Abstract
The large number of experimentally determined molecular structures has led to the development of a new semiotic system in the life sciences, with increasing use of accurate molecular representations. To determine how this change impacts students’ learning, we incorporated image tests into our introductory cell biology course. Groups of students used a single text dealing with signal transduction, which was supplemented with images made in one of three iconographic styles. Typically, we employed realistic renderings, using computer-generated Protein Data Bank (PDB) structures; realistic-schematic renderings, using shapes inspired by PDB structures; or schematic renderings, using simple geometric shapes to represent cellular components. The control group received a list of keywords. When students were asked to draw and describe the process in their own style and to reply to multiple-choice questions, the three iconographic approaches equally improved the overall outcome of the tests (relative to keywords). Students found the three approaches equally useful but, when asked to select a preferred style, they largely favored a realistic-schematic style. When students were asked to annotate “raw” realistic images, both keywords and schematic representations failed to prepare them for this task. We conclude that supplementary images facilitate the comprehension process and despite their visual clutter, realistic representations do not hinder learning in an introductory course.
INTRODUCTION
The recent burst of experimentally determined molecular structures (in excess of 77,000 by 2012; PDB Newsletter, 2011) has led to the development of a new semiotic system, characterized by an increased employment of realistic representations of cellular components in scientific literature, lecture slides, textbooks and, importantly, on the World Wide Web. In contrast with schematic simplifications, such as geometric shapes, we define as realistic those representations of macromolecular objects (ranging from single proteins to subcellular structures such as the ribosome) that resemble the experimentally determined structure. These realistic representations may be the direct product of molecular graphics programs or they may be artists’ impressions of experimentally determined structures (Goodsell and Johnson, 2007; Dahmani et al., 2009) and can include anatomically accurate simplifications, such as ribbon diagrams of protein backbones.
We have adopted the employment of realistic images in our Signal Transduction textbook (Gomperts et al., 2009) and in our cell biology lecture slides (BioScience Image Bank; www.bioscience.heacademy.ac.uk/imagebank, search for “IJsbrand Kramer” and then click on “Agree” to get access to the images). From a number of arguments, both in favor and against the employment of realistic images (listed in Dahmani et al., 2009), we distilled the points of view: 1) many university instructors prefer state-of-the-art representations, because they reflect an expert level of understanding; 2) students will eventually need to be able to provide meaning to realistic representations in order to be able to read and understand current scientific literature (i.e., acquire biomacromolecular three-dimensional literacy); and 3) in a number of cases, structure–function relationships are best illustrated on the basis of real structural information (Nye, 2004). Although the above arguments are all valid from a teacher's point of view, what really matters is how students “handle” the different type of images: to what extent are they able to follow our argumentation? Knowledge about students’ image handling would be helpful to instructors and textbook authors with respect to the iconographic approach best chosen for illustrating learning documents.
Starting in 2007, to assess the effectiveness of realistic iconographic images in cell biology learning materials, we performed a series of image tests in the context of an introductory cell biology course. Typically, we employed realistic renderings, using computer-generated Protein Data Bank (PDB) structures; realistic-schematic renderings, using shapes inspired by PDB structures; and schematic renderings, using simple geometric shapes to represent cellular components. In our previous study (Dahmani et al., 2009), students were instructed to review chapters about the cell membrane and transport mechanisms. In the tests, they were offered images similar to those found in the learning documents but drawn with different iconographic styles. We found that our students had no more difficulty in making sense of the realistic than of the schematic images, and we concluded that despite their complexity, realistic representations are not necessarily intimidating for students.
In the study described here, we approach the question differently in two respects. First, we have increased the number of objects to raise complexity still further. Rather than depicting a single membrane protein, we tested how students deal with different iconographic styles in a signal transduction pathway that comprises 11 different proteins. Second, we have complemented the image-interpretation exercise (in which the students review the subject ahead of time and then must make sense of similar images in different styles and lacking legends and labels [Dahmani et al., 2009]) with a learning exercise. In this exercise, students were given a text supplemented with an explanatory image in different iconographic styles (realistic, realistic-schematic, or schematic). As a control of how images influence text-based learning, one group received a list of keywords only. The students studied the documents and were examined immediately afterward for their knowledge and understanding. We refer to this protocol as “learning with/without image.” The image tests were part of the formal knowledge assessment program of the cell biology course.
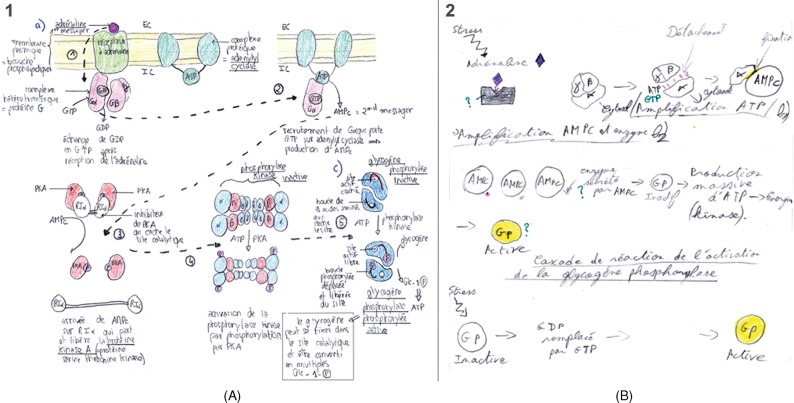
We chose the adrenaline-to-glycogen phosphorylase pathway for this study. It holds an important place in our introductory cell biology course, because of its historical and Nobel background (Krebs and Fischer, 1956; Rall et al., 1957; Sprang et al., 1988), its employment of diverse signaling mechanisms, and its easy integration with other course subjects (muscle contraction, the nicotinic acetylcholine receptor, and ATP production by mitochondria). We have explored two aspects of the pathway: 1) the cascade of signaling events starting with binding of adrenaline to its receptor and ending with the activation of glycogen phosphorylase (tests 1 and 3); and 2) a more focused aspect, the regulation of activation of protein kinase A (PKA; test 2).
Employing the learning with/without images protocol (test 1), we aimed to answer the following two questions: Do supplementary images facilitate the text-based, comprehension process (Mayer, 1997; Carney and Levin, 2002; Schnotz, 2002; Ormrod, 2008)? If so, which of the above-described iconographic styles is most efficient? We anticipated that realistic representations of signaling cascades, because of their visual clutter, would be off-putting to students and hinder learning (Sweller, 2005; Schnotz and Kürschner, 2007). Furthermore, employing the image-interpretation protocol, we aimed to answer a second set of questions, namely: Are schematic representations of proteins, combined with verbal instructions about structural details, sufficient to provide insight into structural aspects of proteins (test 2)? Do realistic representations, because of their elevated level of detail, have more intrinsic explanatory power (i.e., act as a better storyboard) when students are asked to recall information on the basis of a raw image (test 3)?
Our results show that the signal transduction pathway images do indeed support learning relative to keywords, but we find no significant differences in the overall test outcomes between different iconographic approaches. Furthermore, we show that a raw realistic representation of the adrenaline pathway, despite abundant molecular detail, has no higher intrinsic explanatory power than a raw schematic representation. These results confirm our previous findings that, despite the large number of proteins presented, students can handle realistic representations (Dahmani et al., 2009). Moreover, we extend our findings by showing that image supplements, independent of their iconographic approach, always need clear verbal instructions to be meaningful to novice students. We conclude that instructors have quite a bit of freedom in choosing among iconographic approaches, as long as they are clear about the iconographic and scientific codes. We end by providing a number of arguments why realistic or realistic-schematic representations might be more suitable for an introductory cell biology course.
METHODS
Description of the Test Group
We performed the tests at the University of Bordeaux with first-year biology students in their second semester of the life and environmental sciences program (designated SVE, for Science de la Vie et l'Environnement). It is a cohort of roughly 300 students, who are divided into groups for different teaching purposes; three groups for lectures (taught separately), 10 groups for practicals (laboratory exercises), and 20 groups for tutorials. The current Cell Biology course comprises 16 h of lectures (12 sessions), 15 h of practicals (four sessions), and 4 h of student-led tutorials (three sessions). Knowledge is assessed in four 80-min tests (a weighting of 0.075 each), one 90-min in-course exam (weighting: 0.3), and a 90-min final exam (weighting: 0.4). One of the 80-min tests occurred in the lecture theater (large groups) and was used for this report; the other three occurred during the tutorials (small groups). The students attended a 60-h general chemistry course and a 30-h biochemistry course in the preceding semester. In these courses, they were taught about the composition of biological molecules and the characteristics of chemical bonds, as well as the different types of molecular interactions. The tests described here were performed over a period of 5 yr (Spring 2008 to Spring 2012).
Format of the Image Tests
To test how perceptual images aid in learning, we chose a viable learning environment, rather than an artificial experimental setup (for a discussion on this subject, see De Jong, 2010). By viable learning environment, we mean a highly relevant subject (cell biology) and a detailed text supplemented with an image that reiterates the text instructions (as is a common practice in textbooks; Schnotz, 2002, Carney and Levin, 2002), sufficient reading time, and a summative assessment at the end of the exercise. We reasoned that the results of such a study should be meaningful to a wide range of biology instructors. We distinguish two types of tests we employed for testing our hypotheses; these are outlined below.
Learning with/without Image Test Protocol (Test 1).
We employed this protocol to find out whether images aid learning of text-based instructions and, if so, which of the iconographic styles is most efficient. Students were given one text about the adrenaline-to-glycogen phosphorylase pathway plus one of three different renderings of a graphic representation of the same pathway (image a, b, or c) or a list of keywords (image d). Simultaneously, a slide was projected that showed the students what activities were expected from them during the assessment phase of the test and what type of questions they were going to be asked. In short, the slide informed them that they had to draw and describe the signaling pathway in their own style and that they had to answer multiple-choice questions (MCQs). It also showed a list of subjects (glycogen metabolism, role of ATP in activation of proteins, etc.) that were going to be dealt with in the MCQs. We put up this slide because our students feel more confident when they have a clear idea of what we expect from them. Moreover, the instructions give them information regarding what to look for in the text. Both confidence and guidance tend to augment their engagement in learning (particularly for weaker students). The documents were removed after 40 min and were replaced with the question sheet, for which we permitted a roughly 25-min student answer period (for detailed information, see Section I: Test 1 in the Supplemental Material).
The students were not informed about the subject of the test ahead of time, they had not received any formal teaching about the adrenaline-to-glycogen phosphorylase pathway, and they were not instructed to review the subject. Students who participated in this test had not participated in either of the other two tests. The students had, however, received brief instructions in an earlier tutorial session about how the addition of a phosphate (through phosphorylation) and the exchange of GDP for GTP affect protein conformation and activity.
Image-Interpretation Test Protocol (Tests 2 and 3).
We have developed this protocol with two questions in mind: Do schematic images of proteins, combined with a highly coherent text dealing with structural modifications, prepare for insight into structural aspects of the same proteins (tests 2 and 3)? Do realistic representations, because of their elevated level of detail, have more intrinsic explanatory power (i.e., act as a better storyboard) when students are asked to recall information on the basis of a raw image (test 3)? In test 2, students were given one text about the different activation states of protein kinase A (PKA), plus one of two supplemental images of the same subject rendered in different iconographic styles (image a, realistic-schematic, or b, schematic). The role of the inhibitor RI-α and how cAMP is required to separate it from the catalytic subunit were mentioned in the text but not shown in the concomitant image during the reading phase (for detailed information, see Section I: Test 2 in the Supplemental Material). A third group of students received a sheet with a list of keywords (image c). Again, a slide was projected that depicted a rough outline on the questions of the test. After 40 min of reading, the documents were removed, and students were given a question sheet plus a “raw” (unlabeled) realistic rendering of the same activation states, which this time included the inhibitor RI-α (acting as pseudosubstrate). The students were not informed about the subject of the test ahead of time, they had not received any formal teaching about the PKA activation states, and they were not instructed to review the subject. Students who participated in this test had not participated in either of the other two tests.
In test 3, the learning and image-interpretation activities were separated over a number of weeks, and the test was performed twice over a period of 2 yr. In year 1, students were taught with a schematic representation of the adrenaline-to-glycogen phosphorylase pathway. After the receptor signaling section was finished, their knowledge was assessed 4 wk later in an image-interpretation test (lasting for 40 min). In this test, half the students (n = 42) received a “raw” schematic image, whereas the other half (n = 44) received a “raw” realistic one. They all received the same questions sheet. To reduce ambiguity in the identity of the pathway, the labels for glycogen and adrenaline were added to the raw image. In year 2, students were taught with a realistic representation of the pathway and tested in the same way described above. Both the course slides and handout were available to the students during the review periods (for detailed information, see Section I: Test 3 in the Supplemental Material). Students who participated in this test had not participated either of the other two tests.
Assessing Students’ Starting Level and Testing the Image Tests
Pilot tests were performed in student-led tutorials (supervised by I.K.) in preceding years. These pilot tests allowed us to determine the preparation time needed for the tests and allowed us to make improvements to the reading materials and adjust the difficulty of the questions. They also served to verify possible ambiguities in the learning documents and, importantly, they served to assess whether students learned from the documents, because we included additional pretests in the tutorials to reveal student knowledge level prior to the preparation phase (assessment of the starting level). The pretests were similar to the image tests employed in the study presented here.
Grading of the Tests
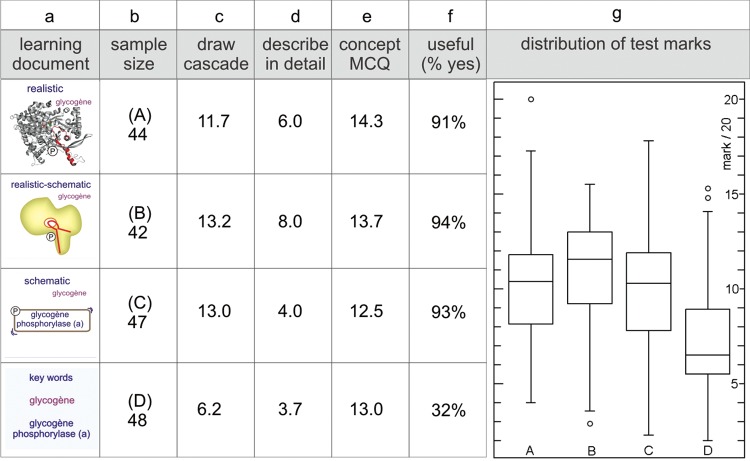
A detailed description of how we graded the tests is provided in the Supplemental Material (Section I). With respect to the drawing shown later in Figure 4, we divided the pathway in five sections and offered 2 points for each step; partially correct answers were given a single point per step. Points were awarded for depicting binding of adrenaline to its receptor and exchange of GDP against GTP (2 points), dissociation of heterotrimeric complex and Gα–GTP-subunit binding to adenylyl cyclase (2 points), production of cAMP leading to the removal of the inhibitor (RIα) and subsequent activation of PKA (2 points), phosphorylation and activation of phoshorylase kinase (2 points), and phosphorylation and activation of glycogen phosphorylase (2 points). Two points were reserved for carefulness of drawing, use of colors or other aspects that make the drawing appealing. All grades for the figures were converted to a mark out of 20 (mark/20).
Figure 4.
Two examples of student drawings. (A) A drawing given a high grade (12/12) from a student who had access to the realistic-schematic representation. (B) A drawing given a low grade (4/12) from a student who had access to the schematic representation.
The Unfolding Exercise
The aim of this exercise, performed with a “raw” realistic representation of the adrenaline-to-glycogen phosphorylase pathway (test 3), is to let students unfold what they read in the image. It serves two intertwined purposes. The first is to address the hypothesis that realistic images may hinder learning due to their visual clutter. The second is to find out where students go wrong in the interpretation of the image. Do they make mistakes in following the sequence of events? Do they make mistakes in interpreting the iconographic code, that is, do they not distinguish a protein from a nucleotide? Or do they make mistakes in the scientific codes, that is, do they recognize the object as a protein kinase but fail to understand what a kinase reaction is all about and how it affects the activity of the substrate protein? Fifteen volunteers were interviewed by tutors 1 wk after the image test with a realistic image supplement. Students were informed about the purpose of the interview; however, to avoid the possibility of prior rehearsal, the subject was not mentioned. In this type of exercise, it is important that the interviewer does not push the students toward a unique (and “correct”) interpretation; in other words, the questions have no wrong or right answers. Student replies were written down (see Figure S1 and Table S1 in the Supplemental Material).
Compensation of Test Grades
Students were informed about the objectives of the tests and about the adjustment procedure we applied if the different images had lead to different average marks. To compensate, we multiplied students’ grades with coefficients (with a ceiling of 20/20) so that all the group averages leveled to the average of the “best-performing image.”
Organization of the Test Sessions
High instructor vigilance is essential to minimize cheating. To minimize students’ “visual access” to images with other iconographic styles during the test session, we separated the lecture theater into three or four sections, each with a different image. Each image section (20–30 students) was monitored by one instructor, whereas a separate instructor answered questions and distributed/removed documents for the whole of the lecture theater. Each test was performed with one lecture group at a time (85–115 students). Because test subjects are immediately communicated among peers, we could not repeat the same test with other groups during the same year. Had we reused a test, the students would be prepared (read up on the subject and study images from the Web or other sources), and from the tutorial pilot tests, we learned that this precludes any distinction in test outcomes between the control group (keywords) and the group with image supplements (they no longer need perceptual images to succeed in the test). For this reason, we had to prepare two image-interpretation tests, one dealing with adrenaline-to-glycogen phosphorylase and the other with the activation states of PKA.
Preparation of the Images
Images were prepared with CorelDraw (Insight Technology Solutions, Velizy, France) and PyMol (www.pymol.org). With the exception of the schematic image, protein shapes were based on information from the PDB (Berman et al., 2000). We prepared three versions: 1) realistic, using a maximum number of PDB structures; 2) realistic-schematic, using shapes inspired by PDB structures; and 3) schematic, using boxes or circles to represent proteins (Figures S1–S3, S5, S6, and S8-S10 in the Supplemental Material). The control group received a list of keywords that, in order to provide some visual distraction, were presented in (colorful) boxes (Figures S4 and S7 in the Supplemental Material). We employed the following PDB coordinates: glycogen phosphorylase, 1gpy (tense state, little active, glucose-6-phosphate–bound) and 1gpa (relaxed state, active, pSer-14); β2-adrenergic receptor, 2rh1; GTP-binding protein (Gi-α), 1gg2; adenylyl cyclase, 1cul; and PKA plus inhibitor RI-α, 2qcs. We used 1hck (Cdk2) as a representative of the inactive state of a serine/threonine protein kinase. With respect to phosphorylase kinase, we based our graphic on the images shown in Venien-Bryan et al. (2002, 2009).
Depiction of Numerical Data and Statistical Analysis
The outcomes of the different sections of the image tests (drawing the cascade, describing the events, and MCQ) are shown as average marks, out of 20, without SD, so that the figures/tables are easier to read. To provide an impression of the distribution of marks, we depict the overall test results in box-and-whisker plots. These indicate the lowest mark (lower bar); the lower, median, and upper quartile (the box); and the highest mark (upper bar). An analysis of variance (ANOVA) statistical analysis was performed to check whether the image type influenced students’ performances on tests. A Student's t test was employed to estimate whether differences between two data sets were statistically significant. We used a Web-based calculation program for both of these analyses (Kirkman, 1996).
RESULTS
Students Learn from the Documents Associated with the Tests
An essential condition for studying a possible impact of iconographic styles is that students learn from the provided documents. Because of practical and time constraints, we could not measure the starting level of the students in the lecture theater. However, from the numerous pilot tests in the tutorials over two preceding years, we learned that the starting level is generally very low. When the adrenaline-to-glycogen phosphorylase learning test (test 1) was performed prior to studying the learning documents (pretest), students (n = 197) obtained an average score of 3.87/20 ± 1.96 (mean ± SD; Figure 1). The average score for the same test after the students (n = 188) had studied the learning documents was 9.66/20 ± 3.55 (Student's t test p value < 0.001). For test 2, in which students learned about the different activation states of PKA, the average score of the pretest in the tutorials was 3.11/20 ± 2.16 (n = 102), and we obtained an average score of 10.1 ± 3.65 (n = 118) for the test described in this study (Student's t test p value < 0.001). The incremental gain in average grades between pretests and the data presented in this article (posttests) indicate that the learning documents promote learning. We therefore conclude that if differences in test results occur, they can be attributed to differences in learning efficacy of the iconographic styles in question. These pretest results also served as a control for test 3, as we employed an identical text and identical images but in a different experimental setting.
Figure 1.
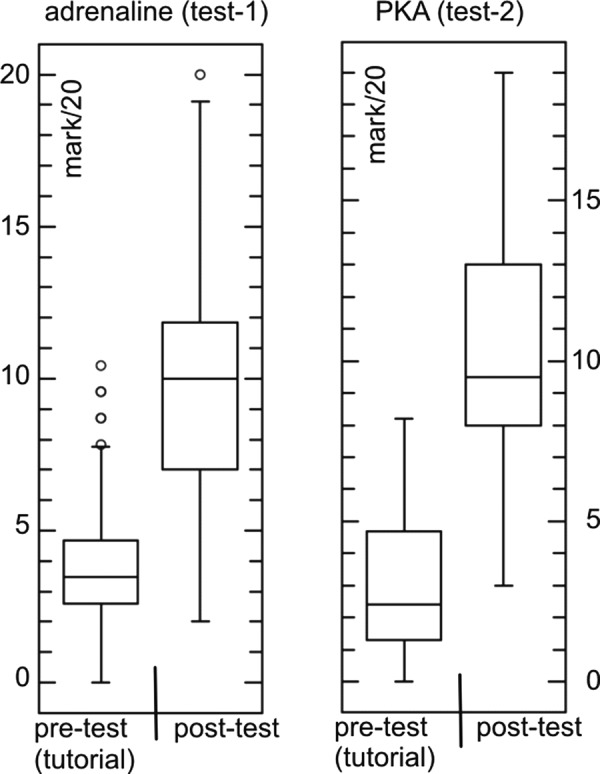
Average score of the image tests, irrespective of the image supplement, before (pretest) and after (posttest) students studied the learning documents. Students were given the pretest during tutorials in the year prior to the experiments described in this article. The posttest represents the results from the image tests held in the lecture theater. The content of the pre- and posttests were similar. Left, results from the adrenaline-to-glycogen phosphorylase image test (test 1); right, the result of the PKA activation states image test (test 2). Score increments are significant for both situations, with a p < 0.001.
Images Aid Learning, But Different Iconographic Approaches Appear to Have Equally Positive Influence
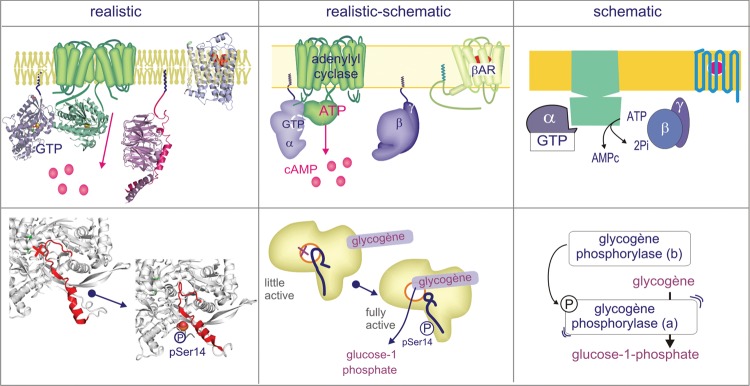
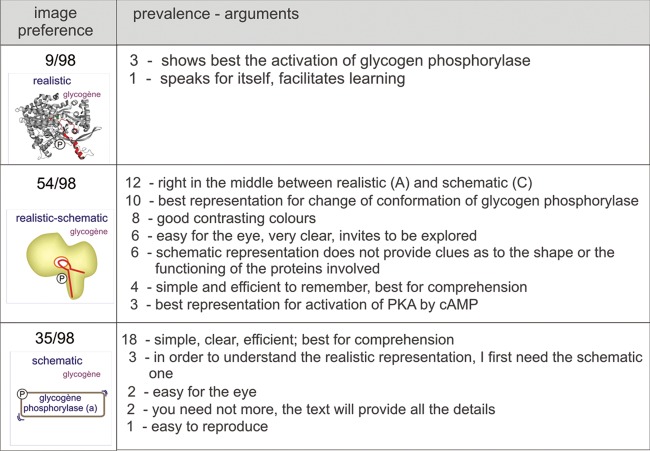
To find out whether images impact learning and if so, which of the different iconographic styles is most efficient (first set of questions), we offered students one text and one of three renderings of the adrenaline-to-glycogen phosphorylase pathway (Test 1; see Figure 2 for examples of the iconographic approach). The control group received the same text, but it was only supplemented with a set of keywords. Images and text covered the same subject (Carney and Levin, 2002), as is common practice in textbook illustrations. The questions were not particularly focused on the images; rather, they were meant to assess text-based knowledge, as we would do in assessments in the first year. Students scored significantly worse when supplied only with keywords. However, the different iconographic approaches appear to have equally positive influence on students’ overall performance (Figure 3g, box-and-whisker plot). This outcome is also reflected in the evaluation scores, for which students were asked to write down on their answer sheet at the end of the test whether or not their image supplement was “useful.” Whereas 32% of the students judged the keywords useful, all the images scored high, in the range of 91–94% (Figure 3f). The images strongly influenced the students’ drawings of the signaling cascade and the detailed descriptions of the activation mechanism of glycogen phosphorylase (phosphorylation of Ser-14) but had less effect on the results for the MCQs. With respect to MCQs, students given the image test with the realistic image scored significantly better (14.3) than students supplied with other supplements, but we found no significant differences among students supplied realistic-schematic, schematic, or keywords (Figure 3, compare c, d, and e). Two examples of student drawings of the signaling cascade are shown in Figure 4; one with a high (12/12) score, from a student who had been given the schematic-realistic image, and the second with a low (4/12) score, from a student who had been given the schematic image. Importantly, both the schematic style and keywords led to an equivalent low mark for the description of the activation mechanism of glycogen phosphorylase (4.0/20 and 3.7/20, respectively; Figure 3d). After the image test and after having received all three image supplements, students were asked which image they preferred; the realistic-schematic image came out best, followed by the schematic, and only 9.2% of the students selected the realistic image. A list of arguments is provided in Figure 5. In short, students have an eye for molecular detail (relation of structure and function), and they appreciate the colors, but at the same time, they are attracted by the clearness of the schematic representation.
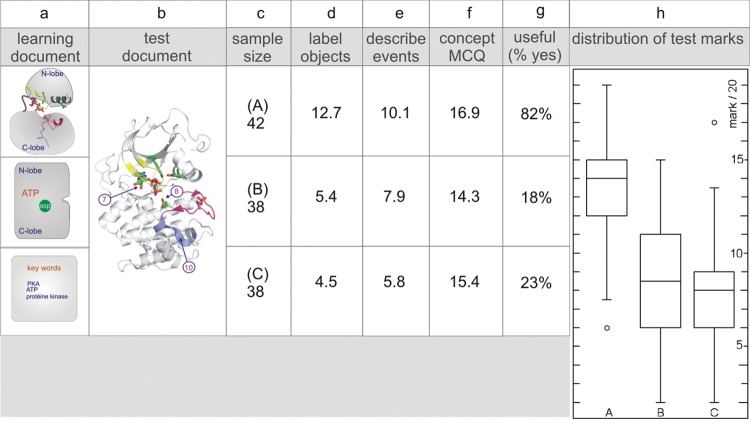
Figure 2.
Iconographic styles employed throughout the study. The images cover the same subject and feature the same components but represent quite different renderings. Only a selection of the components of the adrenaline signaling cascade is presented.
Figure 3.
Outcome of the image test employing different iconographic styles for the representation of the adrenaline-to-glycogen phosphorylase signaling pathway. In an 80-min test protocol, students were provided with one text and one of three different renderings (A–C) of the adrenaline-to-glycogen phosphorylase pathway, ranging from realistic to schematic. The control group (D) received a list of keywords (icons in a). After roughly 45 min of preparation, the learning documents were removed and replaced with a test in which students had to draw the pathway (average results in c), describe in detail the mechanism of activation of glycogen phosphorylase (average results in d), and reply to MCQs (average results in e). At the end, students were asked to indicate whether they regarded the image (or keywords) useful or not (percentage of “yes” replies, f). Notice that with respect to the overall test score (box-and-whisker plot presentation in g), students scored significantly lower when the text was supplemented with a list of keywords only (D). However, the different iconographic styles had equal impact (compare A, B, and C), and the students also found them equally useful. Each test group comprised about 45 subjects (b). Marks were out of 20. There are significant differences between the groups (ANOVA, F = 14.57 and p < 0.0001). The image supplement, independent of its iconographic style, significantly increases the test outcome, but there are no significant differences between the different renderings (A–C).
Figure 5.
Students’ image preferences and arguments for their choices. After the image test, students were given all three renderings of the adrenaline-to-glycogen phosphorylase pathway and were asked to select their favorite and provide arguments. The prevalence column indicates the number of students who made the particular comment (not all students provided arguments). Notice a strong preference for the realistic-schematic representation.
Students Have No Difficulty with the Iconic Code of the Realistic Rendering
In a separate setting, in the week following the test, 15 volunteers were asked to “unfold” what they read in the realistic image (for detailed information, see Section II, Table S1 and Figure S10 in the Supplemental Material). We discovered that students do well with respect to the recognition of the flow of information (going from event (a) to (b) to (c), etc.); they understand the iconic codes of dotted arrows (movement and binding) and active sites (three red concentric lines); and they easily recognize cellular components, such as the plasma membrane, proteins (by shape, size, and ribbon representation), nucleotides, and phosphate. However, about one-third of the students make serious errors in the description of the role of ATP and how, through a phosphorylation reaction, it affects catalytic activity. Certain students have difficulty in understanding how enzymes, catalytic sites, and substrates relate to one another (in both composition and function). From the image test and the student descriptions of the signaling events, we conclude that supplementary images facilitate the comprehension process and, despite their visual clutter, realistic representations do not hinder learning.
Keywords or Schematic Images Do Not Prepare Students for Computer-Aided Renderings of Protein Structures
Next, we asked whether schematic images of proteins, combined with a highly coherent text that provides structural detail, are sufficient to provide students with insight into structural aspects of the same proteins (test 2). We also sought to determine whether realistic images have more intrinsic explanatory power (i.e., act as a better storyboard), because of their detail (test 3). To this end, we employed two different tests, one dealing with molecular detail of the activation states of PKA (test 2) and the other dealing with the above-described signaling pathway of adrenaline (test 3). For image test 2, students were offered a single text and one of two renderings of the activation states of PKA (realistic-schematic or schematic) or a set of keywords (control group). After removal of the learning documents, students received a “raw” realistic presentation of the same subject and a set of questions; they were asked to label molecular structures (N-lobe, activation segment, catalytic site, ATP, etc), describe the different activation states, and reply to a small number of MCQs. Perhaps not surprisingly, students scored significantly lower when supplied with a schematic presentation or keywords, and this corresponds with their low evaluation score for these supplements (18 and 23%, respectively; Figure 6). Only the realistic-schematic representation prepared them for labeling and for a description of the activation states. Again, the images had less influence on students’ replies to MCQs (Figure 6f). The realistic-schematic image led to a significantly higher score (16.9 out of 20) than did the other supplements (14.3 for the schematic image and 15.4 for the keywords).
Figure 6.
Keywords and a schematic representation of the activation states of PKA do not prepare for a realistic test image. In an 80-min test protocol, students were provided with one text and one of two different renderings (A or B) of the activation states of PKA (realistic-schematic or schematic). The control group (C) received a list of keywords (icons in a). After roughly 45 min of preparation, the learning documents were removed and replaced with a test in which students were given a realistic version of the same events (icon in b) and were asked to: label molecular structures (activation segment, ATP, catalytic site, etc.; average results in d), describe the events (average results in e), and reply to MCQs (average results in f). At the end of the test, students were asked to indicate whether they regarded the image (or keywords) useful or not (percentage of “yes” replies in g). Notice the dramatic drop in the overall test score (box-and-whisker plot presentation in h) when students had prepared the subject with a schematic representation (B) or a list of keywords (C). Each test group comprised about 40 subjects (c). Marks were out of 20. There are significant differences between the groups (ANOVA, F = 47.82 and p < 0.0001), the impact of the realistic-schematic image is significantly distinguishable from that of a schematic image or a keywords supplement.
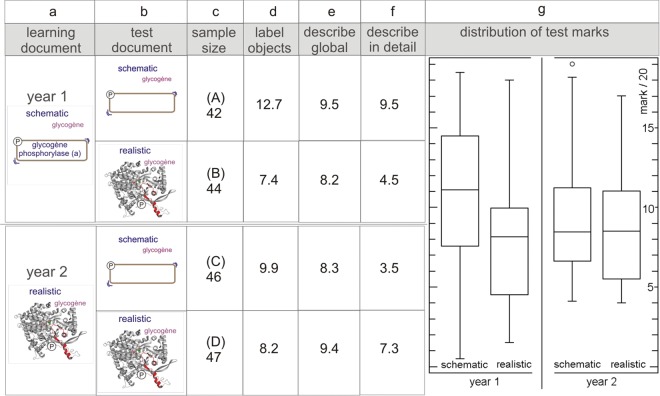
The second test (test 3), dealing with adrenaline-mediated activation of glycogen phosphorylase, had a different experimental setup and was divided over two cohorts in two different years. In year 1, students were taught with a schematic image (in lecture slides, handouts, and multimedia resource) and were tested for their capacity to label and annotate a “raw” realistic or a schematic image. In year 2, they were taught with a realistic image and subjected to the same test (see Figure 7). When taught with a schematic image, students scored significantly lower when they were confronted with a realistic test image (Figure 7g, year 1). Both labeling of the cascade components and description of events were inferior (Figure 7, d–f). The situation reversed, but only in part, in the cohort instructed with a realistic image in year 2. Students now scored better in the description of events when confronted with the realistic image, but the schematic image still proved to be a superior support for the labeling of the pathway components (Figure 7, d–f). Because labeling counted for 50% of the overall test mark, the two sections (“description” versus “labeling”) balance out, and the two groups no longer differ significantly (Figure 7g, year 2). Collectively, these image-annotation experiments show that students are properly prepared for raw images (lacking verbal instructions) only if they have studied them ahead of time in the context of precise verbal instructions. In other words, raw images, whether detailed and realistic or not, have no intrinsic explanatory power. These results confirm previous findings that images carry no information when students have no precise expectations of what to make of them; raw images always need “procedures” to extract the relevant information (Cook et al., 2008a,b; Crisp et al., 2008; Orgil and Crippen, 2010; Palmer, 1978). With respect to gaining structural insight, this means that students must be taught with the appropriate realistic or realistic-schematic image. When it comes to rote learning of components of a signaling cascade, the schematic representation seems by far superior.
Figure 7.
Novice students can only make sense of images if they have studied them ahead of time in the context of precise verbal instructions. In year 1, students had been instructed (in lecture slides and handouts) with a schematic representation of the adrenaline-to-glycogen phosphorylase signaling pathway (icons in a). In an in-course assessment (1 mo later, following instructions to study the signaling chapter), they either received an unlabeled (“raw”) schematic or a realistic representation (icons in b) of the same process. The words adrenaline and glycogen were added to both images. Students were asked to: label the components (β2-adrenergic receptor, G-protein, adenylyl cyclase, etc.; average results in d), globally describe the pathway (receptor occupation leads to an exchange of GDP for GTP in the α-subunit of Gs, etc.; average results in e), and describe in more detail how phosphorylation changes the activity of glycogen phosphorylase (average result in f). Despite having been provided with abundant details, students were not at all prepared to make sense of the realistic image (g, box-and-whisker plot for year 1); they scored much better in all aspects with the familiar schematic representation (significant difference between the two groups: p < 0.00001). We applied the same protocol for year 2, except that students were taught with a realistic representation. This reversed the outcome of the test with respect to description of the events. Students performed much better with the familiar realistic representation (compare average results in e and f). However, they were still more effective in labeling the components of the signaling cascade in the schematic image (average results in d). Because of this, the overall marks do not differ between the two groups (g, box-and-whisker plot for year 2; p = 0.67). Each group comprised about 45 subjects (d). Marks are out of 20.
DISCUSSION
In this study we aimed to answer two sets of questions, the first set being: Do supplementary images facilitate the text-based comprehension process (Mayer, 1997; Carney and Levin, 2002; Schnotz, 2002; Ormrod, 2008)? If so, which of the above-described iconographic styles is most efficient? The second set is: Are schematic images of proteins, combined with a highly coherent text that provides structural detail, sufficient to provide students with insight into structural aspects of the same proteins? Do realistic images have more intrinsic explanatory power because of their detail, thus acting as a better storyboard?
With reference to the first set of questions, we conclude that indeed supplementary images facilitate the comprehension process. However, we find no difference in learning efficacy between the different iconographic styles and conclude that realistic renderings of signaling pathways do not necessarily hinder learning and are not too distracting for novice students. We therefore confirm and extend the findings reported in our previous article, in which we showed that students interpret realistic images as effectively as schematic ones and when given a choice, students do not necessarily select the least complex image (Dahmani et al., 2009). Images have the most impact on the drawing and the (detailed) description of signaling events but have much less influence on the outcome of MCQs. This suggests that for a number of subjects normally assessed in our course, image support is less vital. This of course does not necessarily exclude a role for images in cell biology, as students at this stage must have a large collection of relevant mental representations to which they can make reference (Kosslyn et al., 2006). With respect to the second set of questions, we show that despite detailed verbal instructions, the schematic representation of PKA does not prepare students for insight into protein structure, that is, schematic representations do not habituate students to computer-generated renderings of PDB structures. Finally, an important finding is that the raw realistic representation of the signaling pathway, despite abundant molecular detail, has no higher intrinsic explanatory power; novice students only can make full sense of raw images, irrespective of their iconographic style, when they are instructed in advance with images linked with precise, text-based instructions.
Regarding the question of how to illustrate a cell biology learning document, we draw four lessons from our work and that of others. The first is that instructors actually have a good deal of freedom with respect to choosing between iconographic styles, because, with the appropriate instruction, students quite readily adapt to iconic codes (this object represents a protein, etc.). One should, however, keep in mind that due to their simplicity, schematic images do not necessarily make cell biology easier to understand. Independent of their iconographic style, cell biology images always require a thorough understanding of numerous iconic and scientific codes (the protein is composed of amino acids, each with particular characteristics, and folded in a particular way, etc.; Johnstone, 1991). This is why written instructions are of utmost importance, and they have to be highly coherent for novice students (Mayer, 1989; McNamara et al., 1996). It therefore is essential that instructors are at ease with their choice, because their verbal instructions must correspond with the information carried by the image (Seufert and Brünken, 2006; Bartholomé and Bromme, 2009); and, vice versa, the prompt must prompt coherent verbal instructions (Jucks et al., 2007; Runde et al., 2007). It is our experience that instructors also need images, perhaps more than students when in a lecture setting. A second lesson is that students generally appreciate the realistic-schematic representations. They find this type of image most appropriate (see also Dahmani et al., 2009; Ormrod, 2008), and this may increase their curiosity and motivation, and, as a consequence, act as a tool to aid their thinking (Ainsworth, 1999). A third and important outcome of our study is that if iconic codes are not a problem for students, scientific codes are an obstacle. About one-third of the interviewed students could not fully interpret the realistic image, because they failed to master certain molecular concepts. The problem, therefore, lies more upstream, in an earlier stage of biomolecular grounding (learning through perception [icons] and conceptual instructions). How iconographic styles, in both stills and animations, impact on an early stage of biomolecular grounding is an interesting question. Lastly, choosing between iconographic styles is dependent on the teaching objectives (and subsequent assessment). For instance, the outcome of our MCQs is much less affected by the presence or absence of images. On the contrary, when assessing insight into structural aspects of PKA, using a “raw” ribbon representation, students only scored well when the text was supplemented by a realistic-schematic image and not a schematic one.
We take the point of view that our first-year students should acquire a novice level of biomacromolecular three-dimensional literacy (Craig and Bateman, 2010), and we are gradually building this objective into our cell biology teaching and assessment program (rather than leaving this task to the biochemistry course). One of the arguments is that our teaching experience has taught us that more realistic iconographic styles have the important advantage of being more amenable to an accompanying hands-on molecular modeling practical (“biocomputing” practical); there is less discrepancy between the shapes students obtain on their computer screen and the ones depicted in their teaching documents. Linking lecture content with hands-on molecular modeling, seeing and doing, enhances students’ understanding (Ealy, 2004; Harris et al., 2009) and general visual literacy (Schönborn and Anderson, 2009). This in turn facilitates a natural (and early) access to richly annotated, databases, such as the PDB, ExPASy, the National Center for Biotechnology Information, and others, thus providing students with the necessary skills for Internet-based “information foraging.”
Supplementary Material
ACKNOWLEDGMENTS
We thank our student-tutors, who have helped develop (proofreading and translation into French) and test the feasibility of image tests in their tutorials: Arthur Delmares, Laurence Hiroux, Lionel Tocaven, Pierre Feydel, Romain Carmeille, Cecilia Saubusse, Antoine Bremond, Loic Charles, Dylan Coutou-Picard, Priscilla David, Céline Faucart, William Froger, Emmanuel Esperou, Guillaume Hervot, Romain Mazeau, and Khalil Rouibi. We thank Elisabeth Génot, Thomas Daubon, Isabel Egana, Filipa Curado, and Paolo Ciufici, all members of the Signal Transduction lab at the Institut Européen de Chimie et Biologie (IECB), Pessac, for monitoring the test sessions. Lena Tibell (University of Linkoping, Sweden) is acknowledged for her help with the test protocol. We thank Graham Johnson (University of California at San Francisco, San Francisco, CA), Paul Craig (Rochester Institute of Technology, Rochester NY), and Andrew Goldsborough (IECB, Pessac, France) for helpful comments and text corrections. We thank Rainer Bromme (University of Muenster, Germany), Ton de Jong (University of Twente, Netherlands), and Rob Goldstone (Indiana University, Bloomington, IN) for their stimulating email discussions.
REFERENCES
- Ainsworth S. The functions of multiple representations. Comput Educ. 1999;33:131–152. [Google Scholar]
- Bartholomé T, Bromme R. Coherence formation when learning from text and pictures: what kind of support for whom? J Educ Psychol. 2009;10:282–293. [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney RN, Levin JR. Pictorial illustrations still improve students’ learning from text. Educ Psychol Rev. 2002;14:5–26. [Google Scholar]
- Cook M, Wiebe EN, Carter G. The influence of prior knowledge on viewing and interpreting graphics with macroscopic and molecular representations. Sci Educ. 2008a;92:848–867. [Google Scholar]
- Cook M, Wiebe EN, Carter G. The interpretation of cellular transport graphics by students with low and high prior knowledge. Int J Sci Educ. 2008b;30:239–261. [Google Scholar]
- Craig P, Bateman R. 2010. A proficiency rubric for biomacromole-cular 3D literacy. PDB Newsletter Spring (45). www.rcsb.org/pdb/general_information/news_publications/newsletters/2010q1/education_corner.html (accessed 17 October 2012)
- Crisp V, Sweiry E, Ahmed A, Pollitt A. Tales of the expected: the influence of students’ expectations on exam validity and implications for writing exam questions. Educ Res. 2008;50:95–115. [Google Scholar]
- Dahmani H-R, Schneeberger P, Kramer IM. Analysis of students’ aptitude to provide meaning to images that represent cellular components at the molecular level. CBE Life Sci Educ. 2009;8:226–238. doi: 10.1187/cbe.09-03-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong T. Cognitive load theory, educational research, and instructional design: some food for thought. Instr Sci. 2010;38:105–134. [Google Scholar]
- Ealy JB. Students's understanding is enhanced through molecular modeling. J Sci Educ Technol. 2004;13:461–471. [Google Scholar]
- Gomperts, Kramer, Tatham . Signal Transduction. 2nd ed. New York: Academic Press/Elsevier; 2009. [Google Scholar]
- Goodsell DS, Johnson GT. Filling the gaps: artistic licence in education and outreach. PLoS Biol. 2007;12:e308. doi: 10.1371/journal.pbio.0050308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Peck RF, Colton S, Morris J, Chaibub E, Kallio J. A combination of hand-held models and computer imaging programs helps students answer oral questions about molecular structure and function: a controlled investigation of student learning. CBE Life Sci Educ. 2009;8:29–43. doi: 10.1187/cbe.08-07-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone AH. Why is science difficult to learn? Things are seldom what they seem. J Comput Assist Learn. 1991;7:75–83. [Google Scholar]
- Jucks R, Bromme R, Runde A. Explaining with nonshared illustrations: how they constrain explanations. Learn Instr. 2007;17:204–218. [Google Scholar]
- Kirkman TW. 1996 Statistics to Use. www.physics.csbsju.edu/stats (accessed 17 October 2012) [Google Scholar]
- Kosslyn SM, Ganis G, Thompson WL. Mental imagery and the human brain. In: Jing Q, Rosenzweig MR, d'Ydewalle G, Zhang H, Chen H-C, Zhang K, editors. Progress in Psychological Science around the World, vol. 1, Neural, Cognitive and Developmental Issues. New York: Psychology Press; 2006. pp. 195–209. [Google Scholar]
- Krebs EG, Fischer EH. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochem Biophys Acta. 1956;20:150–157. doi: 10.1016/0006-3002(56)90273-6. [DOI] [PubMed] [Google Scholar]
- Mayer RE. Systematic thinking fostered by illustrations in scientific text. J Educ Psychol. 1989;81:240–246. [Google Scholar]
- Mayer RE. Multimedia learning: are we asking the right questions? Educ Psychol. 1997;32:1–19. [Google Scholar]
- McNamara DS, Kintsch E, Butler-Songer N, Kintsch W. Are good texts always better? Interactions of text coherence, background knowledge, and levels of text understanding in learning from text. Cogn Instr. 1996;14:1–43. [Google Scholar]
- Nye LS. The minds’ eye: biomedical visualization: the most powerful tool in science. Biochem Mol Biol Educ. 2004;32:123–131. doi: 10.1002/bmb.2004.494032020337. [DOI] [PubMed] [Google Scholar]
- Orgil M-K, Crippen K. Teaching with external representations: the case of a common energy-level diagram in chemistry. J Coll Sci Teach. 2010;40:78–84. [Google Scholar]
- Ormrod JE. Human Learning, 5th ed. Upper Saddle River, NJ:: Prentice Hall, chap. 12, 350–390; 2008. Metacognition, self-regulated learning and study strategies. [Google Scholar]
- Palmer SE. Fundamental aspects of cognitive representation. In: Rosch E, Lloyd BB, editors. Cognition and Categorization. Hillsdale, NJ: Erlbaum; 1978. pp. 259–303. [Google Scholar]
- PDB Newsletter 2011. From 7 to 70,000: the PDB reaches a new milestone, Winter (48)
- Rall TW, Sutherland EW, Berthet J. The relationship of epinephrine and glucagon to liver phosphorylase: effect of epinephrine and glucagon on the reactivation of phosphorylase in liver homogenates. J Biol Chem. 1957;225:463–475. [PubMed] [Google Scholar]
- Runde A, Bromme R, Jucks R. Scripting in net-based medical consultation: The impact of external representations on giving advice and explanations. In: Fischer F, Kollar I, Mandle H, Haake JM, editors. Scripting Computer-Supported Collaborative Learning—Cognitive, Computational, and Educational Perspectives. New York: Springer; 2007. pp. 57–72. [Google Scholar]
- Schnotz W. Towards an integrated view of learning from text and visual displays. Educ Psychol Rev. 2002;14:101–120. [Google Scholar]
- Schnotz W, Kürschner C. A reconsideration of cognitive load theory. Educ Psychol Rev. 2007;19:469–508. [Google Scholar]
- Schönborn KJ, Anderson TR. A model of factors determining students’ ability to interpret external representations in biochemistry. Int J Sci Educ. 2009;31:193–232. [Google Scholar]
- Seufert T, Brünken R. Cognitive load and the format of instructional aids for coherence formation. Appl Cogn Psychol. 2006;20:321–331. [Google Scholar]
- Sprang SR, Acharya KR, Goldsmith EJ, Stuart DI, Varvill K, Fletterick RJ, Madsen NB, Johnson LN. Structural changes in glycogen phosphorylase induced by phosphorylation. Nature. 1988;336:216–221. doi: 10.1038/336215a0. [DOI] [PubMed] [Google Scholar]
- Sweller J. The redundancy principle in multimedia learning. In: Mayer R, editor. The Cambridge Handbook of Multimedia Learning. New York:: Cambridge University Press; 2005. pp. 159–167. chap. 10. [Google Scholar]
- Venien-Bryan C, Jonic S, Skamnaki V, Brown N, Bishler N, Oikonomakos NG, Boisset N, Johnson LN. The structure of phosphorylase kinase holoenzyme at 9.9 Å resolution and location of the catalytic subunit and the substrate glycogen phosphorylase. Structure. 2009;17:117–127. doi: 10.1016/j.str.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venien-Bryan C, Lowe EM, Boisset N, Traxler KW, Johnson LN, Carlson GM. Three-dimensional structure of phosphorylase kinase at 22 Å resolution and its complex with glycogen phosphorylase b. Structure. 2002;10:33–41. doi: 10.1016/s0969-2126(01)00691-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.