the endogenous thiolate ligand in ferric cytochrome P-450CAM.
FIGURE 4.
Heme iron coordination in PgDps. A, formation of a hyperporphyrin spectrum with a split Soret upon addition of TCEP to holo-PgDps. Excess apo-PgDps was used to saturate free ferric heme in solution, pH 8.0. Incremental additions of TCEP were then titrated into the protein-heme complex at molar ratios of TCEP/PgDps from 1:1 to up to 1000:1. Inset shows changes in the Q bands. B, UV-visible spectra of heme Strep-tagged PgDps. Inset shows in detail the Q band region. C, UV-visible absorption spectra of hemin incubated with PgDps and PgDps + dithionite. Inset shows changes in the Q bands. Peaks in the reduced spectra (PgDps + dithionite) are as labeled.
Additional evidence for thiolate ligation of the ferric heme in PgDps comes from our observation that the Strep-tagged PgDps displayed a spectrum consistent with a low spin six-coordinate ferric heme with an axial ligand consisting of a thiolate ligand trans to a neutral donor such as histidine, as observed in cystathionine β-synthases (43, 44) or in the M80A ferric mutant of P-450CAM + imidazole (45). Key spectral signature includes a δ peak at 360 nm, an intense Soret peak at 420 nm, coupled with an α band shoulder at 568 nm, a more intense β peak at 540 nm, and a weak charge transfer band at 640 nm (Fig. 4B). We therefore hypothesized that the histidine residue within the Strep-tag (WSHPQFEK) provides the proximal neutral ligand trans to the endogenous thiolate ligation of the ferric PgDps to give a typical histidine-thiolate hexa-coordinated ferric heme protein. Coupled with the SDS-PAGE results (Fig. 3B), these data suggest that the only cysteine residue (Cys-101) in PgDps coordinates the ferric heme iron as a thiolate ligand at the fifth axial position.
To further explore the heme coordination status, ferric holo-PgDps was subsequently reduced with excess dithionite. As shown in Fig. 4C, upon reduction, the Soret peak shifted to 414 nm with a prominent Q band at 555 nm with a flanking shoulder at 535 nm. These spectral patterns are similar to that observed for neutral thiol as a proximal ligand to ferrous heme in other five-coordinated ferrous heme proteins such as the ferrous H175C/D235L mutant of cytochrome c peroxidase (46) or the tetrahydrothiophene adduct of the myoglobin H93G cavity mutant (47). Thus, the spectra seem to indicate protonation of the cysteine upon reduction by dithionite to result in neutral thiol coordination of the ferrous heme.
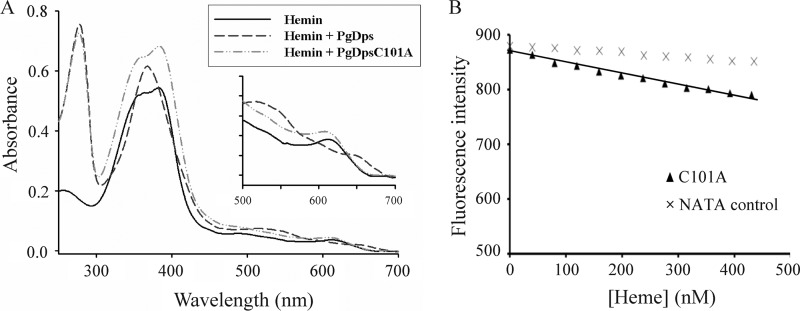
To confirm the role of the single cysteine in PgDps in coordinating the heme iron, a Cys-101 to alanine PgDps mutant (PgDpsC101A) was constructed and assayed for heme binding. As expected, titration of PgDpsC101A into heme did not alter the heme spectral characteristics (Fig. 5A). Furthermore, tryptophan fluorescence quenching data of PgDpsC101A titrated with heme suggest nonsaturable, nonspecific binding (Fig. 5B). These results confirm that Cys-101 is a critical ligand for PgDps heme coordination.
FIGURE 5.
Cysteine-101 of PgDps coordinates the heme iron. A, binding of ferric heme to PgDpsC101A gave similar spectral features to hemin alone, in contrast to the spectral changes induced by native PgDps. B, maximal tryptophan fluorescence quenching of apo-PgDpsC101A at 358 nm, pH 8.0, by sequential titrations of heme. Background quenching was obtained using N-acetyltryptophanamide (NATA) under the same conditions.
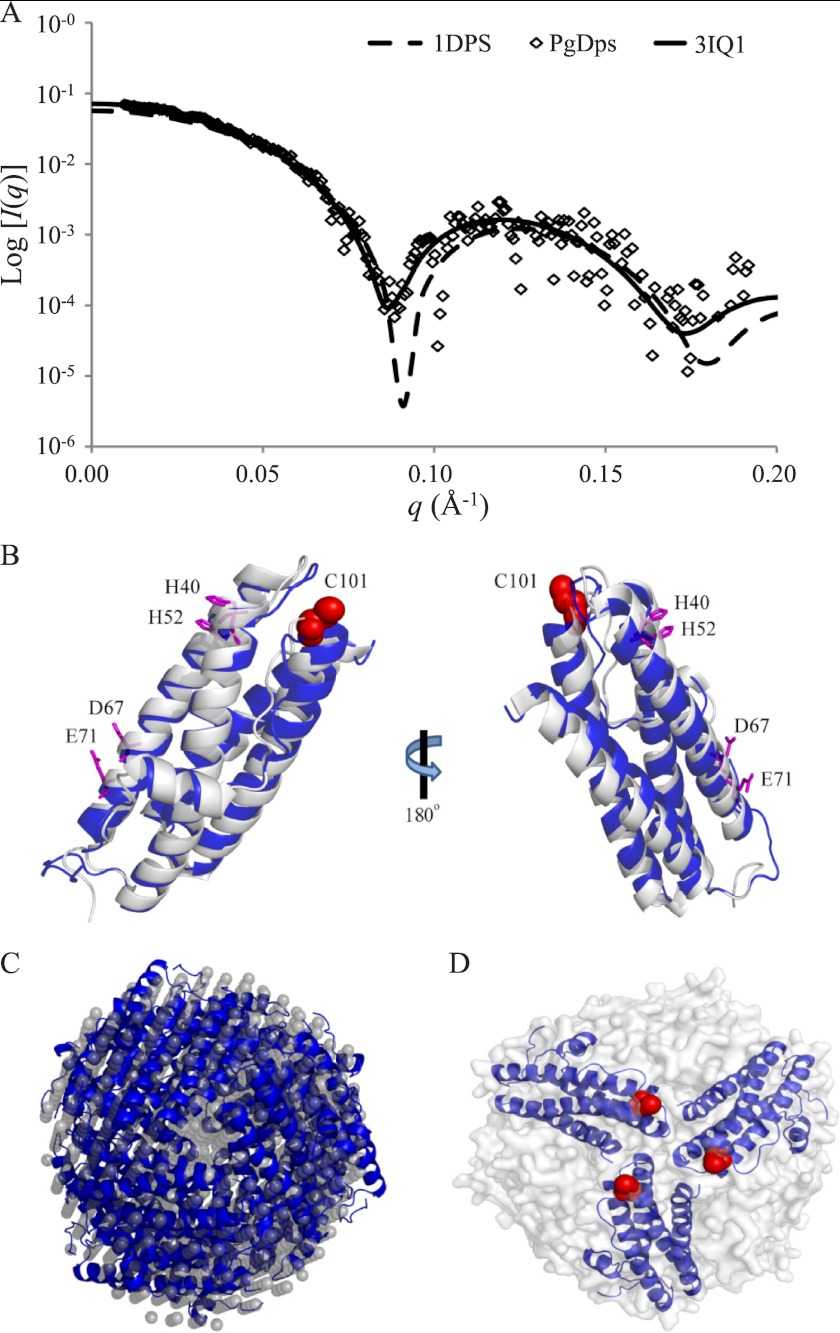
Homology Model of PgDps Dodecamer
A sequence BLAST search revealed that PgDps has the highest sequence identity (50%) to Dps from Vibrio cholerae O1 (PDB code 3IQ1) (E value 5.105 × 10−33, 71% positive). Using the Phyre protein structure prediction server (27), a homology model for monomeric PgDps was generated. The top scoring homology model (E value of 5.196 × 10−23 and an estimated precision of 100%) returned by the Phyre calculations was generated from alignments with the crystal structure of Agrobacterium tumefaciens Dps protein (PDB code 1O9R) (48). Interestingly, the crystal structures of these two Dps proteins (PDB code 1O9R and 3IQ1) are very similar, with a root-mean-square deviation of 1.37 for Cα positions. Furthermore, the subunit arrangements for the dodecamer in these two structures are equivalent (data not shown). In contrast, PgDps modeling based on the crystal structure of E. coli Dps protein (PDB code 1DPS) does not agree well with the experimental scattering profile of PgDps (Fig. 6A). The homology model predicts that PgDps monomer folds in a four-helical bundle typical of Dps proteins and ferritins. Superimposition of the homology model of monomeric PgDps with the monomer structure of Dps protein from Vibrio cholera O1 suggests that the two structures are also very similar with a root-mean-square deviation of 1.37 for Cα positions (Fig. 6B). To model the PgDps dodecamer, 12 monomer homology models were assembled following the same subunit arrangements and 3-fold symmetry as in Protein Data Bank code 3IQ1. The resulting homology model of the PgDps dodecamer fits very well to the experimental de-smeared scattering profile of PgDps (Fig. 2A), and it also fits well into the shape reconstruction model from SAXS (Fig. 6C). Hence, it is likely that the PgDps dodecamer may have a similar structure to Dps proteins from V. cholerae O1 or A. tumefaciens. The PgDps dodecamer model is composed of four repeats of identical trimers. This trimeric subunit arrangement has been seen in other members of the Dps family (for instance, Mycobacterium smegmatis) (49). In such a trimeric arrangement, the single cysteine residue (Cys-101) from each PgDps monomer is exposed partially on the surface of the dodecamer (Fig. 6D), and the conserved residues of the ferroxidase center (i.e. His-40, His-52, Asp-67, and Glu-71) are located in the central core of each subunit protein (Fig. 6B).
FIGURE 6.
Homology model of dodecameric PgDps. A, superimposition of experimental scattering data of PgDps with simulated scattering profiles of two Dps proteins (PDB code 1DPS and 3IQ1) using CRYSOL. B, homology model of a PgDps monomer (blue ribbon) based on the crystal structure of V. cholerae O1 Dps (gray ribbon; PDB code 3IQ1). Residues from the highly conserved ferroxidase center (His-40, His-52, Asp-67, and Glu-71) are shown as magenta sticks along with the location of the heme-coordinating residue Cys-101 (C101; red spheres). C, superimposition of ab initio shape reconstruction from Fig. 2D (in gray spheres) and homology model of dodecameric PgDps (in blue ribbons). D, within the PgDps dodecamer (shown as gray surface), partially exposed cysteine residues (red spheres) from each of three PgDps monomers form a potential heme-binding core.
Extensive PSI-BLAST searches uncovered significant sequence similarities between PgDps and other Dps proteins in a wide array of bacteria (data not shown). However, the heme-coordinating cysteine of PgDps was found mostly in species such as Prevotella denticola, Prevotella melaninogenica, and Bacteriodes clarus, which belong to the order Bacteroidales (Fig. 7). Interestingly, a majority of these Bacteroidales species have been described as potential oral or intestinal anaerobic pathogens.
FIGURE 7.
Multiple sequence alignment of PgDps and orthologs from the order Bacteriodales. Alignment was generated by ClustalW, and the conserved heme-coordinating cysteine is boxed. The full names of the host organisms are as follows: Bster, Bacteroides stercoris; Bclar, Bacteroides clarus; Begge, Bacteroides eggerthii; Bflux, Bacteroides fluxus; Binte, Bacteroides intestinalis; Bcell, Bacteroides cellulosilyticus; Bunif, Bacteroides uniformis; Bhelc, Bacteroides helcogenes; Coral, Capnocytophaga oral strain; Parab, Parabacteroides sp. D13; Pdist, Parabacteroides distasonis; Pjohn, Parabateroides johnsonii; Aindi, Alistipes indistinctus; Ppall, Prevotella pallens; Pnigr, Prevotella nigrescens; Pdisi, Prevotella disiens; Pinte, Prevotella intermedia; Pvero, Prevotella veroralis; Pmult, Prevotella multiformis; Pmel, Prevotella melaninogenica; Pbucc, Prevotella buccae; Pdent, P. dentalis; Pmars, Prevotella marshii; Pging, P. gingivalis.
Heme-induced Oligomerization of PgDps Dodecamers
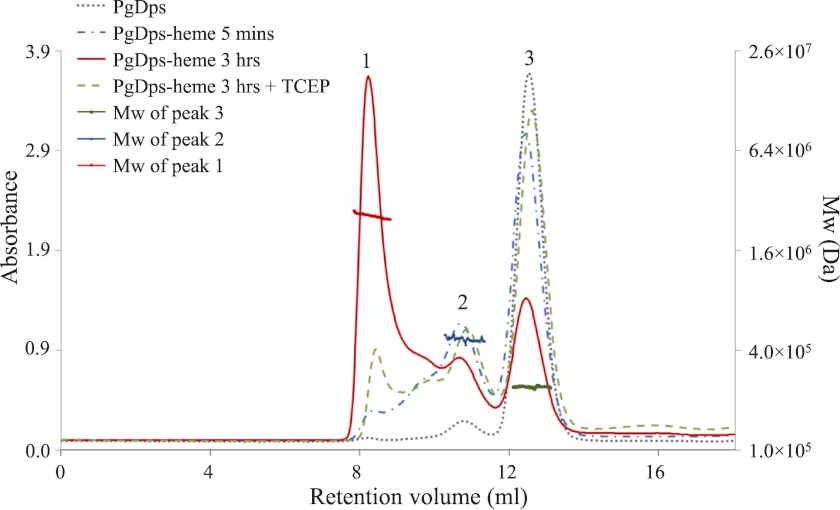
Surface location of the heme-chelating Cys-101 suggests that the heme-binding site in PgDps will be on the surface of the dodecamer. SEC-MALLS was employed to examine the association states of PgDps after incubation with hemin (ferric heme). In the absence of heme, native PgDps self-assembled as a stable dodecamer in solution (Fig. 1B). In the presence of hemin to PgDps in a molar ratio of 1:1, PgDps dodecamers were found to oligomerize into higher molecular mass species, and these were observed to increase with the time of incubation. After a 5-min incubation with hemin, the main population remained as a single dodecameric form with a molecular mass of ∼244 kDa, but a second peak started to emerge at twice the dodecameric mass of ∼494 kDa (Fig. 8). After a 3-h incubation, the major population shifted to a higher oligomeric state with a molecular mass of ∼2471 kDa, equivalent to ∼10 units of dodecameric PgDps. Of note, these high oligomeric species are not monodisperse suggesting multiple species. Furthermore, addition of TCEP to holo-PgDps caused dissociation of the higher order oligomers back to the single dodecameric form (Fig. 8).
FIGURE 8.
Heme-induced oligomerization of PgDps dodecamers. The oligomeric state of PgDps bound to heme in solution was determined by SEC-MALLS. PgDps was pre-incubated with heme at the molar ratio of 1:1 in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl at 37 °C. Molecular mass values of three main elution peaks were calculated using a dn/dc value of 0.185. Heme induced PgDps to form progressively higher oligomeric species over time. Molecular mass of the oligomeric species eluted as peak 1 was calculated at 2471 kDa, which is approximately equal to the molecular mass of 10 dodecamers. The higher oligomeric species could be dissociated back to the predominant dodecameric form after incubation with 2 mm TCEP.
PgDps Is Critical for P. gingivalis Tolerance of Excess Heme under Iron-depleted Conditions
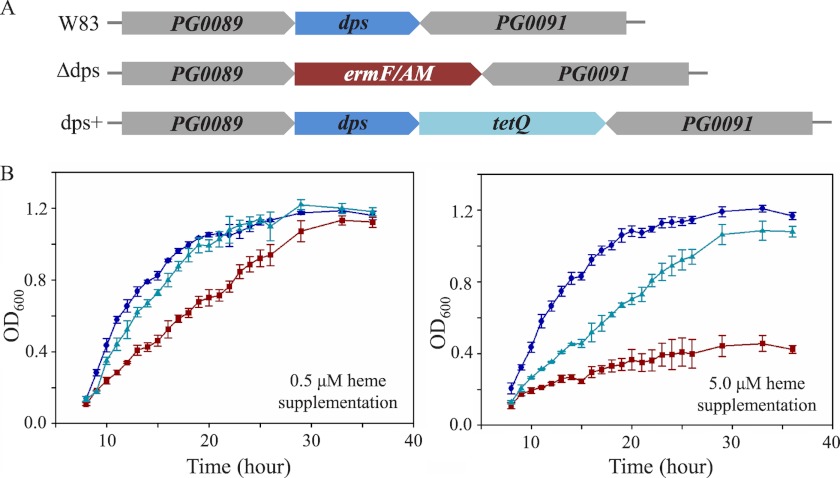
Dps family proteins are known to participate in intracellular iron storage and DNA protection from redox stress. To examine whether the novel heme binding properties of PgDps correlate with similar stress protection functions, especially stress attributed to heme toxicity, growth of wild-type P. gingivalis strain W83, dps-deleted mutant (Δdps), and a complementation mutant of dps deletion (dps+) was investigated over a range of heme concentrations in batch culture. P. gingivalis cultures were passaged once in heme-free medium prior to being inoculated into medium supplemented with different concentrations of heme. Furthermore, to mimic the physiological scarcity of ionic iron, which in vivo is estimated at 10−18 m (50), the iron chelator dipyridyl was added to the growth medium. As shown in Fig. 9, all strains grew robustly when supplemented with 0.5 μm heme as the sole iron source. However, under heme-replete conditions (5 μm), growth of Δdps mutant was noticeably inhibited compared with the wild-type and complemented mutant.
FIGURE 9.
PgDps is critical for P. gingivalis to grow under heme-replete conditions. A, gene map of wild-type strain W83, dps deletion mutant (Δdps), and complemented mutant (dps+) showing the position of the erythromycin resistance cassette (ermF/AM) and tetracycline resistance cassette (tetQ), respectively. B, growth of P. gingivalis wild-type W83 (●), Δdps mutant (■), and dps+ mutant (▴) in iron-depleted medium supplemented with 0.5 μm heme or 5 μm heme.
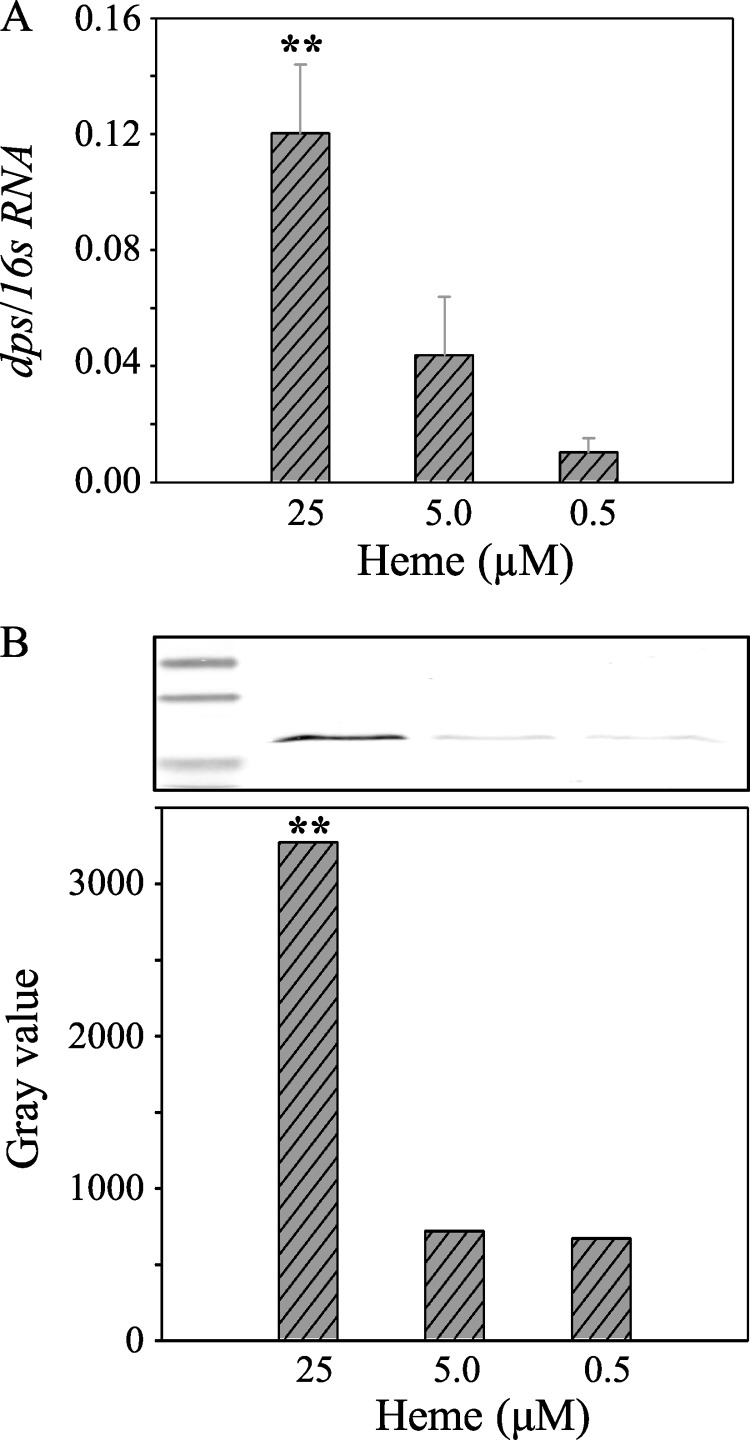
Dps Is Up-regulated in Response to Excess Heme
In the wild-type strain, dps transcription was found to be 10-fold higher under heme-excess conditions (25 μm) than under heme-limited conditions (0.5 μm; Fig. 10A). This result correlated well with PgDps production as quantified via densitometry measurements of Western blot using anti-Dps antibody against the whole culture (Fig. 10B). Taken together with growth defects of the Δdps mutant in heme-replete conditions, we propose that PgDps is up-regulated to sequester heme and neutralize potential heme toxicity.
FIGURE 10.
Differential regulation of PgDps in response to environmental heme concentrations under iron-depleted conditions. A, relative transcription of dps mRNA normalized against the housekeeping 16S RNA in P. gingivalis cultured under various levels of heme supplementation as the only iron source. Results represent mean value of three independent experiments in triplicate ± S.E. B, PgDps protein production by P. gingivalis grown under different heme supplementation was also quantified by densitometry of Western blot bands. Whole culture samples were normalized to A600 of 1.0 before being separated on SDS-PAGE. Polyclonal anti-Dps antibody was used in the immunoblot assay along with alkaline phosphatase-conjugated secondary antibody. ** denotes p < 0.01.
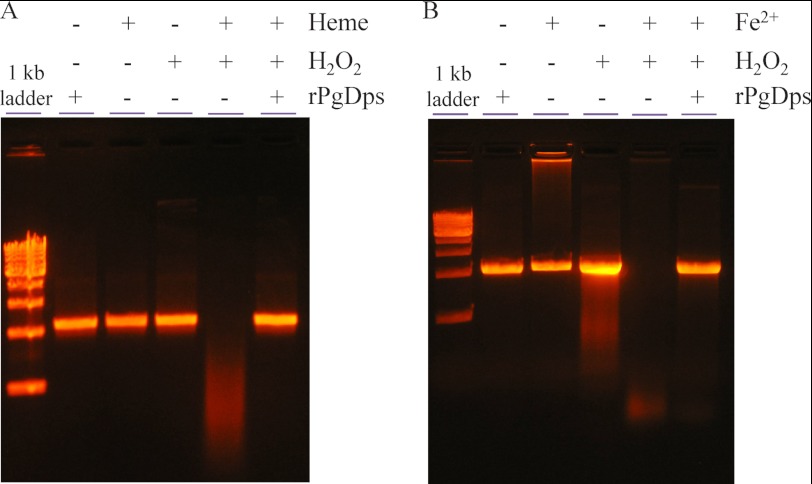
PgDps Protects DNA against Heme-induced Scission and Hydroxyl Radical Damage Generated by Ferrous Iron
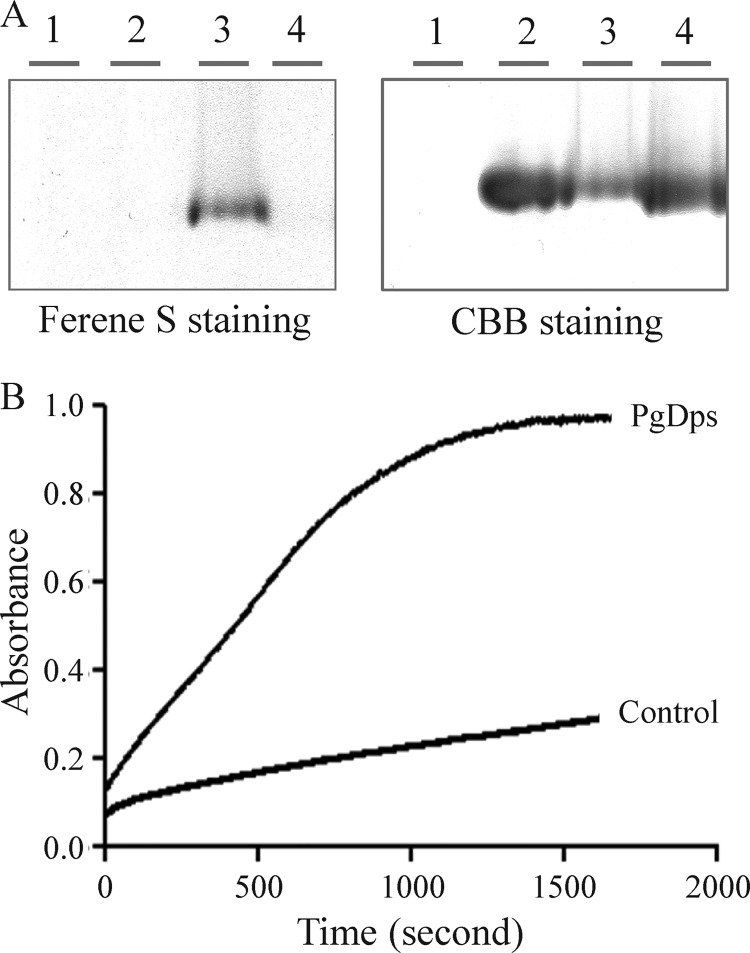
Free heme can cause nicking and subsequent degradation of DNA in the presence of a reducing agent (51). In this process, ferric iron in heme is first reduced to the ferrous form, which reacts with molecular oxygen to generate reactive species that cause DNA hydrolysis. In this respect, the Δdps mutant has been reported to be sensitive to hydrogen peroxide under heme-replete conditions (7.7 μm) (18). Consequently, heme toxicity may be related to DNA strand scission in the presence of H2O2. As shown in Fig. 11A, DNA was more vulnerable to degradation by free heme and H2O2 in vitro, but in the presence of PgDps, DNA was protected from degradation. Similarly, PgDps protects DNA from Fe2+/H2O2-mediated degradation (Fig. 11B). DNA protection could be achieved via two possible mechanisms as follows: (i) PgDps directly interacts with DNA and acts as a physical barrier against free radicals, or (ii) PgDps sequesters free Fe2+ ions and heme to prevent free radical formation. Although a number of Dps from other organisms have been described to be able to bind DNA nonspecifically using gel shift experiments (31) and a previous report described impairment of DNA migration in agarose gel by PgDps at 4 °C in the presence of ferrous ammonium sulfate (18), we were unable to detect nonspecific DNA binding of PgDps either at physiological 37 °C or at 4 °C (data not shown). We did, however, detect impairment of DNA migration in the presence of Fe2+ alone (Fig. 11B), which may explain the previous observation of “DNA binding” by PgDps in the presence of Fe2+, as a similar control was not included (18). Failure of PgDps to bind DNA was confirmed by the observation that iron-loaded PgDps that has been purified through a desalting column to remove unbound ferrous ions was unable to retard DNA migration through agarose gel (data not shown). However, similar to other reported Dps, PgDps does have iron binding capacity as shown by positive ferene S staining for iron after incubation of apo-PgDps with ferrous ion (Fig. 12A). Furthermore, the presence of a putative ferroxidase center, inferred from multiple sequence alignments against other Dps family proteins, suggests that PgDps may retain the ability to oxidize ferrous iron (Fig. 6B). Indeed, PgDps was found to be able to catalyze the conversion of Fe2+ to Fe3+ in the presence of atmospheric oxygen (Fig. 12B). Thus, iron binding and ferroxidase activity of PgDps coupled with heme sequestration may contribute to hydrogen peroxide resistance of the organism.
FIGURE 11.
PgDps protects DNA from H2O2-mediated degradation in the presence of heme or ferrous iron. Linear DNA (10 nm) was pre-incubated with 30 μm PgDps and 10 μm heme (A) or 10 mm fresh ferrous iron (B) at 37 °C anaerobically for 1 h in the dark prior to being exposed to 10 mm H2O2 for 15 min anaerobically at 37 °C in the dark. DNA was separated on 1% agarose in TAE buffer and visualized with ethidium bromide staining.
FIGURE 12.
Ferroxidase activity of PgDps. A, iron binding activity of PgDps. Lane 1, ferrous iron only; lane 2, PgDps only; lane 3, PgDps + ferrous iron; lane 4, PgDps + heme. Samples were separated on 7% native gel, then stained with ferene S and re-stained with Coomassie Brilliant Blue G-250 (CBB). B, time-dependent conversion of 300 μm Fe2+ to Fe3+ in the presence of 5 μm PgDps as monitored at 305 nm at 25 °C. Slow increase in absorbance of the control sample is due to auto-oxidation by atmospheric oxygen.
DISCUSSION
This study on PgDps reveals a new aspect to the Dps family proteins as multiple stress response proteins in bacteria. Dps proteins are recognized to have evolved from iron-sequestering ferritins, a ubiquitous protein superfamily. This superfamily contains three groups of proteins as follows: (i) the large 24-mer ferritins, (ii) heme-containing bacterioferritins, and (iii) the smaller 12-mer mini-ferritin Dps family (52). These families are distantly related and thus have retained a number of structural and functional similarities. In this study, we were able to demonstrate that PgDps is structurally similar to a number of existing Dps species and is composed of 12 identical monomeric subunits that assemble into a spherical shape with a hollow core while retaining iron sequestration and ferroxidase activity of ferritins. However, PgDps also has properties of bacterioferritin whereby it can bind ferric heme, albeit at a different site of binding and in a different mode of heme coordination. In E. coli bacterioferritin, heme was found to intercalate laterally between two subunits by bismethionine axial ligation involving the inter-subunit methionine 52 (53, 54). By contrast, in PgDps, heme iron is penta-coordinated by a thiolate ligand from a surface-located Cys-101 residue. The bound heme, in turn, triggered dodecameric PgDps to assemble into a higher oligomeric species. It is proposed that dimerization of surface-bound ferric heme under physiological conditions, either as μ-oxo or π-π* forms (37, 55), may be the main driver of this higher oligomerization state, and the addition of the phosphine TCEP to coordinate the sixth site of the ferric PgDps leads to dissociation of the bisheme species and, subsequently, dissociation of the higher order oligomers. This theory for dodecamer oligomerization is also compatible with the observation that holo-Strep-tagged PgDps dodecamers do not aggregate to any significant extent even after 3 h of incubation (data not shown), in contrast to detagged PgDps. This is probably due to the six-coordinated heme in Strep-tagged PgDps preventing heme stacking/dimerization. However, we could not discount the possibility that the surface-bound heme itself may introduce additional interaction with a neighboring oligomer to cause their aggregation. These questions will require further structural characterization of holo-PgDps.
The unique heme-binding characteristic of PgDps suggests that it could be a divergent member of the bacterioferritin/ferritin superfamily. One previous report has described a heme-binding member of the Dps family protein from a cyanobacterial Synechococcus sp., but detailed investigation into this phenomenon was not carried out (56). Multiple sequence alignments with other Dps homologs revealed the conservation of the heme-coordinating cysteine mostly in the Bacteriodales class of organisms, suggesting that other members of this group of anaerobic bacteria may utilize Dps to bind heme in a similar manner (Fig. 7).
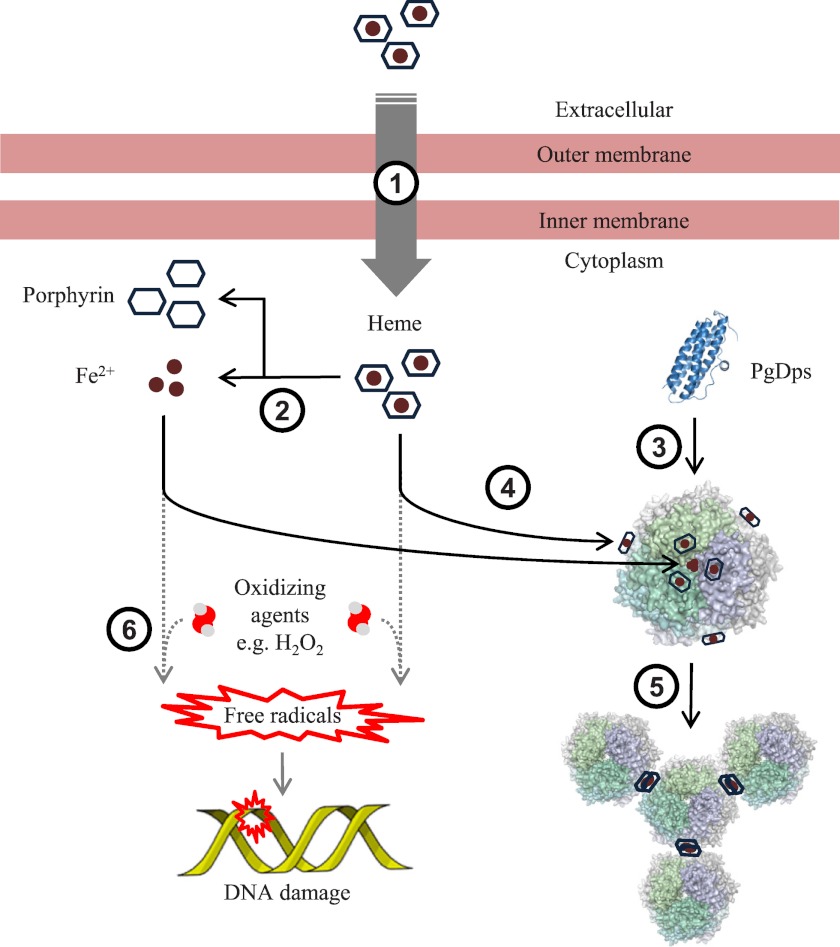
The effective heme binding of PgDps could be employed by the organism to store excess heme and function as an intracellular heme reservoir. However, the simple heme storage function of PgDps cannot explain the growth inhibition of Δdps mutants in heme-replete conditions. In P. gingivalis, iron starvation, such as that encountered in the environments of the mammalian host, induces up-regulation of a number of known heme uptake systems (57). However, the influx of heme acquired from host hemoproteins may present a potential danger to the bacteria. During colonization of the oral tissues, P. gingivalis is exposed to oxygenated environments as well as to other oxidizing agents such as hydrogen peroxide produced by other bacteria in dental plaque biofilms and by host immune cells (58, 59). The interaction of these molecules with heme can generate highly reactive free radicals (60). Accordingly, P. gingivalis must possess the capability to control these toxic effects as the organism is a successful pathogen in a number of mammalian hosts. To counter oxidative insults, P. gingivalis possesses a number of detoxifying systems such as thiol peroxidase, alkyl hydroperoxide reductase, and superoxide dismutase. However, these systems have been shown to be down-regulated in heme-replete conditions (61, 62). In this regard, PgDps was found to protect DNA against heme and Fe2+-mediated hydrolysis in the presence of H2O2. This protective role is supported by the finding that dps expression was up-regulated in response to increasing heme concentrations. Therefore, we propose that PgDps could be an important factor in the management of the intracellular heme and iron pool to avoid oxidative damage. This concept is summarized in a schematic diagram in Fig. 13.
FIGURE 13.
Proposed function of PgDps. In in vivo conditions when iron is scarce and heme is the only iron source, various heme transport receptors are up-regulated and extracellular heme is imported (1) into the cell. The fate of intracellular heme in P. gingivalis is currently not known, but most likely the iron and porphyrin will be separated (2) for utilization in other cellular pathways. In response to the presence of intracellular heme and iron, PgDps is expressed and self-assembled (3) into dodecameric forms. Intracellular ferrous iron is converted to ferric iron by PgDps and is sequestered within the hollow core of the dodecamer, whereas heme is sequestered (4) on the surface of the complex. Binding of heme onto PgDps subsequently induces aggregation (5) of PgDps dodecamers, possibly via heme stacking/dimerization. Without the ferroxidase and sequestration function of PgDps, unbound ferrous iron and heme can produce free radicals in the presence of an oxidizing agent (6) leading to DNA damage.
At present, it is not clear whether there are additional functions of the bound heme in PgDps. Protection of DNA against H2O2-mediated hydrolysis in the presence of heme or Fe2+ may indicate a potential peroxidase activity. Indeed, holo-Dps from Synechococcus sp. has been reported to exhibit a weak peroxidase activity, but the authors concluded the Km value was too low to be physiologically relevant (56). In PgDps, we could not detect a clear peroxidase activity in vitro (data not shown). By way of contrast, thiolate-coordinated heme has been reported to be important in monooxygenase activity of the cytochrome P450 superfamily due to its greater electron donor potential (63). These enzymes are present in all forms of life and partake in a wide array of oxidative reactions, from oxidation of organic compounds to the generation of nitric oxide from l-arginine and halogenation by chloroperoxidases (63). Whether holo-PgDps possesses any of these functions has yet to be determined.
Finally, PgDps was found not to be able to bind DNA nonspecifically as reported previously (18). We were unable to detect impairment of DNA migration in agarose gels in the presence of apo-PgDps or purified Fe2+-loaded PgDps at 4 or 37 °C. We did notice, however, that in the presence of Fe2+ only, some DNA precipitated in the sample wells of the agarose gel (Fig. 11B), analogous to the observation reported previously for PgDps/Fe2+ and interpreted as DNA binding activity (18). The inability to bind DNA is not unique to PgDps, as other Dps family proteins have been reported to share this property (48, 64–67). Furthermore, failure to bind DNA may explain the previous observation that the Δdps mutant was no more sensitive to DNA-damaging reagents such as mitomycin C or metronidazole than the wild-type strain (18). In addition, the observation that the Δdps mutant was more sensitive to H2O2 challenge than the wild type (18) was probably due to PgDps sequestration of heme and Fe2+ and its ferroxidase activity as shown in our current work rather than through DNA binding per se.
In conclusion, our characterization of the functional properties of P. gingivalis Dps demonstrates an important new aspect to the protective function of the Dps protein family. The unique heme-binding property of PgDps was shown to contribute to P. gingivalis resistance to heme toxicity, and conservation of the heme iron-coordinating cysteine within Dps from the Bacteroidales order of bacteria suggests that this protective function may be shared among members of this family.
Acknowledgments
We thank Dr. Catherine Rathsam and Dr. Xiaoyan Zhou for assistance with various experiments and Dr. Mikio Shoji and Prof. Koji Nakayama for generously providing the anti-Dps antibody. We also thank Dr. Ann Kwan for assistance with the analysis of fluorescence data.
This work was supported, in whole or in part, by National Institutes of Health Grant DE 09761 (to Jan Potempa and subcontracted to K.-A. N.). This work was also supported in part by Grant LP0562660 from the Australian Research Council (to N. H.) and Establishment Grant RN38/08 from the Ramaciotti Foundation, Australia (to K.-A. N.).
- Dps
- DNA-protective protein from starved cells
- δ-ALA
- δ-aminolevulinic acid
- eTSB
- enriched tryptic soy broth
- PgDps
- P. gingivalis DNA-protective protein from starved cells
- SAXS
- Small-angle X-ray Scattering
- SEC-MALLS
- size-exclusion chromatography-multiangle laser light scattering
- TCEP
- Tris(2-carboxyethyl)phosphine
- THT
- tetrahydrothiophene
- TMBZ
- tetramethylbenzidine
- PDB
- Protein Data Bank.
REFERENCES
- 1. Ezzo P. J., Cutler C. W. (2003) Microorganisms as risk indicators for periodontal disease. Periodontol. 2000 32, 24–35 [DOI] [PubMed] [Google Scholar]
- 2. Gibson F. C., 3rd, Hong C., Chou H. H., Yumoto H., Chen J., Lien E., Wong J., Genco C. A. (2004) Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 109, 2801–2806 [DOI] [PubMed] [Google Scholar]
- 3. Wegner N., Wait R., Sroka A., Eick S., Nguyen K. A., Lundberg K., Kinloch A., Culshaw S., Potempa J., Venables P. J. (2010) Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase. Implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62, 2662–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Seymour G. J., Ford P. J., Cullinan M. P., Leishman S., Yamazaki K. (2007) Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 13, 3–10 [DOI] [PubMed] [Google Scholar]
- 5. Roper J. M., Raux E., Brindley A. A., Schubert H. L., Gharbia S. E., Shah H. N., Warren M. J. (2000) The enigma of cobalamin (vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J. Biol. Chem. 275, 40316–40323 [DOI] [PubMed] [Google Scholar]
- 6. Gao J. L., Nguyen K. A., Hunter N. (2010) Characterization of a hemophore-like protein from Porphyromonas gingivalis. J. Biol. Chem. 285, 40028–40038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slakeski N., Dashper S. G., Cook P., Poon C., Moore C., Reynolds E. C. (2000) A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol. Immunol. 15, 388–392 [DOI] [PubMed] [Google Scholar]
- 8. Smalley J. W., Byrne D. P., Birss A. J., Wojtowicz H., Sroka A., Potempa J., Olczak T. (2011) HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis. PLoS ONE 6, e17182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dashper S. G., Hendtlass A., Slakeski N., Jackson C., Cross K. J., Brownfield L., Hamilton R., Barr I., Reynolds E. C. (2000) Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J. Bacteriol. 182, 6456–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simpson W., Olczak T., Genco C. A. (2000) Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182, 5737–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anzaldi L. L., Skaar E. P. (2010) Overcoming the heme paradox. Heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78, 4977–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ladan H., Nitzan Y., Malik Z. (1993) The antibacterial activity of haemin compared with cobalt, zinc, and magnesium protoporphyrin and its effect on potassium loss and ultrastructure of Staphylococcus aureus. FEMS Microbiol. Lett. 112, 173–177 [DOI] [PubMed] [Google Scholar]
- 13. Nitzan Y., Wexler H. M., Finegold S. M. (1994) Inactivation of anaerobic bacteria by various photosensitized porphyrins or by hemin. Curr. Microbiol. 29, 125–131 [DOI] [PubMed] [Google Scholar]
- 14. Lewis J. P., Plata K., Yu F., Rosato A., Anaya C. (2006) Transcriptional organization, regulation, and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology 152, 3367–3382 [DOI] [PubMed] [Google Scholar]
- 15. Ratliff M., Zhu W., Deshmukh R., Wilks A., Stojiljkovic I. (2001) Homologues of neisserial heme oxygenase in Gram-negative bacteria. Degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skaar E. P., Gaspar A. H., Schneewind O. (2004) IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443 [DOI] [PubMed] [Google Scholar]
- 17. Zhu W., Wilks A., Stojiljkovic I. (2000) Degradation of heme in Gram-negative bacteria. The product of the hemO gene of Neisseriae is a heme oxygenase. J. Bacteriol. 182, 6783–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ueshima J., Shoji M., Ratnayake D. B., Abe K., Yoshida S., Yamamoto K., Nakayama K. (2003) Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71, 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiancone E., Ceci P. (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions. Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 1800, 798–805 [DOI] [PubMed] [Google Scholar]
- 20. Fletcher H. M., Schenkein H. A., Morgan R. M., Bailey K. A., Berry C. R., Macrina F. L. (1995) Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63, 1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nikolich M. P., Shoemaker N. B., Salyers A. A. (1992) A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob. Agents Chemother. 36, 1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiu J., March P. E., Lee R., Tillett D. (2004) Site-directed, ligase-independent mutagenesis (SLIM). A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen K. A., Zylicz J., Szczesny P., Sroka A., Hunter N., Potempa J. (2009) Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155, 328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jeffries C. M., Lu Y., Hynson R. M., Taylor J. E., Ballesteros M., Kwan A. H., Trewhella J. (2011) Human cardiac myosin binding protein C. Structural flexibility within an extended modular architecture. J. Mol. Biol. 414, 735–748 [DOI] [PubMed] [Google Scholar]
- 25. Franke D., Svergun D. I. (2009) DAMMIF, a program for rapid ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jeffries C. M., Whitten A. E., Harris S. P., Trewhella J. (2008) Small-angle x-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J. Mol. Biol. 377, 1186–1199 [DOI] [PubMed] [Google Scholar]
- 27. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 28. Svergun D., Barberato C., Koch M. H. (1995) CRYSOL–a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 29. Bagshaw C. R., Harris D. A. (1987) in Spectrophotometry and Spectrofluorometry: a Practical Approach (Harris D. A., Bashford C. L., eds) pp. 91–113, IRL Press, Washington, D. C [Google Scholar]
- 30. Shepherd M., Heath M. D., Poole R. K. (2007) NikA binds heme. A new role for an Escherichia coli periplasmic nickel-binding protein. Biochemistry 46, 5030–5037 [DOI] [PubMed] [Google Scholar]
- 31. Calhoun L. N., Kwon Y. M. (2011) Structure, function, and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J. Appl. Microbiol. 110, 375–386 [DOI] [PubMed] [Google Scholar]
- 32. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 33. Koch M. H., Vachette P., Svergun D. I. (2003) Small-angle scattering. A view on the properties, structures, and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 36, 147–227 [DOI] [PubMed] [Google Scholar]
- 34. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) ATSAS 2.1–toward automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 40, S223-S228 [Google Scholar]
- 35. Orthaber D., Bergmann A., Glatter O. (2000) SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallog. 33, 218–225 [Google Scholar]
- 36. Rivera M., Walker F. A. (1995) Biosynthetic preparation of isotopically labeled heme. Anal. Biochem. 230, 295–302 [DOI] [PubMed] [Google Scholar]
- 37. Asher C., de Villiers K. A., Egan T. J. (2009) Speciation of ferriprotoporphyrin IX in aqueous and mixed aqueous solution is controlled by solvent identity, pH, and salt concentration. Inorg. Chem. 48, 7994–8003 [DOI] [PubMed] [Google Scholar]
- 38. Zhang L., Guarente L. (1995) Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang T. J., McCoubrey W. K., Jr., Maines M. D. (2001) Heme oxygenase-2 interaction with metalloporphyrins: function of heme regulatory motifs. Antioxid. Redox Signal. 3, 685–696 [DOI] [PubMed] [Google Scholar]
- 40. Ruf H. H., Wende P., Ullrich V. (1979) Models for ferric cytochrome P450. Characterization of hemin mercaptide complexes by electronic and ESR spectra. J. Inorg. Biochem. 11, 189–204 [DOI] [PubMed] [Google Scholar]
- 41. Knight D. A., Deschamps J. R., Butcher R. J., Simmers C., Chang E. L. (2008) A new coordination mode for tris(2-carboxyethyl)phosphine. Synthesis, crystal structure, and characterization of the mixed-valence Co(III)/Co(II)/Co(III) complex [Co{P(CH2CH2COO)2-(CH2CH2COOH)}2]2[Co(H2O)4][Na2(H2O)4]Cl2·6H2O. Polyhedron 27, 1795–1801 [Google Scholar]
- 42. Sono M., Dawson J. H., Hager L. P. (1985) Phosphine binding as a structural probe of the chloroperoxidase active site. Spectroscopic evidence for endogenous thiolate ligation to the heme iron. Inorg. Chem. 24, 4339–4343 [Google Scholar]
- 43. Kery V., Bukovska G., Kraus J. P. (1994) Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate. Cystathionine β-synthase is a heme protein. J. Biol. Chem. 269, 25283–25288 [PubMed] [Google Scholar]
- 44. Meier M., Janosik M., Kery V., Kraus J. P., Burkhard P. (2001) Structure of human cystathionine β-synthase. A unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 20, 3910–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu Y., Casimiro D. R., Bren K. L., Richards J. H., Gray H. B. (1993) Structurally engineered cytochromes with unusual ligand-binding properties. Expression of Saccharomyces cerevisiae Met-80 → Ala iso-1-cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 90, 11456–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sigman J. A., Pond A. E., Dawson J. H., Lu Y. (1999) Engineering cytochrome c peroxidase into cytochrome P450. A proximal effect on heme-thiolate ligation. Biochemistry 38, 11122–11129 [DOI] [PubMed] [Google Scholar]
- 47. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., Dawson J. H. (2003) Neutral thiol as a proximal ligand to ferrous heme iron. Implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ceci P., Ilari A., Falvo E., Chiancone E. (2003) The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage. X-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 278, 20319–20326 [DOI] [PubMed] [Google Scholar]
- 49. Chowdhury R. P., Vijayabaskar M. S., Vishveshwara S., Chatterji D. (2008) Molecular mechanism of in vitro oligomerization of Dps from Mycobacterium smegmatis. Mutations of the residues identified by “interface cluster” analysis. Biochemistry 47, 11110–11117 [DOI] [PubMed] [Google Scholar]
- 50. Bullen J. J. (1981) The significance of iron in infection. Rev. Infect. Dis. 3, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 51. Aft R. L., Mueller G. C. (1983) Hemin-mediated DNA strand scission. J. Biol. Chem. 258, 12069–12072 [PubMed] [Google Scholar]
- 52. Smith J. L. (2004) The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30, 173–185 [DOI] [PubMed] [Google Scholar]
- 53. Andrews S. C., Le Brun N. E., Barynin V., Thomson A. J., Moore G. R., Guest J. R., Harrison P. M. (1995) Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 270, 23268–23274 [DOI] [PubMed] [Google Scholar]
- 54. Carrondo M. A. (2003) Ferritins, iron uptake, and storage from the bacterioferritin viewpoint. EMBO J. 22, 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. de Villiers K. A., Kaschula C. H., Egan T. J., Marques H. M. (2007) Speciation and structure of ferriprotoporphyrin IX in aqueous solution: spectroscopic and diffusion measurements demonstrate dimerization, but not μ-oxo dimer formation. J. Biol. Inorg. Chem. 12, 101–117 [DOI] [PubMed] [Google Scholar]
- 56. Peña M. M., Bullerjahn G. S. (1995) The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J. Biol. Chem. 270, 22478–22482 [DOI] [PubMed] [Google Scholar]
- 57. Lewis J. P. (2010) Metal uptake in host-pathogen interactions: role of iron in Porphyromonas gingivalis interactions with host organisms. Periodontol. 2000 52, 94–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barnard J. P., Stinson M. W. (1999) Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect. Immun. 67, 6558–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chapple I. L. (1996) Role of free radicals and antioxidants in the pathogenesis of the inflammatory periodontal diseases. Clin. Mol. Pathol. 49, M247–M255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Svistunenko D. A. (2005) Reaction of haem containing proteins and enzymes with hydroperoxides. The radical view. Biochim. Biophys. Acta 1707, 127–155 [DOI] [PubMed] [Google Scholar]
- 61. Lynch M. C., Kuramitsu H. K. (1999) Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect. Immun. 67, 3367–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dashper S. G., Ang C. S., Veith P. D., Mitchell H. L., Lo A. W., Seers C. A., Walsh K. A., Slakeski N., Chen D., Lissel J. P., Butler C. A., O'Brien-Simpson N. M., Barr I. G., Reynolds E. C. (2009) Response of Porphyromonas gingivalis to heme limitation in continuous culture. J. Bacteriol. 191, 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Omura T. (2005) Heme-thiolate proteins. Biochem. Biophys. Res. Commun. 338, 404–409 [DOI] [PubMed] [Google Scholar]
- 64. Su M., Cavallo S., Stefanini S., Chiancone E., Chasteen N. D. (2005) The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry 44, 5572–5578 [DOI] [PubMed] [Google Scholar]
- 65. Liu X., Kim K., Leighton T., Theil E. C. (2006) Paired Bacillus anthracis Dps (mini-ferritin) have different reactivities with peroxide. J. Biol. Chem. 281, 27827–27835 [DOI] [PubMed] [Google Scholar]
- 66. Ishikawa T., Mizunoe Y., Kawabata S., Takade A., Harada M., Wai S. N., Yoshida S. (2003) The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185, 1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haikarainen T., Papageorgiou A. C. (2010) Dps-like proteins. Structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 67, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]