Background: Ca2+ binding to the thin filament protein troponin strictly regulates muscle contraction.
Results: Ca2+ affects measured and mapped dynamics extensively within separated areas of cardiac troponin. in a structurally revealing manner.
Conclusion: Troponin dynamics are critical to its function.
Significance: Mechanistic discovery regarding troponin action improves understanding of normal, adaptive, and pathological cardiac contraction, including heritable cardiomyopathy.
Keywords: Calcium-binding Proteins, Cardiomyopathy, Molecular Motors, Muscle, Protein Dynamics, Protein Folding, Troponin
Abstract
Ca2+ dissociation from troponin causes cessation of muscle contraction by incompletely understood structural mechanisms. To investigate this process, regulatory site Ca2+ binding in the NH2-lobe of subunit troponin C (TnC) was abolished by mutagenesis, and effects on cardiac troponin dynamics were mapped by hydrogen-deuterium exchange (HDX)-MS. The findings demonstrate the interrelationships among troponin's detailed dynamics, troponin's regulatory actions, and the pathogenesis of cardiomyopathy linked to troponin mutations. Ca2+ slowed HDX up to 2 orders of magnitude within the NH2-lobe and the NH2-lobe-associated TnI switch helix, implying that Ca2+ greatly stabilizes this troponin regulatory region. HDX of the TnI COOH terminus indicated that its known role in regulation involves a partially folded rather than unfolded structure in the absence of Ca2+ and actin. Ca2+-triggered stabilization extended beyond the known direct regulatory regions: to the start of the nearby TnI helix 1 and to the COOH terminus of the TnT-TnI coiled-coil. Ca2+ destabilized rather than stabilized specific TnI segments within the coiled-coil and destabilized a region not previously implicated in Ca2+-mediated regulation: the coiled-coil's NH2-terminal base plus the preceding TnI loop with which the base interacts. Cardiomyopathy-linked mutations clustered almost entirely within influentially dynamic regions of troponin, and many sites were Ca2+-sensitive. Overall, the findings demonstrate highly selective effects of regulatory site Ca2+, including opposite changes in protein dynamics at opposite ends of the troponin core domain. Ca2+ release triggers an intramolecular switching mechanism that propagates extensively within the extended troponin structure, suggests specific movements of the TnI inhibitory regions, and prominently involves troponin's dynamic features.
Introduction
The principal direct trigger for muscle relaxation in striated muscles, including the heart, is the dissociation of Ca2+ from the thin filament protein, troponin (1–6). More specifically, it is Ca2+ dissociation from specific site(s) of the troponin subunit TnC that is essential for relaxation. If troponin is removed experimentally, muscles contract regardless of Ca2+ (7). With troponin present, contraction is strictly dependent upon Ca2+ binding to TnC. To investigate this central, partially understood (8–14) regulatory mechanism, the present study widely examines and quantitatively maps the effects of regulatory site II Ca2+ on cardiac troponin's dynamics.
Peptide backbone NH hydrogens exchange with H2O solvent hydrogens or exchange instead with solvent deuterium when a protein is immersed in D2O. The hydrogen bonding of protein folding suppresses, very greatly, the rate of such hydrogen-deuterium exchange in secondary structures. Hydrogen-deuterium exchange rates vary greatly within and between proteins, quantitatively mirroring the great variation in flexibility and local dynamics that is characteristic of proteins. Measurements of the local rates of exchange across a molecule provide a quantitative dynamic map of that protein's many distinct regions (15–19).
In 2010, we reported (20) first application to troponin of the approach again used here: hydrogen-deuterium exchange-mass spectrometry (HDX2-MS). HDX-MS was used to obtain dynamic maps of troponin at 25 °C, in the presence of either saturating or subsaturating Ca2+ concentration. This earlier report found troponin to be highly dynamic generally, undergoing rapid HDX in many regions, and yet also to contain regions of high stability that exhibited very slow HDX. We also observed that troponin's dynamics were affected by the free Ca2+ concentration, in a complex manner. However, the effects of Ca2+ could not be assessed comprehensively, because HDX rates were so fast that 50% of exchange preceded the first time point. Furthermore, site II was not fully emptied in the lower Ca2+ concentration condition.
We later reported (21) a considerably more incisive dynamic map for Ca2+-saturated troponin, obtained by using 10 °C rather than 25 °C HDX conditions. Using a lower temperature to slow the intrinsic HDX rate, it was possible to characterize many troponin residues for the first time and to assess others very much better than previously.
In the present report, these lower temperature conditions are used to map the effect of regulatory site Ca2+ on troponin dynamics. To accomplish this, we developed an approach in which very high Ca2+ occupancy of sites III and IV in the TnC COOH-lobe was assured and unaffected, whereas site II in the NH2-lobe was completely and specifically emptied. To keep sites III and IV unaffected, we employed 1 mm CaCl2, the same concentration used previously for HDX of Ca2+-saturated troponin. To empty site II, we used Ala mutations in selected residues of the metal-coordinating EF-hand loop (22, 23).
As shown below, the results provide a new picture of troponin dynamics in relation to regulatory function. Among the new findings is the demonstration that Ca2+ highly selectively alters troponin's dynamic behavior. Troponin consistently is stabilized locally near the Ca2+ site and consistently is destabilized in a region at the farthest end of the troponin core domain. The mechanistic implications of this and many other HDX findings are discussed. Finally, the data comprehensively and quantitatively validate the concept that cardiomyopathy-inducing mutations cluster in dynamic regions of troponin.
EXPERIMENTAL PROCEDURES
Protein Preparation
Human cardiac CBMII troponin core domain was prepared from bacterially expressed TnI, D65A/E66A TnC, and COOH-terminal TnT fragment 183–288. The three subunits were mixed in a 1:1:1 ratio and then reconstituted into a ternary complex by serial dialysis (20). The complex was isolated by ion exchange chromatography using a Resource S column and an AKTA system (GE Healthcare), concentrated by a Centricon apparatus (Millipore), and stored at −80 °C before use.
HDX and Exchange Analysis by HPLC-Electrospray Ionization Fourier Transform-Ion Cyclotron Resonance MS
Exchange was performed as described previously (20, 21). Briefly, thawed protein samples were placed into exchange buffer (10 mm NaH2PO4, pH 7.0, 0.1 m NaCl) by a spin column. HDX was initiated by mixing with 9 parts of the same, 10 °C buffer but in D2O. Aliquots were removed at different time points to be acid-quenched with pH 2.5 phosphate buffer and flash frozen in liquid nitrogen. Samples were stored at −80 °C.
For analysis, quickly thawed troponin samples were digested with an equal mass of pepsin for 5 min on ice. They were then immediately injected into a micropeptide trap (Microchrom), which was attached to a C18 HPLC column, which was connected to the Fourier Transform-Ion Cyclotron Resonance MS. Peptide envelopes were easily recognizable by correspondence to the possible postexchange m/z of pre-HDX-identified peptides. The centroid mass of each peptide was determined using MagTran (24). The fragments are the same as those described previously for unmodified troponin examined under the same HDX conditions (21), with three exceptions. Time course measurements for TnT 264–288, TnI 79–85, and TnC 81–97 were substituted for TnT 263–288, TnI 78–88, and TnC 82–97, respectively, because data for the latter set were not consistently detected in the CBMII troponin MS spectra.
Curve Fitting of Exchange Kinetic Data
Before curve fitting, corrections for backward D/H exchange were applied as reported previously (21), averaging 23%. Transition sizes and rate constants were determined by non-linear least squares curve fitting of exponential peptide mass increases over time, using Scientist (Micromath). These parameter and error estimate determinations were performed either simply on individual peptide data or globally on grouped data from peptides wherever there was peptide overlap. The globally fit groups of peptides for CBMII troponin were: TnC (28–57, 36–56, 48–56) and (118–132, 122–130, 122–132); TnI (27–53, 29–49, 29–53), (54–66, 54–78, 62–85, 79–85), (97–109, 97–116), (125–134, 125–152), (156–169, 162–169), and (191–197, 191–210); TnT (224–242/225–243 (which are effectively indistinguishable after exchange), 230–243, 237–243) and (243–263/244–263 (which are effectively indistinguishable after exchange), 244–250, 249–263, 251–263), where peptide groups are enclosed in parentheses.
As previously (21), transition magnitudes averaged half the length of the assignable regions in which they were detected. Correspondingly, most assignable regions contained more than one exchange transition. From HDX data alone, one cannot determine the location of a transition within a somewhat larger assignable region that is defined by the size of the peptide(s). The resulting, ∼5-residue uncertainty in localization was addressed as previously (22) to produce the assignments in the present study. In other words, the different transitions were ordered within an assignable region to correspond as much as possible with the troponin high resolution structural data (25). For example, faster exchange was assigned where crystallographic B-factors were higher or particularly where no structure was detected by crystallography. The latter consideration produces an unambiguously justified assignment for HDX occurring prior to 5 s (i.e. without protection).
Calculation of Protection Factors
Measured exchange rates were converted to protection factors, which indicate the degree of HDX slowing relative to rates expected if troponin were unfolded. Protection factors reflect local folding stability, measured in the context of the globally folded molecule. More specifically, protection factors were calculated by comparison of measured HDX rates with the geometric mean of the model peptide unprotected HDX rates (26) for each residue in the assignable region. Values for these means ranged from 0.4 to 1.8 s−1. Because this is a narrow range relative to the much wider variation in HDX rates, Fig. 5 would be altered little if exchange rate values were mapped instead of protection factors.
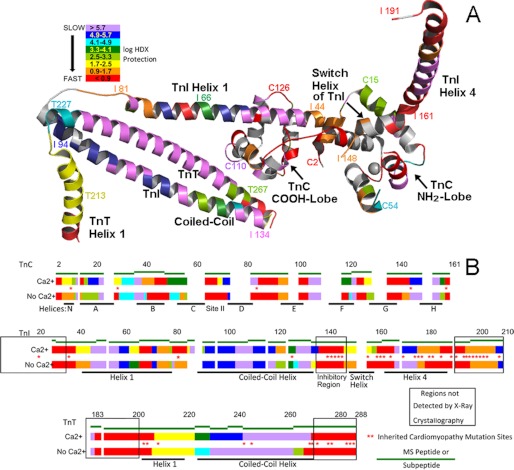
FIGURE 5.
Dynamic maps of cardiac troponin in the absence and presence of site II Ca2+. Shown is a summary of all of the results, using a logarithmic scale to convey the wide range of observed dynamic behavior. A and B show the same color coding. Red, highly dynamic unprotected regions, generally undergoing HDX in <5 s. Violet, highly stable regions that have slow dynamics. Intermediate dynamics are shown according to the degree of protection from HDX, which was calculated from the measured HDX rates (Table 1). A, CBMII troponin (Ca2+-free) findings mapped onto the cardiac troponin three-dimensional structure. Residue number annotation sites were chosen for ease of illustration. Orientation is the same as in Figs. 1–4. In B, the same CBMII troponin data (indicated by No Ca2+) as well as data for unmodified troponin (indicated by Ca2+) are shown mapped onto the linear amino acid sequence positions within each troponin subunit. Green line segments indicate the examined peptides or subpeptides, providing a lower limit (i.e. maximum error) to the spatial resolution of the assignments. Within each green line segment, the Ca2+ dynamic regions are positioned to minimize the differences from the No Ca2+ map. Asterisks indicate sites of cardiomyopathy-inducing missense mutations. Note that they cluster almost entirely within the more dynamic regions of troponin and that many sites are Ca2+-sensitive.
RESULTS
Troponin causes cessation of cardiac contraction when Ca2+ is dissociated from one specific site: site II in the TnC subunit's NH2-lobe. To investigate the effects of regulatory Ca2+ with the highest specificity, the present study compares HDX of unmodified troponin with HDX of troponin with D65A/E66A TnC. Such “CBMII” mutation prevents Ca2+ binding and closely reproduces the normal apo state structure, as shown by detailed NMR study of a very similar NH2-lobe construct (27). CBMII and its skeletal muscle counterpart potently inhibit thin filaments in fibers and in solution. They have been examined in many biochemical and physiological experiments (22, 23, 28–35) because they abrogate Ca2+ binding to the NH2-lobe without altering Ca2+ binding to the TnC COOH-lobe.
Effects of Site II Ca2+ on the Dynamic Properties of Troponin's Regulatory Head Region
The regulatory head region of troponin (25) contains two components: (i) the NH2-lobe of TnC and (ii) the COOH-region of subunit TnI. The TnI COOH-region interacts with the NH2-lobe in a strictly Ca2+-dependent manner (25, 36, 37). This reversible interaction is the initial, immediate vicinity aspect of the regulatory Ca2+ switch that alters thin filament structure so as to control contraction.
Fig. 1 shows HDX as a function of time for peptides derived from the regulatory head region. In each panel, HDX data and best fit curves are shown for both CBMII and unmodified troponin. Measured time points spanned from 5 s to 48 h, allowing measurement of a wide range of faster and slower dynamics. The graphed increases in mass over time report hydrogen-deuterium exchange at peptide backbone NH groups. (Side chain hydrogens either do not exchange or else revert to hydrogen before mass measurement.)
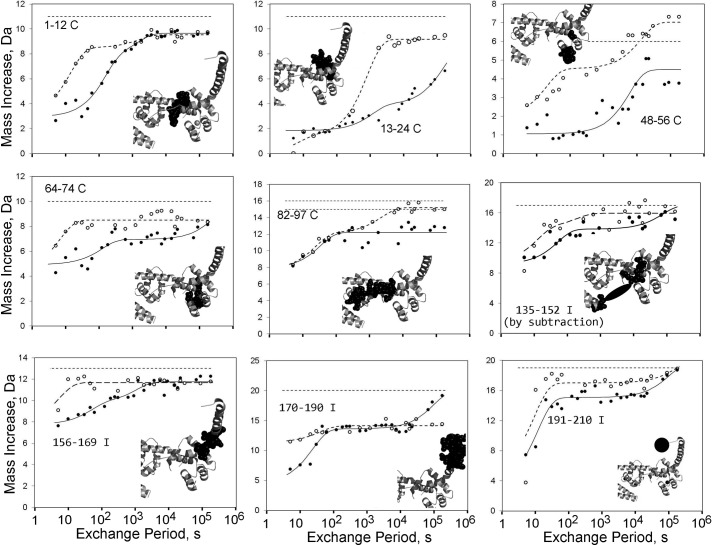
FIGURE 1.
HDX kinetics within troponin's regulatory head region. Troponin was exposed to D2O for times varying between seconds and days. Hydrogen-deuterium exchange resulted in the graphed increases in the masses of fragments generated by pepsin digestion performed post-D2O exposure. Open circles, data for CBMII troponin with inactivated Ca2+ site II. Filled circles, data from Ref. 21 for unmodified troponin, with Ca2+ bound at site II. Dashed and solid lines, best fit curves. In each panel, the location of the graphed peptide within troponin is shown in black in a cropped thumbnail image. (Fig. 5A shows the same orientation with annotations.) The faster TnC transitions for CBMII troponin in each panel, compared with HDX for unmodified troponin, indicate the absence of a large stabilizing effect that occurs when site II is occupied by Ca2+. The illustrated peptides from TnI have a similar effect, but the shifts are smaller in magnitude. Also, note that no TnI peptide is fully exchanged before the first time point, indicating some degree of protection from exchange in each peptide. For CBMII troponin, graphed data were acquired successfully for TnC 81–97 rather than TnC 82–97, which was assessed for unmodified troponin. TnI 135–152 results were obtained by subtraction of the data points and fitted curves shown in Fig. 4 for TnI 125–152 and TnI 125–134. Dotted lines at the top of each panel indicate maximal possible exchange for the graphed peptide(s).
Three broad generalizations are helpful for interpreting the graphs. First, high values at 5 s generally indicate unprotected NH groups. Hydrogen exchanges to deuterium prior to the first time point at sites that have no protein folding-mediated protection from exchange. Second, the time required for an observed HDX transition reflects the relative folding stability of that region. The faster the rate, the weaker the local stability and the faster are the dynamics. Finally, failure to complete HDX by the last, 48 h measurement indicates the most locally stable, least dynamic property that can be characterized.
HDX of TnC NH2-lobe peptides are shown in five of the Fig. 1 panels. The feature most immediately evident is that many NH hydrogens exchanged more slowly when Ca2+ was bound to TnC. Each of these five panels contains at least one transition that occurred earlier when CBMII troponin was studied (open circles) than for unmodified troponin (filled circles). Ca2+ binding to an intact, unmodified site II slowed many HDX transition rates, and no TnC NH2-lobe region had a statistically significant increase in dynamics in the presence of site II Ca2+. Thus, Ca2+ stabilized many portions of the TnC NH2-lobe. The time domain shifts were 1–2 orders of magnitude, indicating that this effect is a large, prominent aspect of regulation. In fact, the 2-order of magnitude shifts in the dynamics of these peptides may be reporting the fractional occupancy of site II with Ca2+. This inference comes from the fact that the experimental 1 mm CaCl2 concentration is about 100-fold greater than the site II Kd for unmodified troponin under these D2O conditions (20).
HDX rates in the TnC NH2-lobe are influenced not only by Ca2+ directly but also by the Ca2+-dependent association of the ∼10-residue TnI switch helix to the NH2-lobe. HDX for the switch helix itself is shown within Fig. 1. In short, the switch helix is stabilized by Ca2+, undergoing faster HDX in the CBMII context than in unmodified troponin.
This is shown in two of the Fig. 1 panels (TnI 156–169 and TnI 135–152). Both peptides include parts of the TnI 150–160 switch helix. CBMII data for each included a pre-20 s HDX transition that was faster than in unmodified (i.e. Ca2+-binding) troponin. It may at first seem puzzling how this transition can be assigned to the switch helix. However, HDX of TnI 162–169 is very fast, occurring before 5 s, regardless of Ca2+ (not shown). Also, the troponin atomic structure indicates that TnI 137–147 is dynamic and unfolded (25). Once these residues are assigned to undergo HDX in <5 s, one can infer that the switch helix is the primary location of the Ca2+-stabilized residues in both 135–152 and 156–169.
Complex, Ca2+-affected dynamics for the COOH-terminal 40 residues of TnI are shown in Fig. 1 TnI panels 170–190 and 191–210. In (Ca2+-free) CBMII troponin this region is not tethered to the TnC NH2-lobe by the switch helix. Interestingly, this untethered state does not yield a fully unfolded structure for the TnI COOH terminus. Rather, half of TnI 170–190 exhibits rather high protection from HDX. Also, much of TnI 191–210 exhibits protection, albeit weakly.
These findings indicate that at 10 °C in the absence of both actin and Ca2+, a large part of the TnI COOH terminus is dynamically folded rather than completely unfolded. The ability of this region to form specific structure is consistent with observations that many alternative point mutations in the TnI COOH terminus induce cardiomyopathy (38–41). A specific but weakly folded structure is consistent with NMR data (42, 43) but is distinct from the very interesting proposal that the region functions as an intrinsically disordered domain (44).
Small Effect of Site II Ca2+ on the Dynamics of the TnC COOH-lobe
Unlike the NH2-lobe, the TnC COOH-lobe was not prominently stabilized by site II Ca2+. Rather, peptides describing HDX of this region were very similar for CBMII troponin and for unmodified troponin (Fig. 2). In each peptide shown, about half of the NH groups exchanged prior to the first 5 s time point; they were unprotected. Also for each peptide, many of the NH hydrogens remained unexchanged after the last, 48 h time point; they were highly protected from exchange. Small transitions were in some cases seen, with effects of Ca2+ not reaching statistical significance. The results imply the absence of any large scale effects of site II on the COOH-lobe.
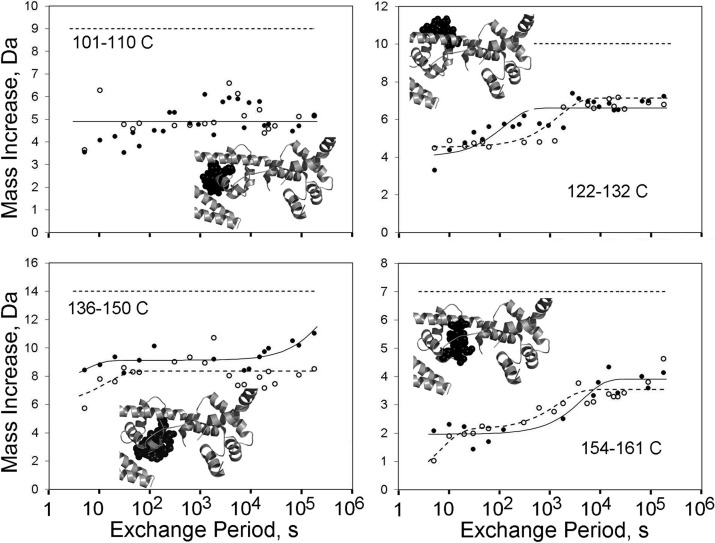
FIGURE 2.
HDX kinetics within the TnC COOH-lobe. As in Fig. 1, the replacement of NH hydrogens by deuterium was measured after variable durations of troponin exposure to D2O. Ca2+ had little effect in these experiments on the HDX occurring in peptides derived from the TnC COOH-lobe. Regardless of Ca2+ at site II, most HDX occurred either before 5 s or after 48 h. Symbols, lines, and thumbnail images are the same as in Fig. 1.
However, these data do not exclude the possibility of limited or localized effects of site II on COOH-lobe dynamics. In fact, there is reason to believe that one portion of the TnC COOH-lobe is stabilized by Ca2+. When studied at 25 °C, TnC 154–161 has an HDX transition that is slower in the presence of millimolar as opposed to micromolar Ca2+ (20). This transition does not appear in the current data set shown in Fig. 2, because it requires longer than 48 h to occur. The importance of this part of TnC is highlighted by the observation that the TnC G159D mutation causes human cardiomyopathy (45), contractile dysfunction (46), and altered interactions with TnI (47–49). Finally, these data do not address how troponin may be affected by site III and IV occupancy by Mg2+ rather than Ca2+.
Selective Effects of Site II Ca2+ on the Dynamics of Core Domain TnT
HDX patterns within TnT were revealing in general and also in relationship to site II. TnT helix 1, comprising residues 204–220, is the most NH2-terminal portion of TnT known clearly to have a well folded structure in the absence of other thin filament proteins. TnT peptide 188–224 contains helix 1 and had an observable HDX transition that was finished within 5 min (Fig. 3). This indicates moderately weak folding of helix 1. The log of the protection factor (Table 1) is in the range 1.76–1.92, providing an estimate of the local protein folding constant; it was slightly less than 100. Site II Ca2+ had no statistically significant effect on the HDX rate of TnT helix 1. However, its fast dynamics are consistent with some other role in regulation, as previously demonstrated by studies of both phosphorylation and cardiomyopathy-linked mutation (50–52).
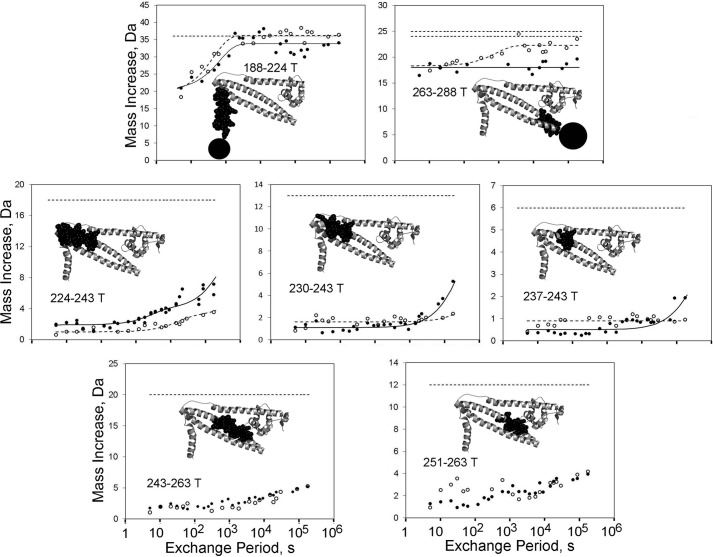
FIGURE 3.
HDX kinetics within TnT. TnT HDX data are shown from the same experiments shown in Figs. 1, 2, and 4. TnT helix 1 was relatively dynamic and unaffected by Ca2+. The TnT strand of the coiled-coil was highly stable, with very slow exchange, and with variable effects of Ca2+ depending upon the region of the coiled-coil. Ca2+ increased the dynamics of the base of the coiled-coil, had no effect for the middle, and stabilized the COOH terminus of the coiled-coil TnT strand. Symbols, lines, and thumbnail images are the same as in Fig. 1. For CBMII troponin, data were acquired successfully for TnT 264–288 rather than data for TnT 263–288, which were acquired for unmodified troponin.
TABLE 1.
HDX rates and assignments
In an unanticipated result, the base of the TnT strand of the TnT-TnI coiled-coil was sensitive to Ca2+; it was stabilized by CBMII. This action can be seen in the late time points of TnT 224–243 and its subpeptide TnT 230–243 (Fig. 3) and to a smaller extent in the HDX data for subpeptide 237–243. Despite the structurally remote location of TnT region 224–237 relative to the NH2-lobe of TnC, site II affected the HDX rate. A conservative extrapolation of the 224–243 and 230–243 data would indicate that half of the NH groups were stabilized by the absence of Ca2+ binding to site II. As shown below, this observation was validated by similar results in TnI.
The above data do not reflect a global effect on the TnT strand, however. This is demonstrated by results for other peptides. Specifically, TnT 243–263 and 251–263 showed identical, very low HDX for 48 h regardless of Ca2+. TnT 237–243 was mostly similar (no effect of Ca2+ except for a 1-Da difference at 48 h). Thus, the mid- and post-mid-portions of the TnT coiled-coil strand do not appear to be affected by Ca2+.
Interestingly, there is an opposite effect of Ca2+ on the dynamics of the coiled-coil's other end, its COOH terminus. Here, as seen in Fig. 3 data for TnT 263–288, Ca2+ stabilized rather than destabilized the TnT strand. For unmodified troponin containing site II Ca2+, HDX of TnT 264–288 never reached completion after 48 h in D2O. This implies that the end of the coiled-coil (which extends to ∼TnT 271) was highly stable. For CBMII troponin, on the other hand, the end of the coiled-coil was not as stable; HDX of virtually the entire 263–288 peptide finished within 1 h in D2O.
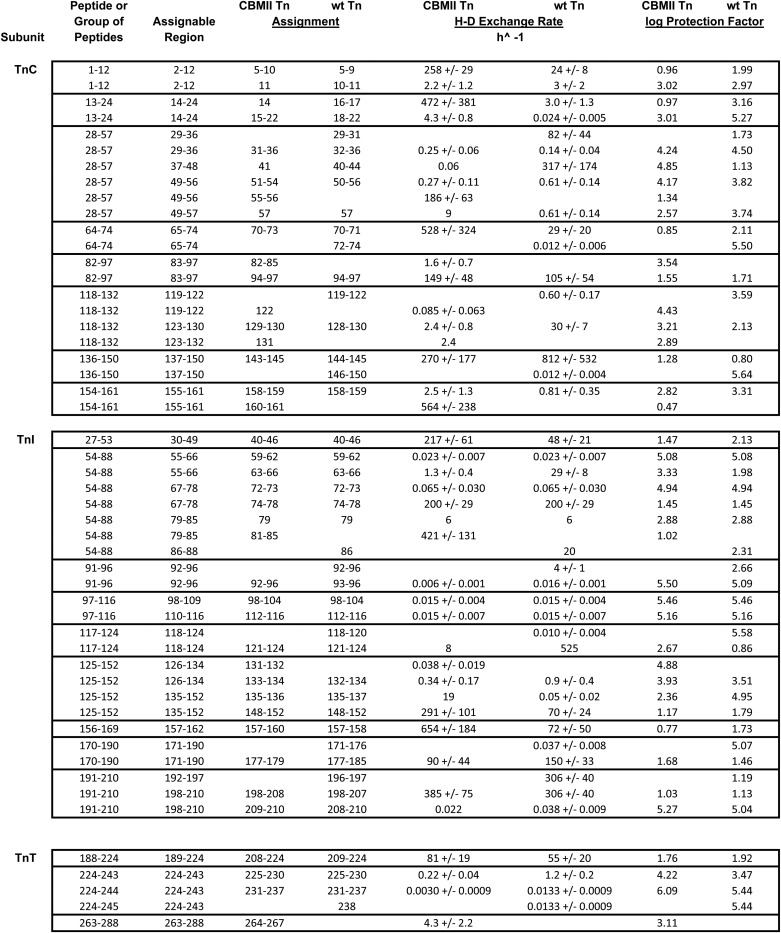
Selective Effects of Site II Ca2+ on the Dynamics of TnI
TnI 91–96 (Fig. 4) showed faster exchange in unmodified troponin than in CBMII troponin. Thus, the TnI strand at the NH2-terminal base of the coiled-coil was stabilized by Ca2+ release. This finding parallels data for the corresponding TnT region, as described above. In a separate but additional similarity to TnT, the immediately succeeding part of the coiled-coil strand (TnI 97–116) was Ca2+-insenstive, stable, and very slowly exchanging.
FIGURE 4.
HDX kinetics within TnI peptides outside the regulatory head region. Shown are HDX data for additional TnI peptides, derived from the experiments also shown in Figs. 1–3. Abolishing Ca2+ binding (CBMII troponin) destabilized the TnI helix 1 NH2 terminus (TnI 29–49 data) and had selective effects on specific other regions of TnI. Many regions were unaffected, but in other regions, CBMII acted to slow HDX, indicating local stabilization in the absence of Ca2+. See “Results.” Symbols, lines, and thumbnail images are the same as in Fig. 1.
In other parts of the coiled-coil, however, the TnI and TnT strands differed. For the TnI strand but not the TnT strand, Ca2+ release-mediated stabilization extended to regions other than the coiled-coil base. Specifically, both TnI 117–124 and TnI 125–134 had HDX transitions that were slower in the CBMII context than in unmodified troponin. The corresponding TnT region did not exhibit this behavior.
In describing the structure of troponin for the first time, Takeda et al. (25) noted that the base of the coiled coil has a hydrophobic interaction with the preceding TnI loop. Also, a hydrogen bond network links TnI Arg-79 to the coiled-coil (53). TnI 62–85 HDX data in Fig. 4 show that CBMII stabilized this TnI loop. The relatively fast, Ca2+-slowed transition in 62–85 is not present in 54–78, so it must reside in loop residues TnI 79–85. Consequently, it can be concluded that site II Ca2+ release has the same, stabilizing effect on all three of the interacting components: both TnT and TnI near the coiled-coil base and also the preceding TnI loop.
Finally, portions of TnI helix 1 were Ca2+-sensitive. A Ca2+ effect on the helix's NH2 terminus was demonstrated by the HDX data for TnI 29–49. The data for this peptide indicated two regions, with different kinetics from each other. One region exchanged before 5 s, regardless of Ca2+. This fast HDX can be attributed to the first half of the peptide, which was not well ordered by crystallography. More interestingly, TnI 29–49 contained a measurable pre-60 s HDX transition that was slowed (i.e. stabilized) by Ca2+. This Ca2+-stabilized region must represent the second half of the peptide, which is the NH2-start of TnI helix 1. The possible structural basis for this stabilization is discussed below.
Most of the remainder of TnI helix 1 was unaffected by Ca2+. HDX data for the long helix 1 peptide TnI 54–78 was similar for CBMII troponin and unmodified troponin. However, one part of helix 1 was stabilized by Ca2+ release. Close examination of the results for TnI 54–66, 54–78, and 62–85 indicate that the same, 3–4-Da, Ca2+-shifted transition was present for all three peptides. Thus, residues 63–66, which comprise the only segment present in the data for all three peptides, became more dynamic when Ca2+ was bound to TnC site II. HDX of this part of TnI helix 1 was slower in the CBMII complex (i.e. in the absence of site II Ca2+).
Comprehensive Map of the Dynamics of Ca2+-free, CBMII Troponin
The processed HDX kinetic results for all regions of troponin are compiled in Table 1 and illustrated in Fig. 5. In addition to HDX rates, Table 1 indicates what those observed kinetics imply about HDX slowing. Specifically, the rate constants were used to calculate how much HDX was slowed compared with HDX rates for unfolded, unprotected model peptides (26). The log of the fold slowing yields the listed protection factors, which are illustrated by color coding in both panels of Fig. 5.
In Fig. 5A, the (Ca2+-free) CBMII troponin results were mapped onto the only high resolution structure available for cardiac troponin, the Ca2+-saturated core domain (25). (The TnI switch helix and helix 4 should be assumed to have a different structure than shown; they would be dissociated from the TnC NH-lobe in CBMII troponin.) Fig. 5B shows the same results (No Ca2+ represents CBMII) mapped onto the linear amino acid sequence. Additionally, Fig. 5B shows findings (Ca2+) derived from our published HDX data (21) for Ca2+-saturated troponin.
Fig. 5A shows that in the absence of Ca2+, the most stable portions of troponin (violet and blue coloring) are the first half of the coiled-coil, parts of the TnC COOH-lobe, and COOH-lobe-associated segments of TnI helix 1. This generalization parallels HDX dynamic mapping results obtained in the presence of Ca2+ (21); the most stable regions remain the same, regardless of site II. Other portions are more dynamic and in fact tend to be very much more dynamic.
Many regions of troponin in Fig. 5 are colored red, indicating very flexible, loosely folded, or unfolded structure. This particularly includes those regions not identified by x-ray crystallography, which are enclosed by large black rectangles in Fig. 5B. The crystallographically undetected TnI COOH terminus is a notable exception. Much of it is weakly folded (orange) rather than unfolded (red).
Violet-shaded regions are much more stable, and they exchanged little hydrogen for deuterium after 48 h of immersion in D2O. This slow exchange, indicating slow dynamics, was particularly evident in the coiled-coil region. Interestingly, the dynamics of the coiled-coil were not the same for TnT and TnI. Part of TnI was more dynamic. This demonstrates that the TnI strand has flexibility that is independent of any unfolding of the TnT strand.
The paired with (+) versus without (−) Ca2+ linear representation of the HDX data in Fig. 5B helps to visualize what parts of each troponin subunit have Ca2+-influenced dynamics. Color differences (i.e. dynamic differences) are seen especially but not exclusively in the TnC NH2-lobe. In general, the changes in this domain are clear measurable effects, as established by the large curve shifts in Fig. 1 with supporting error estimates in Table 1. Fig. 5B also shows that Ca2+ affected several other distinct locations within troponin, outside the TnC NH2-lobe. Finally, it illustrates that the dynamics of many other regions were in fact Ca2+-independent; site II had either a small effect or no effect on dynamics for many sections of each subunit.
Dynamic Map of Troponin Mutation Sites Linked to Inherited Cardiomyopathy
The asterisks in Fig. 5B show the locations (from recent reviews (54–56)) of missense mutations that have been linked to heritable cardiomyopathies. Numerous mutations lie within regions shown to have dynamics that were affected by Ca2+. However, this was not a consistent pattern. Many other mutation sites were not demonstrably Ca2+-sensitive by HDX.
Nevertheless, there was indeed a striking pattern that the comprehensive map in Fig. 5B illustrates. There was a conspicuous tendency for disease-linked mutations to fall within dynamic parts of this dynamic protein. Previous reports (57–60) and reviews (54, 55) have noted the connection between troponin flexibility and cardiomyopathy mutation sites; most cardiomyopathy-inducing troponin mutations occur in regions that are thought to be dynamic. Here this concept was comprehensively and quantitatively validated. Like very few other methods, HDX makes it possible to measure, compare, and map dynamics of an entire molecule, regardless of any gaps in high resolution structural information. The troponin HDX data show that a full 94% of the 50 mapped mutations lay within peptides that were dominated by relatively fast dynamics, indicated by red, orange, or yellow. Furthermore, two-thirds of the mapped mutations occurred in regions (in red) that had no detectable protection (i.e. no detectable folded structure) under at least one condition: in the presence of Ca2+, the absence of Ca2+, or both.
Map of Troponin Regions Stabilized by Regulatory Site Ca2+
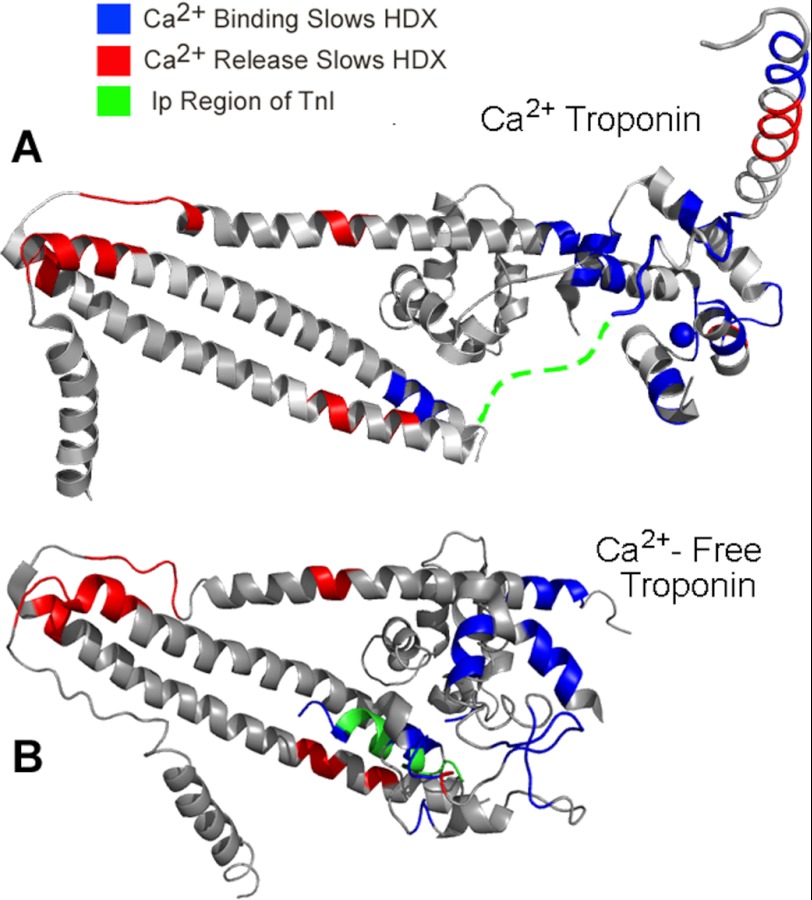
Fig. 6 shows a more binary representation of the effects of Ca2+ on the dynamics of troponin, across the full span of the core domain's extended structure. Regions stabilized by Ca2+ binding (blue) or by Ca2+ release (red) or not changed by Ca2+ (gray) are mapped onto the atomic structure of troponin. Fig. 6A shows changes as mapped onto cardiac Ca2+-saturated troponin. Fig. 6B shows mapping of the same changes onto homologous residues of the skeletal muscle troponin Mg2+-EGTA atomic model (37).
FIGURE 6.
Selective, opposite effects of Ca2+ on the dynamics of different parts of troponin. The figure shows the estimated locations of the areas that were more stable and less dynamic in either the Ca2+ (blue) or Ca2+-free (red) condition. A, results mapped onto the cardiac troponin Ca2+ structure. B, the same results mapped onto the skeletal muscle troponin Mg2+-EGTA structure. TnI 137–147 is shown as a green dashed line in A. Note the greatly altered position of this region in B, where the corresponding residues are again shown in green. In A, unaffected areas are shown in gray, and unassessed areas are shown in white. Ip Region, inhibitory peptide region.
When illustrated in this manner, the specificity of the effects is clear, as is the directionality of the observed differences. The blue regions to the right within Fig. 6A indicate Ca2+-induced stabilization within the regulatory TnC NH2-lobe to which Ca2+ binds as well as stabilization in the associating switch helix. (See Fig. 5 for annotation of the switch helix location). These findings indicate relatively local actions of Ca2+ within the regulatory head region. They are relatively proximate effects and are direct features of the regulatory switching mechanism.
Near to the regulatory head but not included within it, two other regions are stabilized by Ca2+: the NH2-end of TnI helix 1 and the COOH-end of the TnT coiled-coil strand. Each of these effects is located proximate to the regulatory head region of troponin, close enough to suggest that direct contacts of some sort explain the observations.
Two direct interactions can be invoked that have the potential to explain the HDX-detected, Ca2+-dependent stabilization of helix 1. First, the HDX findings may reflect weak interactions between helix 1 and the TnC NH2-lobe that were noted in the crystallography study (25). Second, the 30-residue, cardiac-specific NH2 terminus of TnI is believed to attach to the Ca2+-TnC NH2-lobe (61–64). Such tethering could stabilize the start of TnI helix 1.
The functional importance of the end of helix 1 to the muscle Ca2+ switch is supported not only by Fig. 6 but also by independent results. K36Q mutation, immediately preceding the helix, diminishes sensitivity to Ca2+ and causes dilated cardiomyopathy. Also, protein kinase C phosphorylation of two residues near the helix 1 NH2 terminus alters contractile regulatory function (65–67).
Ca2+-dependent direct contacts could, in principle, explain the stabilization of the COOH-end of the coiled-coil TnT strand. However, no specific structural foundation is known to support such a possibility. Whatever the mechanism, it is unclear whether this coiled-coil end stabilization applies to both strands of the coiled-coil or only to TnT. The HDX data of the relevant TnI peptides leave too much ambiguity in kinetic assignments to define the behavior of the TnI strand's COOH terminus.
Map of Troponin Regions Stabilized by Ca2+ Release
Troponin regions that were stabilized by abolishing Ca2+ binding are indicated in red in Fig. 6. To understand the structural context for these observations, the cardiac HDX findings were mapped in Fig. 6B onto the only available structure for troponin lacking regulatory site Ca2+: the atomic model of skeletal muscle troponin crystallized in Mg2+-EGTA conditions. In this structural model, the TnI switch helix and helix 4 were not detected. Panels A and B of Fig. 6 look qualitatively different from each other primarily because these parts of TnI are absent in B and also because the TnC NH2-lobe is in a rather different location from the cardiac result, above the plane of the page.
Fig. 6B shows that the green shaded TnI inhibitory peptide region (Ip) corresponding to the disordered (dashed line) cardiac TnI 137–147 of Fig. 6A, is folded back above and almost anti-parallel to the coiled-coil. Because Ca2+-free cardiac troponin may have a similar structure, this skeletal muscle troponin model is critical for considering the present results. One immediate implication would be that such a structure could underlie the stabilizing effect of Ca2+ removal on portions of the COOH-half of the coiled-coil (seen in red just below the green helix in Fig. 6B).
Furthermore, this orientation of the TnI inhibitory peptide region in Fig. 6B suggests an explanation for the local stabilization indicated in red at the base of the coiled-coil in the Ca2+-free state. We propose that the effect results from contacts with the TnI COOH terminus that occur once Ca2+ is released from site II. It is striking that the most COOH-terminal portion of TnI shown in the model has folded back in the needed direction for this to occur, anti-parallel to the coiled-coil. Beyond this point, ∼60 COOH-terminal residues are absent in the atomic model. We propose that they interact with this region at the base of the coiled-coil to an extent sufficient to produce the observed slowing of HDX in CBMII troponin. The alternative possibility is that the effects at the base of the coiled-coil are due to transmission of the site II Ca2+ signal along the coiled-coil. This seems somewhat less likely, because there are intervening sequence regions that are Ca2+-insensitive.
Similar structural possibilities apply for the observed effects of site II on TnI 63–66 within helix 1. One cannot say from these data whether the observed Ca2+ sensitivity indicates a direct contact or is mediated by subtle changes in the nearby TnC COOH-lobe. In either case, two lines of evidence indicate that this region is indeed part of the regulatory site Ca2+ switch mechanism despite its location far from site II. First, the observation is robust. The same Ca2+-shifted transition kinetics were observed in three peptides. Second, this precise location is functionally implicated by a previous experiment: mutation A65H shifts the pCa50 for tension development and alters cross-bridge-dependent activation of contraction (68).
DISCUSSION
HDX rates are functionally significant; they quantitatively reflect local folding stability and dynamics. They are not mere markers of solvent accessibility. Therefore, the troponin data in this report have considerable implications both for the details and also for the broad mechanism of troponin's regulatory function.
The details include the finding that Ca2+ stabilizes the troponin regulatory head region, especially the TnC NH2-lobe, but also the switch helix, the nearby end of TnI helix 1, and the coiled-coil terminus. Other regions of the core domain were stabilized not by Ca2+ binding but instead by Ca2+ release. Other details include the observation that the TnI COOH terminus carries out its known inhibitory actions by adopting a partially folded structure in the absence of Ca2+ and actin.
The identified regions of HDX rate Ca2+-sensitivity are part of the regulatory mechanism, in so far as the mechanism can be described within troponin itself. They have altered structural energetics and can be considered part of the Ca2+ switch. Furthermore, previous, particular experimental results serve to validate this interpretation. Phosphorylation and/or mutational studies show effects on regulation for each Ca2+-sensitive HDX region that is now newly identified outside the regulatory head.
The detailed findings point toward several broad mechanistic implications. The Ca2+-sensitive regions are widely rather than narrowly distributed within the core domain. Thus, Ca2+ release triggers an intramolecular switching mechanism that propagates extensively within troponin's extended structure. Also, the current study proves that the mechanistically important regions of troponin are weakly or even reversibly folded. Little energy would be needed for any local changes in structure, including any folding-unfolding transitions, that accompany their Ca2+-regulated binding to targets (actin, tropomyosin, intratroponin targets, or Ca2+ itself). The regulatory switching mechanism prominently involves troponin's dynamic properties.
Via comprehensive, quantitative maps, the data validate the concept that cardiomyopathy-inducing mutations cluster in dynamic regions of troponin. Inherited heart disease due to troponin mutations takes the form of any of the more general cardiomyopathic syndromes: hypertrophic, dilated, or restrictive cardiomyopathy. Detailed study of disease-linked mutations yields functionally complex, mutation-specific results (69). In this highly diverse context, any consistency is notable. Furthermore, the strong clustering of mutations within dynamic regions provides a general clue for the pathogenesis of troponin-linked cardiomyopathies.
One possibility is that the dynamic regions contain the mutation sites by default; the more stable regions cannot tolerate mutations because they are too disruptive to structure and function. Knock-out alleles are not observed, and mutation-induced disease severity usually allows carriers to survive well into adulthood. As a generalization for why mutations cluster in dynamic regions, however, we suggest the opposite inference. The observed clustering occurs because sequence changes in the relevant regions tend to cause the largest functional effects. They tend to be the most, rather than the least, sensitive regions of troponin. Sequence changes in these regions are more likely to be sufficiently disruptive to cause disease rather than to become clinically silent alleles.
As stated above, the mechanistically important regions of troponin are weakly folded, with low stability and low folding energy. Therefore, point mutations in these regions can easily disrupt structure sufficiently to prevent their normal participation in the regulatory mechanism. This is consistent with the observations that any of multiple mutations in the same region may cause disease. In this sense, these regions are weak links in the structural chain required for normal troponin function.
These considerations imply that the clustering of cardiomyopathy-inducing mutations in dynamic regions has two interrelated origins. First, the mutations tend to cluster in areas of troponin that have functional roles in the regulatory mechanism, and such areas of troponin are dynamic. Also, the weak folding of these regions makes them particularly vulnerable to local, mutation-induced unfolding and structural change.
Finally, future studies will be needed to establish what mechanism mediates the observed stabilization of the base of the TnT-TnI coiled-coil in the Ca2+-free state. As suggested above, one possibility is localization of the TnI COOH terminus at the base of the coiled-coil (i.e. stabilization by an intratroponin binding event). The COOH-terminal region of TnI is a key mediator of troponin's ability to control tropomyosin position on actin so that tropomyosin sterically interferes with strong myosin attachment in relaxed muscle (70). More generally, the TnI COOH terminus is a key mediator of troponin's ability to shut off muscle contraction in the absence of Ca2+ (71–74). Therefore, our testable proposal is potentially critical for the structural basis of the regulatory mechanism.
Alternatively, if the base of the coiled-coil is Ca2+-sensitive by a different mechanism, such as structural effects transmitted along the coiled-coil, the implications for regulation still could be important. Troponin-mediated regulation is a remarkable example of action at a distance. Troponin is a very asymmetrical, extended molecule. In addition to the core domain, it includes a tail region that attaches to the actin filament at a point several actins distant from the bulk of the core domain. The troponin tail has many cardiomyopathy mutation sites (55) and is poorly folded unless attached to the thin filament. It is unknown how troponin's elongated structure is involved in regulation. However, a Ca2+-sensitive region as detected and shown in Fig. 6 may act, in the thin filament context, either directly to alter troponin's interactions with actin-tropomyosin (75, 76) or else as part of other transmission of the regulatory signal (77).
In summary, the findings indicate the many mechanistic connections among the details of troponin's fast structural dynamics, troponin's function as the primary on-off switch that regulates muscle contraction, and troponin's identification as a cause of heritable cardiomyopathy.
This work was supported, in whole or in part, by National Institutes of Health Grants HL063774 and UL1TR000050. This work was also supported by the CBC/UIC Proteomics and Informatics Facility, established by a grant from the Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
- HDX
- hydrogen-deuterium exchange
- TnI
- TnC, and TnT, troponin I, C, and T, respectively
- CBMII
- calcium binding mutant (TnC site II).
REFERENCES
- 1. Ebashi S., Endo M., Otsuki I. (1969) Control of muscle contraction. Q. Rev. Biophys. 2, 351–384 [DOI] [PubMed] [Google Scholar]
- 2. Potter J. D., Gergely J. (1974) Troponin, tropomyosin, and actin interactions in the Ca2+ regulation of muscle contraction. Biochemistry 13, 2697–2703 [DOI] [PubMed] [Google Scholar]
- 3. Tobacman L. S. (1996) Thin filament-mediated regulation of cardiac contraction. Annu. Rev. Physiol. 58, 447–481 [DOI] [PubMed] [Google Scholar]
- 4. Gordon A. M., Homsher E., Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi T., Solaro R. J. (2005) Calcium, thin filaments, and the integrative biology of cardiac contractility. Annu. Rev. Physiol. 67, 39–67 [DOI] [PubMed] [Google Scholar]
- 6. Metzger J. M., Westfall M. V. (2004) Covalent and noncovalent modification of thin filament action. The essential role of troponin in cardiac muscle regulation. Circ. Res. 94, 146–158 [DOI] [PubMed] [Google Scholar]
- 7. Moss R. L., Allen J. D., Greaser M. L. (1986) Effects of partial extraction of troponin complex upon the tension-pCa relation in rabbit skeletal muscle. Further evidence that tension development involves cooperative effects within the thin filament. J. Gen. Physiol. 87, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vibert P., Craig R., Lehman W. (1997) Steric-model for activation of muscle thin filaments. J. Mol. Biol. 266, 8–14 [DOI] [PubMed] [Google Scholar]
- 9. Poole K. J., Lorenz M., Evans G., Rosenbaum G., Pirani A., Craig R., Tobacman L. S., Lehman W., Holmes K. C. (2006) A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle x-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 155, 273–284 [DOI] [PubMed] [Google Scholar]
- 10. Heeley D. H., Belknap B., White H. D. (2002) Mechanism of regulation of phosphate dissociation from actomyosin-ADP-Pi by thin filament proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 16731–16736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pirani A., Vinogradova M. V., Curmi P. M., King W. A., Fletterick R. J., Craig R., Tobacman L. S., Xu C., Hatch V., Lehman W. (2006) An atomic model of the thin filament in the relaxed and Ca2+-activated states. J. Mol. Biol. 357, 707–717 [DOI] [PubMed] [Google Scholar]
- 12. Maytum R., Lehrer S. S., Geeves M. A. (1999) Cooperativity and switching within the three-state model of muscle regulation. Biochemistry 38, 1102–1110 [DOI] [PubMed] [Google Scholar]
- 13. Behrmann E., Müller M., Penczek P. A., Mannherz H. G., Manstein D. J., Raunser S. (2012) Structure of the rigor actin-tropomyosin-myosin complex. Cell 150, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Z., Li K. L., Rieck D., Ouyang Y., Chandra M., Dong W. J. (2012) Structural dynamics of C-domain of cardiac troponin I protein in reconstituted thin filament. J. Biol. Chem. 287, 7661–7674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith D. L. (1998) Local Structure and dynamics in proteins characterized by hydrogen exchange and mass spectrometry. Biochemistry 63, 285–293 [PubMed] [Google Scholar]
- 16. Englander S. W. (2000) Protein folding intermediates and pathways studied by hydrogen exchange. Annu. Rev. Biophys. Biomol. Struct. 29, 213–238 [DOI] [PubMed] [Google Scholar]
- 17. Arrington C. B., Robertson A. D. (2000) Kinetics and thermodynamics of conformational equilibria in native proteins by hydrogen exchange. Methods Enzymol. 323, 104–124 [DOI] [PubMed] [Google Scholar]
- 18. Huyghues-Despointes B. M., Scholtz J. M., Pace C. N. (1999) Protein conformational stabilities can be determined from hydrogen exchange rates. Nat. Struct. Biol. 6, 910–912 [DOI] [PubMed] [Google Scholar]
- 19. Kim K. S., Fuchs J. A., Woodward C. K. (1993) Hydrogen exchange identifies native-state motional domains important in protein folding. Biochemistry 32, 9600–9608 [DOI] [PubMed] [Google Scholar]
- 20. Kowlessur D., Tobacman L. S. (2010) Troponin regulatory function and dynamics revealed by H/D exchange-mass spectrometry. J. Biol. Chem. 285, 2686–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kowlessur D., Tobacman L. S. (2010) Low temperature dynamic mapping reveals unexpected order and disorder in troponin. J. Biol. Chem. 285, 38978–38986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Putkey J. A., Sweeney H. L., Campbell S. T. (1989) Site-directed mutation of the trigger calcium-binding sites in cardiac troponin C. J. Biol. Chem. 264, 12370–12378 [PubMed] [Google Scholar]
- 23. Huynh Q., Butters C. A., Leiden J. M., Tobacman L. S. (1996) Effects of cardiac thin filament Ca2+. Statistical mechanical analysis of a troponin C site II mutant. Biophys. J. 70, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z., Marshall A. G. (1998) A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 9, 225–233 [DOI] [PubMed] [Google Scholar]
- 25. Takeda S., Yamashita A., Maeda K., Maéda Y. (2003) Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature 424, 35–41 [DOI] [PubMed] [Google Scholar]
- 26. Bai Y., Milne J. S., Mayne L., Englander S. W. (1993) Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brito R. M., Putkey J. A., Strynadka N. C., James M. N., Rosevear P. R. (1991) Comparative NMR studies on cardiac troponin C and a mutant incapable of binding calcium at site II. Biochemistry 30, 10236–10245 [DOI] [PubMed] [Google Scholar]
- 28. Gillis T. E., Martyn D. A., Rivera A. J., Regnier M. (2007) Investigation of thin filament near-neighbour regulatory unit interactions during force development in skinned cardiac and skeletal muscle. J. Physiol 580, 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreno-Gonzalez A., Gillis T. E., Rivera A. J., Chase P. B., Martyn D. A., Regnier M. (2007) Thin-filament regulation of force redevelopment kinetics in rabbit skeletal muscle fibres. J. Physiol. 579, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Regnier M., Rivera A. J., Wang C. K., Bates M. A., Chase P. B., Gordon A. M. (2002) Thin filament near-neighbour regulatory unit interactions affect rabbit skeletal muscle steady-state force-Ca2+ relations. J. Physiol. 540, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Homsher E., Lee D. M., Morris C., Pavlov D., Tobacman L. S. (2000) Regulation of force and unloaded sliding speed in single thin filaments. Effects of regulatory proteins and calcium. J. Physiol. 524, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morris C. A., Tobacman L. S., Homsher E. (2001) Modulation of contractile activation in skeletal muscle by a calcium-insensitive troponin C mutant. J. Biol. Chem. 276, 20245–20251 [DOI] [PubMed] [Google Scholar]
- 33. Morris C. A., Tobacman L. S., Homsher E. (2003) Thin filament activation and unloaded shortening velocity of rabbit skinned muscle fibres. J. Physiol. 550, 205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Butters C. A., Tobacman J. B., Tobacman L. S. (1997) Cooperative effect of calcium binding to adjacent troponin molecules on the thin filament-myosin subfragment 1 MgATPase rate. J. Biol. Chem. 272, 13196–13202 [DOI] [PubMed] [Google Scholar]
- 35. Cammarato A., Hatch V., Saide J., Craig R., Sparrow J. C., Tobacman L. S., Lehman W. (2004) Drosophila muscle regulation characterized by electron microscopy and 3D reconstrucuction of thin filament mutants. Biophys. J. 86, 1618–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M. X., Spyracopoulos L., Sykes B. D. (1999) Binding of cardiac troponin-I147–163 induces a structural opening in human cardiac troponin-C. Biochemistry 38, 8289–8298 [DOI] [PubMed] [Google Scholar]
- 37. Vinogradova M. V., Stone D. B., Malanina G. G., Karatzaferi C., Cooke R., Mendelson R. A., Fletterick R. J. (2005) Ca2+-regulated structural changes in troponin. Proc. Natl. Acad. Sci. U.S.A. 102, 5038–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura A., Harada H., Park J. E., Nishi H., Satoh M., Takahashi M., Hiroi S., Sasaoka T., Ohbuchi N., Nakamura T., Koyanagi T., Hwang T. H., Choo J. A., Chung K. S., Hasegawa A., Nagai R., Okazaki O., Nakamura H., Matsuzaki M., Sakamoto T., Toshima H., Koga Y., Imaizumi T., Sasazuki T. (1997) Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat. Genet. 16, 379–382 [DOI] [PubMed] [Google Scholar]
- 39. Carballo S., Robinson P., Otway R., Fatkin D., Jongbloed J. D., de Jonge N., Blair E., van Tintelen J. P., Redwood C., Watkins H. (2009) Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ. Res. 105, 375–382 [DOI] [PubMed] [Google Scholar]
- 40. Mogensen J., Murphy R. T., Kubo T., Bahl A., Moon J. C., Klausen I. C., Elliott P. M., McKenna W. J. (2004) Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 44, 2315–2325 [DOI] [PubMed] [Google Scholar]
- 41. Takahashi-Yanaga F., Morimoto S., Harada K., Minakami R., Shiraishi F., Ohta M., Lu Q. W., Sasaguri T., Ohtsuki I. (2001) Functional consequences of the mutations in human cardiac troponin I gene found infamilial hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 33, 2095–2107 [DOI] [PubMed] [Google Scholar]
- 42. Blumenschein T. M., Stone D. B., Fletterick R. J., Mendelson R. A., Sykes B. D. (2006) Dynamics of the C-terminal region of TnI in the troponin complex in solution. Biophys. J. 90, 2436–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murakami K., Yumoto F., Ohki S. Y., Yasunaga T., Tanokura M., Wakabayashi T. (2005) Structural basis for Ca2+-regulated muscle relaxation at interaction sites of troponin with actin and tropomyosin. J. Mol. Biol. 352, 178–201 [DOI] [PubMed] [Google Scholar]
- 44. Hoffman R. M., Blumenschein T. M., Sykes B. D. (2006) An interplay between protein disorder and structure confers the Ca2+ regulation of striated muscle. J. Mol. Biol. 361, 625–633 [DOI] [PubMed] [Google Scholar]
- 45. Mogensen J., Murphy R. T., Shaw T., Bahl A., Redwood C., Watkins H., Burke M., Elliott P. M., McKenna W. J. (2004) Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 44, 2033–2040 [DOI] [PubMed] [Google Scholar]
- 46. Preston L. C., Lipscomb S., Robinson P., Mogensen J., McKenna W. J., Watkins H., Ashley C. C., Redwood C. S. (2007) Functional effects of the DCM mutant Gly159Asp troponin C in skinned muscle fibres. Pflugers Arch. 453, 771–776 [DOI] [PubMed] [Google Scholar]
- 47. Baryshnikova O. K., Robertson I. M., Mercier P., Sykes B. D. (2008) The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I. Biochemistry 47, 10950–10960 [DOI] [PubMed] [Google Scholar]
- 48. Biesiadecki B. J., Kobayashi T., Walker J. S., John Solaro R., de Tombe P. P. (2007) The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ. Res. 100, 1486–1493 [DOI] [PubMed] [Google Scholar]
- 49. Preston L. C., Ashley C. C., Redwood C. S. (2007) DCM troponin C mutant Gly159Asp blunts the response to troponin phosphorylation. Biochem. Biophys. Res. Commun. 360, 27–32 [DOI] [PubMed] [Google Scholar]
- 50. Morimoto S., Lu Q. W., Harada K., Takahashi-Yanaga F., Minakami R., Ohta M., Sasaguri T., Ohtsuki I. (2002) Ca2+-desensitizing effect of a deletion mutation DK210 in cardiac troponin T that causes familial dilated cardiomyopathy. Proc. Natl. Acad. Sci., U.S.A. 99, 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sumandea M. P., Pyle W. G., Kobayashi T., de Tombe P. P., Solaro R. J. (2003) Identification of a functionally critical protein kinase C phosphorylation residue of cardiac troponin T. J. Biol. Chem. 278, 35135–35144 [DOI] [PubMed] [Google Scholar]
- 52. Robinson P., Mirza M., Knott A., Abdulrazzak H., Willott R., Marston S., Watkins H., Redwood C. (2002) Alterations in thin filament regulation induced by a human cardiac troponin T mutant that causes dilated cardiomyopathy are distinct from those induced by troponin T mutants that cause hypertrophic cardiomyopathy. J. Biol. Chem. 277, 40710–40716 [DOI] [PubMed] [Google Scholar]
- 53. Matsumoto F., Maeda K., Chatake T., Maéda Y., Fujiwara S. (2009) Functional aberration of myofibrils by cardiomyopathy-causing mutations in the coiled-coil region of the troponin-core domain. Biochem. Biophys. Res. Commun. 382, 205–209 [DOI] [PubMed] [Google Scholar]
- 54. Willott R. H., Gomes A. V., Chang A. N., Parvatiyar M. S., Pinto J. R., Potter J. D. (2010) Mutations in troponin that cause HCM, DCM AND RCM. What can we learn about thin filament function? J. Mol. Cell Cardiol. 48, 882–892 [DOI] [PubMed] [Google Scholar]
- 55. Tardiff J. C. (2011) Thin filament mutations. Developing an integrative approach to a complex disorder. Circ. Res. 108, 765–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohtsuki I., Morimoto S. (2008) Troponin. Regulatory function and disorders. Biochem. Biophys. Res. Commun. 369, 62–73 [DOI] [PubMed] [Google Scholar]
- 57. Hinkle A., Tobacman L. S. (2003) Folding and function of the troponin tail domain. Effects of cardiomyopathic troponin T mutations. J. Biol. Chem. 278, 506–513 [DOI] [PubMed] [Google Scholar]
- 58. Palm T., Graboski S., Hitchcock-DeGregori S. E., Greenfield N. J. (2001) Disease-causing mutations in cardiac troponin T. Identification of a critical tropomyosin binding region. Biophys. J. 81, 2827–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manning E. P., Guinto P. J., Tardiff J. C. (2012) Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy. An integrative in silico/in vitro approach. J. Biol. Chem. 287, 14515–14523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu B., Tikunova S. B., Kline K. P., Siddiqui J. K., Davis J. P. (2012) Disease-related cardiac troponins alter thin filament Ca2+ association and dissociation rates. PLoS. One. 7, e38259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sadayappan S., Finley N., Howarth J. W., Osinska H., Klevitsky R., Lorenz J. N., Rosevear P. R., Robbins J. (2008) Role of the acidic N′ region of cardiac troponin I in regulating myocardial function. FASEB J. 22, 1246–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Howarth J. W., Meller J., Solaro R. J., Trewhella J., Rosevear P. R. (2007) Phosphorylation-dependent conformational transition of the cardiac specific N-extension of troponin I in cardiac troponin. J. Mol. Biol. 373, 706–722 [DOI] [PubMed] [Google Scholar]
- 63. Baryshnikova O. K., Li M. X., Sykes B. D. (2008) Modulation of cardiac troponin C function by the cardiac-specific N-terminus of troponin I. Influence of PKA phosphorylation and involvement in cardiomyopathies. J. Mol. Biol. 375, 735–751 [DOI] [PubMed] [Google Scholar]
- 64. Westfall M. V., Borton A. R. (2003) Role of troponin I phosphorylation in protein kinase C-mediated enhanced contractile performance of rat myocytes. J. Biol. Chem. 278, 33694–33700 [DOI] [PubMed] [Google Scholar]
- 65. Noland T. A., Jr., Guo X., Raynor R. L., Jideama N. M., Averyhart-Fullard V., Solaro R. J., Kuo J. F. (1995) Cardiac troponin I mutants. Phosphorylation by protein kinases C and A and regulation of Ca2+-stimulated MgATPase of reconstituted actomyosin S-1. J. Biol. Chem. 270, 25445–25454 [DOI] [PubMed] [Google Scholar]
- 66. Burkart E. M., Sumandea M. P., Kobayashi T., Nili M., Martin A. F., Homsher E., Solaro R. J. (2003) Phosphorylation or glutamic acid substitution at protein kinase C sites on cardiac troponin I differentially depress myofilament tension and shortening velocity. J. Biol. Chem. 278, 11265–11272 [DOI] [PubMed] [Google Scholar]
- 67. Pyle W. G., Sumandea M. P., Solaro R. J., De Tombe P. P. (2002) Troponin I serines 43/45 and regulation of cardiac myofilament function. Am. J. Physiol. Heart Circ. Physiol. 283, H1215–H1224 [DOI] [PubMed] [Google Scholar]
- 68. Engel P. L., Kobayashi T., Biesiadecki B., Davis J., Tikunova S., Wu S., Solaro R. J. (2007) Identification of a region of troponin I important in signaling cross-bridge-dependent activation of cardiac myofilaments. J. Biol. Chem. 282, 183–193 [DOI] [PubMed] [Google Scholar]
- 69. Seidman C. E., Seidman J. G. (2011) Identifying sarcomere gene mutations in hypertrophic cardiomyopathy. A personal history. Circ. Res. 108, 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Galińska-Rakoczy A., Engel P., Xu C., Jung H., Craig R., Tobacman L. S., Lehman W. (2008) Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J. Mol. Biol. 379, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tripet B., Van Eyk J. E., Hodges R. S. (1997) Mapping of a second actin-tropomyosin and a second troponin C binding site within the C-terminus of troponin I, and their importance in the Ca2+-dependent regulation of muscle contraction. J. Mol. Biol. 271, 728–750 [DOI] [PubMed] [Google Scholar]
- 72. Rarick H. M., Tu X. H., Solaro R. J., Martin A. F. (1997) The C terminus of cardiac troponin I is essential for full inhibitory activity and Ca2+ sensitivity of rat myofibrils. J. Biol. Chem. 272, 26887–26892 [DOI] [PubMed] [Google Scholar]
- 73. Jin J. P., Yang F. W., Yu Z. B., Ruse C. I., Bond M., Chen A. (2001) The highly conserved COOH terminus of troponin I forms a Ca2+-modulated allosteric domain in the troponin complex. Biochemistry 40, 2623–2631 [DOI] [PubMed] [Google Scholar]
- 74. Ramos C. H. (1999) Mapping subdomains in the C-terminal region of troponin I involved in its binding to troponin C and to thin filament. J. Biol. Chem. 274, 18189–18195 [DOI] [PubMed] [Google Scholar]
- 75. Galińska A., Hatch V., Craig R., Murphy A. M., Van Eyk J. E., Wang C. L., Lehman W., Foster D. B. (2010) The C terminus of cardiac troponin I stabilizes the Ca2+-activated state of tropomyosin on actin filaments. Circ. Res. 106, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Knowles A. C., Irving M., Sun Y. B. (2012) Conformation of the troponin core complex in the thin filaments of skeletal muscle during relaxation and active contraction. J. Mol. Biol. 421, 125–137 [DOI] [PubMed] [Google Scholar]
- 77. Manning E. P., Tardiff J. C., Schwartz S. D. (2011) A model of calcium activation of the cardiac thin filament. Biochemistry 50, 7405–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]