Abstract
Objective: To assess the effect of varying microbubble (MB) and DNA doses on the overall and comparative efficiencies of ultrasound (US)-mediated gene delivery (UMGD) to murine hindlimb skeletal muscle using cationic versus neutral MBs.
Materials and Methods: Cationic and control neutral MBs were characterized for size, charge, plasmid DNA binding, and ability to protect DNA against endonuclease degradation. UMGD of a codon optimized firefly luciferase (Fluc) reporter plasmid to endothelial cells (1 MHz, 1 W/cm², 20% duty cycle, 1 min) was performed in cell culture using cationic, neutral, or no MBs. In vivo UMGD to mouse hindlimb muscle was performed by insonation (1 MHz, 2 W/cm², 50% duty cycle, 5 min) after intravenous administration of Fluc combined with cationic, neutral, or no MBs. Gene delivery efficiency was assessed by serial in vivo bioluminescence imaging. Efficiency of in vivo UMGD with cationic versus neutral MBs was systematically evaluated by varying plasmid DNA dose (10, 17.5, 25, 37.5, and 50 µg) while maintaining a constant MB dose of 1x108 MBs and by changing MB dose (1x107, 5x107, 1x108, or 5x108 MBs) while keeping a constant DNA dose of 50 µg.
Results: Cationic and size-matched control neutral MBs differed significantly in zeta potential with cationic MBs being able to bind plasmid DNA (binding capacity of 0.03 pg/MB) and partially protect DNA from nuclease degradation while neutral MBs could not. Cationic MBs enhanced UMGD compared to neutral MBs as well as no MB and no US controls both in cell culture (P < 0.001) and in vivo (P < 0.05). Regardless of MB type, in vivo UMGD efficiency increased dose-dependently with DNA dose and showed overall maximum transfection with 50 µg DNA. However, there was an inverse correlation (ρ = -0.90; P = 0.02) between DNA dose and the degree of enhanced UMGD efficiency observed with using cationic MBs instead of neutral MBs. The delivery efficiency advantage associated with cationic MBs was most prominent at the lowest investigated DNA dose (7.5-fold increase with cationic versus neutral MBs at a DNA dose of 10 µg; P = 0.02) compared to only a 1.4-fold increase at a DNA dose of 50 µg (P < 0.01). With increasing MB dose, overall in vivo UMGD efficiency increased dose-dependently with a maximum reached at a dose of 1x108 MBs with no further significant increase with 5x108 MBs (P = 0.97). However, compared to neutral MBs, cationic MBs enhanced UMGD efficiency the most at low MB doses. Relative enhancement of UMGD efficiency using cationic over neutral MBs decreased from a factor of 27 for 1x107 MBs (P = 0.02) to a factor of 1.4 for 1x108 MBs (P < 0.01) and no significant difference for 5x108 MBs.
Conclusions: Cationic MBs enhance UMGD to mouse skeletal muscle relative to neutral MBs but this is dependent on MB and DNA dose. The enhancement effect of cationic MBs on UMGD efficiency is more evident when lower doses of MBs or DNA are used, whereas the advantage of cationic MBs over neutral MBs is substantially reduced in the presence of excess MBs or DNA.
Keywords: Ultrasound, gene therapy, ultrasound-mediated gene delivery, microbubble, sonoporation.
INTRODUCTION
Ultrasound (US)-mediated gene delivery (UMGD) is emerging as a promising noninvasive and non-viral strategy for gene therapy 1, 2. In contrast to viral vectors, which bear the risk of immunotoxicity, insertional mutagenesis, and nonspecific gene delivery 3, UMGD is considered to be a safer approach which provides remarkable spatial and temporal control of gene transfer 4. UMGD facilitates cellular uptake of DNA by the phenomenon of sonoporation, which refers to the formation of transient cell membrane microperforations induced by cavitational US bioeffects 5-7. Several studies have shown that sonoporation is markedly enhanced in the presence of microbubbles (MBs) 8, which are lipid-encapsulated gas-filled intravascular contrast agents best known for their role in diagnostic US imaging 9-11 but also function as nuclei for acoustic cavitation in UMGD 8, 12. Exposed to certain acoustic fields, MBs oscillate vigorously and eventually collapse, amplifying the acoustic energy and causing secondary convective bioeffects that thereby promote cell membrane poration and gene uptake 5, 6, 13.
In most published studies of in vivo UMGD, plasmid DNA was co-administered with neutral or negative surface charge MBs. To further enhance gene delivery efficiency, recent studies have employed MBs with positively-charged shells that electrostatically bind to anionic DNA, enabling them to become direct gene carriers for intravascular transportation 14-18. Moreover, it has been shown that plasmid DNA charge-coupled onto the surface of cationic MBs are protected against endonuclease degradation 19, which are subsequently released with preserved function by US-induced MB destruction 14. The overall end effect is a local increase in gene concentration at the selected site of UMGD 1.
It has been recently demonstrated in an in vivo tumor model that UMGD efficiency is substantially increased when using cationic MBs compared to control neutral MBs 20. However, it remains unknown whether this gene transfer advantage afforded by cationic MBs is limited to certain experimental conditions. While US parameters (such as frequency, peak negative pressure, duration of insonation, duty cycle, etc), MB characteristics (such as composition of microbubble shell, size, etc), and the in vivo target tissue have been identified as important factors on the overall success rate of acoustically-mediated gene delivery 21-24, MB and DNA doses are likely to play major roles specifically on the relative efficiencies of in vivo US and MB-mediated gene delivery when directly comparing the use of cationic versus neutral MBs. Both MB and DNA doses directly affect the concentration of both free and bound DNA at the site of tissue transfection - a key factor for efficient gene delivery 25 - and are thus likely to impact the transfection-enhancing mechanisms of cationic MBs.
We hypothesized that the increased gene delivery efficiency afforded by cationic MBs over neutral MBs may depend on certain combinations of MB and DNA doses and that excess MBs or DNA may attenuate the enhancing effects of cationic MBs on in vivo UMGD. To the best of our knowledge, a direct comparison of in vivo UMGD efficiency using cationic versus neutral MBs at varying MB and DNA doses has not been previously reported.
The purpose of this study was to determine the effect of varying MB and plasmid DNA dose on in vivo UMGD efficiencies comparing the use of cationic MBs versus neutral MBs.
MATERIALS AND METHODS
Firefly Luciferase Reporter Plasmid Preparation
A reporter plasmid encoding for a mammalian codon-optimized firefly luciferase (Fluc) driven by a cytomegalovirus (CMV) promoter was constructed in the pcDNA 3.1-puromycin(+) vector backbone (Invitrogen, Carlsbad, CA). Using the pGL 4.10 vector (Promega, Madison, WI) as template, the Fluc gene fragment was PCR amplified (Triple Master PCR System; Eppendorf, Hauppauge, NY) using the forward primer ATAGCTAGCATGGAAGATGCCAAAAACATT containing a Nhe I restriction enzyme site (underlined) and the reverse primer GGCCTCGAGTTACACGGCGATCAAGCCGCCCTT with a Xho I restriction enzyme site (underlined). The Fluc gene fragment was then ligated into the pcDNA 3.1(+) vector. The firefly luciferase plasmid construct (pFluc) was amplified with the Endotoxin Free Maxiprep plasmid ampification kit (Qiagen, Valencia, CA) according to manufacturer instructions and resuspended and stored in Tris-EDTA buffer (10mM TRIS, 1mM EDTA, pH 8.0; EMD Chemicals, Gibbstown, NJ). DNA concentration was measured with a spectrophotometer (Agilent, Santa Clara, CA) by measuring the absorbance at 260nm.
Cell Line Preparation and Maintenance for Cell Culture Experiments
Since MBs remain in the vascular component and interact with vascular endothelial cells, mouse endothelial SVR cells (ATCC, Manassas, VA) were used for cell culture experiments. SVR cells were grown in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) with high concentrations of glucose (4.5 g/L) and L-glutamine supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). At 70% to 80% confluence, cells were collected by trypsinization.
Microbubble Preparation and Characterization
Both cationic and neutral MBs (Bracco Research; Geneva, Switzerland) were composed of a perfluorobutane core encapsulated by a PEGylated lipid shell and prepared as previously reported 26. The shell of neutral MBs was assembled using 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC; Avanti Polar Lipids, Alabaster, AL), 1 mol% of 1,2-dipalmitoyl-sn-glycero-3-phosphoethanol-amine-N-[methoxy(polyethylene glycol)-5000] (DPPE-PEG5000; Avanti Polar Lipids), and 20 mol% of palmitic acid (Fluka, Buchs, Switzerland). Cationic MBs were designed to have a positive surface charge by incorporating 20 mol% 1,2-distearoyl-3-trimethylammonium-propane (DSTAP; Avanti Polar Lipids), a cationic phospholipid, into the otherwise identically composed lipid shell.
Neutral and cationic MBs were stored as lyophilisates in septum-sealed vials filled with perfluorobutane and nitrogen gas and were resuspended in sterile 0.9% saline directly before all experiments. Size distribution analysis of both MB types was performed by electrozone sensing using a Multisizer 3 Coulter Counter (Beckman Coulter SA, Nyon, Switzerland). Additionally, the zeta potentials of cationic and neutral MBs were analyzed by laser Doppler electrophoresis (ZetaSizer 3000; Malvern Instruments, Malvern, UK).
Assessment of DNA Binding to Microbubbles
To quantify the amount of plasmid DNA that can bind to each type of MB, 5x108 cationic or control neutral MBs were incubated with varying doses of pFluc (5, 10, 20, 40, 60, or 80 µg) in 0.9% saline for 15 min to allow for spontaneous electrostatic charge coupling between the anionic DNA and cationic MB shells. Samples were then centrifuged in a 3 ml-syringe barrel for 8 min at 200 g to separate DNA bound to gas-filled buoyant MBs from nonadherent DNA. Unbound DNA was quantified by collecting 200 µl of the subnatant beneath the floated MB layer and subsequent purification using 0.45 µm centrifugation filters (Millipore, Billerica, MA) and high speed centrifugation at 14,000 g for 5 min to remove MB shell contaminants. Next, free DNA in the filtrate was collected and concentration was quantified by spectrophotometry (see above). The total amount of nonconjugated DNA was calculated by multiplying the measured DNA concentration in the subnatant by the total sample volume. The amount of DNA bound to MBs was calculated by subtracting the amount of unbound DNA from the total amount of DNA added to the initial mixture. Given increasing amounts of initial DNA, the maximum amount of bound DNA across DNA doses was determined to be the binding capacity for 5x108 MBs. All experiments were performed in triplicate.
Plasmid DNA-cationic MB binding was demonstrated visually by confocal fluorescence microscopy. Five micrograms of pFluc was labeled with YOYO-1 fluorescent dye (Invitrogen, Carlsbad, CA) according to manufacturer instructions and incubated with 5x108 cationic or neutral MBs in 0.9% saline for 15 min. Samples were then diluted 1:10 and wet mounted on non-charged microscope slides for imaging in fluorescence and brightfield modes at 1,000x magnification using a confocal laser scanning microscope (LSM 510; Carl Zeiss MicroImaging, Thornwood, NY) with a 488 nm argon laser. All experiments were performed in triplicate.
DNase Protection Assay
To compare the protective properties of cationic versus neutral MBs from DNA degradation by endonucleases, mixtures of plasmid DNA and cationic or neutral MBs were incubated with DNase I (New England Biolabs, Ipswich, MA) and the extent of DNA degradation was assessed by gel electrophoresis.
To allow for plasmid DNA-MB interactions, 10 µg of pFluc was incubated with 1x108 cationic or neutral MBs in 100 µl 1mM NaCl for 15 min at room temperature. After dilution with DNase reaction buffer (New England Biolabs, Ipswich, MA) to a total reaction volume of 200 µl, pFluc-MB mixtures were exposed to 0.02 units of DNase I for 15 min at 37º C 19. The reaction was stopped by adding ethylenediaminetetraacetic acid disodium salt solution (EDTA) to a final concentration of 5 mM (Sigma Aldrich, St. Louis, MO). Filter centrifugation (0.45 µm filters centrifuged at 200 g for 40 min) was used to separate unbound plasmid DNA from DNA bound to MBs, which remained above the filter membrane. The charge interactions between bound plasmid DNA and MBs were disrupted by addition of 200 µl of 5 M NaCl and subsequent filter centrifugation (as above). The DNA fraction released from the MBs was recovered from the high-salt solution using DNA purification columns per manufacturer instructions (QIAquick; Qiagen, Valencia, CA). Additional samples of plasmid DNA without MBs served as controls. DNA gel electrophoresis (0.8% Seakem agarose; Fisher Scientific, Pittsburgh, PA) was performed to determine the extent of DNA degradation. DNA bands were stained with ethidium bromide (0.5 µg/ml; Bio-Rad, Hercules, CA) and digitally imaged under ultraviolet light (2UV Transilluminator; UVP, Upland, CA). This assay was performed in triplicate.
Gene Delivery with Ultrasound and Microbubbles in Cell Culture
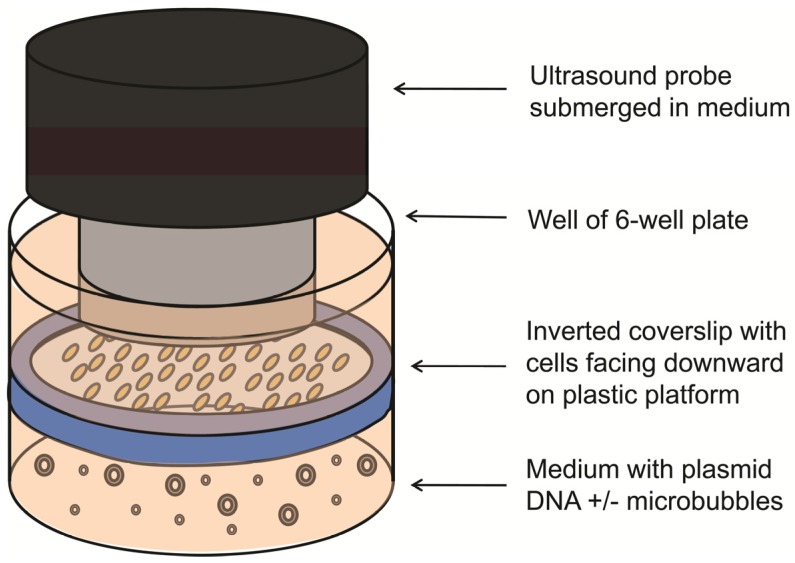
The feasibility of UMGD with cationic and neutral MBs using the Fluc reporter gene plasmid was first assessed in cell culture. SVR cells (1x105) were trypsinized, plated on sterilized glass coverslips (25 mm diameter, VWR Scientific, West Chester, PA), and incubated for 24 hours. Transfection of pFluc in cell culture was performed in triplicate as follows: Four micrograms of pFluc was incubated with 5x107 cationic, neutral, or no MBs for 15 min to allow for cationic MB-plasmid DNA binding (n=3 per group per experiment). An inverted setup (Figure 1) was used to promote transfection by providing close contact between buoyant MBs and cells. First, a sterilized plastic ring was inserted in the well and OptiMem medium (6 ml; Invitrogen, Carlsbad, CA) was added to fill the well up to the top of the ring. Then, the pFluc-MB mixture was added to the medium, and a coverslip of SVR cells (pre-rinsed with phosphate-buffered saline) was placed on the platform with the cells facing the pFluc-MB mixture. Additional OptiMem medium was added on top of the coverslip to aid US transmission. To minimize the generation of standing waves, the 6 well plates were placed on top of a platform and partially submerged within a degassed water bath with an US-absorbing rubber pad placed underneath 27. After MBs were allowed 5 min to float and contact the surfaces of the inverted cells, US was applied at 1 MHz, 1 W/cm², and 20% duty cycle (DC) for 1 min using the Sonitron 2000 US machine (Rich-Mar, Chattanooga, TN) and a 20 mm transducer submerged in the medium above the cover slips. Control groups included UMGD performed without MBs and SVR cells exposed to pFluc-cationic MB mixtures in identical fashion but without application of US. All experiments were performed in triplicate.
FIGURE 1.
Experimental setup for UMGD in cell culture. Vascular endothelial cells grown on a glass coverslip were inverted with cells facing downward on a plastic ring platform in a well of a 6-well plate. Mixtures of pFluc with cationic, neutral, or no MBs were added to medium under the coverslip. US was applied from above the coverslip.
Bioluminescence Quantification of Transfection in Cell Culture
Functional reporter gene expression was quantified 24 hours post transfection using a commercial Firefly luciferase assay system (Promega, Madison, WI) and luminometer (20/20n Luminometer; Turner Biosystems, Sunnyvale, CA) according to manufacturer instructions. For each sample, Fluc activity, measured in relative light units (RLU), was normalized to total protein (BCA Protein Assay-RAC; Pierce, Rockford, IL).
Animal Model for In Vivo Transfection
All experimental procedures involving animals were approved by the Institutional Administrative Panel on Laboratory Animal Care. In vivo UMGD studies were performed on the hindlimb skeletal muscle of 6-8-week-old female nude mice (Charles River Laboratories, Wilmington, MA). Hindlimb skeletal muscle was chosen as the target delivery tissue as it has been previously used for dose optimization studies of UMGD and has been found to be a robust and reproducible model 14, 27-30. For all animal studies, mice were anesthetized with 2% isoflurane (Aerrane, Baxter, Deerfield, IL) in oxygen administered at a rate of 2L/min.
In vivo Gene Delivery with Ultrasound and Microbubbles
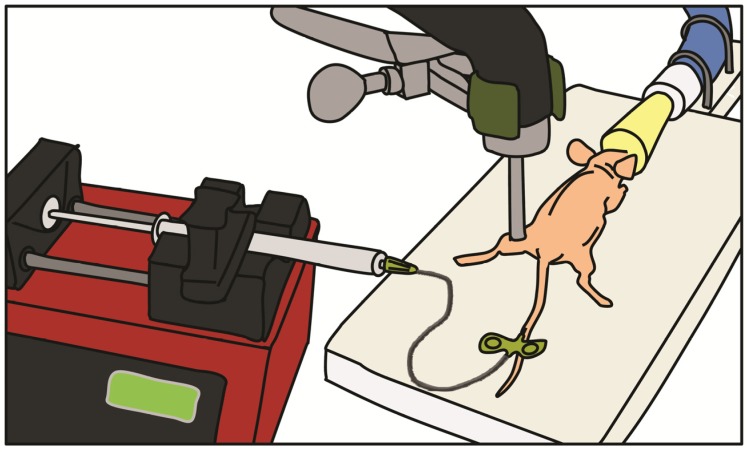
In vivo UMGD to mouse hindlimb skeletal muscle was performed in all conditions (detailed below) using a standardized setup illustrated in Figure 2. After pFluc was incubated with cationic, neutral or no MBs for 15 min to allow for cationic MB-plasmid binding, solutions (400 µl total volume) were administered intravenously via a catheter (MicroMarker Tail Vein Access Cannulation kit; VisualSonics, Toronto, Ontario) placed into a lateral tail vein at a constant rate of 100 µl/min using an automated infusion pump (GeniePlus, Kent Scientific, Torrington, CT). Using the Sonitron 2000 sonoporation system (Rich-Mar, Chattanooga, TN) and a 5 mm transducer positioned directly above the center of the mouse hindlimb muscle, US was applied using the following settings: 1 MHz, 2 W/cm2, 50% DC, and total insonation time of 5 min. An US-absorbing rubber was placed under the hindlimb during insonation to minimize additional energy deposition from reflected waves 27.
FIGURE 2.
Experimental setup for in vivo UMGD. Mixtures of pFluc with cationic, neutral, or no MBs were administered intravenously via a catheter inserted in the tail vein of anesthetized nude mice. The mixtures were infused with a syringe infusion pump at a constant rate. US was applied with an ultrasound probe placed on top of the mouse hindlimb muscle.
To demonstrate an in vivo UMGD efficiency advantage with cationic MBs over neutral MBs and control conditions, the initial animal experiments compared four different conditions (n=6 each): 1) UMGD with 1x108 cationic MBs and 50 µg pFluc; 2) UMGD with 1x108 neutral MBs and 50 µg pFluc; 3) UMGD with no MBs and 50 µg plasmid DNA; and, 4) intravenous administration of 1x108 cationic MBs and 50 µg pFluc but without application of US.
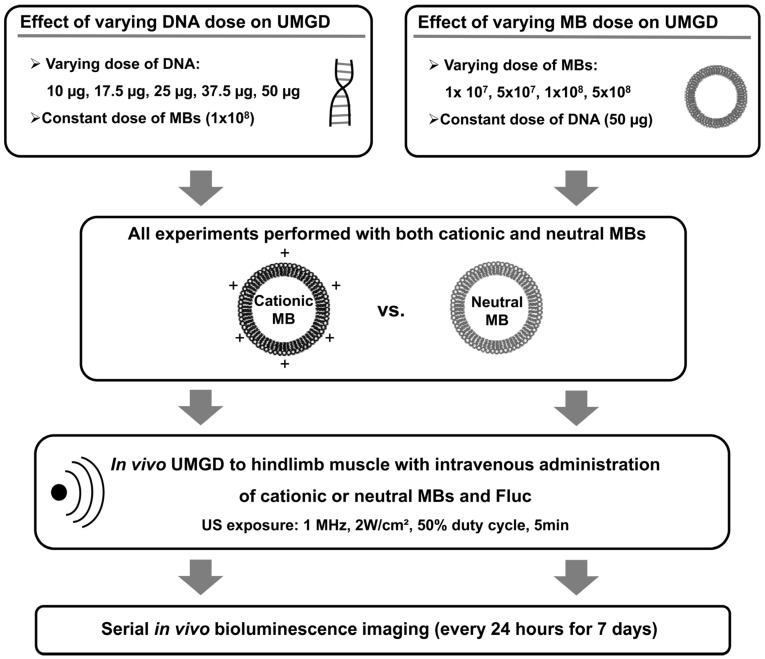
In subsequent experiments, the roles of DNA and MB dose on the relative efficiencies of in vivo UMGD with cationic versus neutral MBs were investigated by systematically varying either DNA or MB dose while maintaining all other variables constant (Figure 3). First, in vivo UMGD was performed (n=6 for each condition) with varying doses of pFluc (10, 17.5, 25, 37.5, or 50 µg) and a constant number of cationic or neutral MBs (1x108 MBs). Second, in vivo UMGD was performed (n=6 for each condition) using a variable number of cationic or neutral MBs (1x107, 5x107, 1x108, or 5x108) and a constant dose of plasmid DNA (50 µg pFluc). Mice were randomly assigned to these experimental groups.
FIGURE 3.
Experimental plan for testing the influence of varying DNA and MB doses on in vivo UMGD efficiency in murine hindlimb skeletal muscles. Different combinations of MB and DNA doses were evaluated for both cationic and neutral MBs. Transgene expression was measured every 24 hours for 7 days using in vivo bioluminescence imaging.
Quantification of In Vivo pFluc Transfection
Efficiency of in vivo delivery of pFluc was assessed by in vivo bioluminescence imaging of Fluc activity with the IVIS-200 imaging system (Caliper Life Sciences, Hopkinton, MA). Fluc substrate, D-luciferin (150 mg/kg; Biosynth AG, Staad, Switzerland), was injected intraperitoneally in treated mice, and images were acquired until peak signal was obtained. Bioluminescence imaging was performed starting at 24 hours post-transfection and continued every 24 hours for seven days. Peak signal intensities for each mouse were used to compare the transfection efficiencies among all conditions tested. Image analysis was performed offline using Living Image 3.2 software (Caliper Life Sciences) by one reader blinded to the treatment conditions. A region of interest of constant shape and size (0.53 cm2) was drawn over the transfected hindlimbs of all animals. Bioluminescence signal in the region of interest was quantified using the maximum radiance measured in photons/sec/cm2/steradian (sr) 31.
Statistical Analysis
All continuous values were expressed as mean ± standard deviation. DNA binding was analyzed by comparing plateau values of the binding curves of each microbubble (MB) type using a Wilcoxon rank sum test. For gene delivery experiments in cell culture, the Wilcoxon rank sum test was used for comparisons between different experimental groups. In vivo UMGD efficiency of cationic versus neutral MBs was compared using a stratified Wilcoxon rank sum test. The degree of gene delivery efficiency advantage that was afforded by using cationic MBs in in vivo UMGD was assessed by calculating the ratio of in vivo bioluminescence signals for the cationic and neutral MB groups (C:N) for each DNA and MB dose tested. For overall comparison of gene delivery with cationic versus neutral MBs and control groups, the test was stratified by the experiment time and MB number. For all other comparisons, the test was stratified by experiment time to remove the batch effect. The trend of signal increase over time was tested by simple linear regression analysis. The estimated rank correlation coefficient (ρ) between DNA dose and C:N was calculated. This correlation could not be performed for MB dose and C:N due to the limited number of doses tested. All statistical analyses were performed with a statistical analysis software (R.2.6.1, http://www.r-project.org). P values of less than 0.05 were considered to indicate a significant difference.
RESULTS
Microbubble Characterization
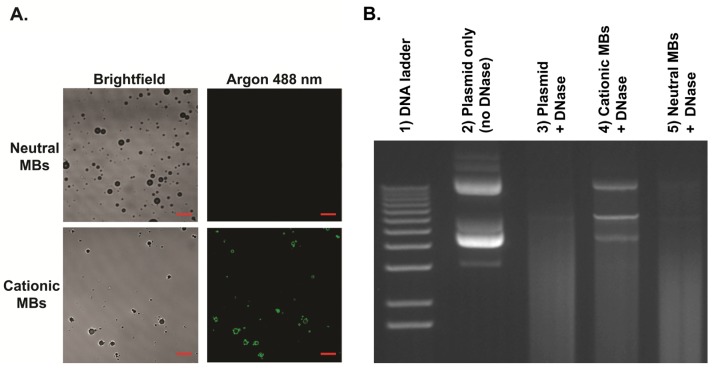
MB characteristics are summarized in Table 1. Size distribution analyses showed no significant differences in mean diameters between cationic (diameter in number (Dn) 1.4 ± 0.1 µm, diameter in volume (Dv) 3.8 ± 0.5 µm) and neutral (Dn 1.4 ± 0.1 µm, Dv 3.9 ± 0.5 µm) MBs. Zeta potential for cationic MBs (+15.8 ± 2.0 mV) was significantly higher than that of neutral MBs (-0.3 ± 0.3 mV; P < 0.01). Quantitative assessment of DNA binding to MBs showed a significantly higher (P < 0.01) loading capacity of 14.9 ± 1.7 µg of DNA per 5x108 MBs for cationic MBs (equal to 0.03 pg per cationic MB) compared to essentially no binding to an equivalent number of neutral MBs (0.27 ± 1.5 µg). Plasmid DNA and cationic MB binding was confirmed visually by confocal microscopy (Figure 4A), demonstrating charge-coupling of fluorescence-labeled DNA to cationic MBs but not neutral MBs. DNase protection assay showed plasmid DNA mixed with (and not charge-coupled to) neutral MBs to be completely degraded after DNase treatment while DNA bound to cationic MBs were partially protected from DNase degradation (Figure 4B).
TABLE 1.
Summary of microbubble characteristics.
| Dn (µm) | Dv (µm) | Zeta potential (mV) | |
|---|---|---|---|
| Cationic microbubbles | 1.4 ± 0.1 | 3.8 ± 0.5 | +15.8 ± 2.0 |
| Neutral microbubbles | 1.4 ± 0.1 | 3.9 ± 0.5 | -0.3 ± 0.3 |
FIGURE 4.
Binding of plasmid DNA to cationic MBs and DNase protection assay. A, Direct binding of YOYO-1-labeled plasmid DNA (10 µg) to cationic MBs (1x108) but not neutral MBs (1x108) was confirmed visually by confocal microscopy (1000x, scale bar = 10 µm). B, Gel electrophoresis confirmed partial protection of plasmid DNA bound to cationic MBs (lane 4) from DNase degradation. Plasmid DNA mixed with and not bound to neutral MBs (lane 5) were completed degraded after DNase treatment. Lane 2 represents control samples containing only plasmid DNA without MBs that were not exposed to DNase. Lane 3 shows complete DNase degradation of plasmid DNA not incubated with either MB type.
Quantification of Ultrasound-Mediated Transfection in Cell Culture
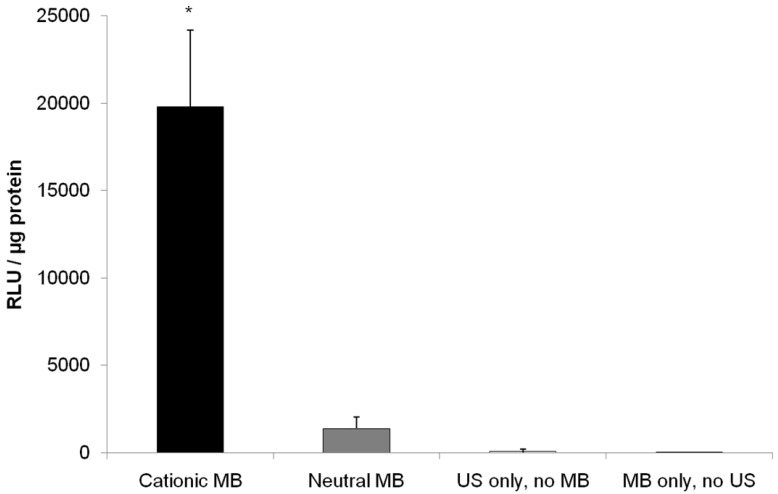
US-mediated transfection of pFluc to vascular endothelial cells yielded luciferase signal 14.5-fold higher (P < 0.001) with cationic MBs (19799 ± 4401 RLU/µg protein) compared to neutral MBs (1361 ± 680 RLU/µg protein) (Figure 5). Fluc activity after UMGD to vascular endothelial cells with either cationic or neutral MBs was significantly higher (P < 0.001) than UMGD without MBs (91 ± 109 RLU/µg protein), indicating that the presence of MBs markedly enhanced overall transfection efficiency. The negative control group of vascular endothelial cells exposure to DNA coupled to cationic MBs but without US application showed background signal (5 ± 2 RLU/µg protein).
FIGURE 5.
Firefly luciferase activity of SVR after in vitro US-mediated delivery of 4 µg pFluc with 5x107 cationic, neutral, or no MBs (n=9 per group). Transgene expression was greatly enhanced with cationic MBs compared to neutral MBs (14.6-fold, P < 0.001). Units are average RLU per µg of protein measured for luciferase acitivity; error bars represent standard deviation. * P < 0.001 relative to the neutral MB, no MB, and no US groups.
Assessment of In Vivo Ultrasound-Mediated Gene Delivery
UMGD with cationic MBs versus neutral MBs, no MBs, and no US
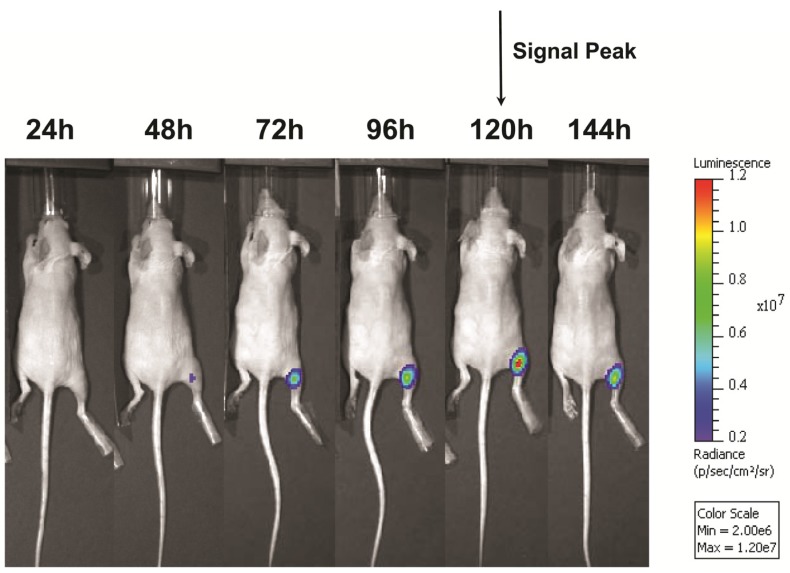
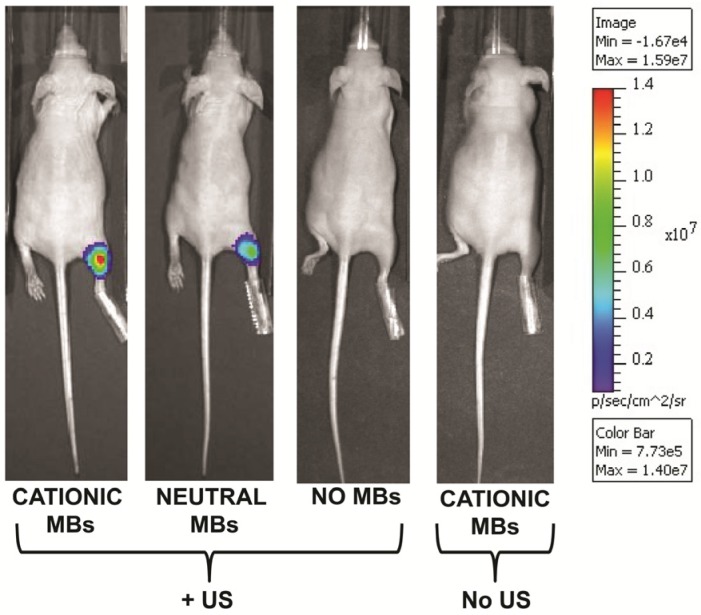
Following UMGD with 50 µg of pFluc and 1x108 of either cationic, neutral, or no MBs, serial in vivo bioluminescence imaging of Fluc expression by murine hindlimb skeletal muscle demonstrated increasing signal over time (P < 0.01) with maximum signal for each mouse observed between 96 and 144 hours post-transfection (Figure 6). In contrast, longitudinal bioluminescence signal remained persistently low for both groups of mice injected with pFluc without MBs and treated with US (P = 0.47) as well as mice injected with pFluc and cationic MBs but US was not applied (P = 0.75) (Figure 7).
FIGURE 6.
Representative serial in vivo bioluminescence images of a mouse obtained every 24 hours for up to 144 hours following UMGD with 1x108 cationic MBs and 50 µg pFluc to mouse hindlimb skeletal muscle. In this mouse, maximum bioluminescence signal (photons/sec/cm2/sr) as a measure of transfection efficiency occurred 120 hours after gene delivery.
FIGURE 7.
Representative in vivo bioluminescence images of four different mice obtained 120 hours after in vivo UMGD with 1x108 MBs and 50 µg pFluc to the right hindlimb muscle using (from left to right) cationic, neutral, or no MBs. The last mouse on the right was treated with systemic administration of cationic MBs and pFluc but US was not applied. In vivo bioluminescence imaging signal (photons/sec/cm2/sr) as a measure of delivery efficiency was higher in the cationic MB mouse compared to the neutral MB mouse. Only background signal was observed in the mice treated with UMGD without MBs and systemic exposure to cationic MBs and pFluc but no US.
In comparing the two UMGD MB groups using this combination of MB and DNA dose, mean bioluminescence signal was significantly higher (P = 0.005) when using cationic MBs (1.5 ± 0.7 x 107 photons/sec/cm2/sr) instead of neutral MBs (1.1 ± 0.4 x 107 photons/sec/cm2/sr). Mean Fluc activity was markedly increased when using either cationic (4969-fold, P < 0.001) or neutral MBs (3654-fold, P < 0.001) in UMGD compared to sonoporation without MBs (3.1 ± 1.1 x 103 photons/sec/cm2/sr), indicating that the presence of MBs was required for detectable gene delivery and transgene expression. There was no significant difference (P = 0.31) in Fluc activity between the no MB UMGD group and the negative control group where mice received pFluc-cationic MB solutions but no US was applied (1.0 ± 1.6 x 104 photons/sec/cm2/sr).
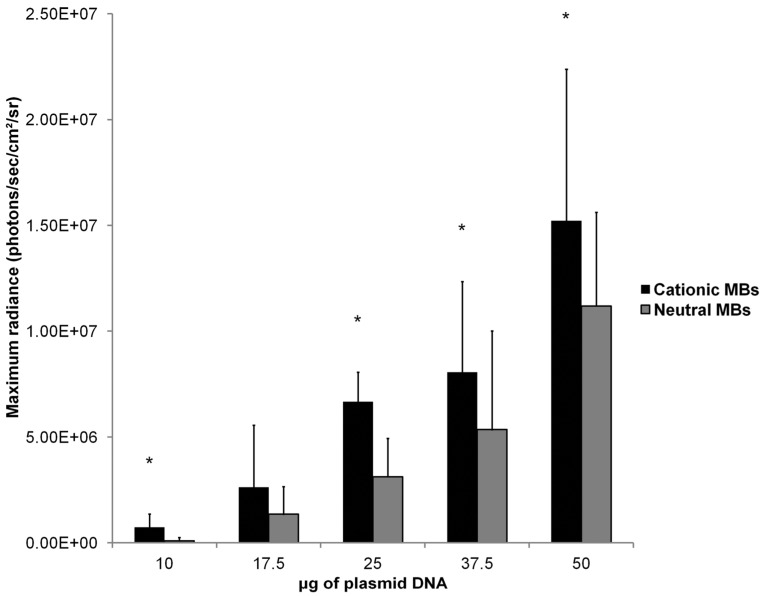
Effect of DNA Dose on In Vivo UMGD Using Cationic Versus Neutral MBs
Mean in vivo bioluminescence signals measured in hindlimb skeletal muscle following in vivo UMGD using 1x108 cationic or neutral MBs and varying doses of pFluc are shown in Figure 8. In general, mean in vivo bioluminescence signal rose with increasing DNA dose for both MB types. Maximum peak Fluc activity was found with a DNA dose of 50 µg for both cationic (1.5 ± 0.7 x 107 photons/sec/cm2/sr, P = 0.04 vs. 37.5 µg) and neutral MBs (1.1 ± 0.4 x 107 photons/sec/cm2/sr, P = 0.047 vs. 37.5 µg).
FIGURE 8.
Effect of varying DNA dose on the efficiency of in vivo UMGD using cationic or neutral MBs. Overall, gene delivery increased with increased DNA dose with maximum observed at the highest DNA dose of 50 µg. However, the degree of enhancement in UMGD efficiency with using cationic MBs over neutral MBs (C:N) was most prominent at the lowest DNA dose. Bars are means of the maximum radiance at individual peak time points ± standard deviations (in photons/sec/cm2/sr, n=6 per group). * P < 0.05 comparing cationic and neutral MB groups.
Mean in vivo bioluminescence signals were significantly higher when cationic MBs were used versus neutral MBs for DNA doses of 10, 25, 37.5, and 50 µg (P < 0.05). For the DNA dose of 17.5 µg, the difference in peak mean bioluminescence signal using cationic MBs (2.6 ± 2.9 x 106 photons/sec/cm2/sr) versus neutral MBs (1.4 ± 1.3 x 106 photons/sec/cm2/sr) did not reach statistical significance (P = 0.14). Increased gene delivery efficiency observed with the use of cationic versus neutral MBs correlated inversely with DNA dose (ρ = -0.90; P = 0.02). The degree of UMGD efficiency enhancement was highest at the lowest plasmid DNA dose of 10 µg (C:N of 7.5) and lowest at the highest DNA dose of 50 µg (C:N of 1.4).
Effect of MB Dose on In Vivo UMGD Using Cationic Versus Neutral MBs
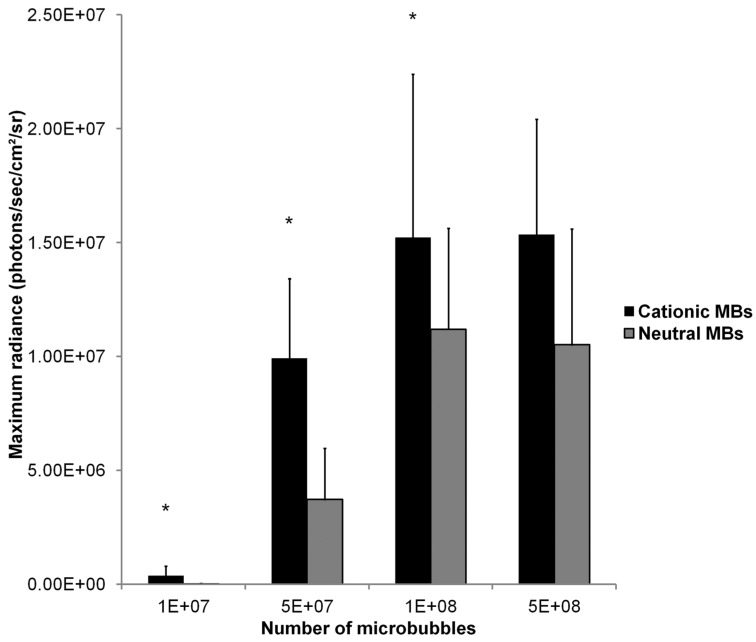
Mean in vivo bioluminescence signals measured in hindlimb skeletal muscle following in vivo UMGD using 50 µg of pFluc and varying doses of cationic or neutral MBs are summarized in Figure 9. Overall, increasing MB numbers resulted in higher in vivo bioluminescence signal for both cationic and neutral MBs with maximum signals observed with 1x108 and 5x108 MBs. There were no significant differences in mean bioluminescence signals comparing the 1x108 and 5x108 MB dose groups for both cationic (P = 0.97) and neutral MBs (P = 0.81).
FIGURE 9.
Effect of varying MB dose on the efficiency of in vivo UMGD using cationic or neutral MBs. In vivo gene transfer increased with increasing MB dose for both cationic and neutral MBs with a plateau at 1x108 MBs. Advantages in UMGD efficiency in using cationic over neutral MBs (C:N) was highest at the lowest MB dose. Bars are means of the maximum radiance at individual peak time points ± standard deviations (in photons/sec/cm2/sr, n=6 per group). * P < 0.05 comparing cationic and neutral MB groups.
A direct comparison of in vivo UMGD efficiency with cationic versus neutral MBs showed significantly higher in vivo Fluc activity when cationic MBs were used for 1x107, 5x107, and 1x108 MBs (P < 0.05), but not for the highest MB dose of 5x108 (P = 0.35). The gene delivery efficiency advantage revealed in using cationic over neutral MBs was most prominent at the lowest MB dose of 1x107 (C:N of 27) but much attenuated when MB dose was increased to 1x108 (C:N of 1.4).
DISCUSSION
Ultrasound and MB-mediated gene delivery has garnered significant attention as a potential method for safe, noninvasive, and regionally-specific gene therapy 4, 21. As with all non-viral gene transfer platforms, potential clinical translation of this technology requires improvements in delivery efficiency 3. By drawing upon decades of experience with cationic lipid-based vectors for non-viral gene therapy 32, recent efforts towards this goal have led to the engineering of MBs with positively-charged lipid shells. In the context of in vivo UMGD using systemically administered MBs, this functionalizes MBs to serve not only as cavitation nuclei for sonoporation but also intravascular transporters of genetic cargo by direct electrostatic binding of plasmid DNA or other nucleotide-based agents. It has been hypothesized that the use of systemically administered cationic MBs that bind and transport DNA in UMGD potentiates gene delivery efficiency by protection of the genetic cargo against endonuclease degradation resulting in an increase in local gene concentration at the site of sonoporation 1, 19, 33.
In a direct head-to-head comparison study between cationic and control neutral MBs, it has been recently shown both in cell culture and in a mouse hindlimb tumor model that UMGD efficiency can be substantially enhanced using cationic MBs 20. However, it remains controversial in the literature whether the use of cationic MBs over conventional neutral or anionic MBs improves transfection efficiency as very few direct comparison studies have been published. In two recent cell culture UMGD studies directly comparing cationic and neutral MBs, Tlaxca et al. 17 showed no significant difference while Nomikou et al. 18 demonstrated superiority with cationic MBs (although no P value was reported). In the only other direct MB comparison UMGD study performed in vivo, no significant difference in gene transfer efficiency between cationic and neutral MBs was found 18. However, it is important to highlight that UMGD in this study was performed with direct intramuscular injection of MB-DNA mixtures rather than systemic administration 18. With these results and the assumption that UMGD efficiency is determined by a multitude of factors, we hypothesized that a transfection superiority seen with cationic over neutral MBs in in vivo UMGD may not be universal but could be maximized under certain conditions. In our study, we focused on DNA and MB dose as key determinants and investigated their roles on in vivo UMGD efficiency overall and the relative efficiency differences between cationic and neutral MBs.
We first characterized the MBs used in this study. Cationic and control neutral MBs were size-matched and synthesized using the same lipid shell platform except for the addition of a positively-charged lipid for the cationic MBs. Saturation binding experiments demonstrated that positively-charged MBs could bind up to 0.03 pg of plasmid DNA per MB, which is within range of previous reports 14, while neutral MBs bound essentially no DNA. Adherence of fluorescence-labeled plasmid DNA to cationic MBs but not neutral MBs was confirmed visually by confocal microscopy. DNAse protection assay demonstrated that direct conjugation of plasmid DNA to cationic MB partially protected bound DNA from nuclease degradation, providing a mechanism for how cationic MBs may increase local DNA concentration at the selected site of sonoporation and thus enhance UMGD in the in vivo setting as systemically administered free plasmid DNA is rapidly degraded by endonucleases 19, 34.
Next, in a cell culture model, we directly compared the efficiencies of pFluc transfection in using cationic, neutral, or no MBs in UMGD. Vascular endothelial cells were chosen for in vitro gene delivery as MBs are intravascular gene carriers and interact primarily with endothelial cells in sonoporation 1. Relative to neutral MBs, use of cationic MBs enhanced UMGD 14.5-fold, which is in contradistinction to the results reported by Tlaxca et al. 17. This is likely due to differences in experimental setup. By placing MB-DNA mixtures in medium below downward facing cells, cationic MBs with plasmid DNA electrostatically conjugated to their shells allowed for transportation of the genetic payload to the cell surfaces as the buoyant MBs floated, concentrating the administered DNA at the site of sonoporation. In contrast, neutral MBs mixed with nonbound plasmid DNA left the free DNA to distribute throughout the volume of medium below the cells. Additionally, relative to sonoporation without MBs, UMGD in the presence of either MB type substantially amplified transfection (15-fold for neutral MBs, 219-fold for cationic MBs), confirming prior reports that MBs potentiate sonoporation 8. These studies were performed using fixed DNA and MB doses because several in vitro optimization studies have previously investigated the effects of varying DNA and MB dose on UMGD efficiency 17, 18, 27, 29, 35, 36. These published studies generally showed improved gene delivery with more DNA 17, 29, 35-37 and more MBs 17, 18, 27, 38 although some reported saturation at higher levels 17, 39. Moreover, results from such cell culture studies do not necessarily translate to facilitating optimal gene transfer in vivo.
To systematically evaluate the role of DNA and MB dose on in vivo UMGD efficiency, we chose mouse skeletal muscle as the target tissue because it was the model used for several published UMGD studies 14, 27-30, allowing comparison of our results with previous reports. Furthermore, this model allows better reproducibility as opposed to a tumor model where tumor growth and vascularity can vary. Either DNA or MB dose were varied while all other parameters were kept constant. It should be emphasized that MBs and plasmids were administered intravenously and not by invasive direct intramuscular injection where low levels of gene delivery can occur by injection alone 40, 41.
To begin, reporter gene expression was assessed by serial in vivo bioluminescence imaging after in vivo UMGD using one combination of DNA and MB dose (50 µg of pFluc, 1x108 of either cationic or neutral MBs) and compared to no MB and no US controls. In contrast to the cell culture experiment results, there was essentially no detectable Fluc activity when no MBs were used for sonoporation-mediated gene delivery. These results emphasize the differences between in vitro and in vivo conditions and suggest that MBs are required for substantial in vivo UMGD.
In examining the influence of DNA and MB dose on overall in vivo UMGD efficiency regardless of MB type, we showed dose dependent enhancement of gene delivery efficiency with more DNA and more MBs. Maximum gene delivery was observed with the highest pFluc dose of 50 µg and the two highest MB doses of 1x108 and 5x108 MBs. In two published investigations on in vivo UMGD to the heart that administered MBs and plasmid DNA intravenously, one found a proportional increase in efficiency with increased MB concentration 42 while the other showed improved efficiency with higher DNA doses 43. This correlation between increased DNA dose and improved gene delivery is not surprising given that DNA is the agent of intended intracellular delivery. Luo et al. 25 have demonstrated that methods which increase local DNA concentration particularly at the target cell surface augment transfection efficiency. A high administered DNA dose alone can achieve this by increasing the probability of plasmid DNA being in close proximity to the target cell membrane. Similarly, administering a large number of MBs may potentiate UMGD by increasing the probability of MBs being close to the target cell membrane, thereby increasing the number of sonoporation-enhancing cavitation nuclei available to assist in the formation of more transient cell membrane microperforations 42.
While maintaining a constant pFluc dose of 50 µg but varying MB dose, a saturation in UMGD efficiency was reached with 1x108 MBs for both cationic and neutral MBs; increasing MB dose to 5x108 MBs failed to yield a relative increase in in vivo gene transfer. Alter et al. 42, using the commercially available Sonazoid MB, also showed a saturation point at 3.6x108 MBs. At the higher MB doses employed in our study, it is possible that the 50 µg of pFluc initially administered was insufficient to take advantage of the enhancement associated with using more MBs and thus limited the potential for a further increase in gene delivery, suggesting that an optimal ratio of MB and plasmid DNA likely exists. Alternatively, an excessive number of intravascular MBs may prevent a further rise in transfection by causing irreversible microperforations and cellular damage 17, 24, 44. With the range of DNA doses examined, although no plateau was reached, one may have been found if higher DNA doses were tested. A saturation point in in vivo UMGD with the highest DNA doses provided was shown by Chen at al. 43 Testing DNA doses greater than 50 µg is warranted in future studies. Intravenous administration of up to 200 µg of DNA has been reported in previous murine UMGD studies 42.
We also systematically analyzed how varying DNA and MB dose affects relative gene delivery efficiencies in directly comparing cationic and control neutral MBs. Among the DNA doses tested, significant transfection superiority was found with cationic over neutral MBs for all DNA doses (10, 25, 37.5 and 50 µg) except for 17.5 µg. When 17.5 µg of pFluc was used, peak mean in vivo bioluminescence signal for the cationic MB group was 1.9-fold that of the neutral MB group; however, statistical significance was not achieved most likely due to high variability as reflected by the associated high standard deviation. For the four MB doses tested, use of cationic MBs resulted in enhanced gene delivery efficiency relative to neutral MBs for 1x107, 5x107, and 1x108 MBs but not 5x108 MBs. However, this cannot be explained by variability alone. Evaluation of trends show an inverse relationship between DNA and MB dose with C:N, where the degree of efficiency enhancement associated with use of cationic MBs was maximal at both the lowest DNA and MB doses and minimal at the highest doses (or nonexistent at 5x108 MBs). Interestingly, in a MB dose variation study of UMGD in cell culture, Nomikou et al. 18 found only a minimal relative increase in gene expression for cationic MBs compared to neutral MBs, which may not have reached statistical significance (no P value reported), at the highest concentration of MBs tested.
Overall, our results suggest that cationic MBs enhance in vivo UMGD efficiency the most when low DNA and/or MB doses are used. Conversely, systemic administration of a relative excess of plasmid DNA and/or MBs may override the efficiency advantages associated with use of cationic MBs in UMGD. Mechanistically, this also may be attributed to the increased probability of DNA or MBs being in close proximity to the cell membrane with high doses of either.
Our MB and DNA dose variation study data provide considerable insights into how best to exploit the gene delivery efficiency advantages afforded by using cationic MBs in in vivo UMGD. The implications for clinical translation of this technology are several-fold. While our results showed that increased MB and DNA dose improve gene delivery efficiency regardless of MB type, there will likely be limitations to the number of MBs and amount of DNA that can be safely administered systemically in patients. Use of high DNA and MB doses would also render this treatment costly. To scale up this gene delivery platform from mice to large animals and eventually humans will likely require a substantial increase in MB and DNA dose to accommodate markedly higher total blood volumes. Therefore, cationic MBs may play a critical role in limiting the dose of DNA and MBs while enhancing sonoporation efficiency enough to achieve a suitable level of gene transfer.
There are recognized limitations of this study. The potential for toxicities, particularly with the high MB and DNA doses, was not specifically evaluated in the in vivo studies. Furthermore, it is unknown whether the use of cationic over neutral MBs leads to any differences in potential adverse effects. Also, the findings described may be specific to the cationic and neutral MBs employed and conditions utilized and thus may not be generalizable to other MB designs, other US parameter settings, or other target in vivo tissue such as tumor models.
CONCLUSION
In conclusion, our results suggest that regardless of MB type (cationic versus neutral), increased MB or DNA dose increases in vivo UMGD efficiency with a saturation point reached with the highest two MB doses tested. However, upon examining how DNA and MB dose affect relative efficiencies in directly comparing cationic and control neutral MBs in in vivo UMGD, the efficiency advantage afforded by using cationic MBs was maximal at the lowest DNA and MB doses, reduced with higher doses, and abrogated with the highest MB dose tested. The use of cationic MBs for in vivo UMGD can therefore be considered a promising approach to reduce the required doses for successful gene delivery. Future studies will need to determine the optimal ratio of MB and DNA dose that best exploits the advantages of cationic MBs and to confirm that enhanced gene delivery with cationic MBs also results in improved treatment efficiency when using therapeutic genes.
Acknowledgments
This work has been supported by the German National Academic Foundation fellowship (C.M.P.), RSNA Research and Education Foundation Research Resident grant RR0823 (D.S.W.), RSNA Seed grant RSD0809 (J.K.W.), the SMIS NIH fellowship program (M.A.P.), and the Howard S. Stern Research Grant of the Society of Gastrointestinal Radiologists (J.K.W.).
References
- 1.Hernot S, Klibanov AL. Microbubbles in ultrasound-triggered drug and gene delivery. Adv Drug Deliv Rev. 2008;60:1153–66. doi: 10.1016/j.addr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaneko OF, Willmann JK. Ultrasound for molecular imaging and therapy in cancer. Quant Imaging Med Surg. 2012;2:87–97. doi: 10.3978/j.issn.2223-4292.2012.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glover DJ, Lipps HJ, Jans DA. Towards safe, non-viral therapeutic gene expression in humans. Nat Rev Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- 4.Newman CM, Bettinger T. Gene therapy progress and prospects: ultrasound for gene transfer. Gene Ther. 2007;14:465–75. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- 5.Schlicher RK, Radhakrishna H, Tolentino TP. et al. Mechanism of intracellular delivery by acoustic cavitation. Ultrasound Med Biol. 2006;32:915–24. doi: 10.1016/j.ultrasmedbio.2006.02.1416. [DOI] [PubMed] [Google Scholar]
- 6.Kimmel E. Cavitation bioeffects. Crit Rev Biomed Eng. 2006;34:105–61. doi: 10.1615/critrevbiomedeng.v34.i2.10. [DOI] [PubMed] [Google Scholar]
- 7.Miller DL, Pislaru SV, Greenleaf JE. Sonoporation: mechanical DNA delivery by ultrasonic cavitation. Somat Cell Mol Genet. 2002;27:115–34. doi: 10.1023/a:1022983907223. [DOI] [PubMed] [Google Scholar]
- 8.Greenleaf WJ, Bolander ME, Sarkar G. et al. Artificial cavitation nuclei significantly enhance acoustically induced cell transfection. Ultrasound Med Biol. 1998;24:587–95. doi: 10.1016/s0301-5629(98)00003-9. [DOI] [PubMed] [Google Scholar]
- 9.Pysz MA, Willmann JK. Targeted contrast-enhanced ultrasound: an emerging technology in abdominal and pelvic imaging. Gastroenterology. 2011;140:785–90. doi: 10.1053/j.gastro.2011.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kircher MF, Willmann JK. Molecular body imaging: MR imaging, CT, and US. part I. principles. Radiology. 2012;263:633–43. doi: 10.1148/radiol.12102394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher MF, Willmann JK. Molecular body imaging: MR imaging, CT, and US. Part II. Applications. Radiology. 2012;264:349–68. doi: 10.1148/radiol.12111703. [DOI] [PubMed] [Google Scholar]
- 12.Miller MW, Miller DL, Brayman AA. A review of in vitro bioeffects of inertial ultrasonic cavitation from a mechanistic perspective. Ultrasound Med Biol. 1996;22:1131–54. doi: 10.1016/s0301-5629(96)00089-0. [DOI] [PubMed] [Google Scholar]
- 13.Collis J, Manasseh R, Liovic P. et al. Cavitation microstreaming and stress fields created by microbubbles. Ultrasonics. 2010;50:273–9. doi: 10.1016/j.ultras.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Christiansen JP, French BA, Klibanov AL. et al. Targeted tissue transfection with ultrasound destruction of plasmid-bearing cationic microbubbles. Ultrasound Med Biol. 2003;29:1759–67. doi: 10.1016/s0301-5629(03)00976-1. [DOI] [PubMed] [Google Scholar]
- 15.Haag P, Frauscher F, Gradl J. et al. Microbubble-enhanced ultrasound to deliver an antisense oligodeoxynucleotide targeting the human androgen receptor into prostate tumours. J Steroid Biochem Mol Biol. 2006;102:103–13. doi: 10.1016/j.jsbmb.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Carson AR, McTiernan CF, Lavery L. et al. Gene therapy of carcinoma using ultrasound-targeted microbubble destruction. Ultrasound Med Biol. 2011;37:393–402. doi: 10.1016/j.ultrasmedbio.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tlaxca JL, Anderson CR, Klibanov AL. et al. Analysis of in vitro transfection by sonoporation using cationic and neutral microbubbles. Ultrasound Med Biol. 2010;36:1907–18. doi: 10.1016/j.ultrasmedbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomikou N, Tiwari P, Trehan T. et al. Studies on neutral, cationic and biotinylated cationic microbubbles in enhancing ultrasound-mediated gene delivery in vitro and in vivo. Acta Biomater. 2011;8:1273–80. doi: 10.1016/j.actbio.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Lentacker I, De Geest BG, Vandenbroucke RE. et al. Ultrasound-responsive polymer-coated microbubbles that bind and protect DNA. Langmuir. 2006;22:7273–8. doi: 10.1021/la0603828. [DOI] [PubMed] [Google Scholar]
- 20.Wang DS, Panje C, Pysz MA. et al. Cationic versus neutral microbubbles for ultrasound-mediated gene delivery in cancer. Radiology. 2012;264:721–32. doi: 10.1148/radiol.12112368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrara K, Pollard R, Borden M. Ultrasound microbubble contrast agents: fundamentals and application to gene and drug delivery. Annu Rev Biomed Eng. 2007;9:415–47. doi: 10.1146/annurev.bioeng.8.061505.095852. [DOI] [PubMed] [Google Scholar]
- 22.Liang HD, Tang J, Halliwell M. Sonoporation, drug delivery, and gene therapy. Proc Inst Mech Eng H. 2010;224:343–61. doi: 10.1243/09544119JEIM565. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, Qi H, Huang L. et al. Factors controlling the efficiency of cationic lipid-mediated transfection in vivo via intravenous administration. Gene Ther. 1997;4:517–23. doi: 10.1038/sj.gt.3300424. [DOI] [PubMed] [Google Scholar]
- 24.Zarnitsyn VG, Prausnitz MR. Physical parameters influencing optimization of ultrasound-mediated DNA transfection. Ultrasound Med Biol. 2004;30:527–38. doi: 10.1016/j.ultrasmedbio.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18:893–5. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 26.Schneider M, Anantharam B, Arditi M. et al. BR38, a new ultrasound blood pool agent. Invest Radiol. 2011;46:486–94. doi: 10.1097/RLI.0b013e318217b821. [DOI] [PubMed] [Google Scholar]
- 27.Li YS, Davidson E, Reid CN. et al. Optimising ultrasound-mediated gene transfer (sonoporation) in vitro and prolonged expression of a transgene in vivo: potential applications for gene therapy of cancer. Cancer Lett. 2009;273:62–9. doi: 10.1016/j.canlet.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Liang HD, Dong B. et al. Gene transfer with microbubble ultrasound and plasmid DNA into skeletal muscle of mice: comparison between commercially available microbubble contrast agents. Radiology. 2005;237:224–9. doi: 10.1148/radiol.2371040805. [DOI] [PubMed] [Google Scholar]
- 29.Liang HD, Lu QL, Xue SA. et al. Optimisation of ultrasound-mediated gene transfer (sonoporation) in skeletal muscle cells. Ultrasound Med Biol. 2004;30:1523–9. doi: 10.1016/j.ultrasmedbio.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 30.Lu QL, Liang HD, Partridge T. et al. Microbubble ultrasound improves the efficiency of gene transduction in skeletal muscle in vivo with reduced tissue damage. Gene Ther. 2003;10:396–405. doi: 10.1038/sj.gt.3301913. [DOI] [PubMed] [Google Scholar]
- 31.Ray P, Gambhir SS. Noninvasive imaging of molecular events with bioluminescent reporter genes in living subjects. Methods Mol Biol. 2007;411:131–44. doi: 10.1007/978-1-59745-549-7_10. [DOI] [PubMed] [Google Scholar]
- 32.Wasungu L, Hoekstra D. Cationic lipids, lipoplexes and intracellular delivery of genes. J Control Release. 2006;116:255–64. doi: 10.1016/j.jconrel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Tinkov S, Bekeredjian R, Winter G. et al. Microbubbles as ultrasound triggered drug carriers. J Pharm Sci. 2009;98:1935–61. doi: 10.1002/jps.21571. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Shollenberger LM, Conwell CC. et al. Mechanism of naked DNA clearance after intravenous injection. J Gene Med. 2007;9:613–9. doi: 10.1002/jgm.1054. [DOI] [PubMed] [Google Scholar]
- 35.Duvshani-Eshet M, Machluf M. Therapeutic ultrasound optimization for gene delivery: a key factor achieving nuclear DNA localization. J Control Release. 2005;108:513–28. doi: 10.1016/j.jconrel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 36.Meijering BD, Henning RH, Van Gilst WH. et al. Optimization of ultrasound and microbubbles targeted gene delivery to cultured primary endothelial cells. J Drug Target. 2007;15:664–71. doi: 10.1080/10611860701605088. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ, Greenleaf JF, Kinnick RR. et al. Ultrasound-mediated transfection of mammalian cells. Hum Gene Ther. 1996;7:1339–46. doi: 10.1089/hum.1996.7.11-1339. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe A, Otake R, Nozaki T. et al. Effects of microbubbles on ultrasound-mediated gene transfer in human prostate cancer PC3 cells: comparison among Levovist, YM454, and MRX-815H. Cancer Lett. 2008;265:107–12. doi: 10.1016/j.canlet.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Mehier-Humbert S, Yan F, Frinking P. et al. Ultrasound-mediated gene delivery: influence of contrast agent on transfection. Bioconjug Chem. 2007;18:652–62. doi: 10.1021/bc0602432. [DOI] [PubMed] [Google Scholar]
- 40.Danko I, Williams P, Herweijer H. et al. High expression of naked plasmid DNA in muscles of young rodents. Hum Mol Genet. 1997;6:1435–43. doi: 10.1093/hmg/6.9.1435. [DOI] [PubMed] [Google Scholar]
- 41.Doh SG, Vahlsing HL, Hartikka J. et al. Spatial-temporal patterns of gene expression in mouse skeletal muscle after injection of lacZ plasmid DNA. Gene Ther. 1997;4:648–63. doi: 10.1038/sj.gt.3300460. [DOI] [PubMed] [Google Scholar]
- 42.Alter J, Sennoga CA, Lopes DM. et al. Microbubble stability is a major determinant of the efficiency of ultrasound and microbubble mediated in vivo gene transfer. Ultrasound Med Biol. 2009;35:976–84. doi: 10.1016/j.ultrasmedbio.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Chen S, Shohet RV, Bekeredjian R. et al. Optimization of ultrasound parameters for cardiac gene delivery of adenoviral or plasmid deoxyribonucleic acid by ultrasound-targeted microbubble destruction. J Am Coll Cardiol. 2003;42:301–8. doi: 10.1016/s0735-1097(03)00627-2. [DOI] [PubMed] [Google Scholar]
- 44.Bao S, Thrall BD, Miller DL. Transfection of a reporter plasmid into cultured cells by sonoporation in vitro. Ultrasound Med Biol. 1997;23:953–9. doi: 10.1016/s0301-5629(97)00025-2. [DOI] [PubMed] [Google Scholar]