Abstract
Histo-blood group antigens (HBGAs) are a source of antigenic variation between individuals that modulates resistance and susceptibility to pathogens and is a barrier to the spread of enveloped viruses. HBGAs are also produced by a few prokaryotes where they are synthesized by glycosyltransferases (GTs) related to human HBGA synthases. Here we report the first structure of a bacterial GT of this family, from an intestinal resident, Bacteroides ovatus. Unlike its mammalian homologues and other GTs with similar folds, this protein lacks a metal-binding Asp-X-Asp motif and is fully active in the absence of divalent metal ions, yet is strikingly similar in structure and in its interactions with substrates to structurally characterized mammalian metal-dependent mammalian homologues. This shows how an apparently major divergence in catalytic properties can be accommodated by minor structural adjustments and illustrates the structural underpinnings of horizontal transfer of a functional gene from prokaryotes to vertebrates.
Mammalian genomes encode five GTs from Carbohydrate Active Enzymes (CAZy) family 6, a group of enzymes that catalyze the transfer of α-galactose (α-Gal) or α-N-acetyl-galactosamine (α-GalNAc) to the 3-OH group of a β-linked Gal or GalNAc in an acceptor substrate. These GTs include the histo-blood group (HBG) A and B synthases (known as GTA and GTB), an α-1,3-galactosyltransferase (α3GT) that catalyzes the synthesis of the xenoantigen or α-gal epitope, isogloboside 3 synthase (iGb3S) and Forssman glycolipid synthase (FS). α3GT, iGb3S and FS are inactive in humans because of missense and frame-shift mutations in their genes, while allelic variation in blood group genes results the production of various combinations of O, A and B antigens (HBGA) in different individuals. The lack of active α3GT and iGb3S results in the absence of α-Gal epitopes from human tissues, and presence of antibodies (about 1–3 % of the IgG) against the α-Gal epitope in the circulations of all humans while the lack of A and/or B antigen in individuals is associated with the presence of antibodies against the absent antigen. The production of antibodies against glycans produced by family 6 GTs is thought to result from exposure to these antigens on bacteria and enveloped viruses (reviewed by Brew et al1).
Vertebrate GT6 members are multi-domain membrane proteins with small N-terminal cytosolic domains, a transmembrane helix, a stem and a C-terminal catalytic domain. Their catalytic activities are completely dependent on divalent metal ions, particularly Mn2+. Crystallographic structures have been determined for the catalytic domains of human GTA and GTB2,3,4,5 and bovine α3GT in free states and in complexes with substrates and inhibitors6,7,8,9; structure-function relationships in these enzymes have also been investigated by extensive mutational studies10,11,12. GT6 members have a GT-A fold, one of the two predominant folds among GTs (GT-A and GT-B) and share, with most other GT-A fold GTs, a Asp-X-Asp (D-X-D) sequence motif that interacts with the metal cofactor and donor substrate (UDP-GalNAc or UDP-Gal).
BoGT6a is one of two GT6 paralogs encoded by the B. ovatus genome and is a representative of the GT6s that are found in at least 25 other Gram-negative bacterial species from four phyla (Firmicutes, Deferribacteres, Proteobacteria and Bacteroidetes) and genes from various unidentified bacterial species in the human gut metagenome. The bacterial GT6 have a N-terminal catalytic domain and a C-terminal membrane-association basic-hydrophobic-basic (BHB) domain1 suggesting that they associate with the bacterial cell membrane to facilitate their roles in the synthesis of the lipopolysaccharide O-antigen13. We have previously expressed a C-terminally truncated form of BoGT6a, lacking the membrane-binding region that is less prone to aggregation than the full-length protein14. Enzymatic characterization of this form of BoGT6a showed that it catalyzes the transfer of GalNAc from UDP-GalNAc to 2′-fucosyllactose (FAL) and similar molecules. Most significantly, BoGT6a was found to be fully active in the presence of EDTA and not stimulated by Mn2+ or Mg2+14. Since its homologues from other bacteria, with one exception, also have an Asn-X-Asn (N-X-N) sequence in place of the metal-binding D-X-D motif, it seems likely that these are also metal-independent. The exception is the GT6 from a member of the phylum Chlamydiae, Parachlamydia acanthamoebae, which has a D-X-D motif, lacks a BHB domain and is similar in size and sequence to a GT6 from a bacteriophage, the cyanophage PSSM-2. Numerous genes that encode GT6 homologues that resemble PSSM-2 and appear to be from other bacteriophages or marine bacteria are present in the Marine Metagenome database.
Here we present the crystal structure of the C-terminally truncated form of the metal-independent BoGT6a at high resolution and in a complex with an acceptor substrate 2′-fucosyllactose (FAL). The structure exhibits remarkable similarity to the catalytic domains of human GTA and GTB. Also, we have used isothermal calorimetry (ITC) to investigate the binding of the donor and substrates, UDP-GalNAc and FAL, respectively, and have developed a model of the complex of UDP-GalNAc with BoGT6a. P. acanthamoebae GT6 was expressed and found to have a similar substrate specificity to BoGT6a but requires divalent metal ions for activity. Finally, based on our experimental data, we discuss the role of the metal ion in members of the GT6 family and the evolutionary significance of metal dependence and independence.
Results
Overall structure of apo BoGT6a
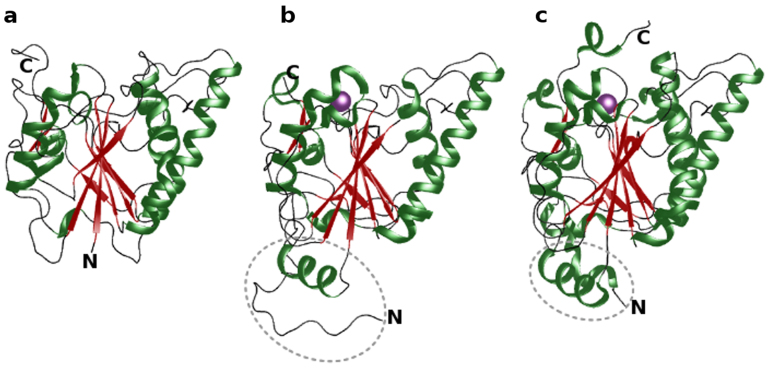
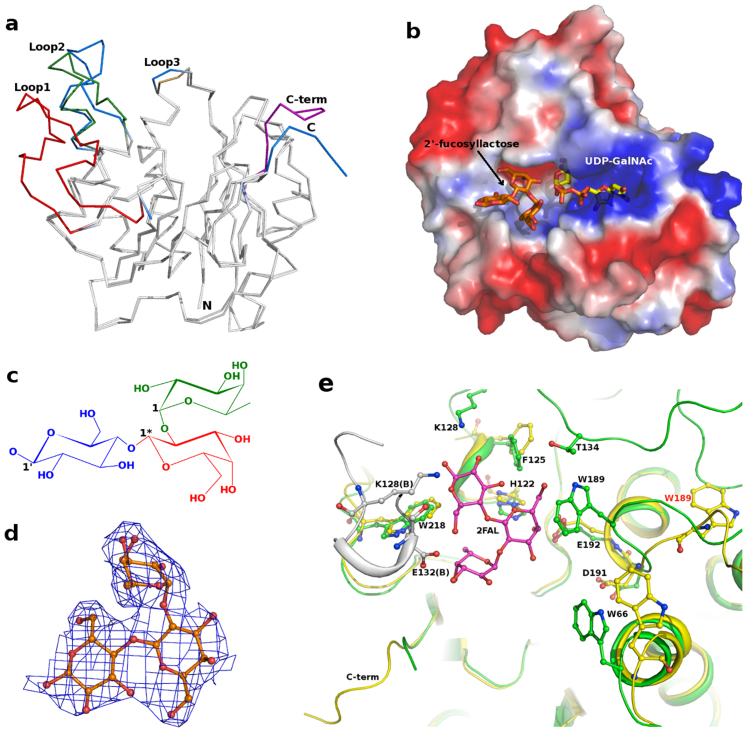
The structure of native apo-BoGT6a encompasses 10 β-strands, 4 α-helices and three 310 helices calculated using STRIDE15. The overall topology of BoGT6a is strikingly similar to those of characterized mammalian members of GT6 family with sequence similarity of 49% to the catalytic domains of ABO(H) blood group enzymes – GTA, GTB and α3GT (Fig. 1). It is truncated at the N-terminus relative to the catalytic domains of the mammalian enzymes by about 46 residues, a region that has been thought to be necessary for correct folding of α3GT16. The native structure of BoGT6a has a disordered region (residues 126–151) that could not be modeled because of lack of visible electron density. However, in the substrate-bound form (BoGT6a-FAL), electron density for this region is clearly visible and its structure is stabilized by inter- and intra-molecular interactions that were not observed in the native structure (discussed below). The root mean square deviation (r.m.s.d. calculated using TMALIGN17) for Cα atoms and all atoms between native BoGT6a and the four molecules (A, B, C and D) of the BoGT6a-FAL complex are 0.77–0.87 and 0.99–1.3 Å respectively. The flexible C-terminal segment beyond residue Ile228 undergoes a significant conformational change on binding the acceptor when compared with the native structure (Fig. 2a). Similarly there are conformational rearrangements of residues 66–67 and 175–190 associated with FAL binding. For further discussion, these regions are designated Loop1 (126–151), Loop2 (175–190), Loop3 (66–67) and C-term (228–236) (Figs. 2a, Fig. S1).
Figure 1. Cartoon representation of secondary structural elements of (a) BoGT6a (present study), (b) GTA (PDB id: 1ZI1) and c) α3GT (PDB id: 1GX4).
N- and C- terminus of the protein molecules are labelled. The N-terminal extension in GTA and α3GT are marked. The manganese ion in GTA and α3GT are shown as purple spheres.
Figure 2.
(a) Cα superposition of BoGT6a and BoGT6a-FAL structures.Structural variations in loop regions Loop1 (126–151) in red, Loop2 (175–190) in forest green, Loop3 (66–67) in sand, C-term (228–234/236) in purple of BoGT6a-FAL are shown. BoGT6a loops and C-term are coloured in marine blue; (b) Surface potential charge representation of BoGT6a with the bound FAL and modelled UDP-GalNAc molecule; (c) Chemical structure of FAL. Only the 1st carbon atom positions are labelled for each of the monomeric unit; (d) Observed electron density [(2Fo-Fc) map contoured at 1.0σ] for the bound FAL in the structure of the complex; (e) Acceptor binding site of BoGT6a with interacting residues and ligand FAL (2FAL) shown as ball-and-stick model. The position of Trp189 both in native BoGT6a (in yellow) and BoGT6a-FAL complex (in green) is shown and labeled in red (BoGT6a) and in black (BoGT6a-2′-fucosyllactose). The reorientation of this loop (Loop2) stabilizes Loop1 that shares hydrogen bonding interactions with the bound FAL. Residues interacting with 2′-fucosyllactose from molecule B are coloured in grey.
The native structure of BoGT6a showed the presence of three ions (presumably from the crystallization buffer), a Ca2+ ion located close to the C-terminus of the protein, interacting with Glu216, Asp230 and Asn232, a Cl− ion bound by a group of solvent molecules in a loop between Met29 and Phe34. The ions are distant from the N-X-N motif (~19 and 14 Å for Cl− and Ca2+ ions respectively). In addition, one bound HEPES molecule and some 200 water molecules were identified in the structure (Table 1).
Table 1. Crystallographic data for BoGT6a and BoGT6a-2′-fucosyllactose (FAL) complex.
| BoGT6a | BoGT6a-FAL | |
|---|---|---|
| Space group | P4322 | P21 |
| Number of molecules per asymmetric unit | 1 | 4 |
| Cell dimensions | a = 41.23 Å, b = 41.23 Å, c = 282.9 Å | a = 70.85 Å, b = 93.87 Å, c = 75.51 Å |
| α = β = γ = 90° | β = 93.8° | |
| Resolution range (Å) | 33.32–1.91† | 70.69–3.00 |
| Rsymma (outer shell) | 0.075 (0.14)†† | 0.13 (0.49) |
| I/σI (outer shell) | 18.1 (4.5) | 9.3 (2.6) |
| Completeness (outer shell) % | 77.9 (22.2) | 94.1 (93.7) |
| Total no. of reflections | 600206 | 72365 |
| Unique no. of reflections | 20120 | 18641 |
| Redundancy (outer shell) | 4.3 (1.6) | 3.9 (3.8) |
| Wilson B-factor (Å2) | 20.0 | 47.78 |
| Rcrystb/Rfreec | 17.91/23.19 | 18.07/26.19 |
| Average B-factor (Å2) | ||
| Overall | 14.0 | 36.6 |
| Protein | 10.9 | A: 30.5, B: 35.2, C: 41.7, D: 38.8 |
| Ligand | 27.1 (HEPES); Cl− (10.8); Ca2+ (13.3) | 36.2 (FAL) |
| Water | 19.1 | — |
| RMSD values | ||
| bond length (Å) | 0.008 | 0.009 |
| bond angle (°) | 1.114 | 1.429 |
| Ramachandran plot statistics (%) | ||
| Favoured | 98.6 | 96.7 |
| Additionally allowed | 1.4 | 3.3 |
| PDB code | 4AYL | 4AYJ |
aRsymm = ΣhΣi|I(h) − i(h)|/ΣhΣiIi(h), where Ii(h) and I(h) are the ith and the mean measurements of the intensity of reflection h, respectively.
bRcryst = Σh|Fo − Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h, respectively.
cRfree is equal to Rcryst for a randomly selected 5.0% subset of reflections not used in the refinement.
†Reflections up to 1.91 Å resolution were used in all crystallographic calculations.
††Due to low Rsymm value, resolution cutoff of 1.91 Å was used and the crystal diffracted up to 1.7 Å. Data completeness figures at 2.1 Å are – overall and last shell (2.18–2.1 Å) are 90.5 and 67.6% respectively.
Comparison of BoGT6a with other members of the GT6 family
BoGT6a is closely similar in structure to GTA (PDB id: 1ZI118), GTB (PDB id: 1R7U19) and α3GT (PDB id: 1GX46) (Fig. 1) with overall r.m.s.d.17 values of 1.96 (179 Cα atoms), 1.96 (180 Cα atoms) and 2.15 Å (200 Cα atoms) respectively. At the sequence level BoGT6a is 49%, 48% and 48% similar to GTA, GTB and α3GT respectively. The four molecules of the BoGT6a-FAL structure superimpose on GTA and GTB with an overall r.m.s.d. value of 1.6 Å and with α3GT, the r.m.s.d. values range from 1.75 to 2.0 Å. The principle differences in r.m.s.d. values between BoGT6a and the BoGT6a-FAL complex and their mammalian homologues are mostly attributable to the conformational changes in Loop2, Loop3, and the C-terminus of the molecule.
Conformational changes associated with acceptor (FAL) binding and implications for acceptor specificity
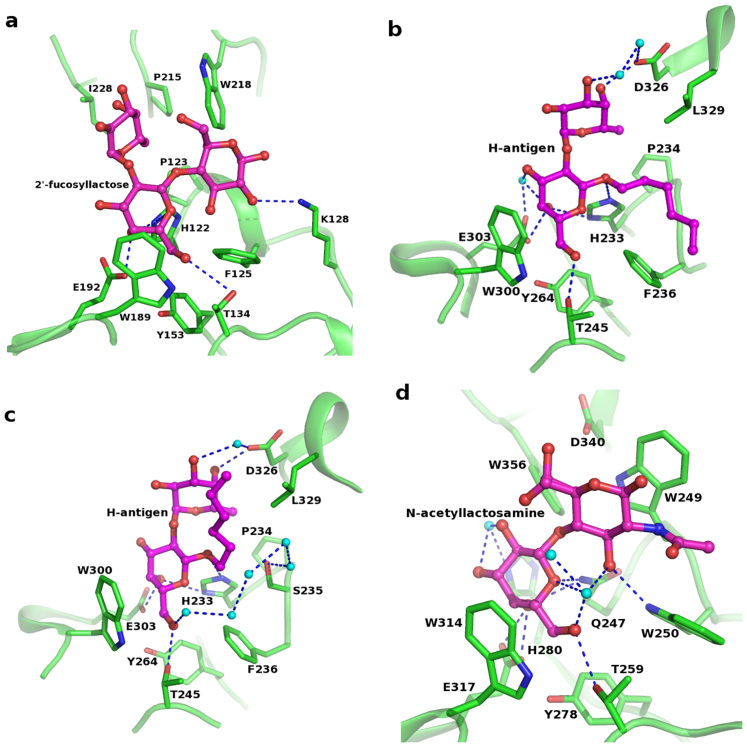
In the complex of BoGT6a with FAL, the acceptor substrate binds in a pocket (Figs. 2b–c) and interacts with residues in the C-terminal half of the polypeptide chain. The complex is highly ordered, as indicated by the clear electron density for the substrate (Fig. 2d), and includes a major conformational change in the C-terminal region of BoGT6a (compared with the native), which adopts a ‘partially closed’ conformation (Fig. 2e). The acceptor molecule interacts with the protein through a network of H-bonds with His122, Thr134 and Glu192 (from molecule A) and Lys128 and Glu132 (from molecule B) (Fig. 2e, Table 2). Corresponding residues from molecules C and D are also involved with FAL binding but display subtle variations in their interactions. This indicates that the interaction of molecule B with the ligand bound to molecule A (and molecule C and D) of BoGT6a is a crystallographic artifact. Hydrophobic interactions, particularly with the aromatic side chains of Phe125, Trp189 and Trp218, are also important for FAL binding. The amino acid sequence alignment in Fig. S1 shows the locations of the flexible loops and residues that interact with substrates and highlights the high levels of sequence identity between the prokaryotic and mammalian GT6. Several residues that interact with FAL are conserved between BoGT6a and its closest mammalian homologues GTA and GTB including His122, Thr134, Tyr153, Trp189 and Glu192 (Fig. S1 and Fig. 3). In α3GT which, unlike GTA/B and BoGT6a, binds acceptor substrates that lack a 2′-fucosyl moiety, there are substitutions for acceptor-binding residues that can be linked to its specificity: His122 is substituted by Gln and both Gly 124 and Ile 228 by Trp (residues marked as A1, A2 and A7 in Fig. S1). Trp249 (A2) in α3GT interacts with the non-polar B-face of glucose in lactose, undergoing a large conformational change to accommodate the acceptor and being stabilized in its new location by a H-bond with Asp340. A similar conformational change is not required in either BoGT6a or GTA/B as the corresponding residue is Gly in BoGT6a and GTA, and Ser in GTB (Fig. S1 and Fig. 3a–c). The residue at position A7 is also a key determinant of acceptor substrate specificity; the smaller side chains in GTA/B and BoGT6a (Ala and Ile, respectively) allow the accommodation of the 2′-fucose of the acceptor substrate (Fig. 3a–c) but Trp356 of α3GT does not (Fig. 3d).
Table 2. Hydrogen bonding interactions between BoGT6a and FAL. The distances given are the range observed in all four molecules of BoGT6a-FAL. Protein atom labels italicised are from a symmetry related molecule. Atom labels are as shown in Figure 2.
| Protein Atom (BoGT6a) | Ligand Atom (2′-fucosyllactose) | Distance (Å) |
|---|---|---|
| O5* | 3.28–3.41 | |
| His 122 NE2 | O4* | 2.31–2.83 |
| Lys 128 NZ | O2′ | 3.06–3.39 |
| Thr 134 OG1 | O6* | 3.28–3.45 |
| Trp 189 NE1 | O6* | 2.84–3.33 |
| Glu 192 OE1 | O4* | 2.41–3.21 |
| Glu 192 OE2 | O4* | 2.64–3.21 |
| O1′ | 2.56–3.46 | |
| Lys 128 NZ | O2′ | 3.27–3.39 |
| O5′ | 3.13–3.43 | |
| Glu 132 OE2 | O6′ | 2.71–3.20 |
Figure 3. Acceptor binding pocket among GT6 family members showing the conserved residues and interactions.
(a) BoGT6a in complex with FAL; (b) GTA in complex with H-antigen (PDB id: 1LZI); (c) GTB in complex with H-antigen (PDB id: 1LZJ); (d) α3GT in complex with LacNAc (PDB id: 1GX4). Water molecules are shown as cyan spheres.
Glu192 (marked A5 in Fig. S1) forms H-bonds with FAL and is projected to also H-bond with the donor substrate. It is conserved in all known members of the GT6 family and has been shown to have a key role in catalysis in both metal-dependent and metal-independent GT6. Previous studies have shown that substitution of Glu192 by Gln in BoGT6a results in a 2 × 104 reduction in kcat for GalNAc transfer to FAL14. The highly conserved Trp189 (Fig. S1), interacts with the acceptor substrate in all structurally characterized GT6 members (Fig. 3).
Overall, on binding the acceptor substrate, Loop1 is stabilized by at least three hydrogen bonds through residues Lys128 and Thr134. Loop2 undergoes a major shift, affecting Trp189, that forms a stacking interaction with the bound FAL.
The C-terminal region of BoGT6a, like the C-terminus of α3GT undergoes a major conformational change (between open and closed conformations) upon ligand binding9. Lys231 of the C-terminal region of BoGT6a in one of the four molecules in the asymmetric unit interacts with the fucose moiety of FAL through hydrogen bonds. The equivalent residue in α3GT is Lys359, which directly interacts with the phosphate moiety of the bound product, UDP (Fig. S2), in the closed state and through a water mediated hydrogen bonding interaction with the acceptor substrate, N-acetyllactosamine6. Mutational and structural studies have shown that Lys359 and Arg365 of α3GT play an important role in catalysis6,9 and Lys231 of BoGT6a is also functionally important since the Lys231Ala mutation in BoGT6a generates a >25-fold reduction in kcat14.
Donor substrate (UDP-GalNAc) interactions
We have unsuccessfully attempted to grow crystals of BoGT6a and its low-activity Glu192Gln mutant in a complex with UDP-GalNAc. However, we were able to investigate the binding of the substrates, UDP-GalNAc and FAL by isothermal titration calorimetry (ITC) in HEPES buffer, pH 7.5 containing 1 mM DTT and 0.2 M NaCl. The heat released was measured as aliquots of ligand were added successively to solutions of the enzyme (1.433 ml, 13 μM) in the stirred reaction cell at 25°C. A binding isotherm was generated by plotting the integral of the heat of binding against the concentration of ligand which can be analyzed as described to calculate the association constant (Ka), enthalpy of binding (ΔHobs), stoichiometry of binding (N) and entropy of binding (ΔS), assuming a 1:1 stoichiometry of binding (see Methods for discussion and details).
The ITC studies with wild-type BoGT6a show that it binds UDP-GalNAc with high affinity in the absence of metal ions and H-antigen (Table 3, Fig. S3). Analysis of the ITC data, with the assumption of 1:1 stoichiometry, gives a Kd value for UDP-GalNAc (59 μM) in good agreement with the Ki value determined by steady state kinetics (67 μM) supporting the basis of this analysis. The results also show that the binding of UDP-GalNAc is enthalpy-driven with a highly favourable (negative) enthalpy change (−23.5 kcal/mol) partially balanced by an unfavourable entropy change (−TΔS of 17.7 kcal/mol). BoGT6a catalyzes UDP-GalNAc hydrolysis at low rate (<0.1% of transferase activity14), which could contribute to the measured heat of UDP-GalNAc binding. To determine if this could affect the thermodynamic profile for this interaction, we carried out an ITC study of UDP-GalNAc binding to a very low activity mutant of BoGT6a (E192Q14). Although the (negative) enthalpy of binding was less (by about 7 kcal/mol), there was a similar reduction in the negative entropy contribution to binding, resulting in an essentially unchanged Ka and ΔG of binding. This indicates that UDP-GalNAc hydrolysis does not affect the general conclusions regarding UDP-GalNAc binding. ITC studies of FAL binding to wild-type BoGT6a indicated weak, low affinity binding. However, the addition of 3 mM UDP in the protein and ligand solutions (11-fold greater than the Kd) resulted in a stronger signal and affinity, reflected in a Kd of 77 μM (ΔG = −5.6 kcal/mol) and a negative enthalpy change of −15.4 kcal/mol partially compensated by an unfavorable entropy of binding (−TΔS of 9.8 kcal/mol). This suggests that the binding of UDP may increase the strength of FAL binding either by inducing a conformational change or by direct interactions of the β phosphate O- of the bound UDP with the acceptor 3 –OH as in α3GT21. Overall, ITC studies indicate that the binding of donor and acceptor substrates to BoGT6a is not ordered, but appears to be synergistic since binding of a donor substrate analog enhances the binding of acceptor. Along with kinetic studies that indicate a sequential mechanism (both substrates bind prior to catalysis) this suggests (but does not prove conclusively) that BoGT6a may have a random sequential mechanism14.
Table 3. Thermodynamic parameters for the binding of substrates and substrate analogues to BoGT6a. Titrations were conducted at 25°C (298 K) with 13 μM BoGT6a in 20 mM HEPES buffer pH 7.5 containing 0.2 M NaCl and 1 mM DTT.
| Ligand | UDP-GalNAc E192Q variant | UDP-GalNAc WT | FAL | FAL |
|---|---|---|---|---|
| Other ligands | UDP (3 mM) | |||
| Ka (*10−3) | 18.0±0.4 | 17.8±0.5 | 0.81±0.02 | 13.02±0.24 |
| ΔG (kcal/mol) | −5.78±0.02 | −5.80±0.02 | −3.97±0.03 | −5.61±0.01 |
| ΔH (kcal/mol) | −16.4±0.2 | −23.5±0.3 | −13.5±0.3 | −15.4±0.1 |
| TΔS (kcal/mol) | −10.6±0.2 | −17.7±0.3 | −9.6±0.7 | −9.8±0.1 |
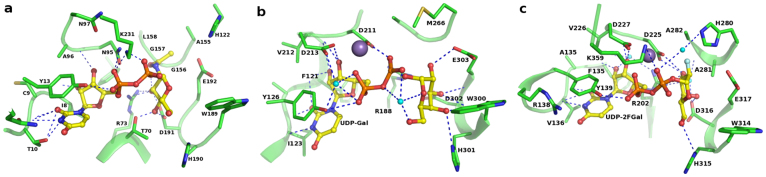
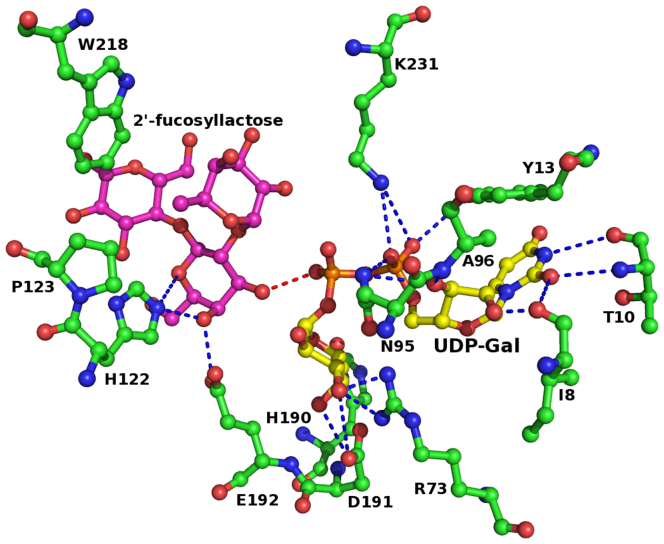
Models of complexes of UDP-Gal and UDP-GalNAc with BoGT6a were generated by molecular docking of UDP-Gal and UDP-GalNAc using α3GT in complex with UDP (PDB: 1GX46), UDP-Gal (PDB: 2VS511), as templates and UDP-GlcNAc extracted from its complex with MurG (PDB: 1NLM22). Models of BoGT6a docked to UDP-Gal and UDP-GalNAc were energy minimized using the CNS software suite23; the modeled complexes with UDP- GalNAc and UDP-Gal shows that the mode of donor substrate binding in BoGT6a is closely similar to those of α3GT and GTA/GTB (Figs. 4,5), emphasizing the high level of conservation of donor substrate binding site, and active site residues in general between the bacterial and mammalian GT6 family members. The residues are shown in ball-and-stick representation in Figure 5 and exhibit H-bonding interactions to the donor substrate through their main-chain or side-chain atoms (Table 4). Mutational studies with BoGT6a have shown that residues A155GGL158 (Fig. 4a) are critical for its specificity for UDP-GalNAc vs UDP-Gal14. The corresponding residues in α3GT (Fig. 4c) are H280AAl283 and screening combinatorial libraries with mutations in this region identified a mutant of α3GT with the sequence A280GGL283 that has greatly enhanced GalNAc transferase activity10. The two substitutions in the corresponding region between GTA and GTB are responsible for their respective specificities for UDP-GalNAc and UDP-Gal24. Other residues of BoGT6a: Ile8, Thr10, Thr70, Arg73, Asn95, His190 and Asp191 appear to form H-bonds with the UDP-GalNAc. Of these residues the main chain atom O of Ile8, the main chain and side chain atoms of Thr10 and side chain atoms OD1 and ND2 of Asn95 directly H-bond with the UDP moiety (Fig. 4a). Residues Thr70, Arg73, His190 and Asp191 interact through multiple H-bonds with the GalNAc or Gal moiety of the donor substrate. Asn95 interacts with the ribose and phosphate moiety of the UDP molecule. Mutation of Asn95 of BoGT6a to Asp has been shown to result in a 36-fold increase in the Km for UDP-GalNAc and a >4000-fold decrease in kcat whereas the substitution of Asp for the Asn97, has only minor effects on the Km and kcat14.
Figure 4. Donor binding sites of (a) BoGT6a – UDP-GalNAc molecule modeled; (b) GTB-UDP-Gal (PDB id: 1ZJ1); (c) α3GT-UDP-2FGal (PDB id: 1GX4).
The manganese ion is shown as a purple sphere and water molecules as small spheres in cyan.
Figure 5. Catalytic site of BoGT6a showing the bound acceptor molecule FAL (magenta) and modeled UDP-Gal (yellow) as ball-and-stick model.
Interacting residues are shown as ball-and-stick model and coloured in green.
Table 4. Potential hydrogen bond interactions between UDP-Gal and BoGT6a in their modeled complex.
| Protein Atom (BoGT6a) | Ligand Atom (UDP-Gal) | Distance (Å) |
|---|---|---|
| Ile8 O | O2 | 3.45 |
| Thr10 O | N3 | 3.23 |
| Thr10 N | O2 | 2.84 |
| Tyr13 OH | O2A | 2.97 |
| Asn69 ND2 | O4 | 3.02 |
| N3 | 3.27 | |
| Arg73 NH2 | O3′ | 2.48 |
| Asn95 OD1 | O3D | 2.85 |
| O1A | 2.61 | |
| Asn95 ND2 | O3′ | 3.09 |
| O2′ | 2.91 | |
| Ala96 N | O3D | 2.72 |
| Gly156 O | O3′ | 3.21 |
| Gly157 N | O2′ | 3.19 |
| His190 NE2 | O6′ | 2.99 |
| Asp191 OD2 | O4′ | 3.41 |
| Asp191 OD1 | O4′ | 2.54 |
| Glu192 N | O4′ | 3.07 |
| K231 NZ | O1A | 3.35 |
| O2A | 3.41 |
GT6 from parachlamydia acanthamoebae, halls coccus (PaGT6)
Based on structures and molecular phylogeny analyses the GT6 family form three clades (1) prokaryotic enzymes with a minimal catalytic domain and a D-X-D motif represented by the cyanophage PSSM-2 enzyme, its Marine Metagenome homologues and PaGT6 (2) the multi-domain vertebrate enzymes and (3) the bacterial enzymes with C-terminal membrane binding BHB domains and N-X-N replacing D-X-D. An array of evidence supports the notion that the evolution of the GT6 family included horizontal gene transfer (HGT) between prokaryotes and an ancestor of the vertebrates1, raising the question about the route(s) of gene transfer. HGT is facilitated by donor and recipient having a shared environment25 as exemplified by the array of GT6-positive bacterial species that are symbionts or pathogens of various vertebrates and the many GT6 sequences in the Marine Metagenome that are probably from uncharacterized cyanophage or cyanobacteria. P. acanthamoebae is an endosymbiont that can infect humans as well as acanthamoebae, and the simpler structure of PaGT6 and other group (2) members identifies them as models for the ancestor of the GT6 family that have characteristics expected in vectors for HGT. To investigate its functional properties we cloned and expressed PaGT6 as a soluble His-tagged Sumo fusion construct. The protein, purified by Ni2+-chelate column chromatography migrated as a single band with an apparent molecular weight of 40 kDa, consistent with the size expected from it sequence (PaGT6 27 kDa plus the Sumo-His tag of 12 kDa).
As in previous studies14 glycosyltransferase activity measurements were conducted using a radiochemical assay with UDP-[3H]-Gal and UDP-[3H]-GalNAc as potential donor substrates and H-antigen trisaccharide (2′-fucosyl-N-acetyl lactosamine), FAL and N-acetyl lactosamine as possible acceptor substrates in the presence and absence of Mn2+ or Mg2+. These showed that PaGT6 catalyzes the transfer of GalNAc from UDP-GalNAc to H-antigen trisaccharide or FAL in the presence of either Mn2+ or Mg2+, with Mg2+ giving higher activity. It was inactive in the absence of metals and with LacNAc as acceptor substrate. A detailed study of its properties will be reported elsewhere.
Discussion
The metal-independent (bacterial) and metal-dependent (vertebrate, phage and Parachlamydia) members of GT6 provide an example of functional divergence in closely similar proteins. The GT6 family consists of retaining GTs that catalyze a reaction in which the α-configuration of the transferred carbohydrate in the donor substrate is also present in the product. Based on structural and other evidence it appears that GTs of this type employ a SNi mechanism involving the formation of a short-lived carbocation transition state in which the incoming nucleophile (acceptor) is activated by the UDP leaving group26; this contrasts with the double displacement mechanism utilized by retaining glycosidases, enzymes that catalyze an analogous reaction. In the better-characterized metal-dependent enzymes the metal ion functions as a Lewis acid catalyst, facilitating the departure of the UDP leaving group, but also functions in donor substrate binding and may have a role in moving the departing group in the active site during catalysis26,27.
In general, GT-A fold GTs from different families are metal-dependent and have degenerate metal binding motifs similar to D-X-D. The exceptions are some sialyltransferases (STs) and the retaining GT, core 2 β-1,6-N-acetylglucosaminyltransferase (C2GnT;C2GnT-L), a member of GT family 1428. STs are structurally heterogeneous and include members of five different GT families that have both GT-A and GT-B folds29; they are all metal-independent but their donor substrates differ in being a monophosphate derivative (a CMP-sialic acid) as opposed to diphosphate (e.g. UDP)29. Crystallographic studies of complexes of core 2 β-1,6-N-acetylglucosaminyltransferase with substrates together with mutational studies suggest that basic residues close to the C-terminus of the catalytic domain function in donor substrate binding and catalysis and may have a similar role to the metal ion in metal-dependent GTA-fold GTs28. Together, these GTs reinforce the perspective that GTs with GTA folds can be metal-dependent or metal-independent, however, the GT6 family differs from other GTA fold families in encompassing both metal-dependent and metal-independent forms.
The close similarity between the active site structure and substrate interactions of BoGT6a and the metal-dependent mammalian GT6 members suggests that they have similar catalytic mechanisms, generating uncertainty regarding the role of the metal ion in catalysis. There is an obvious link between metal binding (and metal dependence) and the D-X-D sequence. Mutational data are consistent with the general features of our model of the UDP-GalNAc complex with BoGT6a, since mutation of Asn95 of the N-A-N motif to Asp, disrupted donor substrate binding and greatly reduced kcat. Metal-independence appears to be linked, at least in part, to the replacement of the aspartates of the D-X-D motif in the metal-dependent GT6s, by asparagine, eliminating charge repulsion between the phosphates of the donor substrate and the aspartyl side chain carboxyls. In the modeled BoGT6a complex with UDP-GalNAc, the amide of Asn95 H-bonds with O2 of the α-phosphate, in keeping with the effects of mutating this residue to Asp. Our mutational studies have shown substitution of Ala for Lys231, of the C-terminus of BoGT6a reduces kcat by 26-fold, whereas the same substitution for Arg229, produces only a 3-fold loss. This supports the interaction of Lys231 with the donor substrate as shown in Fig. 5. Thus, in relation to substrate binding, interactions of the phosphates of the donor substrate with the amide moiety of Asn95 may replace those with the metal ion. The role of the metal ion in stabilizing the UDP leaving group could be replaced by interactions with basic residues near the C-terminus, particularly Lys231. The existence of homologous metal-independent and metal-dependent groups raises the question whether metal dependence and independence has some adaptational significance. There are interesting parallels between the GT6 family and 3-deoxy-D-manno-octulosonate-8-phosphate synthase (KDO8PS), a bacterial enzyme that catalyzes the synthesis of a precursor of the endotoxin of Gram-negative bacteria. There are two subdivisions of the KDO8PS family that are dependent on a divalent metal (Zn2+ or Fe2+) for activity or are metal-independent. The metal-dependent forms bind the metal ion through the side chains of a cysteine, a histidine and a glutamate, whereas in the metal-independent enzymes, the cysteine is replaced by asparagine30. It is interesting that metal- dependence and metal-independence can be conferred by substituting Asn for the metal-binding Cys and vice versa, although full catalytic activity can require up to three additional substitutions31,32,33. It seems possible that in GT6, as well as KDO8PS, the metal ion may function only in substrate binding.
In summary, BoGT6a is surprisingly similar to human GTA in structure, and also utilizes precisely equivalent residues for binding substrates, despite a major functional divergence from vertebrate GT6s in having metal-independent catalytic activity. The results of molecular phylogeny analyses indicate that a metal-dependent prokaryotic GT6, typified by those of P. acanthamoebae GT6 and cyanophage PSSM-2, was the ancestor of the vertebrate clade through horizontal gene transfer1. BoGT6a and its homologues in other bacterial symbionts and pathogens in humans catalyze the synthesis of histo-blood group antigens on the bacterial surface that potentially contribute to the generation of antibodies against non-self histo-blood groups. The GT6 gene products in bacteria and vertebrates have acquired some structural adaptations required for the expression of their biological functions. One is the attachment to membranes, through the acquisition of a C-terminal “BHB” membrane-association domain and N-terminal membrane-insertion and stem domains, respectively. Also, the metal-independence of the bacterial enzymes may be an adaptation to low availability of divalent metals in the cytosol of Gram-negative anaerobes.
Methods
Protein expression and purification
The N-terminally His-tagged form of BoGT6a was expressed in E. coli BL21(DE3) cells and purified as previously reported14 The purified protein was dialyzed against 20 mM Tris-HCl, pH 7.9, containing 0.1 M NaCl and 2 mM dithiothreitol (DTT); 10 mM EDTA was added to the buffer for storage.
P. acanthamoebae genomic DNA was utilized as a template for PCR using primers designed to amplify the GT6 gene with additional sequences on the 5′and 3′ end for cloning into the Expresso T7 SUMO peTite vector (Lucigen).
Forward: 5′-CGCGAACAGATTGGAGGTTTGTTATTTAGCCACTCTCTTTAT G-3′
Reverse: 5′-GTGGCGGCCGCTCTATTATTATCCCTTTCGCATTTCTTCATG-3′
Cloning and transformation into BL21 (DE3) E. coli cells was performed following the instructions of the supplier. BL21 cells were grown to OD600 of 0.5–1.0 and induced with 10 mM IPTG. Soluble protein was isolated and SUMO-HIS-GT6 protein was purified by Ni2+ chelate chromatography using Ni2+-NTA Super Resin (Qiagen) with elution by 150 mM Imidazole. The purified protein (2 mg/litre of culture) was dialyzed against 20 mM Tris, 0.2 M NaCl pH 7.5.
Crystallization, X-ray data collection and processing
Native BoGT6a was concentrated to 6–8 mg/ml prior to crystallization. Both the native and FAL complex were crystallized using the sitting drop vapor diffusion method with a Phoenix crystallization robot on a 96 well Intelli-plate (Art Robbins Instrument). The drops were set up at a 1:1 ratio of protein to mother liquor and incubated at 16°C. Single crystals of native (apo-protein) BoGT6a grew in well solution containing 0.1 M Tris, pH 7.5, 0.2 M calcium acetate and 15% (w/v) PEG 4000 of crystallization screen Clear Strategy Screen 2 (Molecular Dimensions Ltd., UK). To form the BoGT6a-FAL complex, BoGT6a (4 mg/ml) and FAL (10 mM) were incubated overnight at 16°C and crystals were grown in a well solution containing 20% PEG 3350, 0.1 M Bis-Tris propane, pH 8.5 and 0.2 M Na/K tartrate of crystallization screen Pact Premier (Molecular Dimensions Ltd., UK).
Diffraction datasets for native BoGT6a (to 1.91 Å) and the BoGT6a/FAL complex (to 3 Å) were recorded at Diamond Light Source (Didcot, Oxon-UK) on station I02 at 100 K. Cryo cooling was achieved by stabilizing the crystals (prior to X-ray data collection) in 25% PEG 4000 and 30% PEG 3350 for the native and FAL complex crystals respectively. Even though the native crystals diffracted to 1.7 Å resolution, because of the long c-axis the data were recorded using thin oscillation width (ΔΦ) of 0.1° and 0.07°. The crystal was stable in the X-ray beam and over 1000 images were recorded to obtain a complete dataset at high resolution. The native dataset was processed using HKL2000 suite34 in primitive tetragonal space group (P4322, a = b = 41.2 Å, c = 282.9 Å), while the dataset for the BoGT6a-FAL complex was processed using iMosflm35 in primitive monoclinic space group (P21, a = 70.7 Å, b = 93.6 Å, c = 75.4 Å, β = 93.9°) (Table 1).
Structure determination
Initial molecular replacement trials of structural solutions were carried out with PHASER36, MOLREP37 and AMoRe38 software suites from the CCP4 package39 using a BoGT6a homology model built using GTA (PDB: 1WSZ3) and α-1,3-galactosyltransferase (α3GT) (PDB: 2JCO9) as templates, but these failed in all crystal systems.
Further analysis of the data, processed in a primitive triclinic system using the Pointless Suite40, suggested that the crystal form might be primitive tetragonal. Automated model building and refinement trials were performed, for the data processed in primitive tetragonal space group, using MrBump41, which uses CHAINSAW42 and/or MOLREP37 as the model editing tool. The solutions were ranked based on Rfree values, which indicated that the possible space group is either P43212 or P4322. These refined models were selected for input to PHASER36, which gave the best molecular replacement solution with higher log-likelihood gain values for space group P4322 with one molecule per asymmetric unit. However, all the molecular replacement solutions froze at a crystallographic R-factor (Rcryst) value of ~40% during refinement.
A careful analysis of the structure-based sequence alignment of the template against the preliminary refined model indicated that certain loops had higher thermal values that could affect the refinement process. These loops which spanned residues 65–72, 112–116, 124–156, 181–188 and 219–226 were removed from the model. The edited model was fed into ArpWarp classic43 for automated loop building and fitting into electron density map of 2 chains with a total of 206 residues. After several cycles of ArpWarp classic run43 and restrained refinement the Rcryst and Rfree values dropped to 0.235 and 0.303 respectively. Several additional cycles of manual model building and refinement were carried out using COOT44 and PHENIX software45 suites respectively (Table 1).
Phases for the BoGT6a-FAL complex data were calculated by the molecular replacement method (using the fully refined native BoGT6a structure as the starting model) in space group P21 with four molecules in the asymmetric unit. Clear electron density was observed for the acceptor substrate FAL and inserted in to the structure. Further refinement and model building were carried out using PHENIX software45 suite and COOT44 (Table 1). In spite of moderate resolution, the structure is highly ordered and the information about the structural changes due to acceptor molecule recognition is quite evident in all four copies of the asymmetric unit.
Isothermal titration calorimetry (ITC)
Solutions of wild-type BoGT6a and its E192Q mutant14 (12–30 μM) were dialyzed extensively against HEPES buffer (20 mM), pH 7.5 containing 250 mM NaCl and 1 mM DTT and degassed prior. Isothermal titrations were performed at 25°C with ligands dissolved in the same buffer, UDP-GalNAc (3 mM), and FAL in the presence or absence of UDP (3 mM) using a MicroCal VP-ITC micro calorimeter. During each titration, 16 aliquots (10–20 μl) of ligand solution were injected, each over 16 sec, spaced at 300 sec intervals. The stirring speed was 200 rpm. Heats of binding were determined by integrating the signal from the calorimeter, and binding isotherms were generated by plotting the heats of binding against the ratio of enzyme to inhibitor. The software package Origin 5.0 from Microcal Inc. was used to calculate the association constant (Ka) enthalpy change (ΔH) and stoichiometry (N). This analysis presumes that the parameter c ( = [MT]/Kd, where [MT] is the concentration of enzyme) is between 1 and 100020. This condition was not attained for the interactions investigated here (c ≤ 0.35), as in previous studies with bovine α3GT20, because of the limitations of enzyme solubility under these experimental conditions. However, it was possible to increase the concentration of ligand sufficiently to saturate the enzyme. Also, a value of 1.0 for N is supported by structural data for FAL and is reasonable to assume for UDP-GalNAc based on structural data from homologous glycosyltransferases. The free energy of binding (ΔG) and entropy contribution to binding (TΔS) was calculated from the relationships ΔG = −RTLn(Ka) and TΔS = ΔH−ΔG respectively.
Glycosyltransferase assays
Glycosyltransferase activities of PaGT6 (2 μg) were measured using 3H-UDP-GalNAc as the donor substrate in radioassays as previously described14 with the following potential acceptor substrates- a) 1 mM FAL (Accurate Chemical), b) 1 mM blood group H type II trisaccharide (V-labs) and c) 3 mM N-acetyl-lactosamine.
Author Contributions
N.T. and T.T.K.P. carried out all the crystallographic work. A.S. grew the crystals for X-ray diffraction work. P.T. prepared the protein. K.R.A. and N.T. analyzed all the structural data. B.S. and M.L.-W. prepared and purified the proteins. B.S. carried out the ITC studies and data analysis while M.L.-W performed all the enzyme activity measurements and data analysis. K.B. and K.R.A. wrote the manuscript. K.R.A. and K.B. conceived the study and supervised the work. All authors reviewed the manuscript.
Supplementary Material
Supplementary File
Acknowledgments
This work was supported by a Wellcome Trust (UK) equipment grant (number 088464) to K.R.A. We thank Professor Eleanor Dodson (University of York, UK) for help with ‘molecular replacement’ used in the structure determination of apo BoGT6a and the scientists at Diamond Light Source, Didcot, Oxon (UK) for their support during X-ray diffraction data collection. T.T.K.P. wishes to thank Biotechnology Center of Ho Chi Minh City, Vietnam and University of Bath for a post-graduate studentship. K.B. was supported by NIH (USA) grant AR40994 and P.T. and B.S. were partly funded by the National Science Foundation (USA) GK-12 Program.
Accession codes: Atomic coordinates and structure factors have been deposited in the Protein Data Bank – codes 4AYL and 4AYJ for the native protein and its complex with FAL respectively.
References
- Brew K., Tumbale P. & Acharya K. R. Family 6 glycosyltransferases in vertebrates and bacteria: inactivation and horizontal gene transfer may enhance mutualism between vertebrates and bacteria. J Biol Chem 285, 37121–37127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro J. A. et al. ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes. J Biol Chem 283, 10097–10108 (2008). [DOI] [PubMed] [Google Scholar]
- Lee H. J. et al. Structural basis for the inactivity of human blood group O2 glycosyltransferase. J Biol Chem 280, 525–529 (2005). [DOI] [PubMed] [Google Scholar]
- Patenaude S. I. et al. The structural basis for specificity in human ABO(H) blood group biosynthesis. Nat Struct Biol 9, 685–690 (2002). [DOI] [PubMed] [Google Scholar]
- Persson M. et al. Structural effects of naturally occurring human blood group B galactosyltransferase mutations adjacent to the DXD motif. J Biol Chem 282, 9564–9570 (2007). [DOI] [PubMed] [Google Scholar]
- Boix E. et al. Structure of UDP complex of UDP-galactose:beta-galactoside-alpha -1,3-galactosyltransferase at 1.53-A resolution reveals a conformational change in the catalytically important C terminus. J Biol Chem 276, 48608–48614 (2001). [DOI] [PubMed] [Google Scholar]
- Gastinel L. N. et al. Bovine alpha1,3-galactosyltransferase catalytic domain structure and its relationship with ABO histo-blood group and glycosphingolipid glycosyltransferases. EMBO J 20, 638–649 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaluddin H. et al. Crystal structure of alpha-1,3-galactosyltransferase (alpha3GT) in a complex with p-nitrophenyl-beta-galactoside (pNPbetaGal). Biochem Biophys Res Commun 385, 601–604 (2009). [DOI] [PubMed] [Google Scholar]
- Jamaluddin H., Tumbale P., Withers S. G., Acharya K. R. & Brew K. Conformational changes induced by binding UDP-2F-galactose to alpha-1,3 galactosyltransferase- implications for catalysis. J Mol Biol 369, 1270–1281 (2007). [DOI] [PubMed] [Google Scholar]
- Tumbale P., Jamaluddin H., Thiyagarajan N., Acharya K. R. & Brew K. Screening a limited structure-based library identifies UDP-GalNAc-specific mutants of alpha-1,3-galactosyltransferase. Glycobiology 18, 1036–1043 (2008). [DOI] [PubMed] [Google Scholar]
- Tumbale P., Jamaluddin H., Thiyagarajan N., Brew K. & Acharya K. R. Structural basis of UDP-galactose binding by alpha-1,3-galactosyltransferase (alpha3GT): role of negative charge on aspartic acid 316 in structure and activity. Biochemistry 47, 8711–8718 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity. Biochemistry 42, 13512–13521 (2003). [DOI] [PubMed] [Google Scholar]
- Yi W. et al. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J Am Chem Soc 127, 2040–2041 (2005). [DOI] [PubMed] [Google Scholar]
- Tumbale P. & Brew K. Characterization of a metal-independent CAZy family 6 glycosyltransferase from Bacteroides ovatus. J Biol Chem 284, 25126–25134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinig M. & Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res 32, W500–W502 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang P. G. & Brew K. Specificity and mechanism of metal ion activation in UDP-galactose:beta -galactoside-alpha -1,3-galactosyltransferase. J Biol Chem 276, 11567–11574 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang Y. & Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts J. A. et al. Differential recognition of the type I and II H antigen acceptors by the human ABO(H) blood group A and B glycosyltransferases. J Biol Chem 281, 3625–3632 (2006). [DOI] [PubMed] [Google Scholar]
- Nguyen H. P. et al. The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases. J Biol Chem 278, 49191–49195 (2003). [DOI] [PubMed] [Google Scholar]
- Indyk L. & Fisher H. F. Theoretical aspects of isothermal titration calorimetry. Methods Enzymol 295, 350–364 (1998). [DOI] [PubMed] [Google Scholar]
- Boix E., Zhang Y., Swaminathan G. J., Brew K. & Acharya K. R. Structural basis of ordered binding of donor and acceptor substrates to the retaining glycosyltransferase, alpha-1,3-galactosyltransferase. J Biol Chem 277, 28310–28318 (2002). [DOI] [PubMed] [Google Scholar]
- Hu Y. et al. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A 100, 845–849 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- Seto N. O. et al. Donor substrate specificity of recombinant human blood group A, B and hybrid A/B glycosyltransferases expressed in Escherichia coli. Eur J Biochem 259, 770–775 (1999). [DOI] [PubMed] [Google Scholar]
- Andersson J. O., Hirt R. P., Foster P. G. & Roger A. J. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol Biol 6, 27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S. et al. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat Chem Biol 7, 631–638 (2011). [DOI] [PubMed] [Google Scholar]
- Lairson L. L., Henrissat B., Davies G. J. & Withers S. G. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77, 521–555 (2008). [DOI] [PubMed] [Google Scholar]
- Pak J. E., Satkunarajah M., Seetharaman J. & Rini J. M. Structural and mechanistic characterization of leukocyte-type core 2 beta1,6-N-acetylglucosaminyltransferase: a metal-ion-independent GT-A glycosyltransferase. J Mol Biol 414, 798–811 (2011). [DOI] [PubMed] [Google Scholar]
- Rao F. V., Rich J. R., Rakić B., Buddai S., Schwartz M. F., Johnson K., Bowe C., Wakarchuk W. W., Defrees S., Withers S. G. & Strynadka N. C. Structural insight into mammalian sialyltransferases. Nat Struct Mol Biol. 16, 1186–1188 (2009). [DOI] [PubMed] [Google Scholar]
- Duewel H. S. & Woodard R. W. A metal bridge between two enzyme families. 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus requires a divalent metal for activity. J Biol Chem 275, 22824–22831 (2000). [DOI] [PubMed] [Google Scholar]
- Cochrane F. C., Cookson T. V., Jameson G. B. & Parker E. J. Reversing evolution: re-establishing obligate metal ion dependence in a metal-independent KDO8P synthase. J Mol Biol 390, 646–661 (2009). [DOI] [PubMed] [Google Scholar]
- Li J., Wu J., Fleischhacker A. S. & Woodard R. W. Conversion of aquifex aeolicus 3-deoxy-d-manno-octulosonate 8-phosphate synthase, a metalloenzyme, into a nonmetalloenzyme. J Am Chem Soc 126, 7448–7449 (2004). [DOI] [PubMed] [Google Scholar]
- Shulami S. et al. A reciprocal single mutation affects the metal requirement of 3-deoxy-D-manno-2-octulosonate-8-phosphate (KDO8P) synthases from Aquifex pyrophilus and Escherichia coli. J Biol Chem 279, 45110–45120 (2004). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. J. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Battye T. G. G., Kontogiannis L., Johnson O., Powell H. R. & Leslie A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A. & Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J. Apply. Cryst. 30, 1022–1025 (1997). [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994). [Google Scholar]
- CCP4. Collaborative Computational Project, Number 4, "The CCP4 Suite: Programs for Protein Crystallography". Acta Crystallogr D 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- Keegan R. M. & Winn M. D. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr D 64, 119–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 (2008). [Google Scholar]
- Langer G., Cohen S. X., Lamzin V. S. & Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. J. Coot: model-building tools for molecular graphics. Acta Crystallogr D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File