Background: The enzyme synthesizing the pyrimidine moiety (HMP-P synthase or THI5p) of vitamin B1 is poorly characterized.
Results: We determined an atomic model of THI5p and have identified its active site using mutagenesis experiments.
Conclusion: THI5p forms a dimer and uses PLP as a substrate rather than as a cofactor.
Significance: Our study provides seminal information on the enzymatic mechanism of HMP-P synthase.
Keywords: Enzyme Mechanisms, Enzyme Structure, Protein Structure, Pyridoxal Phosphate, Thiamine, X-ray Crystallography, HMP-P Synthase
Abstract
Vitamin B1 is essential for all organisms being well recognized as a necessary cofactor for key metabolic pathways such as glycolysis, and was more recently implicated in DNA damage responses. Little is known about the enzyme responsible for the formation of the pyrimidine moiety (4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P) synthase). We report a structure-function study of the HMP-P synthase from yeast, THI5p. Our crystallographic structure shows that THI5p is a mix between periplasmic binding proteins and pyridoxal 5′-phosphate-dependent enzymes. Mutational and yeast complementation studies identify the key residues for HMP-P biosynthesis as well as the use of pyridoxal 5′-phosphate as a substrate rather than as a cofactor. Furthermore, we could show that iron binding to HMP-P synthase is essential for the reaction.
Introduction
Vitamin B1 (thiamin) is vital in all organisms as a cofactor for key cellular metabolic pathways such as glycolysis, the oxidative pentose phosphate pathway, and the citric acid cycle, as well as the Calvin cycle in plants (1). Vitamin B1 also plays an important role in response to DNA damage and during pathogen attack in plants (2). Although it is an essential compound for all forms of life, only prokaryotes, fungi, and plants are able to perform its de novo biosynthesis. Regulation of the biosynthesis can occur through end product feedback inhibition that involves the direct binding of the vitamin to the mRNA of one of the biosynthesis genes, i.e. riboswitch control (3). Interestingly, although riboswitch control is maintained in bacteria and plants, it does not appear to exist in yeast. Instead, a series of transcription factors are proposed to regulate vitamin B1 biosynthesis in the latter (4, 5). The general mechanism of vitamin B1 biosynthesis involves the separate formation of the thiazole and the pyrimidine heterocycles, which are then condensed to produce thiamin pyrophosphate. In prokaryotes, the process of pyrimidine and thiazole heterocycle formation is relatively well characterized by both structural and mechanistic studies (1). However, in eukaryotes, the formation of these heterocycles is only beginning to be elucidated (6, 7). Indeed, although the formation of the thiazole moiety appears to be conserved between fungi and plants employing the key gene THI4 (annotated THI1 in plants), it is different from the route employed by bacteria. The latter reaction has recently been reconstituted from yeast and employs the THI4 protein in a suicide mechanism using NAD+ as a precursor (8). However, the formation of the pyrimidine moiety is very different even between fungi and plants. In plants, the formation of the pyrimidine moiety, 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P),4 follows the same route as bacteria employing the THIC enzyme and 5-aminoimidazole ribonucleotide as a precursor (7). There is no homolog of THIC in fungi; instead HMP-P is biosynthesized by members of the THI5 gene family (9). Moreover, isotopic labeling studies have indicated that HMP is derived from pyridoxine and histidine in yeast rather than 5-aminoimidazole ribonucleotide (10, 11). Therefore, the last important piece to complete our understanding of de novo thiamin biosynthesis across all organisms is a structural and functional characterization of a member of the THI5 family. Very recently, an important step forward has been made by Lai et al. (12), who have been able to reconstitute the THI5p-dependent HMP-P synthase activity in vitro. However, a detailed structural analysis, incorporating a full characterization of active site residues, an iron-binding site, and the oligomeric state of the protein, has not yet been provided.
Here, we report on the structural and biochemical characterization of THI5p from Saccharomyces cerevisiae. The protein adopts a fold related to the periplasmic binding protein (PBP) family. Interestingly, we observe both PLP and an iron atom bound to the protein as isolated allowing us to suggest numerous residues of the active site necessary for HMP-P biosynthesis. THI5p is a dimer and, although exceptionally using PLP as a substrate, has notable similarities with enzymes dependent on this molecule as a cofactor. Indeed, the proposed mechanism of action has major implications for the regulation of vitamin B1 biosynthesis in yeast.
EXPERIMENTAL PROCEDURES
Molecular Cloning and Protein Purification
The full-length THI5 nucleotide sequence was amplified from the pBG1805-ORF YFL058W plasmid (purchased from Open Biosystems) and cloned into pET-24b for expression either with a C-terminal hexahistidine tag (annotated “tagged” and used for protein crystallization) or without any tag (annotated “untagged” and used for protein biochemical characterization). The THI5p proteins were expressed at 37 °C in the Escherichia coli BL21(DE3) pRARE strain following induction with 0.2 mm isopropyl β-d-thiogalactopyranoside. Purification of the tagged protein was done using a Ni2+-nitrilotriacetic acid column (Qiagen) followed by size exclusion chromatography (Superdex 200 column, GE Healthcare). The untagged version of THI5p was purified using a HiPrep DEAE-Sepharose Fast Flow 16/10 column (GE Healthcare) and size exclusion chromatography as above. Details for the purification buffers are given in the supplemental material. Seleno-methionine (SeMet)-labeled THI5p proteins were expressed in the methionine auxotrophic strain B834(DE3) of E. coli using M9 minimal medium with SeMet and then purified as described above.
Numerous mutants were generated for the biochemical characterization using the QuikChange II site-directed mutagenesis kit (Stratagene) and according to the manufacturer's instructions. The complete list of mutations together with the oligonucleotides used can be found in supplemental Table S1.
Crystallization
THI5p crystals were obtained only with the His tag version of the protein and using the sitting-drop vapor diffusion technique at 18 °C in 96-well crystallization plates (Swiscii). The drops were prepared by mixing 0.4 μl of the solution of the tagged protein (at a concentration of 15–20 mg/ml) with an equal volume of crystallization solution and were equilibrated against 35 μl of crystallization solution. A large number of conditions yielded crystals with very poor diffraction properties. To improve the diffraction properties of these crystals, we engineered multiple surface mutations using the THI5p structural model predicted by the program Phyre (13). The following mutants were generated: E101K/D102S (THI5p-mutant 1); K240S/E241G (THI5p-mutant 2); Q317T (THI5p-mutant 3), K240S/E241G/Q317T (THI5p-mutant 4); and E101K/D102S/Q317T (THI5p-mutant 5). THI5p-mutant 4 and THI5p-mutant 5 gave high diffracting crystals under the following conditions, respectively: 1) 0.2 m lithium sulfate monohydrate, 0.1 m BisTris, pH 5.5, 25% polyethylene glycol 3350 (crystal 1); and 2) 18% 2-propanol, 0.1 m sodium citrate, pH 5.5, 20% polyethylene glycol 4000 (crystal 3). The SeMet derivative of THI5p-mutant 4 also gave high diffracting crystals under the following conditions: 0.2 m potassium thiocyanate, 0.1 m BisTris, pH 5.7, 27% polyethylene glycol 3350, 10% dodecyl-β-d-maltoside (crystal 2). THI5p crystals appeared in a few hours and grew to 50 × 50 × 10 μm. Crystal 3 was soaked with 1 mm PLP for 24 h in the crystallization condition. Crystals were then transferred into cryoprotective buffers (i.e. crystallization conditions supplemented with 10% glycerol) prior to flash freezing in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Diffraction data for THI5p-mutant 4 and THI5p-mutant 5 crystals were collected at Beamline ID14-4 at the European Synchrotron Radiation Facility (Grenoble, France). Single anomalous diffraction datasets of SeMet-containing THI5p-mutant 4 crystals were measured on the PXIII beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland. The data sets belong to the two following space groups: (i) crystal 1 in P21 with unit cell dimensions a = 78.8 Å, b = 188.4 Å, and c = 101.6 Å and eight molecules per asymmetric unit; (ii) crystal 2 in P212121 with unit cell dimensions a = 54.4 Å, b = 109.2 Å, and c = 122.1 Å and two molecules per asymmetric unit; (iii) crystal 3 in P21 with unit cell dimensions a = 79.48 Å, b = 191.84 Å, and c = 101.89 Å and eight molecules per asymmetric unit. Crystallographic data are reported in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
Values in parentheses are for highest resolution shell.
| Crystal 1 | Crystal 2 | Crystal 3 | |
|---|---|---|---|
| Data collection | |||
| Wavelength | 0.98011 Å | 0.97795 Å | 0.93940 Å |
| Space group | P21 | P212121 | P21 |
| Unit cell dimensions | |||
| a, b, c | 78.79, 188.46, 101.67 Å | 54.44, 109.21, 122.10 Å | 79.48, 191.84, 101.89 Å |
| α, β, γ | 90.0, 112.51, 90.0° | 90.0, 90.0, 90.0° | 90.0, 112.72, 90.0° |
| Resolution | 49.142 to 2.7 Å (2.77 to 2.7 Å) | 19.998 to 2.7 (2.77 to 2.7 Å) | 48.190 to 2.9 Å (2.98 to 2.9 Å) |
| No. of unique reflections | 71,456 | 38,761 | 61,787 |
| Redundancy | 1.92 | 19.37 | 4.82 |
| Rmeasa | 5.6 (62.7) | 11.0 (129.3) | 12.3 (89.6) |
| I/σ(I) | 13.68 (1.77) | 25.14 (2.85) | 12.90 (2.30) |
| Completeness | 95.27% (96.5%) | 100% (100%) | 99.1% (98.4%) |
| Refinement | |||
| Rworkb | 20.75% | 21.06% | 22.10% |
| Rfreec | 24.69% | 25.42% | 26.26% |
| No. of atoms | |||
| Protein | 20763 | 4608 | 20804 |
| Ligand (PLP) | 16 | 64 | |
| Solvent | 157 | 109 | |
| Mean B-factors | |||
| Protein | 73 Å2 | 45 Å2 | 61 Å2 |
| Ligand (PLP) | 85 Å2 | 45 Å | |
| Solvent | 53 Å2 | 41 Å2 | |
| Root mean square deviation from ideal geometry | |||
| Bond lengths | 0.012 Å | 0.010 Å | 0.010 Å |
| Bond angles | 1.435° | 1.312° | 1.137° |
| Residues in favored region of the Ramachandran plot | 96.95% | 96.28% | 96.45% |
| Residues in allowed region of the Ramachandran plot | 3.05% | 3.72% | 3.55% |
a with Ih the intensity of reflection and nh the multiplicity and h the reflections indices.
b with Fobs and Fcalc the observed and calculated structure factors, respectively.
c Rfree indicates the cross-validation of Rwork.
The structure of the SeMet derivative of THI5p-mutant 4 was determined by the SAD method using the anomalous signal of the selenium atoms. The dataset was indexed, integrated, and scaled with the XDS package (14). Heavy atom location and phasing were performed with the Shelx suite (15) and the SHARP procedure (16). Furthermore, the program SOLOMON (17) was used for phase improvement by solvent flipping. Model building was done using the program COOT (18). The model was then used for phasing the datasets from the THI5p-mutant 4 and THI5p-mutant 5 protein crystals using the molecular replacement program Phaser from the CCP4 package (19). The atomic models were refined using the program Phenix (20). The refinement process included successive rounds of simulated annealing, minimization, B-factor refinement, calculation of composite omit maps, and difference Fourier maps. Several protein loops could not be built due to the high flexibility that varied from molecule to molecule. The list of absent residues in each molecule for the three reported crystal structures is given in supplemental Table S2. Once we finished building the polypeptide chains, the PLP, sulfate ions, and water molecules were added. The PLP molecules were built within a large electron density peak observed in the Fo − Fc difference density map. The electron density peak protrudes from the Lys-62 residue to the N-terminal loop of helix 4 (supplemental Fig. S1). Molecule B of crystal 1 and molecules A, B, E, and H from crystal 3 had such electron density signals in both the 2Fo − Fc and the Fo − Fc maps. The other molecules from crystals 1 and 3 only showed a smaller Fo − Fc difference density peak near the N-terminal loop of helix 4. In the latter cases, we built a sulfate ion present in the respective crystallization conditions used. Importantly, the architecture of the THI5p active site was practically identical in all molecules (supplemental Fig. S2). As the position of the built PLP molecule is unchanged in all the molecules from crystals 1 and 3, the structural panels in the figures were prepared using the atomic model of THI5p refined against the dataset collected from crystal 1 as it was at the highest resolution (2.7 Å). The final models show good stereochemistry as indicated by the program PROCHECK with no residues in the disallowed regions of the Ramachandran plot (21). Structure factors and atomic models have been deposited in the Protein Data Bank with the following Protein Data Bank codes: crystal 1, 4H67; crystal 2, 4H65, and crystal 3, 4H6D.
Spectrometry and Static Light Scattering Analysis
The spectrophotometric measurements were recorded on a BioTek® Synergy 2 microplate reader using purified untagged or tagged THI5p (0.6 mm) in 50 mm sodium phosphate buffer, pH 7.8, containing 150 mm sodium chloride and 2 mm dithiothreitol in the absence or presence of 0.1 m sodium hydroxide at room temperature. In the case of the electrospray ionization mass spectrometry, the samples were prepared by dialyzing 200 μl of untagged THI5p (200 μm) in the same buffer as above against 0.2% formic acid. Mass spectra were recorded on an AB Sciex 5600 TripleTOF device. For the static light scattering analysis, THI5p (tagged or untagged) (at a concentration of 2 mg·ml−1) was separated by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) employing 50 mm potassium phosphate buffer, pH 7.5, containing 50 mm potassium chloride and 0.01% sodium azide as the buffer, which was connected immediately downstream to a miniDAWN TREOS light scattering instrument (Wyatt Technologies). The data were analyzed using the software ASTRA (Wyatt Technologies). An absorption coefficient of 52112.5 m−1·cm−1 was used for the calculation of the weight average molecular mass.
Ferrozine Assay
Ferrozine is a specific reagent that forms a magenta colored complex with ferrous ions (absorbance at 593 nm (22)). The iron concentration in our purified protein samples was determined according to a method described by Beinert (23). Briefly, 100 nmol of untagged THI5p was treated with 1% HCl (w/v) in a final volume of 100 μl and incubated for 10 min at 80 °C. The sample was then cooled to room temperature before adding in this order: 500 μl of 7.5% (w/v) ammonium acetate, 100 μl of 4% (w/v) ascorbic acid, 100 μl of 2.5% (w/v) SDS, and 100 μl of 1.5% (w/v) ferrozine. After centrifugation at 11,000 × g for 2 min, the absorbance at 593 nm was measured. A standard curve was made in parallel using 2–20 nmol of ferrous ammonium sulfate from a 0.2 mm stock solution. An estimation of the amount of iron released from the protein was extrapolated from the standard curve.
Yeast Complementation
S. cerevisiae strain RWY16 (MATα ade2Δ::hisG his3Δ200 thi13::HIS3 leu2Δ0 thi12::LEU2 lys2Δ0 met15Δ0 trp1D63 thi11::TRP1 ura3Δ0 thi5::KANMX4; see Ref. 9) and SD complete medium, i.e. supplemented with all the amino acids and bases (with the exception of uracil as appropriate, see below), was used throughout. Glucose was added as a carbon source to a final concentration of 2% (w/v), apart from the complementation assay itself, where it was replaced by 2% raffinose + 0.01% glucose for preparation of the overnight culture and then by 2% galactose to achieve maximal expression of THI5p (GAL1 promotor induction) during the complementation. Prior to transformation, RWY16 was cultivated on SD complete medium. All of the yeast DNA transformations were performed with the polyethylene glycol/lithium acetate method (24). The selection of transformants was performed in the absence of uracil by growing at 30 °C for 72 h. The pBG1805-ORF YFL058W plasmid was used as the template for all site-directed mutagenesis manipulations (see supplemental Table S1 for primer sequences). All mutations were subsequently confirmed by DNA sequencing. For the complementation assay, the RWY16 strains harboring pBG1805-ORF YFL058W plasmid with either wild-type or mutant THI5 sequences were cultivated overnight at 30 °C in 5 ml of SD complete medium minus uracil. Cells were washed 10 times with 10 ml of sterile water and resuspended in 10 ml of sterile water. The A600 of each cell suspension was adjusted to 0.2 units by dilution with sterile water in 96-well plates. For each strain, this suspension was diluted 1:10 and 1:100, leading to an A600 of 0.02 and 0.002, respectively. The different dilutions were then spotted onto solid SD complete medium in the presence or absence of thiamin (1.2 μm) and with 2% galactose as the sugar source.
RESULTS AND DISCUSSION
Overall Structure of THI5p from Yeast
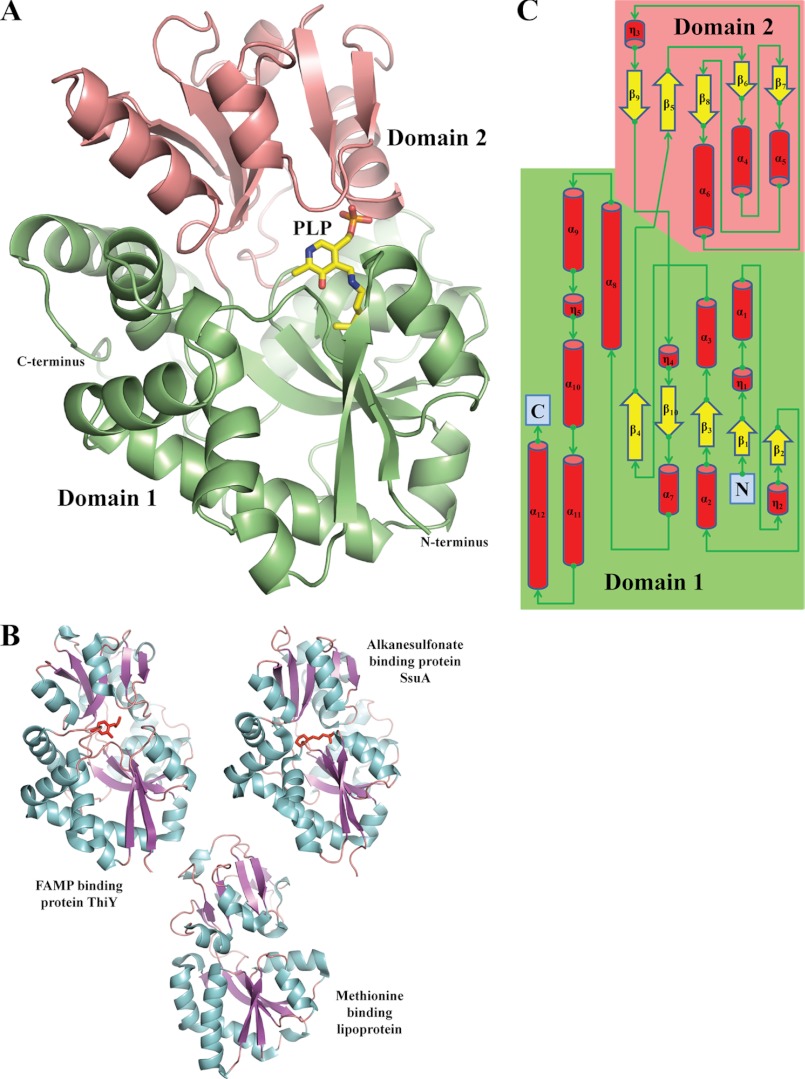
The HMP-P synthase (annotated as THI5p) from S. cerevisiae was cloned and recombinantly expressed in E. coli either without or with a C-terminal hexahistidine tag. The tagged form was used for crystallization studies. THI5p crystallized in two space groups, P21 with eight molecules in the asymmetric unit (crystal 1 and crystal 3) and P212121 with two molecules (crystal 2). Atomic models were subsequently refined to 2.7 Å resolution (crystals 1 and 2) or 2.9 Å resolution (crystal 3; Fig. 1A; see Table 1 for crystallographic and refinement data). THI5p adopts a fold typical of bacterial PBPs (Fig. 1B; 25), e.g. the N-formyl-4-amino-5-(aminomethyl)-2-methylpyrimidine-binding protein, ThiY; the aliphatic sulfonate binding protein, SsuA; or the methionine-binding lipoprotein from Coxiella burnetii, despite its very low sequence identity (less than 20%). Structurally, THI5p is composed of two domains, which have a three-layer αβα architecture (Fig. 1, A and C). The mixed β-sheets of the two domains contain five β-strands with the respective topologies, ↑β4↓β10↑β3↑β1↑β2 (domain 1) and ↑β7↑β6↑β8↓β5↑β9 (domain 2). The two domains are connected by crossover loops between strands β4 and β5 and between strand β9 and helix η4. Moreover, domain 2 is surrounded and stabilized by the helices α10 and α12 of domain 1 (see the topology diagram in Fig. 1C). In several cases, the protein fragments (residues 117–130 and residues 187–195) and the C-terminal extremity of THI5p (residues 335–340) were prone to disorder as judged from missing electron density signals (see supplemental Table S2 for the exact list of residues absent in each molecule).
FIGURE 1.
Structure of THI5p. A, overall view of the THI5p monomer showing the two domains (colored in salmon and green) with α-helices, β-strands, and connecting loops (molecule C, crystal 1). The PLP bound to molecule B of crystal 1 is superimposed on molecule C and shown as sticks. B, three representative periplasmic binding protein models, ThiY (3IX1), SsuA (3KSX), and methionine binding lipoprotein (3TQW). Bound substrates are shown as red sticks. C, topology diagram of THI5p. Secondary structure elements are numbered starting from the N terminus. The structural panels in this figure and the following have been prepared with the program PyMOL (available on line).
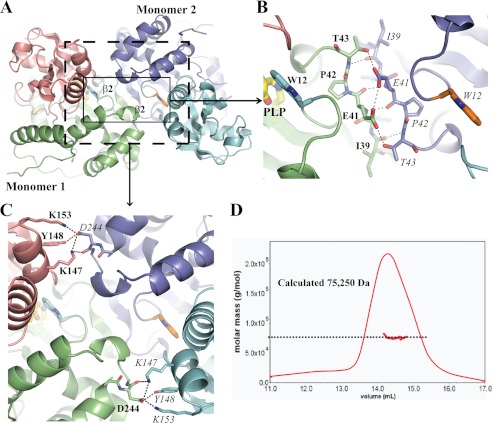
We observed THI5p as a dimer in crystals 1–3, with each crystal containing either 4 (crystal 1 and 3) or 1 (crystal 2) dimer(s) per asymmetric unit. Notably, the dimer is built from the same surface of interaction in the two crystal lattices. Specifically, on the one hand two monomers form a pseudo antiparallel β-sheet with their β2-strands (Fig. 2A), which is further stabilized by multiple hydrogen bonds contributed by the side chain oxygen atoms of residues Glu-41 and Thr-43 (Fig. 2B). A second contact point involves Lys-147, Tyr-148, and Lys-153 in one monomer and Asp-244 in the other monomer (Fig. 2C). Additional contacts are found between hydrophobic residues from both monomers located in domain 1. Altogether, these multiple contact points bury ∼10% of each monomer surface (supplemental Fig. S3) (39). Furthermore, these intermolecular contact points are mirrored on each side of the dimer interface, thus locking the subdomains in a given orientation. The THI5p dimer was also observed in solution as shown by size exclusion chromatography coupled to static light scattering, where the estimated size of the protein (75,250 Da) approaches that expected of a dimer (77,184 Da; Fig. 2D). Recently, the structure of the Candida albicans THI5p homolog was solved by Lai et al. (12). It is unclear from the data presented if they also observed this dimeric organization, likely important for the structural organization of the catalytic site (see below).
FIGURE 2.
THI5p is a dimer. A, ribbon representation of the THI5p dimer (molecule A and B, crystal 1) with Trp-12 highlighted in stick mode. The boxes indicate the contact points shown in more detail in B and C. B, close view of contact point 1 formed upon THI5p dimerization indicating backbone interactions that form a pseudo β-sheet as well as hydrogen bonds (see main text for details). C, contact point 2 showing residues from each monomer that establish multiple hydrogen bonds (see main text for details). D, static light scattering analysis of THI5p. The calculated weight average molecular mass (bold red line) and the Rayleigh ratio (plain red line) were plotted in an elution volume-dependent manner.
Defining the Active Site of HMP-P Synthase
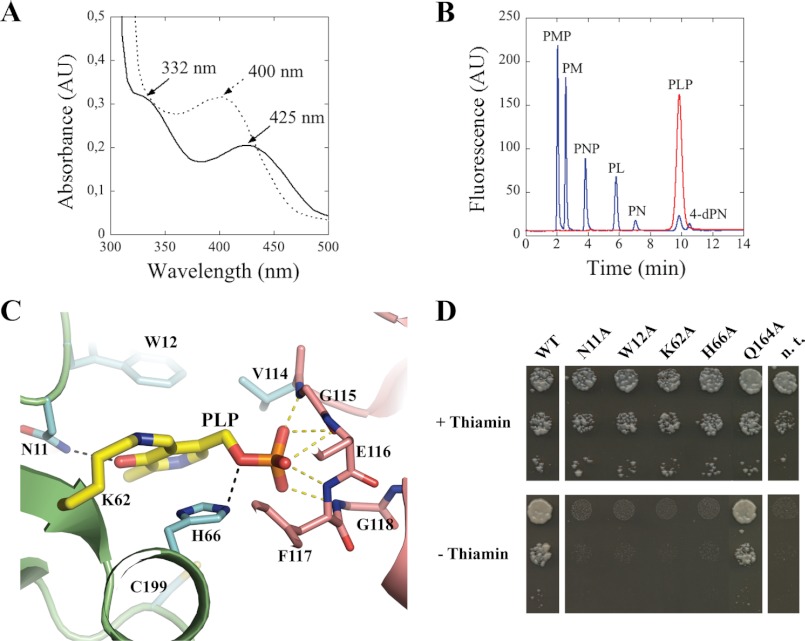
Although it has been proposed that HMP-P synthase from yeast uses a B6 vitamer molecule as a substrate during HMP-P biosynthesis (10, 26), the identity of this vitamer remained to be verified. During the course of the THI5p purification protocol employed in this study, we observed that the protein preparations had a pale yellow color. A UV-visible absorbance spectrum of the purified protein revealed a maximum at 425 nm with a distinct shoulder at 332 nm in addition to the protein peak at 280 nm (Fig. 3A). These absorption bands are characteristic of the keto-enol tautomeric equilibrium of a pyridoxal 5′-phosphate (PLP) molecule when bound to the protein as a Schiff's base (27, 28). PLP is one of nature's most versatile cofactors and is well established as co-purifying with proteins dependent on it as a cofactor. Treatment of the protein with sodium hydroxide induced a hypsochromic shift of the 425 nm band to 400 nm, which is characteristic of the release of free PLP (Fig. 3A) (28). Moreover, an HPLC analysis of the latter compound, as released upon alkali treatment of the protein compared with B6 vitamer standards, confirmed that it was indeed PLP (Fig. 3B). These features were observed with both the untagged and the C-terminal hexahistidine-tagged version of the protein used for the structural study. No other B6 vitamer was observed upon purification and alkali treatment of the protein. Thus, armed with the knowledge that HMP-P synthesis in yeast is dependent on vitamin B6 as a substrate, this strongly suggested that PLP is the vitamer being used by the THI5p protein. At a minimum, this study provides the proof that HMP synthase is covalently bound to PLP.
FIGURE 3.
Defining the active site of THI5p. A, UV-visible spectra of THI5p (0.6 mm) in the presence (dashed line) or absence (black line) of sodium hydroxide (0.1 m). B, HPLC profile of vitamin B6 standards (5 pmol, blue) and the released PLP from THI5p in A (red). PL(P), pyridoxal(phosphate); PM(P), pyridoxamine(phosphate); PN(P), pyridoxine(phosphate); 4-dPN, 4-deoxypyridoxine. AU, absorbance units. C, close view of the active site of THI5p (molecule B, crystal 1). Key residues and PLP are shown in stick mode. Protein domains and atoms are colored as in Fig. 1A. D, yeast complementation assay. Strain RWY16 transformed with wild-type THI5 (WT) or mutants thereof as indicated; n.t., not transformed. The three spots in each column are respective 1:10 serial dilutions.
In this context, it is noteworthy that the two subdomains described for THI5p define a space where the structurally related PBP proteins generally bind their cognate ligand (Fig. 1B). This region contained strong electron density peaks in both the 2Fo − Fc and the Fo − Fc electron density maps. One molecule of crystal 1 and four molecules of crystal 3 showed electron density peaks large enough to build a PLP compound (Fig. 3C; supplemental Fig. S1; see “Experimental Procedures” for details). In all cases, PLP is covalently bound to Lys-62 in THI5p with the phosphate group stabilized by a glycine-rich loop near the N terminus of helix α4 (Fig. 3C). Such a configuration is often seen in PLP-dependent enzymes (29, 30). The imine linkage to Lys-62 was similarly observed in the recent structure of the C. albicans THI5p homolog reported to be in a complex with PLP (12). The molecules with smaller Fo − Fc peaks or with no signal in the 2Fo − Fc electron density map generally showed a difference density peak near the glycine-rich loop, in which we positioned a sulfate ion present in the crystallization buffer, based on the coordination sphere. Interestingly, the pyridine ring of PLP is sandwiched between the indole ring of Trp-12 and the imidazole ring of His-66 (Fig. 3C). Furthermore, the His-66 NE2 atom forms a hydrogen bond with the O4P of PLP, whereas asparagine 11 contacts the O3 position of PLP (Fig. 3C). The PLP O3 position is crucial in the ketoenamine and enolimine tautomeric equilibrium. Such a network of interactions locks the PLP in a precise position and orientation, reinforcing the notion of its role as a substrate rather than as a co-factor (see below for details).
To validate the functionality of the amino acid residues responsible for PLP association, we set up a complementation assay in yeast. The THI5 gene family of S. cerevisiae comprises four highly conserved members named THI5 (YFL058w), THI11 (YJR156c), THI12 (YNL332w), and THI13 (YDL244w). For our purpose, we employed a strain of S. cerevisiae where all four THI5 paralogs have been knocked out (strain RWY16, MATα ade2Δ::hisG his3Δ200 thi13::HIS3 leu2Δ0 thi12::LEU2 lys2Δ0 met15Δ0 trp1D63 thi11::TRP1 ura3Δ0 thi5::KANMX4; Ref. 9). This strain does not grow in the absence of thiamin supplementation; however, reintroduction of the wild-type THI5 gene alone allows the strain to grow in the absence of thiamin (Fig. 3D). Notably, transformation of RWY16 with modified THI5 coding sequences, where any one of the active site residues Asn-11, Trp-12, Lys-62, or His-66 (Fig. 3C) has been mutated to an alanine, abolishes the capacity of the strain to sustain growth in thiamin-free medium (Fig. 3D). As an extra control, introduction of THI5 mutated at glutamine 164, which does not appear to be part of the active site, permits growth identical to the wild type on medium lacking thiamin (Fig. 3D). This study thus validates the assignment of active site residues. It is noteworthy that His-66 has been suggested to be the source of histidine for pyrimidine formation by Lai et al. (12). The mutant forms of THI5p used for crystallization (THI5p-mutant 4 and 5) were also tested for complementation and showed a level of growth similar to that of the wild type (supplemental Fig. S4).
PLP, a Substrate Rather than a Cofactor
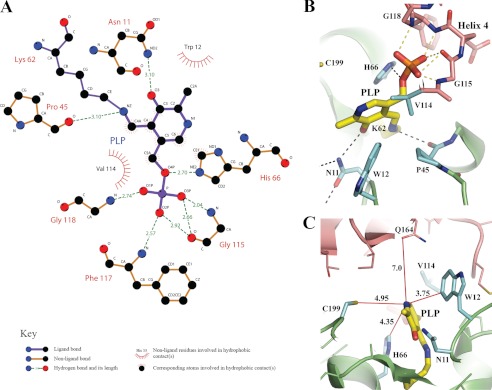
Solving the structure of S. cerevisiae THI5p with PLP bound allows us to perform a direct comparison with PLP-dependent enzymes. PLP-dependent enzymes can be classified into five groups based on their fold (31). However, THI5p is not homologous to any of the PLP-dependent enzyme families based on either sequence identity or structural similarity, because none of the five different folds superimposes with that of THI5p. Nevertheless, a closer comparison of the presently identified active site of THI5p with that of PLP-dependent enzymes demonstrates architectural similarities but also, and more importantly, significant differences in the way the B6 vitamer binds to THI5p (Fig. 4A). One common observation between THI5p and PLP-dependent enzymes is the covalent association of PLP with a lysine residue (Fig. 3, A and C). However, although the reaction center of PLP-dependent enzymes is located around this covalently bound lysine, there is no space to accommodate another molecule near the Lys-62 residue. The O3 atom of PLP, next to the covalently linked lysine NZ atom, plays a critical role in the keto/enol tautomeric equilibrium, the ketoenamine being the reactive form of the co-factor PLP (29). In our structure, this atom is hydrogen bonded to the ND2 position of Asn-11 (Figs. 3C and 4B). Simultaneously, the backbone oxygen of Pro-45 forms a hydrogen bond with the NZ position of PLP (Fig. 4, A and B), a situation unusual for PLP-dependent enzymes. Furthermore, PLP-dependent enzymes establish a salt bridge with the N1 position of the bound PLP to regulate its protonation state and therefore its capacity to increase the positive charge of the NZ atom, again favoring the keto form in the tautomeric equilibrium. In our PLP-bound THI5p structure, the shortest distance between a protein atom and the N1 position is 3.75 Å and corresponds to the carbon atom at CZ3 position of Trp-12. The CE1 position of His-66 and the sulfur group of Cys-199 are 4.35 and 4.95 Å away, respectively (Fig. 4C). Such an environment around the nitrogen atom of the pyridine ring would not favor its electron withdrawing potential necessary for its co-factor function (32). It is likely that a similar situation is found in the C. albicans THI5p homolog structure (12). Moreover, these modifications and the face-to-edge orientation of Trp-12 and His-66 with regards to the PLP molecule likely modify the electronic properties of the PLP π-orbitals. Although the latter are known to be an important determinant for the specificity imparted by particular PLP-mediated reactions like deprotonation/decarboxylation (33), the recently proposed Diels-Alder reaction mechanism of HMP-P biosynthesis is also critically dependent on conjugated π systems (10, 12). The above noted differences would allow the use of PLP as a substrate rather than as a co-factor by modifying its electronic orbitals to favor the PLP reaction with the cyanamide donor to form HMP-P (Figs. 3C and 4B).
FIGURE 4.
PLP/THI5p interaction network. A, schematic diagram of the interaction network formed between PLP and the THI5p active site residues. The LigPlot software (38) was used for plotting with the legend as indicated. B, close view of THI5p-bound PLP with hydrogen bonds to O3 and NZ of PLP indicated (see main text for details). C, a large empty cavity is found around the N1 atom of PLP. Several red lines show the distance to the nearest THI5p atoms. In contrast to PLP-dependent enzymes, no salt bridge was found between N1 of PLP and THI5p. The polypeptide chain used for rendering is molecule B from crystal 1.
Requirement of an Iron Atom for HMP-P Biosynthesis
We proceeded to further characterize THI5p by mass spectrometry analysis. Interestingly, electrospray ionization mass spectrometry (ESI-MS) of the untagged protein revealed a single predominant peak with a mass of 38,513.4 Da (supplemental Fig. S5). This corresponds to the predicted size of the protein (38,460.5 Da) plus an additional mass of 52.9 Da. To assess the possibility that the 52.9-Da additional mass could be due to the presence of an iron ion, a ferrozine assay was performed, specific for iron quantification in biological samples (23, 34), and it clearly shows the presence of an iron atom (Table 2). Indeed, the very recent study of Lai et al. (12) noted that reconstitution of HMP-P synthase activity requires the presence of iron, corroborating our findings. Given that we isolated the protein under aerobic conditions coupled with the observation that oxygen is required for the reaction (12), it can be assumed that Fe(II) is required for the HMP-P synthase reaction to occur.
TABLE 2.
Quantification of iron by the ferrozine assay
The standard errors are the average of three experimental repetitions.
| Ferrous ammonium sulfate | Manganese | Nickel | THI5p-WT | Mutant THI5p |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| K62A | H66A | C196A | C199A | ||||||||
| N (nmol) | 2 | 5 | 10 | 20 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| A593 | 0.032 ± 0.001 | 0.089 ± 0.001 | 0.152 ± 0.001 | 0.346 ± 0.001 | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.041 ± 0.001 | 0.025 ± 0.001 | 0.015 ± 0.001 | 0.009 ± 0.002 | 0.012 ± 0.002 |
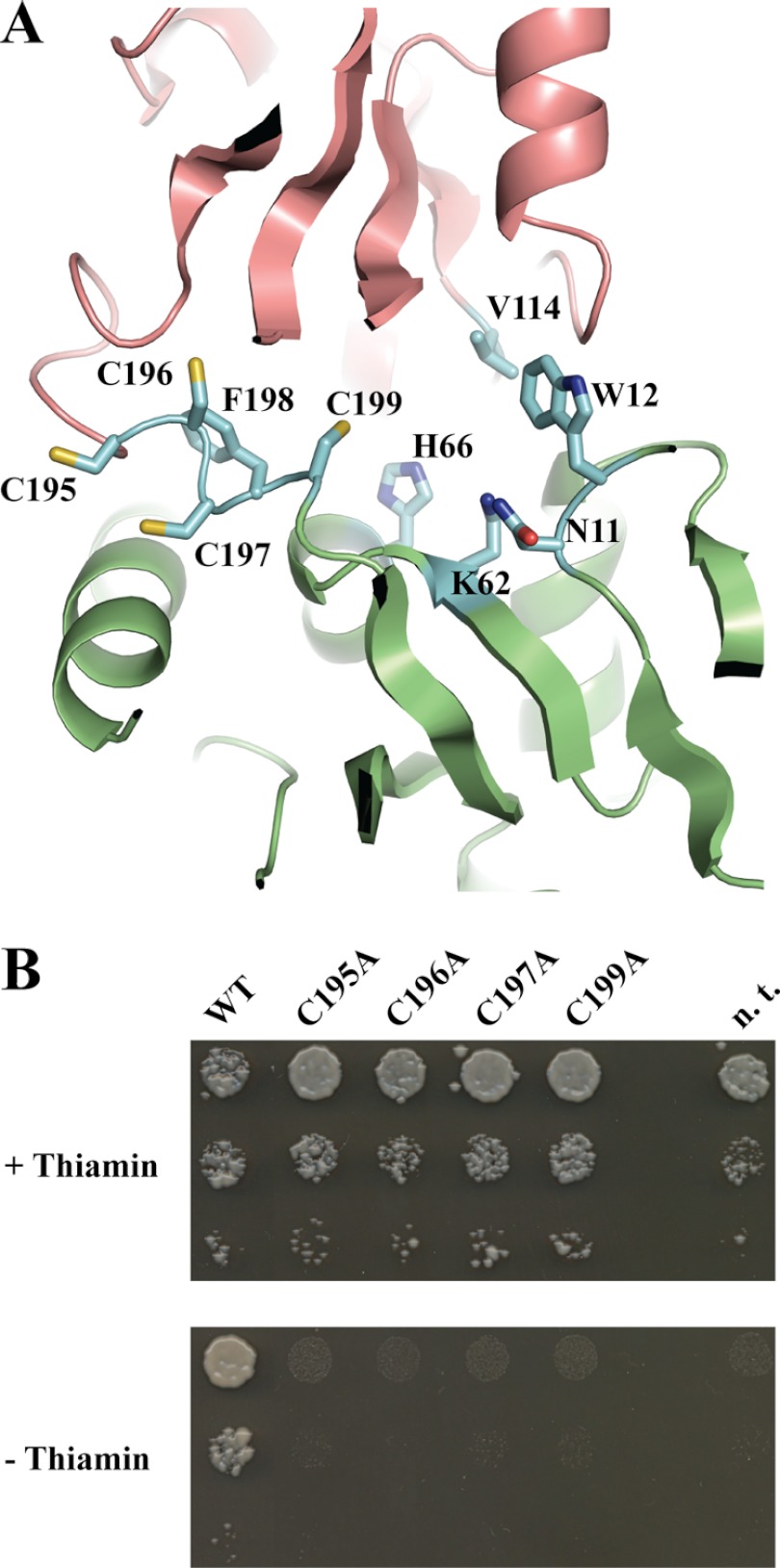
Although we did not observe any iron ion in our x-ray structures of THI5p, the putative binding site of the iron atom can still be proposed. In particular, all THI5p homologs have a strictly conserved CCCFC motif in the middle of their sequence (S. cerevisiae THI5p residue numbers 195–199). This sequence has previously been proposed to accommodate an iron-sulfur cluster based on the structure of the PBP protein, ThiY, although the motif could not be modeled, as it does not exist in the latter (35). This atypical motif was clearly visible in our experimentally phased electron density map (Fig. 5A). It is unclear if the C. albicans THI5p structure had a visible CCCFC motif (12). Nevertheless, the authors did not assess the functional importance of this strictly conserved sequence. This motif does not adopt a particular configuration similar to any known iron-binding fold, perhaps due to the absence of a bound iron in our crystal structure. To validate the functional importance of these cysteine residues during HMP-P synthesis, we mutated each into an alanine residue and performed the complementation assay as before. Notably, the mutation of any of these individual cysteines (Cys-195, Cys-196, Cys-197, or Cys-199) resulted in the abolishment of the growth of RWY16 on thiamin-free medium (Fig. 5B). Moreover, the ferrozine assay revealed a considerable reduction in the absorbance at 593 nm with the recombinantly expressed mutant proteins, indicating an attenuation in the coordination of iron (Table 2). Therefore, we propose that these cysteine residues coordinate an iron atom necessary for the HMP synthesis reaction mediated by THI5p.
FIGURE 5.
A CCCFC motif is essential for THI5p activity. A, conserved iron-binding motif CCCFC (shown in “blue” in stick mode) is located at the interface between domains 1 and 2 within the active site of THI5p. The panel was rendered using molecule C from crystal 1. B, yeast complementation assay demonstrates that each individual cysteine residue is essential. Strain RWY16 transformed with wild-type THI5 (WT) or mutants thereof as indicated; n.t., not transformed. The three spots in each column are respective 1:10 serial dilutions.
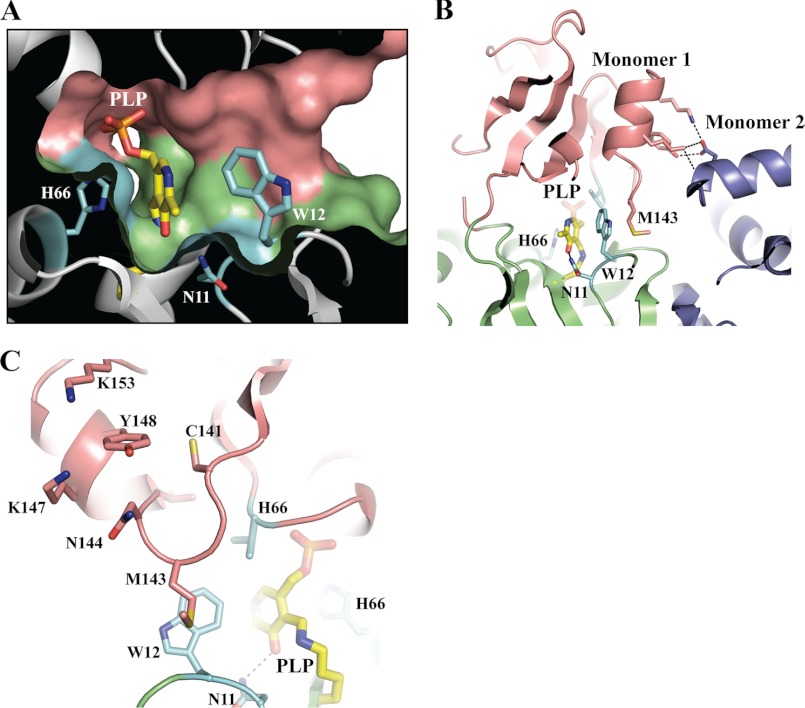
Dimerization Is Likely to Mediate a Gating Mechanism in THI5p
PLP is bound to THI5p within a large internal cavity between domains 1 and 2 (Figs. 1A and 6A). In our x-ray structure, access to this cavity is blocked by Trp-12. Interestingly, the Met-143-containing loop of domain 2 is stacking on Trp-12, locking it in place (Fig. 6B). We propose that the cavity is gated by Trp-12, movement of which is facilitated by the flexible loop containing Met-143. Similar situations have been encountered in PLP-dependent enzymes, where flexible loops have been proposed to act as lids opening and closing the active site (36). In this context, it is noteworthy that the Met-143 residue appears to be held in place by one of the nearby THI5p dimerization contact points as described earlier involving Lys-147, Tyr-148, and Lys-153 (Fig. 6, B and C). PBP proteins typically exist as monomers and undergo open and closed conformations to trap their respective substrate, a mechanism also known as the Venus Flytrap mechanism (37). As THI5p adopts a PBP fold, we believe its domains are able to move as rigid bodies. However, upon dimerization, these movements will be hindered (Figs. 2A and 6B). The intriguing possibility that THI5p utilizes dimerization to facilitate trapping of its substrate provides a fertile area for future work to further explain the enzymatic mechanism of this remarkable enzyme.
FIGURE 6.
Gating mechanism for HMP-P biosynthesis. A, surface representation of THI5p. The view shows the large internal cavity where PLP is bound with residue Trp-12 acting as a gate. His-66 and Asn-11 are shown as colored sticks. B and C, ribbon representations highlighting the THI5p dimerization contacts and the residue Met-143 (domain 2) as possible key components locking Trp-12 (blue, domain 1) and PLP (yellow) in place. The representations are based on molecule B from crystal 1.
Conclusion and Outlook
Although a clear explanation for the use of PLP as a substrate in the biosynthesis of HMP-P has now been provided, one question that intrigued us pertained to the second substrate, histidine. We did not obtain any evidence for the presence of histidine in our x-ray structures. Moreover, we could not see how histidine could be accommodated in the active site of THI5p, according to our structural analyses. The answer to this conundrum has been provided by Lai et al. (12) while our study was being completed, such that the complementary information provided by both studies now provides a comprehensive description of HMP-P synthase. Interestingly, the evidence provided by Lai et al. (12) suggests that it is in fact the active site His-66 residue that is used as the second substrate. This scenario thus mirrors that of the companion protein THI4p, which is required for the biosynthesis of the other heterocycle moiety of thiamin (thiazole), where a backbone cysteine is utilized to provide the sulfur of the thiazole ring (8). The utilization of both THI4p and THI5p in such a single turnover capacity could serve to balance the separate provision of the pyrimidine and thiazole heterocycles, respectively. It is important to note that yeast does not have thiamin pyrophosphate-dependent riboswitches, as observed in bacteria and plants, to shut down the biosynthesis pathway once the supply has been met (3). Important physiological consequences like PLP depletion could occur if THI4p and THI5p activity levels are not controlled properly. Such single-turnover enzymes provide an alternative mechanism to ensure that the PLP pool is not depleted, thus removing the need for riboswitch control of thiamin biosynthesis in yeast.
Acknowledgments
We are indebted to Dr. Peter A. Meacock (University of Leicester) for kindly providing yeast strain RWY16. We thank the Functional Genomics Center Zürich for the mass spectrometry analysis. We are also grateful to Dr. Nicolas Szydlowski and Dr. Michael Moulin (University of Geneva) for assistance and for developing the vitamin B6 HPLC method (to be published elsewhere) and to Dr. Markus Kaufmann (University of Geneva) for performing the static light scattering analysis. We thank all the members of the Thore laboratory for providing excellent feedback. We are grateful to Vincent Olieric and Wang Meitian at the Swiss Light Source and to Andrew McCarthy at the European Synchrotron Radiation Facility, whose outstanding efforts have made these experiments possible. We further acknowledge the financial contributions from the E. Boninchi Foundation, the E. & L. Schmidheiny Foundation, and the Swiss National Science Foundation R'equip Grant 316030-128787 used to set up the crystallography platform.
This work was supported by Swiss National Science Foundation Research Grants PP00A-119186 (to T. B. F.), 31003A-124909 and 31003A-140924 (to S. T.), and the University of Geneva.

This article contains supplemental Figs. S1–S5, Tables S1 and S2, and Experimental Procedures.
The atomic coordinates and structure factors (codes 4H67, 4H65, and 4H6D) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HMP-P
- 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate
- PLP
- pyridoxal 5′-phosphate
- PBP
- periplasmic binding protein
- SeMet
- seleno-methionine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Jurgenson C. T., Begley T. P., Ealick S. E. (2009) The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fitzpatrick T. B., Basset G. J., Borel P., Carrari F., DellaPenna D., Fraser P. D., Hellmann H., Osorio S., Rothan C., Valpuesta V., Caris-Veyrat C., Fernie A. R. (2012) Vitamin deficiencies in humans. Can plant science help? Plant Cell 24, 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winkler W., Nahvi A., Breaker R. R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419, 952–956 [DOI] [PubMed] [Google Scholar]
- 4. Tang C. S., Bueno A., Russell P. (1994) ntf1+ encodes a 6-cysteine zinc finger-containing transcription factor that regulates the nmt1 promoter in fission yeast. J. Biol. Chem. 269, 11921–11926 [PubMed] [Google Scholar]
- 5. Zurlinden A., Schweingruber M. E. (1997) Identification of a DNA element in the fission yeast Schizosaccharomyces pombe nmt1 (thi3) promoter involved in thiamine-regulated gene expression. J. Bacteriol. 179, 5956–5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee A., Jurgenson C. T., Schroeder F. C., Ealick S. E., Begley T. P. (2007) Biosynthesis of thiamin thiazole in eukaryotes. Conversion of NAD to an advanced intermediate. J. Am. Chem. Soc. 129, 2914–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raschke M., Bürkle L., Müller N., Nunes-Nesi A., Fernie A. R., Arigoni D., Amrhein N., Fitzpatrick T. B. (2007) Vitamin B1 biosynthesis in plants requires the essential iron sulfur cluster protein, THIC. Proc. Natl. Acad. Sci. U.S.A. 104, 19637–19642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chatterjee A., Abeydeera N. D., Bale S., Pai P. J., Dorrestein P. C., Russell D. H., Ealick S. E., Begley T. P. (2011) Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wightman R., Meacock P. A. (2003) The THI5 gene family of Saccharomyces cerevisiae. Distribution of homologues among the hemiascomycetes and functional redundancy in the aerobic biosynthesis of thiamin from pyridoxine. Microbiology 149, 1447–1460 [DOI] [PubMed] [Google Scholar]
- 10. Zeidler J., Sayer B. G., Spenser I. D. (2003) Biosynthesis of vitamin B1 in yeast. Derivation of the pyrimidine unit from pyridoxine and histidine. Intermediacy of urocanic acid. J. Am. Chem. Soc. 125, 13094–13105 [DOI] [PubMed] [Google Scholar]
- 11. Ishida S., Tazuya-Murayama K., Kijima Y., Yamada K. (2008) The direct precursor of the pyrimidine moiety of thiamin is not urocanic acid but histidine in Saccharomyces cerevisiae. J. Nutr. Sci. Vitaminol. 54, 7–10 [DOI] [PubMed] [Google Scholar]
- 12. Lai R. Y., Huang S., Fenwick M. K., Hazra A., Zhang Y., Rajashankar K., Philmus B., Kinsland C., Sanders J. M., Ealick S. E., Begley T. P. (2012) Thiamin pyrimidine biosynthesis in Candida albicans. A remarkable reaction between histidine and pyridoxal phosphate. J. Am. Chem. Soc. 134, 9157–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 14. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E. Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation, and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 17. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 18. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laskowski R. A., Moss D. S., Thornton J. M. (1993) Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 22. Stookey L. (1970) Two new spectrophotometric reagents for copper. Talanta 17, 644–647 [DOI] [PubMed] [Google Scholar]
- 23. Beinert H. (1978) Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 54, 435–445 [DOI] [PubMed] [Google Scholar]
- 24. Gietz R. D., Woods R. A. (2006) Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol. Biol. 313, 107–120 [DOI] [PubMed] [Google Scholar]
- 25. Fukami-Kobayashi K., Tateno Y., Nishikawa K. (1999) Domain dislocation. A change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 [DOI] [PubMed] [Google Scholar]
- 26. Tazuya K., Azumi C., Yamada K., Kumaoka H. (1995) Pyrimidine moiety of thiamin is biosynthesized from pyridoxine and histidine in Saccharomyces cerevisiae. Biochem. Mol. Biol. Int. 36, 883–888 [PubMed] [Google Scholar]
- 27. Moore P. S., Dominici P., Voltattorni C. B. (1995) Transaldimination induces coenzyme reorientation in pig kidney dopa decarboxylase. Biochimie 77, 724–728 [DOI] [PubMed] [Google Scholar]
- 28. Mozzarelli A., Bettati S. (2006) Exploring the pyridoxal 5′-phosphate-dependent enzymes. Chem. Rec. 6, 275–287 [DOI] [PubMed] [Google Scholar]
- 29. Eliot A. C., Kirsch J. F. (2004) Pyridoxal phosphate enzymes. Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415 [DOI] [PubMed] [Google Scholar]
- 30. Denesyuk A. I., Denessiouk K. A., Korpela T., Johnson M. S. (2003) Phosphate group binding “cup” of PLP-dependent and non-PLP-dependent enzymes. Leitmotif and variations. Biochim. Biophys. Acta 1647, 234–238 [DOI] [PubMed] [Google Scholar]
- 31. Christen P., Mehta P. K. (2001) From cofactor to enzymes. The molecular evolution of pyridoxal 5′-phosphate-dependent enzymes. Chem. Rec. 1, 436–447 [DOI] [PubMed] [Google Scholar]
- 32. Bisht S., Rajaram V., Bharath S. R., Kalyani J. N., Khan F., Rao A. N., Savithri H. S., Murthy M. R. (2012) Crystal structure of Escherichia coli diaminopropionate ammonia-lyase reveals mechanism of enzyme activation and catalysis. J. Biol. Chem. 287, 20369–20381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunathan H. C. (1966) Conformation and reaction specificity in pyridoxal phosphate enzymes. Proc. Natl. Acad. Sci. U.S.A. 55, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fish W. W. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158, 357–364 [DOI] [PubMed] [Google Scholar]
- 35. Bale S., Rajashankar K. R., Perry K., Begley T. P., Ealick S. E. (2010) HMP binding protein ThiY and HMP-P synthase THI5 are structural homologues. Biochemistry 49, 8929–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matsuda N., Hayashi H., Miyatake S., Kuroiwa T., Kagamiyama H. (2004) Instability of the apo-form of aromatic l-amino acid decarboxylase in vivo and in vitro. Implications for the involvement of the flexible loop that covers the active site. J. Biochem. 135, 33–42 [DOI] [PubMed] [Google Scholar]
- 37. Mao B., Pear M. R., McCammon J. A., Quiocho F. A. (1982) Hinge bending in l-arabinose-binding protein. The “Venus-flytrap” model. J. Biol. Chem. 257, 1131–1133 [PubMed] [Google Scholar]
- 38. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) LIGPLOT. A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 39. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]