Background: The detailed mechanism of PIP2-induced Kir channel gating remains elusive.
Results: Specific mutations increase the flexibility of the cytosolic GH loop and accelerate the PIP2-induced gating kinetics of Kir2 channels.
Conclusion: Interactions of the GH loop with the N terminus regulate the PIP2-induced gating kinetics of Kir2 channels.
Significance: We identify a novel region in Kir channels involved in the control of PIP2-induced gating.
Keywords: Gating, Molecular Dynamics, Site-specific Mutagenesis, Patch Clamp, Potassium Channels, Gating Kinetics, PIP2, Two-electrode Voltage Clamp
Abstract
Inwardly rectifying K+ (Kir) channels set the resting membrane potential and regulate cellular excitability. The activity of Kir channels depends critically on the phospholipid PIP2. The molecular mechanism by which PIP2 regulates Kir channel gating is poorly understood. Here, we utilized a combination of computational and electrophysiological approaches to discern structural elements involved in regulating the PIP2-induced gating kinetics of Kir2 channels. We identify a novel role for the cytosolic GH loop. Mutations that directly or indirectly affect GH loop flexibility (e.g. V223L, E272G, D292G) increase both the on- and especially the off-gating kinetics. These effects are consistent with a model in which competing interactions between the CD and GH loops for the N terminus regulate the gating of the intracellular G loop gate.
Introduction
Inwardly rectifying K+ (Kir) channels play a vital role in many physiological processes, such as control of heart rate, maintenance of the resting membrane potential, hormonal secretion, extracellular K+ buffering in the brain, and K+ secretion in the kidney. These physiological processes are regulated by specific stimuli that control the gating of Kir channels. Examples of stimuli gating Kir channels include intracellular H+ for Kir1 (1), the βγ subunits of G proteins for Kir3 (2), and phosphatidylinositol 4,5-bisphosphate (PIP2)4 for all Kir channels (3, 4). The inwardly rectifying ATP-inhibited K+ (KATP) (Kir6.2) channel was the first reported to depend on PIP2 (5, 6). Subsequently, all known Kir channels were shown to depend directly on phosphoinositides for maintenance of their activity (3, 5, 7–10). In general, Kir channels, as is the case with most ion channels tested, run down when PIP2 is depleted (dephosphorylated by lipid phosphatases) and reactivate when PIP2 is applied from the intracellular side (11). The gating kinetics of Kir channels are quite distinct between different Kir channels, but the underlying reasons are not understood.
Kir channels, like other K+ channels, are homo- or heterotetramers of four subunits. Each subunit shares a common structure consisting of a cytosolic N-terminal domain, an outer transmembrane helix (M1), an ion-selective P loop (selectivity filter (SF)), an inner transmembrane helix (M2) lining most of the permeation pathway, and a cytosolic C terminus controlling ion flow (12, 13) (Fig. 1A). The SF and bundle crossing of the M2 helix are thought to serve as gates, constriction points along the permeation pathway limiting ion flow (12). Mechanisms of gating (i.e. transitions between the closed and open states) for the SF and M2 gates have been studied and proposed (14). The N- and C termini, which are located on the intracellular side of the membrane, form a comparable cytosolic extension to the transmembrane portion of the pore. In addition to the SF and M2 transmembrane gates, a cytosolic constriction (HI or G loop) of the permeation pathway has been proposed as a third gate (9) (Fig. 1A). Specific acidic residues localized in the cytosolic pore are thought to give rise to inward rectification (15). Functional studies and mutagenesis data have identified several residues as potential PIP2-interacting sites in both the N- and C termini of the cytoplasmic domain of the Kir2.1 channel. Many of these residues are basic and thus capable of forming salt bridge interactions with the negatively charged head group of PIP2 (3, 8, 10, 16). Recent crystallographic evidence has confirmed this view (17, 18). Moreover, crystal structures of a Kir3.1 chimera caught the intracellular G loop gate in dilated (open) and constricted (closed) conformations. The movement in the G loop observed in different structures of the cytoplasmic domains is also notable (9, 19). However, it is not known how the G loop gate integrates into channel gating and how it affects the gating kinetics of Kir channels.
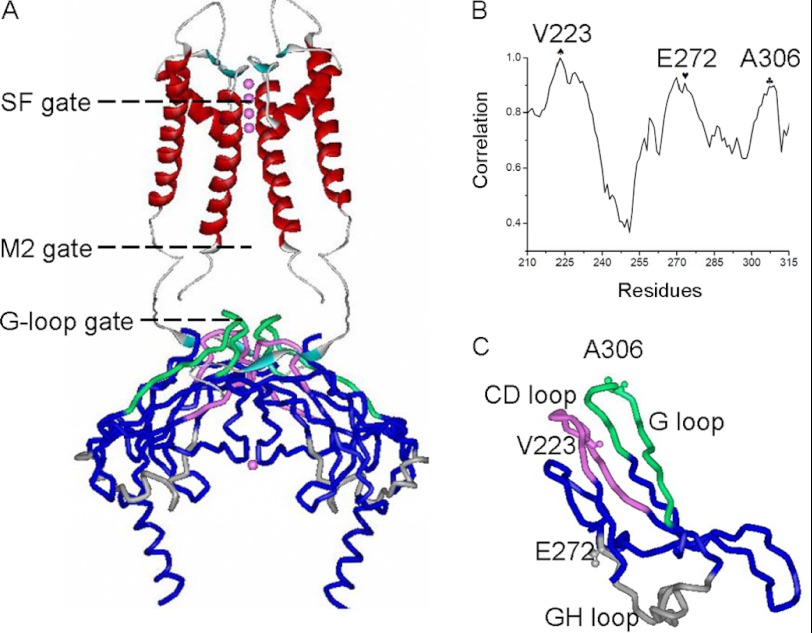
FIGURE 1.
MD simulation performed on the C-terminal domain of Kir2. Shown are highly correlated movements among Val-223, Glu-272, and Ala-306. A, two Kir2.2 subunits shown with the SF, M2 gate, and cytosolic G loop gate indicated. Similarly, the CD and GH loops that regulate the stability of the G loop gate have been highlighted. B, correlation of movements among C-terminal residues of Kir2.1. Val-223, Glu-272, and Ala-306 are shown as ♠, ♥, and ♣, respectively. C, schematic structure of one subunit cytosolic domain where the Kir2.1 residues Val-223, Glu-272, and Ala-306 are shown as small spheres. The pink spheres in A represent the potassium ions in the ion pathway. In A and C, the CD, GH, and G loops are highlighted in pink, gray, and green, respectively.
Although recent crystallographic studies have greatly enhanced our understanding of the three-dimensional structure of Kir channels (13, 17, 20), it has not been possible yet to understand the dynamic gating mechanism of these channels induced by molecules such as PIP2. A recent study of multiple KirBac3.1 structures in either the open or closed conformations has highlighted the importance of intersubunit interactions between the N and C termini and the G loops of adjacent subunits in Kir channel gating (21).
Here we set out to study Kir channel gating kinetics, focusing on the cytosolic channel regions using a voltage-gated phosphatase to control PIP2 levels (22) and computational, mutagenesis, and electrophysiological approaches to probe the role of specific secondary structural elements and specific residues within them. We observed highly correlated movements of the Kir2.1 (Val-223) (of the CD loop) and Glu-272 (of the GH loop) with Ala-306 at the G loop gate (see sequence alignment in supplemental Fig. S1). Mutations, at Val-223 in Kir2.1 or Val-224 in Kir2.2, accelerated both the opening and closing gating kinetics of the channel and increased the flexibility of the GH loop. Similar results were obtained with mutations of key residues within the GH loop. Our results are explained by a model in which PIP2 induces the switching of the N terminus from the CD loop to the GH loop (around the short αG helix) (Fig. 1C). Mutations that decrease the GH loop interactions with the N terminus (e.g. V223L, E272G, D292G) increase the flexibility of the GH loop and allow the N terminus to gate the G loop with faster kinetics.
EXPERIMENTAL PROCEDURES
Chemicals
Phosphatidylinositol-4,5-bisphosphate diC8 PIP2 was purchased from Avanti Lipids and was prepared as described previously (8, 10). All other chemicals were purchased from Sigma.
Homology Modeling
Modeler V9.5 (23) was used to add missing residues to the crystal structures of Kir2.1 (PDB code 1U4F), and the full-length crystal structure of Kir2.2 (PDB code 3JYC). Modeler was also used to create a homology model of the cytosolic domain of Kir2.2. The mutant channels were constructed by substituting the WT side chain with the specified side chains. The models were then subjected to at least 3000 steps of a steepest descent minimization using the CHARMM program with the implicit membrane/solvent Generalized Born (GB) model (24).
Molecular Docking
AUTODOCK (25) was used for the docking studies. We replaced PIP2 with its analog diC1 PIP2, which has two methyl groups. The atomic charges of the PIP2 head group were taken from the ab initio calculations by Lupyan and colleagues (26). A grid map was generated for the Kir2.2 full-length structure using CHNOP (i.e. carbon, hydrogen, nitrogen, oxygen, and phosphor) elements sampled on a uniform grid containing 120 × 120 × 120 points 0.375 Å apart. The center of the grid box was set to the center of known critical PIP2-sensitive residues, i.e. Gln-51, Arg-65, Lys-183, Arg-186, Lys-188, Lys-189, Arg-190, Arg-219, Lys-220, Arg-229, and Arg-313. The Lamarckian Genetic Algorithm (LGA) was selected to identify the binding conformations of the ligands. 100 docking simulations were performed, and the final docked PIP2 analog configurations were selected on the basis of docked binding energies and cluster analysis. The PIP2-Kir2.2 complex was constructed on the basis of the docked PIP2 analog-Kir2.2 complex structure and refined by CHARMM using the same protocol as described above.
Molecular Dynamics (MD) Simulations
The crystal structure of the Kir2.1 cytosolic domain was initialized as follows, solvating the molecule in a rectangular water box of 82 × 104 × 103 Å3 and neutralizing the water box by adding Na+ and Cl− of ∼100 mm. MD simulations were performed using NAMD with CHARMM27 all-atom force field parameters (27). An integration time step of 1 fs, a uniform dielectric constant of 1.0, a scaling factor for 1–4 interactions of 1.0, and periodic boundary conditions were applied in all simulations. A smooth (12–16 Å) cutoff and the Particle Mesh Ewald (PME) (28) were employed to calculate van der Waals forces and full electrostatics, respectively. Prior to the equilibration process, energy minimization (5000 steps with backbone atoms of C-terminal fixed and then another 5000 steps with all atoms free), followed by a heating-up process from 0 to 300 K over 35 ps, were performed. Then, two 5-ns equilibration processes with either all atoms free or partly constrained were performed with the temperature held at 300 K using Langevin dynamics while the pressure was held at 1 atm using the Langevin piston method. An RMSF (root mean squared fluctuation) analysis was performed on the basis of the two equilibration processes.
For the full-length Kir2.2 channel simulation, the channels were immersed in an explicit palmitoyloleoyl-phosphatidylcholine bilayer generated from the visual molecular dynamics membrane package. After being solvated with SPC water molecules, neutralized by Na+ as the counter ions, and including K+ located in the selectivity filter as obtained from the crystal structures, each system involved ∼141,000 atoms in the MD simulations. GROMACS v4.0.5 was used to conduct the simulation with the GROMOS96 53a6 force field. The force field parameters for PIP2 were generated from the Prodrg server (8), and the same atomic charges of the PIP2 head group in the docking step were used in the MD simulations. The lipid parameters were obtained from Dr. Tieleman. Long range electrostatics were calculated using the PME8 method with a 12 Å cut-off. Van der Waals interactions were modeled using Lennard-Jones 6–12 potentials with a 14 Å cut-off. All simulations were conducted at a constant temperature of 300 K using the Berendsen thermostat. The system pressure was coupled at isotropic (X+Y, Z) directions referenced to 1 bar using the Berendsen method9. All bonds were constrained with the LINCS algorithm. The time step was 2 fs, and the neighboring list was updated every 10 time steps.
Prior to production runs, energy minimization of 3000 steps of steepest descent were carried out on each system, followed by a 0.5-ns two-step equilibration process with either all atoms free or partly constrained. An electric field of 0.06 V/nm was applied in this step as well as the production run, along the z axis of the box, to maintain the lower potentials in the intracellular side. The treatment of the electric field has been detailed in Refs. 29, 30. A 100-ns production run was conducted on each system, and coordinates were saved every 10 ps for analysis. Visual molecular dynamics were used for conformational visualization.
Interaction Network Analysis
The interactions of hydrogen bonds, salt bridges, and hydrophobic contacts analysis were calculated using the Simulaid (31–34) program after energy minimization. The Simulaid outputs for interactions were reorganized with in-house scripts for the facility of comparison among the systems.
Molecular Biology
All cDNA constructs were subcloned into the pGEMHE plasmid vector and used as described (10). Point mutants were produced by Pfu mutagenesis with a Quikchange kit (Stratagene). Sequences were confirmed by DNA sequencing. Recombinant Kir2.1, its mutants, and Ci-VSP were expressed in Xenopus laevis oocytes as described previously (10, 35). cRNA was produced with T7 RNA polymerase using a kit (Promega). cRNAs of the various Kir2.1, Kir2.2, their mutants, and of Ci-VSP were injected in the range of 0.5–10 ng/oocyte depending on the functional expression level of the given construct.
Electrophysiology
Recordings in X. laevis oocytes were performed 1 to 2 days after cRNA injection. Whole-oocyte currents were measured by conventional two-electrode voltage clamp (TEVC) using a GeneClamp 500 amplifier (Molecular Devices, CA). Electrodes were filled with 3 m KCl dissolved in 1% agarose to prevent the leakage of KCl into the oocytes. The electrodes had resistances less than 1 mΩ. Oocytes were constantly perfused with a high-potassium solution (ND96K) containing 96 mm KCl, 1 mm NaCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES (pH 7.4 with KOH). A low-potassium solution (ND96) was used in some experiments to inhibit most of the Kir2.1 currents at −80 mV. To activate Ci-VSP, the following voltage protocol was used in TEVC oocyte recordings: 1-s sweeps composed of a 170-ms ramp from −80 to +80 mV followed by an 830-ms step to +80 mV. To deactivate Ci-VSP, the oocytes were held at −80 mV. Sweeps were applied until the resulting currents reached steady state. ND96 contained 96 mm NaCl, 1 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 5 mm HEPES (pH 7.4 with NaOH). Current amplitudes were measured at +80 mV and −80 mV. Data acquisition and analysis were achieved using pClamp9.2 (Molecular Devices, CA) and Origin 7.5 (Microcal, MA) software.
Macropatch channel activity was recorded from devitellinized oocytes under the inside-out mode of standard patch-clamp methods using an Axon 200B patch-clamp amplifier and Clampex 10.0 data acquisition software (Molecular Devices). Electrodes were made from borosilicate glass using a Sutter P-97 microelectrode puller (Sutter Instrument Co., CA) that gave a tip diameter of 5–15 μm and had a resistance of 0.5–1 MΩ when filled with an electrode solution containing 120 mm KCl, 2 mm MgCl2, and 10 mm HEPES (pH 7.4 with KOH). Two bath solutions were used: a FVP solution containing 60 mm KCl, 5 mm EDTA-K, 5 mm KF, 0.1 mm Na3VO4, 10 mm K4P2O7, 10 mm HEPES (pH 7.4 with KOH) and FVP solution with X μm (0–100 μm for dose response and 30 μm for gating kinetics measurements) diC8 PIP2. Data were analyzed using pClamp9.2 (Molecular Devices) and Origin 7.5 (Microcal) software.
HA Tag Expression Experiments
We engineered an HA tag to the extracellular loop between the outer transmembrane helix and the P region of Kir2.1 (8). Oocytes were blocked for 60 min in ND96 with 1% BSA at 4 °C, labeled with 0.5 μg/ml rat monoclonal anti-HA antibody (3F10, Roche) (in 1% BSA overnight at 4 °C), washed at 4 °C, and incubated with HRP-coupled secondary antibody (goat anti-rat) (in 1% BSA for 60 min). Cells were washed (1% BSA, 4 °C) and transferred to ND96 without BSA. Individual oocytes were placed in 55 μl of SuperSignal ELISA (Pierce) and incubated at room temperature for 1 min. Chemiluminescence was quantified in a TD-20E luminometer (Turner Designs, CA). As a control, we used uninjected oocytes.
Data Analysis
The error bars in the figures represent S.E. Each experiment shown or described was performed on five to seven oocytes of the same batch for TEVC recording and three to five oocytes of the same batch for macropatch recordings. A minimum of three to four batches of oocytes was tested for each experiment shown.
RESULTS
The Val-223 and Glu-272 Show High Correlativity with A306 in the G Loop Gate
To explore the mechanism by which the G loop gate relates to the rest of the cytosolic domain during PIP2-induced Kir channel gating, we performed 10-ns MD simulations of the cytosolic domains of the WT Kir2.1 and analyzed the relative movements between C-terminal residues and the G-loop residue Ala-306. From the free MD simulations, we found that the relative movements between Glu-272 (at the beginning of the GH loop), Ala-306 (at the apex of the G loop), and Val-223 (in the CD loop) were highly correlated (Fig. 1B). Fig. 1C shows the location of residues Val-223, Glu-272, and Glu-306 in the cytosolic portion of the crystal structure. Unlike the CD and G loops that are physically proximal and are thought to interact with each other, the GH loop is located quite far from the other two loops (Fig. 1C). We hypothesized that Val-223 and Glu-272 could play important roles in the PIP2-induced Kir channel gating.
Kir2.1(V223L) and Kir2.2(V224L) Affect the PIP2-induced Gating Kinetics through a Common Mechanism
We first examined the effect of mutations at the Kir2.1 223 position from Val to Leu or Met. Fig. 2A shows a right shift of the dose response curve of V223L responding to the stimulatory effect of PIP2 (EC50 for WT, 2.75 ± 0.12 μm and for V223L, 8.57 ± 0.76 μm). However, unlike other mutants reported to decrease the apparent affinity to PIP2, the V223L mutant displayed a 20% increase in its peak currents (Fig. 2B). We next measured membrane surface expression of the WT and V223L mutant using HA-tagged channels. Our data indicated a 42% reduction in cell surface expression for V223L (0.58 ± 0.01 when WT Kir2.1 was normalized to 1). These results suggested that the increased whole-cell currents of the V223L mutant were not due to an increase in available channels on the cell surface but likely to increased activity. To further examine this possibility, we measured the gating kinetics of Kir2.1 and its V223L mutant using either TEVC and the lipid phosphatase Ci-VSP (22) in intact cells or inside-out patches of the patch-clamp technique. Use of Ci-VSP allows rapid and reversible control of phosphatase activity through voltage-clamp recordings, which, in turn, has allowed us to discern the macroscopic closing and opening Kir2 gating kinetics induced by PIP2 dephosphorylation and rephosphorylation.
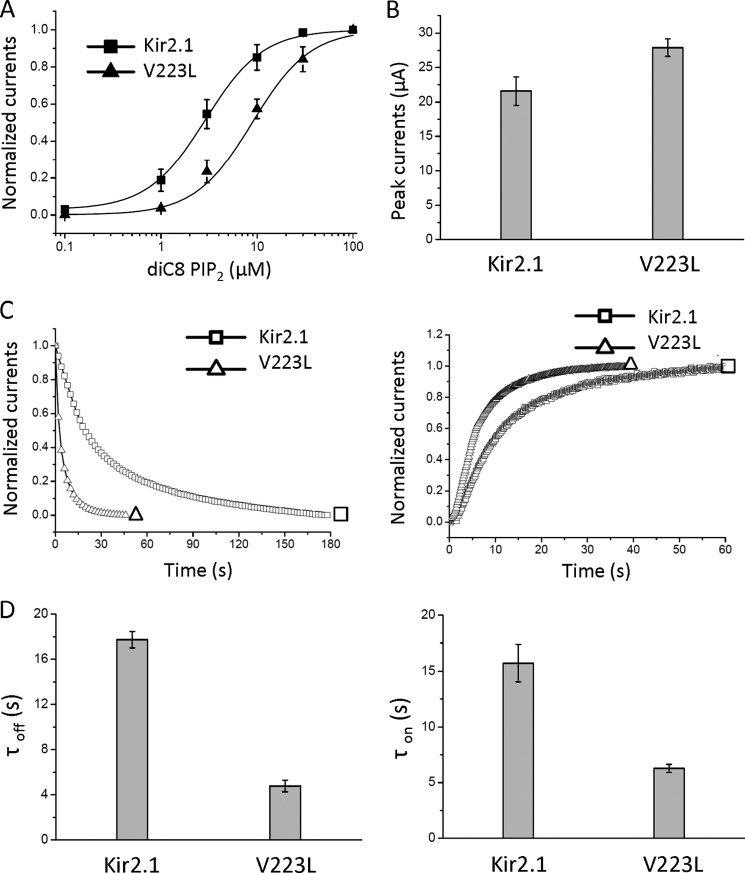
FIGURE 2.
The V223L mutation accelerates the gating kinetics of Kir2. A, dose response of diC8 PIP2 of Kir2.1 and V223L, showing a right shift in the apparent affinity of PIP2 sensitivity as a result of the mutation. B, peak currents of Kir2.1 and V223L do not show a decrease in whole-cell current, as other mutations do, that decrease the apparent channel affinity to PIP2. C, the representative time course of inhibition (PIP2 dephosphorylation induced by Ci-VSP, a 3.7-fold effect) and reactivation (PIP2 rephosphorylation, a 2.5-fold effect) of WT-Kir2.1 and V223L. D, bars are the time constants (mean ± S.E. of at least six experiments) corresponding to C.
Fig. 2, C and D, shows an acceleration of the gating kinetics of the V223L mutant relative to the WT by inhibition (left panels) or activation (right panels) of Ci-VSP (kinetics of current inhibition (τoff) of V223L, 4.78 ± 0.51 s versus WT, 17.73 ± 0.72 s; kinetics of current recovery (τon) of V223L, 6.28 ± 0.36 s versus WT, 15.72 ± 1.66 s). Ci-VSP reduces membrane PIP2 levels when the membrane is depolarized (36). Alternatively, we examined the gating kinetics of activation and deactivation of Kir2.1 WT and its V223L mutant by perfusion and washout of exogenous diC8 PIP2 in inside-out patches. Following patch excision, we perfused the patch with a high-potassium solution (HK) lacking ATP, which results in PIP2 dephosphorylation and rundown of currents. After complete rundown, diC8 PIP2 was applied at the intracellular side of patches expressing WT or V223L to reactivate the channels. The kinetics of both the rundown (τoff) (right panels) and reactivation (τon) (left panels) by diC8 PIP2 were accelerated by the V223L mutation (τoff for WT, 21.97 ± 1.42 s and for V223L, 3.44 ± 2.09 s; τon for WT, 7.25 ± 1.71 s and for V223L, 4.05 ± 0.73 s (supplemental Fig. S2).
We then tested the effects of the V223M mutation on the PIP2-dependent gating kinetics. The apparent affinity of the WT and V223M channels to diC8 PIP2 were indistinguishable (EC50 for WT, 2.75 ± 0.12 μm and for V223M, 2.62 ± 0.46 μm) (supplemental Fig. S3A). V223M also showed similar amplitude currents in TEVC as the WT Kir2.1 (supplemental Fig. S3B). Moreover, the kinetics of Ci-VSP-induced changes in currents were also not significantly different for WT versus the V223M mutant (τoff for WT, 17.73 ± 0.72 s and for V223M, 16.93 ± 0.95 s; τon for WT, 15.72 ± 1.66 s and for V223M, 12.27 ± 0.61 s) (supplemental Fig. S3, C and D). Thus, V223M, unlike the V223L mutant, displayed similar apparent affinity to PIP2, whole-cell current level and gating kinetics as the WT Kir2.1 channel.
We next asked whether other Kir2 channels showed similar effects as the Kir2.1(V223L) mutant did. Kir2.2 also has a Val (Val-224) at the analogous position to the Kir2.1(V223) (supplemental Fig. S4A). The Kir2.2(V224L) mutant, like the Kir2.1(V223L) mutant, accelerated both τoff (left panels) and τon (right panels) (τoff for WT, 42.87 ± 7.45 s and for V224L, 1.36 ± 0.22 s; τon for WT, 176.88 ± 19.42 s and for V224L, 13.53 ± 0.43 s) (supplemental Fig. S4, B and C). Unlike Kir2.1, the apparent affinity of the Kir2.2(V224L) mutant for diC8-PIP2 was left-shifted (EC50 for WT, 6.13 ± 0.43 μm and for V224L, 2.46 ± 0.13 μm) (supplemental Fig. S4D). The WT and the V224L Kir2.2 mutant showed similar whole-cell currents (supplemental Fig. S4E).
V223L and V224L, but not V223M, also Increase the Flexibility of the GH Loop in the Linker between the CD and G Loops
To explore the mechanism by which the V223L and V224L affected the PIP2-induced gating kinetics, we performed 10-ns MD simulations of the cytosolic domains of the WT Kir2.1, Kir2.2 channels and the three mutants (V223L, V223M, and V224L) and analyzed the RMSF, which is a measure of the flexibility of each residue for the WT and mutant channels. Fig. 3A shows significant changes in the flexibility of the GH loop (amino acids 270 to 294, which precisely define the boundaries of the GH loop, see supplemental Fig. S1) that were induced specifically by the V223L or V224L mutation compared with the WT or the V223M mutant, respectively.
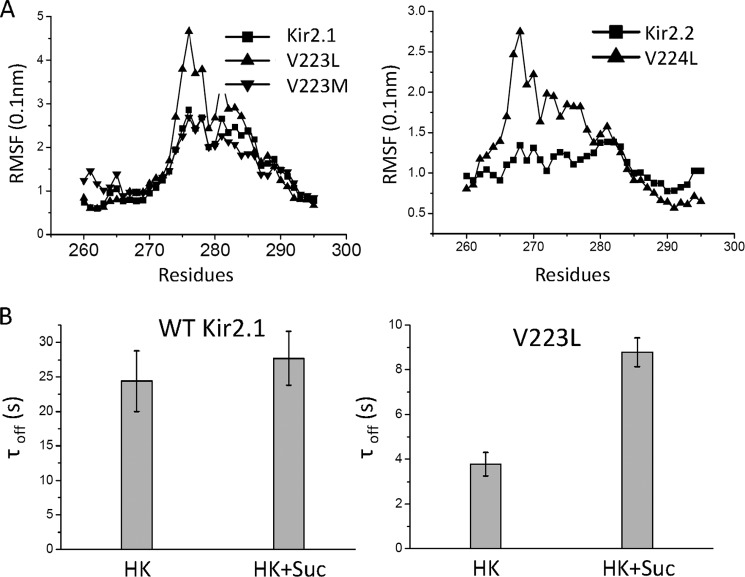
FIGURE 3.
Induction of flexibility of the linker between the CD and G-loops is coupled with changes in the gating kinetics of Kir channels. A, RMSF analysis of the linker between CD and G loops of Kir2.1, Kir2.2, and their mutants. The Val-to-Leu mutations but not the Met mutation cause an increase in the RMSF of Kir2.2 B, time constants (mean ± S.E. of at least three experiments) of rundown (because of diC8 PIP2 washout) of WT-Kir2.1 (left panel) and V223L (right panel) in the presence and absence of sucrose. Only Kir2.1(V223L) shows a significant slowing of the rundown kinetics in the presence of sucrose.
We asked whether this enhanced flexibility induced by the Val to Leu mutation could be related to its effects on the channel gating kinetics. To address this question, we examined the PIP2-induced gating kinetics of the WT Kir2.1 and V223L mutant channels in intracellular solutions with increased viscosity. The dynamics of a protein in solution are intimately coupled to the dynamics of the solvent. The fluctuation amplitudes and relaxation rates, i.e. the flexibility of proteins, can be reduced by increases in solution viscosity (37–40). Previous studies have shown that increases in solution viscosity affect the moving parts of voltage-dependent Na+ channels during gating, causing a decrease in the rate of channel activation (41). We increased the viscosity of the intracellular solution by addition of 2 m sucrose (42) and measured rundown kinetics of currents from inside-out patches expressing the WT Kir2.1 or the V223L mutant channels. Compared with the normal intracellular solution, the solution with the 2 m sucrose showed a non-significant trend of slowing down the time course of rundown in the WT (- sucrose, 24.41 ± 8.40 s; + sucrose, 27.70 ± 7.90 s) (Fig. 3B). In contrast, the rundown kinetics of the V223L mutant were slowed significantly (- sucrose, 3.77 ± 0.53 s; + sucrose, 8.78 ± 0.65 s).
The E272G Mutant Mimics the Effect of the V223L Mutant
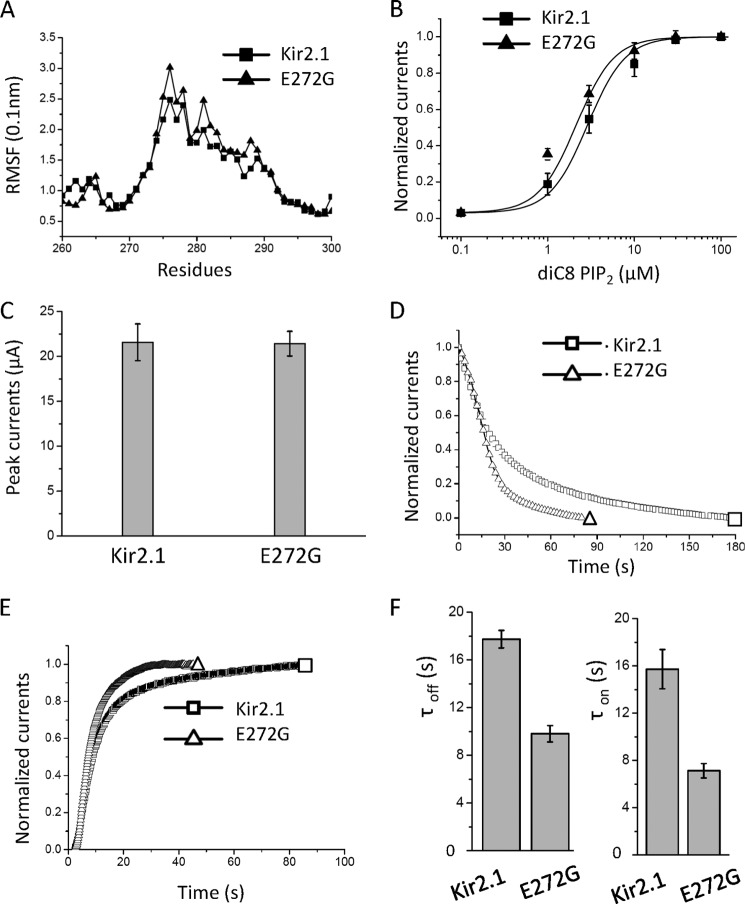
If indeed Val-223 were to be allosterically coupled to the G loop (at Ala-306) via the flexible region (at Glu-272), could an increase in the relative flexibility of Glu-272 mimic the V223L effects on Kir2.1 gating? Gly residues are utilized in protein structures to confer flexibility (e.g. the Gly hinge in the inner helix that forms the permeation pathway) (39, 43, 44). We introduced a glycine at Glu-272 and tested whether it increased local flexibility and whether it mimicked the gating effects of V223L. The RMSF analysis of the E272G mutant indeed showed a significant increase in local flexibility (Fig. 4A). The apparent affinity of the E272G mutant to diC8-PIP2 was slightly left-shifted (EC50 values for WT and the E272G mutant were 2.75 ± 0.12 μm and 2.14 ± 0.21 μm, respectively) (Fig. 4B), whereas the mutant displayed similar peak currents (Fig. 4C). Interestingly, and consistent with V223L, the E272G mutant accelerated the gating kinetics of both channel inhibition and reactivation by Ci-VSP (τoff for WT, 17.73 ± 0.72 s and for E272G, 9.81 ± 0.68 s; τon for WT, 15.72 ± 1.66 s and for E272G, 7.13 ± 0.61 s) (Fig. 4, D–F). A Gly scan of this region (from 266 to 272, supplemental Fig. S5A) revealed that Gly mutations at Thr-268 and Ile-269 in the βG strand were not tolerated well (were either functionally dead or displayed very low activity, respectively), whereas mutations (Pro-266 and Ile-267) on the N-terminal side of the GH loop were without effect. Mutants at the beginning of the flexible GH loop (Val-270, His-271, and Glu-272) showed significant effects on PIP2-induced gating (supplemental Fig. S5, B and C).
FIGURE 4.
The Kir2.1(E272G) also accelerates the gating kinetics albeit to a smaller extent than V223L. A, The RMSF of Kir2.1 and E272G in the GH loop, the linker between the CD and G-loops. The mutant causes an increase in RMSF relative to the control. B, dose-responses of diC8 PIP2 on Kir2.1 and the E272G mutant reveal a small leftward shift in apparent affinity caused by the mutation. C, peak currents of Kir2.1 and the E272G mutant appear comparable. D, the representative time course of current inhibition (PIP2 dephosphorylation induced by Ci-VSP, a 1.8-fold effect) of WT-Kir2.1 and E272G. E, the representative time course of reactivation (PIP2 rephosphorylation, a 2.2-fold effect comparable with the inhibitory effect shown in D) of WT-Kir2.1 and E272G. F, summary bars (left and right) represent the time constants (mean ± S.E. of at least six experiments) corresponding to D and E, respectively.
A Conserved Gating Mechanism Linking the CD and G Loops
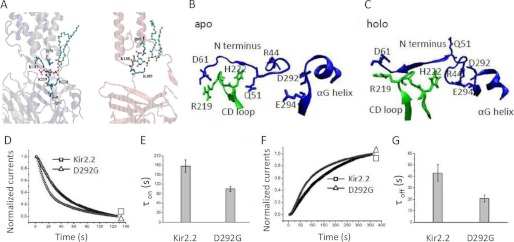
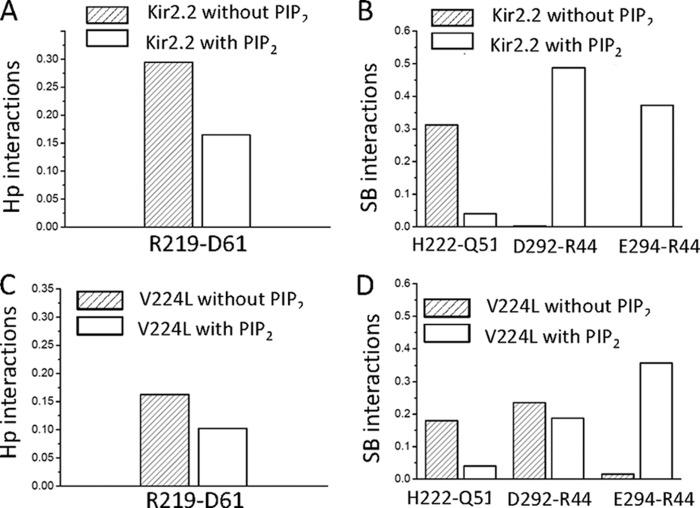
To gain structural insights of how PIP2 might induce channel gating via Val-223 of the CD loop in a manner that involves the GH loop, we performed 100 ns free MD simulations on the WT Kir2.2 and V224L, both in the absence (apo) and presence (holo) of PIP2 using the full-length Kir2.2 crystal structure (13). Supplemental Table S1 shows salt bridge interactions between PIP2 and Kir2.2 during a 20-ns interval in the second half of the simulations. The simulation showed PIP2 to interact with residues Gln-51, Arg-78, Arg-80, Lys-183, Lys-188, Lys-189, Arg-219, and Lys-220 (Fig. 5A and supplemental Table S1) in good agreement with a cocrystal structure of Kir2.2 in complex with PIP2 (17). Comparison of the apo and holo simulations revealed that PIP2 releases the N terminus from the CD loop by weakening the interactions between Arg-219-Asp-61 and His-222-Gln-51 (Fig. 5, B and C, Fig. 6, A and B, and Supplemental Table S2). The N terminus then switches to interact with the αG helix by strengthening the interactions between Asp-292-Arg-44 and Glu-294-Arg-44 (Fig. 5, B and C, and Fig. 6, A and B). The V224L mutant, with an increased flexibility of the GH loop, reduced the intersubunit interactions of the N terminus with the CD loop residues in the apo state (Gln-51-His-222 and Asp-61-Arg-219) and the interactions of the N terminus with the αG helix in the holo state (Asp-292-Arg-44 and Glu-294-Arg-44) compared with the WT (Fig. 6, C and D). Thus, the mutant decreased overall interactions of both the CD and GH loops with the N terminus and increased the flexibility of the GH loop, as if interactions with the N terminus normally constrain the flexibility of the GH loop. To examine the prediction from the simulations of the importance of the Asp-292 interactions, we tested the effects of the D292G mutation on the Kir2.2 gating kinetics. The D292G mutation accelerated the gating kinetics of both channel inhibition and reactivation by Ci-VSP (τoff for WT, 42.87 ± 7.45 s and for D292G, 20.74 ± 2.93s; τon for WT, 176.88 ± 19.42 s and for D292G, 104.82 ± 7.66 s) (Fig. 5, D–G). We also made the E294G mutation, but that caused a significant reduction in whole-cell currents (from 6.56 ± 0.73 μA for the WT to 0.68 ± 0.03 μA for E294G), making it difficult to characterize it further.
FIGURE 5.
Partial crystal structure model of Kir2. The WT shows detailed interactions between the CD-loop, N terminus, and the αG helix. A, the residues that interact with PIP2 from our 100-ns-long MD simulations. PIP2 is shown in a ball-and-stick representation. B and C show the detailed structures of WT Kir2.1 with (holo) and without (apo) PIP2, respectively, to underscore changes in the relationship of the N terminus with the CD and GH loops. D and F are representative trace currents of Kir2.2 WT and D292G, one of the GH loop residues predicted by our simulations to interact with PIP2. PIP2 dephosphorylation of the D292G mutant induced by Ci-VSP (a 2.1-fold effect) and PIP2 rephosphorylation (a 1.7-fold effect) relative to the control, respectively. E and G are summary data (mean ± S.E. of at least six experiments).
FIGURE 6.
PIP2 modulates gating of Kir channels by altering interactions between the CD loop, N terminus, and the αG helix of the GH loop. Shown is the effect of PIP2 on averaged hydrophobic (Hp) interactions between Arg-219-Asp-61 (A) and salt bridge (SB) (B) interactions between His-221-Gln-51, Glu-292-Arg-44, and Glu-294-Arg-44 in the Kir2.2 WT. C, effect of PIP2 on averaged hydrophobic interactions between Arg-219-Asp-61 (C) and salt bridge (D) interactions between His-221-Gln-51, Glu-292-Arg-44, and Glu-294-Arg-44 in Kir2.2 V224L.
DISCUSSION
PIP2-induced channel gating is an intrinsically dynamic process that is difficult to understand by the few available static crystal structures that are missing key transition steps connecting them. Here we apply a combination of computational and electrophysiological approaches to discern the role of important structural elements in the PIP2-induced gating of Kir2 channels, focusing on the cytosolic G-loop gate. We started with MD simulations performed on the crystal structure of the WT Kir2.1 cytosolic domains. Following the MD simulation, we identified that the movements of Ala-306 (of the G loop) correlated highly with Val-223 (of the CD loop) and Glu-272 (of the GH loop).
Mutagenesis of Val-223 and kinetic electrophysiological measurements showed that V223L, but not V223M, right-shifted the diC8-PIP2 dose response, increased currents and caused faster gating kinetics by accelerating both the inhibition and recovery rates. The corresponding mutant in Kir2.2, V224L, showed similar kinetic effects as V223L. Additional MD simulations performed on Kir2.1 (V223L) and Kir2.2 (V224L) showed similar increases in the flexibility of residues corresponding to the GH loop of Kir2 channels (i.e. the 270–294 residues of the Kir2.1 or the 265–294 residues of the Kir2.2).
A number of crystal structures of KirBac3.1 (21) revealed interactions between the N terminus and the CD loop, forming the “latched” or closed state. In a recent computational study with the Kir3.1 chimera, we revealed that PIP2 weakens these interactions, forcing the latched N terminus to transition to an “unlatched” or open state (21, 45). The fact that the N-terminal interactions underlie the transition of the G-loop gate from the closed to the open state in such different channels as the Kir3.1 chimera and KirBac3.1 suggest that the critical role of the N terminus in Kir channel gating has been conserved from prokaryotes to eukaryotes.
Our 100-ns-long MD simulations were performed on the Kir2.2 WT or the V224L channel in the presence or absence of PIP2. There was excellent agreement between the predicted Kir2.2 interacting residues with PIP2 (Gln-51, Arg-78, Arg-80, Lys-183, Lys-188, Lys-189, Arg-219, and Lys-220) and those reported for Kir2.1 from mutagenesis and electrophysiological measurements (8) (Kir2.2. corresponding residues Gln-51, Arg-65, Lys-183, Arg-186, Lys-188, Lys-189, Arg-190, Arg-219, Lys-220, Arg-229, and Arg-313). A similar agreement exists with Kir2.2 residues shown in a recent crystal structure (17) (Arg-65, Arg-78, Trp-79, Arg-80, Lys-183, Arg-186, Lys-188, Lys-189, Arg-190, and Arg-219). Thus, six of the eight residues predicted to interact with PIP2 were shown to affect PIP2 sensitivity experimentally (supplemental Table S1). Other residues shown experimentally to affect PIP2 sensitivity could be doing so in an allosteric manner. The interaction network between the CD loop, the N terminus, and the loop segment around the αG helix of the GH loop revealed that there were appreciable interactions between the N terminus and the CD loop (His-222-Gln-51 and Arg-219-Asp-61) in the absence of PIP2 stabilizing the closed state. PIP2 weakened these two pairs of interactions, interacting with Arg-219 directly and stabilizing interactions between the N terminus and the GH loop: Asp-292-Arg-44 and Glu-294-Arg-44. Interestingly, Asp-292 and Glu-294 are exactly at the other end of the GH loop from Glu-272 that affected PIP2-induced gating kinetics. Thus, the N terminus appears to have switched from stabilizing the CD loop in the closed state to stabilizing the αG helical region within the GH loop in the open state.
The V224L mutation not only stabilized the CD-loop and N-terminal interactions with PIP2 but also weakened all four pairs of interactions between the N terminus and the CD loop or the GH loop. The weakened His-221-Gln-51 and Arg-219-Asp-61 interactions would make it easier for the N terminus to switch from stabilizing the closed to stabilizing the open state, accelerating the activation kinetics of Kir2 channels. The weakened interactions between the N terminus and αG helical region of the GH loop allowed for the increased flexibility of the GH loop, ensuring that the channel can easily transition from the open to the closed state, thus accelerating the deactivation kinetics as well. The flexibility of the GH loop (residues 270 to 294), reflecting the weakened interactions between the N terminus and the GH loop, showed greater effects on the deactivation kinetics (τoff).
Acknowledgments
We thank Heikki Vaananen and Sophia Gruszecki for oocyte isolation and all members of the Logothetis laboratory for critical feedback on this work at every stage of its development.
This work was supported, in whole or in part, by grant National Institutes of Health Grant HL059949 (to D. L.). This work was also supported by NSFC (National Natural Science Foundation of China) Grants 11145003 (to H. A.), 10975045 and C2009000029 (to Y. Z.), and 30730031 (to H. Z.) and by 973 Program Grants 2007CB512100 and C2009001104 (to H. Z.). Computations performed at VCU were supported by the Center for High Performance Computing (CHiPC) and by the National Institutes of Health Grant S10RR027411 (to M. C.).

This article contains supplemental Figs. S1–S5 and Tables 1 and 2.
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- M2
- inner transmembrane helix
- SF
- selectivity filter
- MD
- molecular dynamics
- RMSF
- root mean square fluctuation
- TEVC
- two-electrode voltage clamp
- Ci-VSP
- Ciona intestinalis voltage-sensitive phosphatase.
REFERENCES
- 1. Schulte U., Fakler B. (2000) Gating of inward-rectifier K+ channels by intracellular pH. Eur. J. Biochem. 267, 5837–5841 [DOI] [PubMed] [Google Scholar]
- 2. Logothetis D. E., Kurachi Y., Galper J., Neer E. J., Clapham D. E. (1987) The β γ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 325, 321–326 [DOI] [PubMed] [Google Scholar]
- 3. Huang C. L., Feng S., Hilgemann D. W. (1998) Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature 391, 803–806 [DOI] [PubMed] [Google Scholar]
- 4. Sui J. L., Petit-Jacques J., Logothetis D. E. (1998) Activation of the atrial KACh channel by the βγ subunits of G proteins or intracellular Na+ ions depends on the presence of phosphatidylinositol phosphates. Proc. Natl. Acad. Sci. U.S.A. 95, 1307–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hilgemann D. W., Ball R. (1996) Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science 273, 956–959 [DOI] [PubMed] [Google Scholar]
- 6. Fan Z., Makielski J. C. (1997) Anionic phospholipids activate ATP-sensitive potassium channels. J. Biol. Chem. 272, 5388–5395 [DOI] [PubMed] [Google Scholar]
- 7. Baukrowitz T., Schulte U., Oliver D., Herlitze S., Krauter T., Tucker S. J., Ruppersberg J. P., Fakler B. (1998) PIP2 and PIP as determinants for ATP inhibition of KATP channels. Science 282, 1141–1144 [DOI] [PubMed] [Google Scholar]
- 8. Lopes C. M., Zhang H., Rohacs T., Jin T., Yang J., Logothetis D. E. (2002) Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron 34, 933–944 [DOI] [PubMed] [Google Scholar]
- 9. Pegan S., Arrabit C., Zhou W., Kwiatkowski W., Collins A., Slesinger P. A., Choe S. (2005) Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat. Neurosci. 8, 279–287 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H., He C., Yan X., Mirshahi T., Logothetis D. E. (1999) Activation of inwardly rectifying K+ channels by distinct PtdIns(4,5)P2 interactions. Nat. Cell Biol. 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 11. Logothetis D. E., Jin T., Lupyan D., Rosenhouse-Dantsker A. (2007) Phosphoinositide-mediated gating of inwardly rectifying K(+) channels. Pflugers Arch. 455, 83–95 [DOI] [PubMed] [Google Scholar]
- 12. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) The structure of the potassium channel. Molecular basis of K+ conduction and selectivity. Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 13. Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cuello L. G., Jogini V., Cortes D. M., Pan A. C., Gagnon D. G., Dalmas O., Cordero-Morales J. F., Chakrapani S., Roux B., Perozo E. (2010) Structural basis for the coupling between activation and inactivation gates in K(+) channels. Nature 466, 272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo D., Lu Z. (2003) Interaction mechanisms between polyamines and IRK1 inward rectifier K+ channels. J. Gen. Physiol. 122, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soom M., Schönherr R., Kubo Y., Kirsch C., Klinger R., Heinemann S. H. (2001) Multiple PIP2 binding sites in Kir2.1 inwardly rectifying potassium channels. FEBS Lett. 490, 49–53 [DOI] [PubMed] [Google Scholar]
- 17. Hansen S. B., Tao X., MacKinnon R. (2011) Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whorton M. R., MacKinnon R. (2011) Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inanobe A., Matsuura T., Nakagawa A., Kurachi Y. (2007) Structural diversity in the cytoplasmic region of G protein-gated inward rectifier K+ channels. Channels 1, 39–45 [PubMed] [Google Scholar]
- 20. Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clarke O. B., Caputo A. T., Hill A. P., Vandenberg J. I., Smith B. J., Gulbis J. M. (2010) Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 141, 1018–1029 [DOI] [PubMed] [Google Scholar]
- 22. Horn R. (2005) Electrifying phosphatases. Sci. STKE 307, 50. [DOI] [PubMed] [Google Scholar]
- 23. Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23, 318–326 [DOI] [PubMed] [Google Scholar]
- 24. Im W., Lee M. S., Brooks C. L., 3rd. (2003) Generalized born model with a simple smoothing function. J. Comput. Chem. 24, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 25. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 26. Lupyan D., Mezei M., Logothetis D. E., Osman R. (2010) A molecular dynamics investigation of lipid bilayer perturbation by PIP2. Biophys. J. 98, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacKerell A. D., Brooks C. L., Nilsson L., Roux B., Y. Won M. K. (1998) CHARMM: The energy function and its parameterization with an overview of the program. The Encyclopedia of Computational Chemistry 1, 271–277 [Google Scholar]
- 28. Darden T., York D., Pedersen L. (1993) Particle Mesh Ewald. An N.Log(N) method for ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 29. Roux B. (2008) The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95, 4205–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bjelkmar P., Niemela P. S., Vattulainen I., Lindahl E. (2009) Conformational changes and slow dynamics through microsecond polarized atomistic molecular simulation of an integral Kv1.2 ion channel. PLOS Comput. Biol. 5 e1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mezei M. (1997) Optimal position of solute for simulations. J. Comput. Chem. 18, 812–815 [Google Scholar]
- 32. Mezei M. (2003) A new method for mapping macromolecular topography. J. Mol. Graph. Model. 21, 463–472 [DOI] [PubMed] [Google Scholar]
- 33. Mezei M., Filizola M. (2006) TRAJELIX. A computational tool for the geometric characterization of protein helices during molecular dynamics simulations. J. Comput. Aided Mol. Des. 20, 97–107 [DOI] [PubMed] [Google Scholar]
- 34. Mezei M. (2010) Simulaid. A simulation facilitator and analysis program. J. Comp. Chem. 31, 2658–2668 [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez-Menchaca A. A., Adney S. K., Tang Q. Y., Meng X. Y., Rosenhouse-Dantsker A., Cui M., Logothetis D. E. (2012) PIP2 controls voltage-sensor movement and pore opening of Kv channels through the S4-S5 linker. Proc. Natl. Acad. Sci. U.S.A. 109, E2399–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. (2005) Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243 [DOI] [PubMed] [Google Scholar]
- 37. Beece D., Eisenstein L., Frauenfelder H., Good D., Marden M. C., Reinisch L., Reynolds A. H., Sorensen L. B., Yue K. T. (1980) Solvent viscosity and protein dynamics. Biochemistry 19, 5147–5157 [DOI] [PubMed] [Google Scholar]
- 38. Frauenfelder H., Chen G., Berendzen J., Fenimore P. W., Jansson H., McMahon B. H., Stroe I. R., Swenson J., Young R. D. (2009) A unified model of protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin T., Peng L., Mirshahi T., Rohacs T., Chan K. W., Sanchez R., Logothetis D. E. (2002) The (β)γ subunits of G proteins gate a K(+) channel by pivoted bending of a transmembrane segment. Mol. Cell 10, 469–481 [DOI] [PubMed] [Google Scholar]
- 40. Ding S., Ingleby L., Ahern C. A., Horn R. (2005) Investigating the putative glycine hinge in Shaker potassium channel. J. Gen. Physiol. 126, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kukita F. (2000) Solvent effects on squid sodium channels are attributable to movements of a flexible protein structure in gating currents and to hydration in a pore. J. Physiol. 522, 357–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang J., Krishnamoorthy G., Saxena A., Zhang G., Shi J., Yang H., Delaloye K., Sept D., Cui J. (2010) An epilepsy/dyskinesia-associated mutation enhances BK channel activation by potentiating Ca2+ sensing. Neuron 66, 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alam A., Jiang Y. (2009) High-resolution structure of the open NaK channel. Nat. Struct. Mol. Biol. 16, 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 45. Meng X. Y., Zhang H. X., Logothetis D. E., Cui M. (2012) The molecular mechanism by which PIP(2) opens the intracellular G-loop gate of a Kir3.1 channel. Biophys. J. 102, 2049–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]