Abstract
Malignant glioblastomas are characterized by their ability to infiltrate into normal brain. We previously reported that binding of the multifunctional cytokine TNF-like weak inducer of apoptosis (TWEAK) to its receptor fibroblast growth factor–inducible 14 (Fn14) induces glioblastoma cell invasion via Rac1 activation. Here, we show that Cdc42 plays an essential role in Fn14-mediated activation of Rac1. TWEAK-treated glioma cells display an increased activation of Cdc42, and depletion of Cdc42 using siRNA abolishes TWEAK-induced Rac1 activation and abrogates glioma cell migration and invasion. In contrast, Rac1 depletion does not affect Cdc42 activation by Fn14, showing that Cdc42 mediates TWEAK-stimulated Rac1 activation. Furthermore, we identified two guanine nucleotide exchange factors (GEF), Ect2 and Trio, involved in TWEAK-induced activation of Cdc42 and Rac1, respectively. Depletion of Ect2 abrogates both TWEAK-induced Cdc42 and Rac1 activation, as well as subsequent TWEAK-Fn14–directed glioma cell migration and invasion. In contrast, Trio depletion inhibits TWEAK-induced Rac1 activation but not TWEAK-induced Cdc42 activation. Finally, inappropriate expression of Fn14 or Ect2 in mouse astrocytes in vivo using an RCAS vector system for glial-specific gene transfer in G-tva transgenic mice induces astrocyte migration within the brain, corroborating the in vitro importance of the TWEAK-Fn14 signaling cascade in glioblastoma invasion. Our results suggest that the TWEAK-Fn14 signaling axis stimulates glioma cell migration and invasion through two GEF-GTPase signaling units, Ect2-Cdc42 and Trio-Rac1. Components of the Fn14-Rho GEF-Rho GTPase signaling pathway present innovative drug targets for glioma therapy.
Introduction
Glioblastomas are the most malignant and common primary brain tumor in adults. Glioblastomas are highly infiltrative, leading to a poorly defined tumor mass. As a result, complete resection of the tumor is not feasible without compromising neurologic function, and despite adjuvant chemo- and radiation therapy, the 5-year survival rate is just under 10% (1). The invasive nature of glioblastoma correlates to an increased resistance to current therapeutic strategies, the mechanisms of which are complex and remain to be fully characterized (2).
Glioma cell invasion requires adhesion to extracellular matrix, subsequent degradation, and remodeling of this matrix, as well as signaling initiated by promigratory growth factors, and Rho GTPase–mediated organization and remodeling of the actin cytoskeleton (3). Specifically, the Rho GTPase family members RhoA, Rac1, and Cdc42 are key regulators of cell migration and have been implicated in the formation of stress fibers, induction of lamellipodia, and filopodia protrusion (4). The regulation of Rho GTPase activation is mediated by 3 classes of proteins: guanine nucleotide exchange factors (GEF), which are responsible for activating Rho GTPases to their GTP-bound state; GTPase-activating proteins (GAP), which enact phosphate bond hydrolysis thus inactivating Rho GTPases to a GDP-bound state; and GDP dissociation inhibitors (GDI) which bind to and stabilize Rho GTPases in their inactive GDP-bound form (5). To date, 22 Rho GTPases and 80 Rho GEFs belonging to either the Dbl or DOCK families have been identified (6).
Previous studies have shown that the fibroblast growth factor inducible–14 (Fn14) receptor can signal to induce Rac1 activation (7). Fn14 is a transmembrane receptor belonging to the TNF receptor superfamily (TNFRSF) and serves as the receptor for the multifunctional cytokine TWEAK (8). The Fn14 cytoplasmic tail lacks a death domain but contains TNFR-associated factor (TRAF) binding sites specific for TRAF1, 2, 3, and 5 (9). Fn14 expression is minimal to absent in normal brain tissue but correlates with tumor grade in glioblastoma (10). Activation of Fn14 by TWEAK promotes glioma cell migration, invasion, and survival (7, 10, 11). The TWEAK-Fn14 signaling axis mediates glioma migration and invasion via the Rac1 GTPase and fosters a self-promoting feedback loop whereby Rac1-mediatedTWEAK-Fn14 signaling induces Fn14 gene expression via the NF-κB pathway (7). While Rac1 is ubiquitously expressed among tissue types (12, 13), the levels of Rac1 protein expression in astrocytomas directly correlate with tumor grade in tissue microarray analysis. Furthermore, in glioblastoma, but not in lower grade astrocytomas, prominent plasma membrane staining of Rac1 is observed. These observations indicate that Rac1 is constitutively active in glioblastomas, underlining the importance of Rac1 in these tumors (14). To date, 5 GEFs that can activate Rac1 (Ect2, Vav3, Trio, SWAP-70, and Dock180- ELMO1) have been shown to contribute to the invasive behavior of glioblastoma (14–16). Four of these GEFs have been shown to be overexpressed in glioblastoma versus nonneoplastic brain (Ect2, Vav3, Trio, and SWAP-70; refs. 14, 16), and expression of Dock180 is higher in the tumor rim than in the tumor core.
In this study, we describe a role for TWEAK-Fn14 signaling through a multi-GEF, multi-Rho GTPase signaling pathway that includes Ect2, Trio, Cdc42, and Rac1. We show that Rac1 activation via TWEAK-Fn14 signaling is dependent upon Cdc42 function. We also report that Ect2 mediates Cdc42 activation, whereas Trio mediates Rac1 activation following TWEAK stimulation. Depletion of Ect2, Trio, or Cdc42 by siRNA oligonucleotides suppresses TWEAK-Fn14–induced Rac1 activation and subsequently glioma cell migration and invasion. Finally, we show that the inappropriate expression of either Fn14 or Ect2 in the astrocyte population of glial fibrillary acidic protein (GFAP)-tva transgenic mice induces astrocyte cell motility and proliferation, suggesting that the aberrant expression of Fn14 observed in glioblastoma may play an important role in glioblastoma progression, especially cell invasion.
Materials and Methods
Cell culture conditions
Human astrocytoma cell lines T98G and U118 (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle's Media (DMEM; Gibco) supplemented with 10% heat-inactivated FBS (Gibco) at 37°C with 5% CO2. For all assays with TWEAK treatment, cells were cultured in reduced serum (0.5% FBS) for 16 hours before stimulation with recombinant TWEAK at 100 ng/mL in DMEM + 0.1% bovine serum albumin (BSA) for the indicated time.
Antibodies, reagents, and Western blot analysis
A monoclonal Cdc42 antibody was purchased from Santa Cruz Biotechnology. Anti-myc was purchased from Cell Signaling Technologies. A polyclonal Ect2 antibody and a monoclonal antibody to tubulin were purchased from Millipore. Human recombinant TWEAK was purchased from PeproTech, and laminin from human placenta was obtained from Sigma. Lipofectamine 2000 was purchased from Invitrogen.
For immunoblotting, cells were lysed in 2× SDS sample buffer (0.25 mol/L Tris-HCl, pH 6.8, 10% SDS, 25% glycerol) containing 10 µg/mL aprotinin, 10 µg/mL leupeptin, 20 mmol/L NaF, 2 mmol/L sodium orthovanadate, and 1 mmol/L phenylmethylsulfonylfluoride. Protein concentrations were determined using the BCA Assay (Pierce) with BSA as a standard. Thirty micrograms of total cellular protein was loaded per lane and separated by SDS-PAGE. After transfer at 4°C, the nitrocellulose (Invitrogen) was blocked with either 5% nonfat milk or 5% BSA in TBS, pH 8.0, containing 0.1% Tween-20 (TBST) before addition of primary antibodies and followed with peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG. Protein bands were detected using SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific) with a UVP BioSpectrum 500 Imaging System.
Preparation of recombinant adenoviruses and infection
Adenoviruses expressing myc-tagged Fn14 wild-type protein and the cytoplasmic domain truncation mutant myc- Fn14tCT protein were previously described (7). Cells were infected at matched multiplicity of infections ranging from 5 to 20. For adenoviral infection, 1.5 × 105 cells were plated into a 6-well plate and cultured for 24 hours before infection.
Immunoprecipitation, Rac and Cdc42 activity assays, and nucleotide-free GEF pulldowns
For immunoprecipitation, cells were lysed on ice for 10 minutes in a buffer containing 10 mmol/L Tris-HCl (pH 7.4), 0.5% Nonidet P-40, 150 mmol/L NaCl, 1 mmol/L phenylmethylsulfonylfluoride, 1 mmol/L EDTA, 2 mmol/L sodium orthovanadate, 10 µg/mL aprotinin, and 10 µg/mL leupeptin (Sigma). Equivalent amounts of protein (500 µg) were precleared and immunoprecipitated from the lysates and then washed with lysis buffer followed by S1 buffer [10 mmol/L HEPES (pH 7.4), 0.15 mol/L NaCl, 2 mmol/L EDTA, 1.5% Triton X-100, 0.5% deoxycholate, and 0.2% SDS]. Samples were then resuspended in 2× SDS sample buffer and boiled in the presence of 2-mercaptoethanol (Sigma), separated by SDS-PAGE, transferred to nitrocellulose for 1 hours at 4°C, and proteins were detected using SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific).
Activity assays for Rac1 and Cdc42 were done according to the protocol of the manufacturer (Pierce). Lysates were harvested, and equal concentrations of lysates were assessed for Rac or Cdc42 activity. Affinity pull-down analyses of active GEFs bound to Rho GTPases were conducted using a nucleotide-free Rac1 mutant (G15A) expressed and purified as described (17). Recombinant Rac1 G15A-GST protein was produced in BL21 cells. Cells were lysed in B-PER lysis buffer (Pierce) containing protease inhibitors and purified with glutathione (GSH) beads. Equal amounts of total glutathione S-transferase (GST) fusion protein were then incubated with total cellular protein lysate (1 mg), and precipitated lysates were resolved with SDS-PAGE.
siRNA transfection
siRNA oligonucleotides specific for Rac1, GL2 luciferase (18), Ect2, and Trio were previously described (14). siRNA sequences for Cdc42 are Cdc42-1 (5′–AAAGACTCCTTTCTTGCTT- GT) and Cdc42-2 (5′–AATAACTCACCACTGTCCAAA). Transient transfection of siRNA was conducted using Lipofectamine 2000. Cells were plated at 70% confluency in DMEM + 10% FBS without antibiotics and were transfected within 8 hours of plating. The siRNA and Lipofectamine were diluted in serum-free DMEM. After 5 minutes, the mixtures were combined and incubated for 20 minutes at room temperature to enable complex formation. siRNA oligonucleotides were transfected at 50 nmol/L, and no cell toxicity was observed. Maximum inhibition of mRNA and protein levels was achieved 48 to 72 hours posttransfection.
Radial cell migration assay
Cell migration was quantified as previously described (19). Glioma cells were transfected with siRNA targeting luciferase, Ect2, or Cdc42. After 24 hours, cells were plated onto 10-well glass slides precoated with 10 µg/mL laminin. Cells were cultured in reduced serum (0.5% FBS) for an additional 16 hours before TWEAK addition, and migration rate was assessed over 24 hours.
Lentiviral production
The cDNA encoding murine myc-tagged Fn14 wild-type protein (pSec/Tag2/Fn14wt) or signal-deficient myc- Fn14tCT (7) was excised and ligated into the lentiviral transfer vector pCDH (System Biosciences) that contains a second transcriptional cassette for the expression of GFP. An empty pCDH vector expressing only GFP was used as a control. VSV-G pseudo-typed recombinant lentiviruses were produced by co-transfection of 293 packaging cells with the pCDH construct and the pPACK packaging mix (System Biosystems) according to the manufacturer's directions. For lentiviral transduction, medium containing recombinant lentiviruses was harvested from the packaging cells after 48 hours, concentrated by polyethylene glycol precipitation and centrifugation, and added to subconfluent cultures of T98G and U118 cells together with 8 µg/mL polybrene for 4 to 6 hours. Positively transduced cells were enriched by mass-sorting the GFP-positive cells on a Vantage flow cytometer (BD Biosciences).
Organotypic brain slice invasion assay
Preparation and culture of brain slices was carried out as described previously (20) with minor modification. Glioma cells (T98G and U118) stably expressing GFP were placed bilaterally onto the putamen of 400-µm thick slices of freshly isolated 4- to 6-week-old murine brains. Glioma cell invasion into the brain slices was quantified using the LSM 5 confocal microscope, and depth of invasion (z-axis stacks) was calculated as previously described (20).
Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry
For matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS), protein bands were stained with SYPRO ruby protein staining kit (Bio Rad) according to the manufacturer's protocol. Prominent protein bands present in the nucleotide-free Rac1-G15A mutant lysates were visualized under UV light and isolated. The samples of generated peptides were dissolved in 5 mL of 0.5% trifluoroacetic acid and measured by MALDI-TOF MS analysis on a Voyager reflector instrument (Applied Biosystems) and a Q-STAR mass spectrometer (Perceptive Biosystems) in positive ion mode at the University of Arizona (Tucson, AZ) Proteomic Facility. Data searches were done using the National Center for Biotechnology Information (NCBI) protein data bank with a minimum matching peptide setting of 4, a mass tolerance setting of 50 to 200 ppm, and a single trypsin miss cut setting.
Quantification of lamellipodia formation
Glioma cells were transfected with siRNA targeting luciferase, Rac1, Cdc42, Ect2, or Trio. After 24 hours, cells were plated onto 10-well glass slides precoated with 10 µg/mL laminin. Twenty-four hours later, cells were cultured in reduced serum (0.5% FBS) for an additional 16 hours before TWEAK stimulation for 5 minutes. Subsequently, cells were fixed in 4% formaldehyde/PBS, permeabilized with 0.1% Triton-X100, dissolved in PBS, and incubated with Alexa- Fluor phalloidin (Molecular Probes) to stain for F-actin. Slides were mounted with ProLong Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). Images were collected using a Zeiss LSM 510 microscope, equipped with a 63× objective, ZEN 2009 image analysis software, and Adobe Photoshop CS3. For each experimental condition, 10 images were taken randomly. Lamellipodia were traced using ImageJ software. For each cell, the fraction of the cell perimeter that displayed lamellipodia was calculated.
RCAS astrocyte-specific infection in transgenic mouse models
Immortalized DF-1 chicken fibroblasts (obtained from American Type Culture Collection) were transfected by calcium phosphate precipitation with plasmids in which the cDNAs for alkaline phosphatase (AP), Fn14, Ect2, and hepatocyte growth factor (HGF) were cloned into the ALV-A expression vector, RCAS-Y [a derivative of RCASBP(A); ref. 21]. Transfected cells were cultured in DMEM with 10% FBS and 1× penicillin/streptomycin, and passaged 1:3 every other day to maintain maximal logarithmic growth and viral production. Viral expression was detected by Western blotting using whole-cell lysates.
Mice (G-tva) expressing the TVA receptor protein under control of the GFAP promoter were used (22). Viral-producing DF-1 cells from a confluent T-75 flask were trypsinized, centrifuged, resuspended in 100 µL of medium, and placed on ice. DF-1 cells infected with RCASBP(A)-AP and RCASBP(A)-Fn14, -Ect2, or -HGF were combined in equal proportions and co-injected into the newborn mouse brain. A single intracranial injection of 5 µL was made at the intersection of the coronal and sagittal sutures using a Hamilton Gastight Syringe. The number of wild-type mice per cohort injected with virally producing DF-1 cells was as follows: alkaline phosphatase (n = 2), Fn14 (n = 15), Ect2 (n = 19), and HGF (n = 4). The number of transgenic (TG/+) mice per cohort injected with virally producing DF-1 cells was as follows: alkaline phosphatase (n = 4), Fn14 (n = 14), Ect2 (n = 18), and HGF (n = 10).
Mice injected with RCAS viruses were euthanized 10 weeks postinjection, and whole brains were removed and fixed in 10% neutral-buffered formalin overnight. After fixing, brains were dehydrated in 20% sucrose, 2% glycerol in PBS. Dehydrated tissues used for alkaline phosphatase staining were cut into 60-µm sections on a vibratome. Fixed brain tissue was incubated at 65°C in PBS (pH 9.5) for 1 hour to remove endogenous alkaline phosphatase activity. Sections were then subject to 1-Step NBT/BCIP (Thermo Scientific) until violet staining was visualized.
Statistical analysis
Statistical analyses were done using the 2-sample t test. P < 0.05 was considered significant.
Results
Ect2 binds to the Fn14 receptor and regulates TWEAK-stimulated Rac1 activation
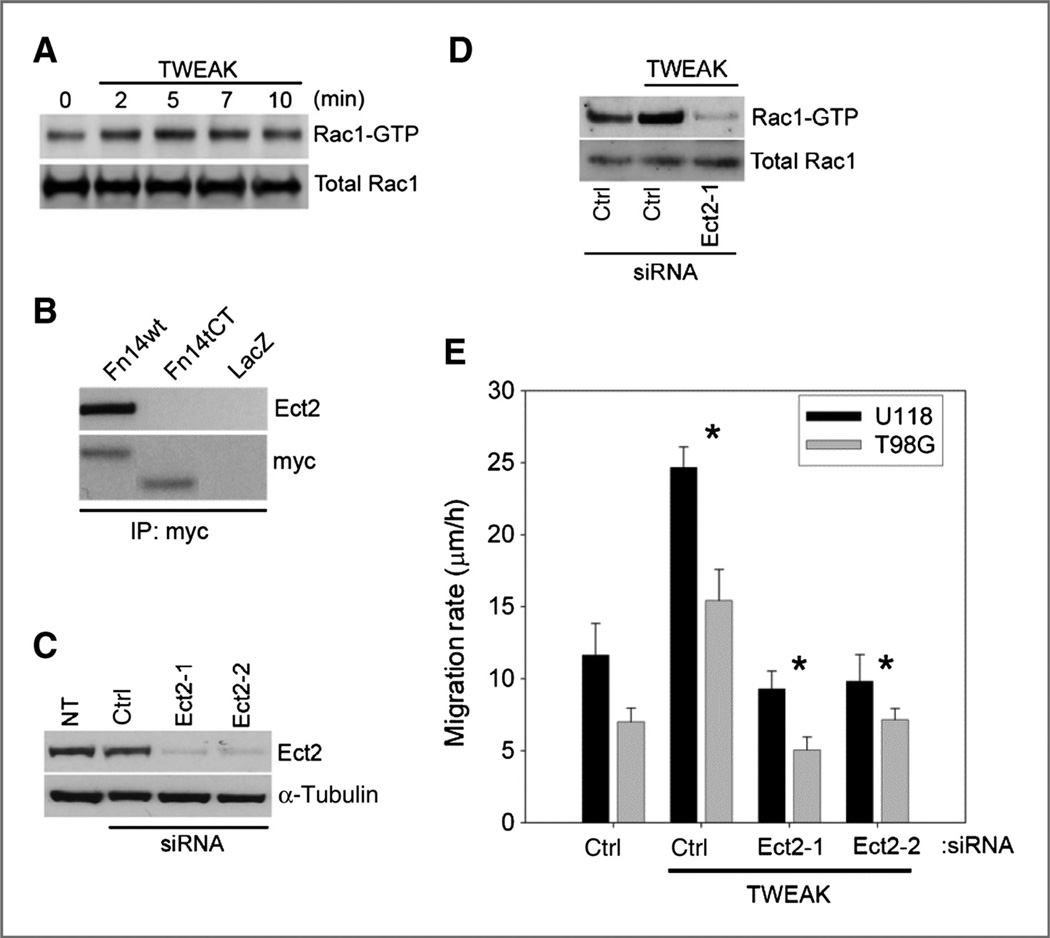
We have previously shown that TWEAK-Fn14 signaling stimulates Rac1 activity (7). Consistent with our previous data, TWEAK stimulation of glioma cells results in a rapid increase of Rac1 activity (Fig. 1A). We also reported that specific Rac1 GEFs (Ect2, Trio, and Vav3) are upregulated in glioblastomas and regulate glioma migration and invasion (14). Here, we examined which GEFs are important in regulating TWEAK-induced Rac1 activation. Co-immunoprecipitation studies indicated that the Fn14 wild-type receptor, but not an Fn14 mutant with a truncated cytoplasmic tail (tCT), interacted with Ect2 (Fig. 1B) but not Trio or Vav3 (data not shown). To determine whether Ect2 is important for TWEAK-induced Rac1 activation, we obtained 2 siRNAs targeting Ect2 and analyzed whether Ect2 depletion affected TWEAK-stimulated Rac1 activation. siRNA-mediated knockdown of Ect2 expression in the glioma cell lines with 2 independent siRNA oligonucleotides was approximately 90% effective (Fig. 1C). Depletion of Ect2 suppressed TWEAK-induced Rac1 activation to levels below the untreated baseline of GTP-bound Rac1 (Fig. 1D) and also suppressed-TWEAK-induced glioma cell migration to levels below baseline (Fig. 1E). These data implicate Ect2 as a nucleotide exchange factor for TWEAK-Fn14–mediated Rac1 activation and cell migration.
Figure 1.
Ect2 binds to Fn14 and regulates TWEAK-induced Rac1 activation and glioma cell migration. A, T98G cells were cultured in reduced serum followed by treatment with 100 ng/mL TWEAK for the indicated times. Whole-cell lysates were assessed for active Rac1 via affinity pull-down with the Pak1-binding domain (PBD). B, T98G cells were infected with adenoviruses expressing myc-tagged Fn14 wild-type (wt), Fn14 C-terminal–truncated (Fn14tCT), or control LacZ. Whole-cell lysates were immunoprecipitated (IP) for myc with subsequent Western blot analysis for Ect2 and myc. C, T98G cells were transfected with 2 siRNA oligonucleotides targeting Ect2 and assessed for the level of shutdown efficiency relative to nontransfected (NT) or ctrl siRNA transfection targeting nonmammalian luciferase (Ctrl) by Western blot analysis. D, T98G cells were cultured in reduced serum followed by treatment with TWEAK (100 ng/mL) for 10 minutes in the presence of either control (ctrl) luciferase–targeted siRNA or siRNA-targeting Ect2 and were assessed by Western blotting. E, glioma cells were transfected with siRNA targeting either control (ctrl) luciferase or 2 independent siRNA oligonucleotides to Ect2 (Ect2-1 and Ect2-2). After 24 hours, cells were cultured in reduced serum medium (0.5% FBS) for 16 hours and were seeded onto 10-well glass slides precoated with 10 µg/mL human laminin. Cells were either left untreated or treated with TWEAK, and glioma cell migration was assessed over 24 hours. Data represent the average of 3 independent experiments (*, P < 0.01).
TWEAK regulation of Rac1 activation is dependent upon Cdc42 function
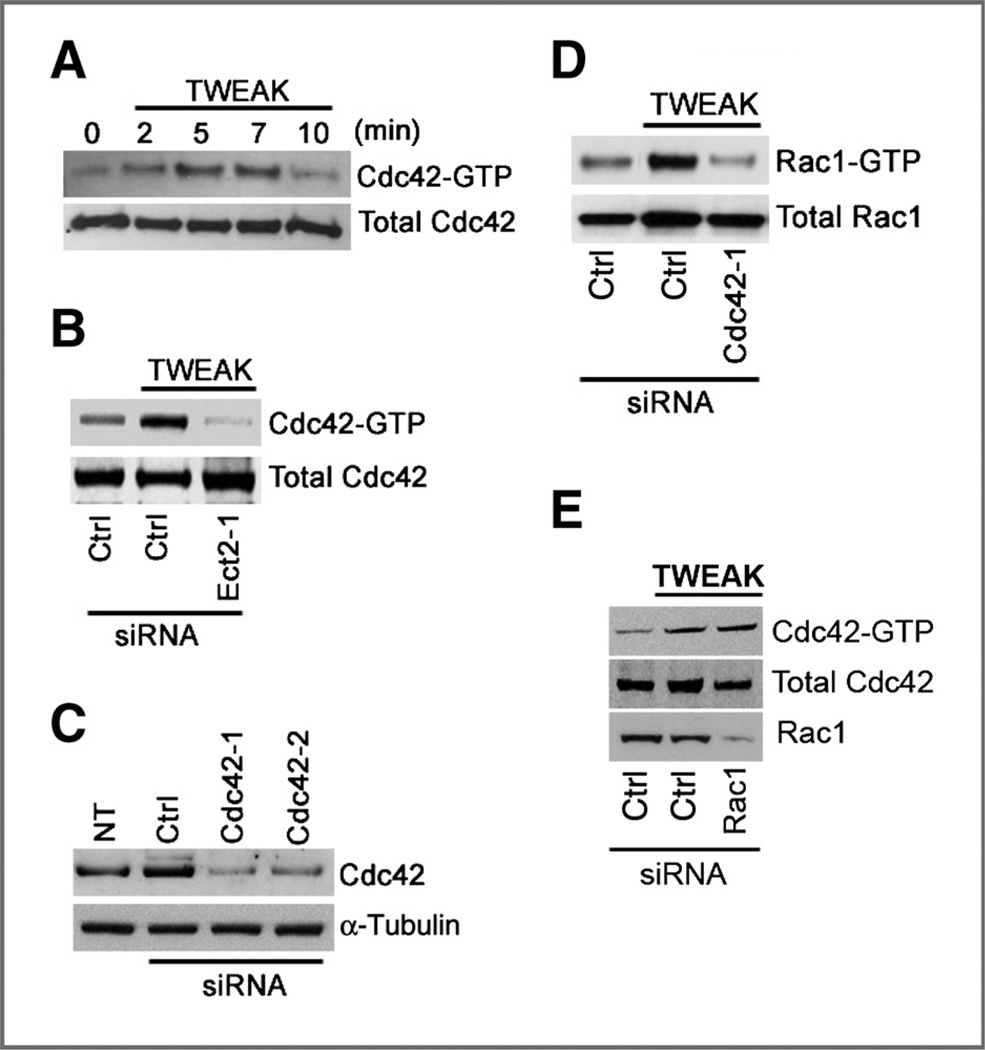
Rac1 has been shown to function downstream of Cdc42 in a number of different settings (23, 24). We therefore examined whether TWEAK also stimulates Cdc42 and whether Cdc42, in turn, mediates TWEAK-induced Rac1 activation. We found that Cdc42 is activated by TWEAK with maximal activation achieved at 5 minutes (Fig. 2A). Because Ect2 can catalyze nucleotide exchange on Cdc42 (18, 25), we also examined whether Cdc42 activation by TWEAK was dependent upon Ect2. Depletion of Ect2 by siRNA oligonucleotides prevents TWEAK-induced Cdc42 activation (Fig. 2B), indicating that Ect2 indeed mediates TWEAK-induced Cdc42 activation. To determine the epistatic relationship between Cdc42 and Rac1 in TWEAK signaling, we examined whether Cdc42 activity was necessary for Rac1 activation and vice versa. Using siRNA directed against Cdc42 (Fig. 2C), we assessed Rac1 activity under depletion of Cdc42. Depletion of Cdc42 expression resulted in the suppression of TWEAK-induced Rac1 activation (Fig. 2D), whereas Rac1 depletion had no effect on TWEAK-induced Cdc42 activation (Fig. 2E). Collectively, these data indicate that Ect2 mediates the activation of Cdc42 downstream of TWEAK-Fn14 and that, in turn, Cdc42 activates Rac1.
Figure 2.
TWEAK stimulation of Rac1 activation is dependent upon Cdc42. A, T98G cells were treated with TWEAK for the indicated times and assayed for Cdc42 activity using the Pak1-binding domain (PBD) assay. B, T98G cells were transfected with Ect2 siRNA oligonucleotides or luciferase control siRNA (Ctrl). Cells were then cultured under reduced serum (0.5% FBS) for 24 hours, followed by treatment with TWEAK for 5 minutes and then assessed for Cdc42 activity. C, T98G cells were either left nontransfected (NT), transfected with 2 independent siRNAs designed against Cdc42 (Cdc42-1 and Cdc42-2), or transfected with control siRNA. Cells were then lysed, and Western blot analysis for Cdc42 was conducted to confirm protein depletion. D, cells transfected with either control or Cdc42-targeting siRNA were cultured under reduced serum for 16 hours. Cells were then either left untreated or treated with TWEAK for 10 minutes and assayed for Rac1 activity. E, cells transfected with either control or Rac1-targeting siRNA were cultured under reduced serum for 16 hours. Cells were then either left untreated or treated with TWEAK for 5 minutes and assessed for Cdc42 activity.
Depletion of Cdc42 or Ect2 by siRNA suppresses TWEAK- or Fn14-induced cell migration and invasion
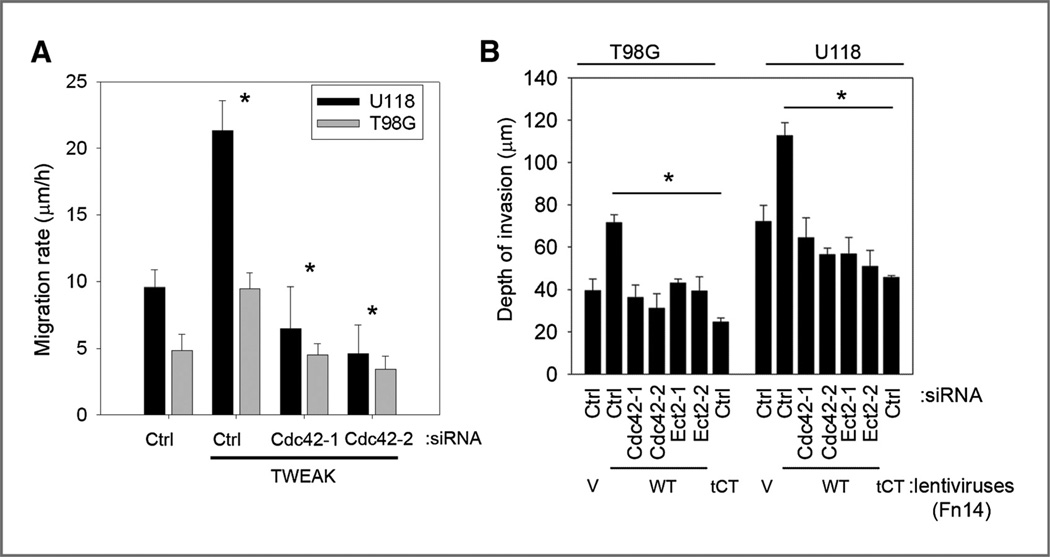
Cdc42 is known to control directional responses to extracellular cues at the front of the cell (4) and has been reported to play an important role in chemotaxis and cell invasion (26–28). Therefore, we investigated the role of Cdc42 function in TWEAK-induced glioma cell migration. TWEAK stimulation of migration was ablated in glioma cell lines under Cdc42-knockdown conditions (Fig. 3A). This level of inhibition of cell migration is similar to that caused by Ect2 depletion (Fig. 1E). We next assessed whether Cdc42 or Ect2 knockdown affects Fn14-stimulated invasion using an ex vivo rat brain slice model as detailed previously (20). Glioma cells infected with recombinant lentiviruses encoding GFP alone (V), wild-type Fn14 (WT), or Fn14 containing a truncation of the cytoplasmic domain (tCT) were transfected with siRNA oligonucleotides targeting either Cdc42 or Ect2 and subsequently assessed for depth of invasion into the brain slices over 48 hours. As reported previously (7), glioma cells overexpressing the Fn14wt receptor have increased invasive activity, whereas cells expressing the Fn14tCT receptor do not (Fig. 3B). siRNA-mediated knockdown of Cdc42 and Ect2 in glioma cells expressing Fn14wt abrogated Fn14-induced cell invasion relative to control nonmammalian- targeted siRNA in Fn14wt cells (Fig. 3B). These data corroborate the cell migration data and further show that Ect2-Cdc42 mediates Fn14-induced glioma cell migration and invasion.
Figure 3.
Cdc42 and Ect2 regulate TWEAK-induced glioma cell migration in vitro and Fn14-induced invasion ex vivo. A, T98G and U118 glioma cells were transfected with siRNA targeting either control nonmammalian luciferase (Ctrl) or 2 independent siRNA oligonucleotides targeting Cdc42 (Cdc42-1 and Cdc42-2). After 24 hours, cells were cultured in reduced serum medium (0.5% FBS) for 16 hours and were seeded onto 10-well glass slides precoated with 10 µg/mL human laminin. Cells were either left untreated or treated with TWEAK, and glioma cell migration was assessed over 24 hours. Data represent the average of 3 independent experiments (*, P < 0.01). B, T98G and U118 glioma cells were stably transduced with lentiviruses expressing GFP alone (V), the Fn14 wild-type (WT), or the cytoplasmic domain–truncated Fn14 receptor (tCT). In certain experiments, cells were either transfected with siRNA oligonucleotides against Cdc42, Ect2 or control luciferase (ctrl). Cells were implanted bilaterally onto the putamen of murine organotypic brain slices and observed at 48 hours. Depth of invasion was then calculated from z-axis images collected by confocal laser scanning microscopy. The mean value of the depth of invasion was obtained from 6 independent experiments (*, P < 0.01).
Trio activates Rac1 downstream of Fn14
Our data show that Ect2 depletion suppresses TWEAK-induced Cdc42 and Rac1 activation. Furthermore, depletion of Cdc42 expression by siRNA oligonucleotides suppresses TWEAK-induced Rac1 activation, which suggests that Ect2 is an indirect exchange factor for Rac1 and argues that an additional GEF(s) may activate Rac1 downstream of TWEAK-Fn14 signaling. To identify such GEFs, we conducted a Rho GEF pull-down assay using a nucleotide- free Rac1 GTPase mutant (G15A-Rac1) GST-fusion protein that can precipitate activated Rac1 GEFs out of cell lysates. T98G glioma cells were treated with TWEAK, subjected to the nucleotide-free Rac1 GEF pull-down assay, and proteins were resolved by SDS-PAGE analysis. Prominent protein bands present in the G15A-Rac1 complex under TWEAK stimulation but absent in the untreated G15A-Rac1 complex were recovered from the gel. Proteins were eluted, trypsin-digested, and MALDI-TOF and tandem mass spectrometric (MS-MS) analysis of the trypsin digests were conducted. A known Rac1-GEF, Trio, was a candidate protein identified by MS in the TWEAK-treated G15A-Rac1 complex. We therefore pursued Trio as a potential Rac1 activator, using 2 siRNAs that significantly reduce Trio expression (Fig. 4A). The increased activity of Rac1 seen upon TWEAK stimulation is eliminated under Trio-knockdown conditions (Fig. 4B), indicating that Trio mediates TWEAK-induced Rac1 activation. Interestingly, depletion of Trio expression by siRNA oligonucleotides had no effect on TWEAK-induced Cdc42 activation (Fig. 4C), suggesting that Trio functions downstream of Cdc42 to induce Rac1 activation. In addition, depletion of Trio inhibited TWEAK-induced glioma cell migration in vitro (Fig. 4D). Likewise, Trio depletion also abrogated Fn14-induced cell invasion in the ex vivo rat brain slice model (Fig. 4E), further supporting the function of Trio in mediating Fn14-induced cell invasion. Therefore, these data suggest that Trio functions downstream of Ect2 and Cdc42 to mediate TWEAK-Fn14–stimulated Rac1 activation.
Figure 4.
Trio activates Rac1 upon TWEAK stimulation or Fn14 overexpression. A, T98G cells were transfected with 2 siRNA oligonucleotides targeting Trio and assessed for the level of shutdown efficiency relative to nontransfected (NT) or ctrl siRNA transfection targeting nonmammalian luciferase (ctrl) by Western blot analysis. Glioma cells transfected with Trio siRNA were cultured under reduced serum for 16 hours followed by treatment with TWEAK for 10 minutes and assessment for Rac1 activity (B) or Cdc42 activity (C). D, T98G and U118 glioma cells were transfected with siRNA targeting either control nonmammalian luciferase (ctrl) or 2 independent siRNA oligonucleotides targeting Trio (Trio-1 and Trio-2). After 24 hours, cells were cultured in reduced serum medium (0.5% FBS) for 16 hours and were seeded onto 10-well glass slides precoated with 10 µg/mL human laminin. Cells were either left untreated or treated with TWEAK, and glioma cell migration was assessed over 24 hours. Data represent the average of 3 independent experiments (*, P < 0.01). E, T98G and U118 glioma cells were stably infected with lentiviruses expressing GFP and either the control vector (V), the Fn14 wild-type (WT), or the cytoplasmic domain–truncated Fn14 receptor (tCT). In certain experiments, cells were either transfected with siRNA oligonucleotides against Trio or control luciferase gene (ctrl). Cells were implanted bilaterally onto the putamen of murine organotypic brain slices and observed at 48 hours. Depth of invasion was then calculated from z-axis images collected by confocal laser scanning microscopy. The mean value of the depth of invasion was obtained from 6 independent experiments (*, P < 0.01).
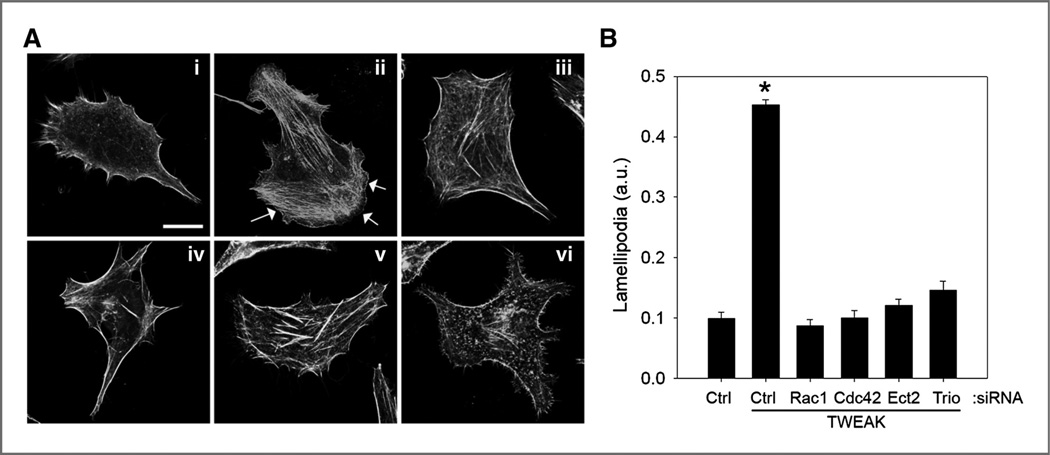
Depletion of Rac1, Cdc42, Ect2, or Trio by siRNA suppresses TWEAK-induced lamellipodia formation
Activated Cdc42 and Rac1 are known to control actin cytoskeleton–based dynamics and promote lamellipodia formation. To determine whether TWEAK activation of Cdc42 and Rac1 signaling confers changes in the actin cytoskeleton, we treated cells with TWEAK and examined filamentous actin using phalloidin staining. We found that TWEAK stimulation of glioma cells strongly stimulates lamellipodia formation [Fig. 5A, (ii, arrows)] as compared with control untreated cells [Fig. 5A, (i)]. Furthermore, treatment with siRNA oligonucleotides targeting Rac1, Cdc42, Ect2, or Trio significantly inhibited TWEAK-stimulated lamellipodia formation (Fig. 5A and B). These data are in line with the migration and invasion data and are consistent with the existence of a signaling cascade comprising Ect2, Cdc42, Trio, and Rac1 that functions downstream of Fn14 to control the invasive behavior of glioblastoma cells.
Figure 5.
TWEAK-induced lamellipodia formation in glioma cells requires the function of Rac1, Cdc42, Ect2, and Trio. T98G glioma cells were transfected with siRNA targeting either control nonmammalian luciferase (ctrl), Rac1, Cdc42, Ect2, or Trio. After 24 hours, cells were seeded onto 10-well glass slides precoated with 10 µg/mL human laminin. Cells were further grown for 24 additional hours and then cultured in reduced serum medium (0.5% FBS) for 16 hours. Cells were either left untreated or treated with TWEAK (5 minutes) and stained for filamentous actin using AlexaFluor-phalloidin. A, representative images of control transfected nontreated (i) or TWEAK-treated (ii) cells, as well as TWEAK-treated cells after depletion of Rac1 (iii), Cdc42 (iv), Ect2 (v), or Trio (vi) are shown. Arrows indicate lamellipodia. Bar, 10 µm. B, quantification of lamellipodia formation. Data represent the average of 10 cells per condition (*, P < 0.001). a.u., arbitrary units.
We also examined focal adhesions in TWEAK-stimulated glioma cells using immunofluorescence staining for vinculin and paxillin. However, we did not observe any significant changes in the number or distribution of focal adhesions upon TWEAK treatment of control cells or TWEAK stimulation of cells depleted of Rac1, Cdc42, Ect2, or Trio (data not shown).
Fn14 and Ect2 promote proliferation and migration of astrocytes in vivo
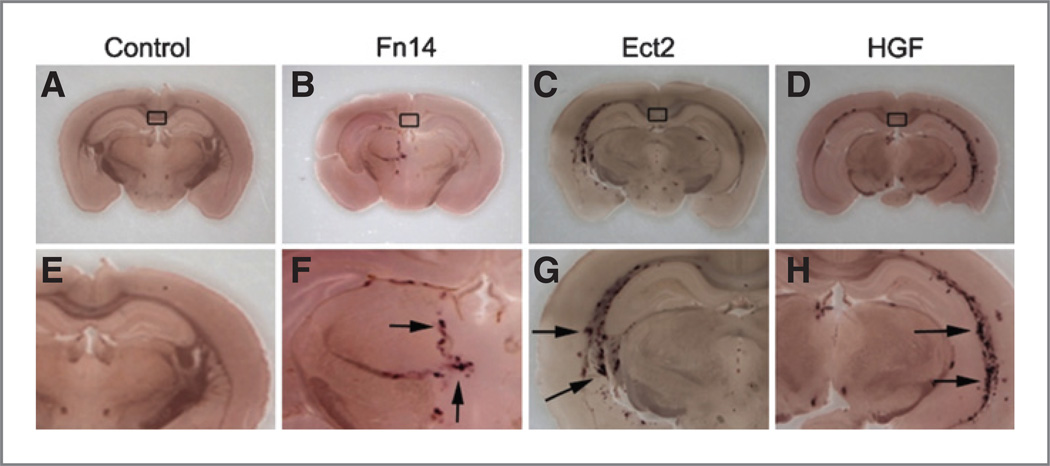
We previously showed that activation of the Fn14 receptor results in glioma cell migration and invasion in vitro (7); furthermore, Ect2 has also been implicated as a regulator of glioma cell migration and invasion (14), a key feature of glioblastoma tumors. To assess the role of Fn14 and Ect2 in vivo, we used transgenic mice engineered to express TVA, the avian cell surface receptor for subgroup A avian leukosis viruses (ALV-A), under control of the astrocyte-specific GFAP promoter. This allows for the selective infection of astrocytes with ALV-A from virus-infected and -producing DF-1 chicken fibroblasts to deliver both the Fn14 or Ect2 and alkaline phosphatase genes (22, 29, 30). It is important to note that the same cells producing alkaline phosphatase– expressing virus are used to infect TVA+ mice with alkaline phosphatase alone or in the combination of alkaline phosphatase and either Fn14-, Ect2-, or HGF-expressing viruses. Co-injection of DF-1 cells with Fn14 or Ect2 expressing and alkaline phosphatase–expressing viral vectors into the brains of the transgenic mice induced astrocytic cell migration along tracts at the intersection of the coronal and sagittal sutures (Fig. 6B and C and F and G). This effect was similar to that caused by co-injection of DF-1 cells infected with avian leucosis viruses expressing alkaline phosphatase and a known robust inducer of cell motility, HGF (ref. 31; Fig. 6D and H). The injection of DF-1 cells producing alkaline phosphatase–expressing viruses alone did not stimulate cell migration (Fig. 6A and E). Given the limited number of cells injected and the lack of selective proliferation or survival advantage under alkaline phosphatase expression alone, it is technically difficult to find these cells after necropsy and staining and thus the injection sites are labeled with a box (Fig. 6). Furthermore, the number of alkaline phosphatase– positive cells in the Fn14 or Ect2 and AP co-injection conditions was clearly increased relative to conditions in which alkaline phosphatase was injected alone, indicating a role for Fn14 and Ect2 in astrocyte proliferation in addition to migration. Co-injection of HGF- and alkaline phosphatase– producing DF-1 cells also resulted in increased numbers of alkaline phosphatase–staining cells. Moreover, analysis of virally infected cells in culture suggests no evidence of toxicity associated with alkaline phosphatase expression. This is consistent with previous studies using alkaline phosphatase expression as a glial cell marker with no evidence of deleterious effects on glial cell proliferation or survival in vitro (32) or in vivo (33) These data suggest that the abnormal expression of Fn14 or Ect2 can be an inducer of migration in situ. Thus, increased expression of Fn14 or Ect2 observed in glioblastoma may play a critical role in the malignant behavior of glioblastoma.
Figure 6.
Proliferation and migration of RCAS-Fn14 and RCAS-Ect2–infected astrocytic cells in vivo. DF-1 chicken fibroblasts producing subgroup A avian leukosis viruses (ALV-A) carrying the RCAS-AP plasmid along with either the RCAS-Fn14 (B and F), RCAS-Ect2 (C and G) or RCAS-HGF (D and H) plasmids were injected at the intersection of the coronal and sagittal sutures of transgenic mice expressing the TVA receptor for ALV-A under control of the GFAP promoter. In certain animals, chicken DF-1 cells producing RCAS-AP ALV-A viruses were injected alone as a negative control (A and E). Rectangles indicate area of injection. Arrows indicate alkaline phosphatase staining 10 weeks postinfection. Images (A–D) and (E–H) represent magnification at ×2 and ×4, respectively.
Discussion
In this article, we describe a signaling cascade comprising Ect2, Cdc42, Trio, and Rac1 that mediates TWEAK-stimulated glioblastoma cell invasion via the TNFRSF member Fn14 (Fig. 7). In addition, we report that ectopic expression of either Fn14 or Ect2 in the astrocytic lineage induces aberrant proliferation and migration of glial cells, underlining the importance of Fn14 and Ect2 in the malignant behavior of glioblastoma.
Figure 7.
Schematic model of TWEAK-Fn14 signaling via Cdc42 and Rac1 to drive glioma migration/invasion. We propose that TWEAK engagement of the Fn14 receptor results in the activation of Cdc42 by the Ect2 GEF. Ect2-Cdc42 signaling results in the activation of Rac1 by the Trio GEF to drive glioma cell migration and invasion. PM, plasma membrane.
Over the past several years, a number of Rho GEFs that contribute to the invasive behavior of glioblastoma cells have been identified: Ect2, Vav3, Trio, SWAP-70, and Dock180 (14–16). All these GEFs can act on the Rac1 GTPase in vitro (6, 25, 34, 35). In addition, siRNA-mediated depletion of these GEFs has been shown to diminish Rac1 activation in glioblastoma cells (14–16). Several of these GEFs can also act on Cdc42 and RhoA, but thus far, the roles of these GTPases in glioblastoma cell invasion have not been investigated. Here, we show that Cdc42 plays a critical role in glioblastoma cell migration and invasion stimulated by TNF family member TWEAK. Cdc42 is best known for its key role in the control of cell polarity (36), but it also has been implicated in the regulation of cell migration and invasion in a variety of different cell types (26, 28).
Our data also place Cdc42 upstream of Rac1 in TWEAK-stimulated cell migration and invasion. Importantly, depletion of Cdc42 completely inhibits TWEAK-stimulated Rac1 activation, indicating that the Ect2-Cdc42 module mediates the main, if not the only, signaling pathway that controls Rac1 activation downstream of TWEAK. We note that Cdc42 has been shown to function upstream of Rac1 in the regulation of a variety of different biologic functions, including cell polarity, cell migration, and glucose-stimulated insulin secretion (37, 38), although several Rac1- independent functions of Cdc42 also have been delineated (23, 24).
We also showed that depletion of the GEF Trio strongly inhibits TWEAK-stimulated activation of Rac1, but not of Cdc42, suggesting that Trio relays Cdc42 activation to that of Rac1. We note however that we cannot exclude the possibility that additional GEFs contribute to the activation of Rac1 by Cdc42. Indeed, in other systems, other GEFs have been shown to mediate Cdc42-induced Rac1 activation, including Cool-2/αPix and Tiam1 (37, 39), suggesting that Cdc42 may couple to Rac1 via different GEFs, depending on the biologic setting. Trio contains 2 GEF domains, which act on Rac1 and RhoA, respectively, but not Cdc42 (40). The role of RhoA in glioblastoma cell invasion remains to be elucidated however. Notably, we previously showed that stimulation of glioblastoma cells with TWEAK leads to a decrease in RhoA activation (7), making it unlikely that RhoA activity contributes to TWEAK-stimulated glioblastoma invasion.
Our data showing that depletion of Ect2 completely inhibits TWEAK-stimulated Cdc42 activation also indicate that Ect2 is the sole GEF that is responsible for Cdc42 activation in this setting. It is interesting to note that Ect2 can also act as an exchange factor for Rac1, both in vitro (25) and in cells (41). However, even though we observed that depletion of Ect2 strongly inhibits TWEAK-induced Rac1 activation, it is unlikely that Ect2 directly acts on Rac1 in TWEAK-stimulated glioblastoma cells, as depletion of Cdc42 completely inhibits TWEAK-stimulated Rac1 activation. Thus, our data support a model in which TWEAK-Fn14 in glioblastoma cells signals via Ect2, leading to Cdc42 and subsequently to Rac1 activation (Fig. 7). In addition, although our results suggest that Trio is a critical mediator of Rac1 activation downstream of TWEAK and Cdc42, in the absence of biochemical data showing that Trio activity is regulated by Cdc42, we cannot exclude the possibility that Trio may be activated via cross-talk with other TWEAK-stimulated signaling events. Interestingly, in lung cancer cells, Ect2 has been shown to mediate the activation of Rac1, but not Cdc42 (41). Moreover, during mitosis, Ect2 activates Cdc42 during metaphase but regulates RhoA during telophase (42, 43). Thus, these findings suggest that Ect2 acts on different GTPases depending on the cellular context. The molecular mechanisms that underlie the substrate specificity dynamics of Ect2 largely remain to be elucidated and may involve the interaction of Ect2 with different binding partners, such as MgcRacGAP and the PKC-Par6 complex (41–44).
The Fn14-Rac1 axis is highly deregulated in glioblastoma tumors. Expression levels of Fn14, Ect2, and Trio increase with astrocytoma grade and correlate with poor patient outcome (7, 14, 45). We also have noted an increase in the degree of cytoplasmic localization of Ect2 in glioblastoma versus low-grade astrocytoma (14). Ect2 localizes to the nucleus in nontransformed cells, and aberrant localization of Ect2 to the cytoplasm correlates with its oncogenic activity (46, 47). Rac1 expression levels also increase with astrocytoma grade (14). In addition, prominent Rac1 plasma membrane localization in a significant subset of glioblastoma tumors indicates that this GTPase is hyperactive in these tumors (14). We also note that the Fn14-Rac1-NF-κB axis contributes to the malignant behavior of glioblastoma cells by promoting cell survival (7, 11).
On the basis of their role in glioma cell migration, we assessed the ability of Fn14 and Ect2 to promote astrocyte migration and proliferation when inappropriately expressed in glial cells of transgenic mice. Previous work from Holland and Varmus showed that basic fibroblast growth factor production in astrocytes induces the proliferation and migration of glial cells, whereas injection of cells producing alkaline phosphatase–expressing virus alone results in only a small number of infected cells near the site of injection that do not migrate (22). Here, we report the ability of both Fn14 and Ect2 to induce cell proliferation and motility with no evidence of toxicity to cells when introduced into the murine astrocyte population.
Thus, the functional observations described in this article and our previous work (7, 11), together with the observations that Fn14 and Ect2 are overexpressed in patient glioblastoma tumors in comparison with non-neoplastic tissue (10, 14), strongly suggest that blocking Fn14 signaling may slow the invasion and progression of the promigratory population of glioblastoma tumors. Furthermore, our finding that Ect2, Cdc42, and Trio are essential signaling components that mediate TWEAK-stimulated activation of Rac1 suggests novel avenues for therapeutic intervention in TWEAK-Fn14–dependent glioblastoma tumors. The recent development of an immunoconjugate containing a highly specific anti-Fn14 monoclonal antibody conjugated to recombinant gelonin (rGel), a cytotoxic ribosome-inactivating N-glycosidase, that can kill Fn14-expressing cells in vitro and induce long-term tumor growth suppression in nude mice bearing T-24 human bladder cancer cell xenografts supports the proposal that Fn14-targeted agents may have antitumor effects (48). Additional therapeutic strategies targeting the TWEAK-Fn14 axis, including small-molecule antagonists of TWEAK trimerization or TWEAK-Fn14– binding and Fn14-Fc fusion proteins, have been recently reviewed (8). There is also an increasing body of literature that supports the feasibility of targeting GTPases and their cognate GEFs as promising anti-cancer therapies (49, 50). Thus, targeting the TWEAK-Fn14 signaling axis presents a unique therapeutic strategy for invasive glioblastoma cells.
Acknowledgments
The authors thank Dr. Keith Burridge (University of North Carolina, Chapel Hill, NC) for a gift of G15A-Rac1-GST plasmid and Dr. Eric Holland (Memorial Sloan- Kettering Cancer Center, New York, NY) for a gift of the transgenic mice.
Grant Support
The work was supported by NIH grants R01 CA130940 (N.L. Tran), R21 NS060023 (M.H. Symons), R01 NS055126 (J.A. Winkles), R01 CA103956 (J.C. Loftus), a VARI-TGen Integration Grant (N.L. Tran and B.O Williams), and the ARCS Foundation Eller Scholarship and Science Foundation Arizona Fellowship (S.P. Fortin).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed.
Authors' Contributions
Conception and design: S.P. Fortin, B.O. Williams, J.T.D. Ross, M.H. Symons, N. L. Tran
Development of methodology: C.R. Zylstra-Diegel, B.O. Williams, J.C. Loftus, M. H. Symons, N.L. Tran
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S.P. Fortin, M.J. Ennis, C.A. Schumacher, J.T.D. Ross, J.A.Winkles
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S.P. Fortin, M.J. Ennis, C.R. Zylstra-Diegel, J.T.D. Ross, M.H. Symons, N.L. Tran
Writing, review, and/or revision of the manuscript: S.P. Fortin, M.J. Ennis, B.O. Williams, J.A. Winkles, J.C. Loftus, M.H. Symons, N.L. Tran
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C.A. Schumacher, C.R. Zylstra-Diegel, J.T.D. Ross
Study supervision: N.L. Tran
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–2422. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 3.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 5.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 7.Tran NL, McDonough WS, Savitch BA, Fortin SP, Winkles JA, Symons M, et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res. 2006;66:9535–9542. doi: 10.1158/0008-5472.CAN-06-0418. [DOI] [PubMed] [Google Scholar]
- 8.Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7:411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown SA, Richards CM, Hanscom HN, Feng SL, Winkles JA. The Fn14 cytoplasmic tail binds tumour-necrosis-factor-receptor-associated factors 1, 2, 3 and 5 and mediates nuclear factor-kappaB activation. Biochem J. 2003;371:395–403. doi: 10.1042/BJ20021730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran NL, McDonough WS, Donohue PJ, Winkles JA, Berens TJ, Ross KR, et al. The human Fn14 receptor gene is up-regulated in migrating glioma cells in vitro and overexpressed in advanced glial tumors. Am J Pathol. 2003;162:1313–1321. doi: 10.1016/S0002-9440(10)63927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran NL, McDonough WS, Savitch BA, Sawyer TF, Winkles JA, Berens ME. The tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-fibroblast growth factor-inducible 14 (Fn14) signaling system regulates glioma cell survival via NFkappaB pathway activation and BCL-XL/BCL-W expression. J Biol Chem. 2005;280:3483–3492. doi: 10.1074/jbc.M409906200. [DOI] [PubMed] [Google Scholar]
- 12.Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. rac, a novel ras-related family of proteins that are botulinum toxin substrates. J Biol Chem. 1989;264:16378–16382. [PubMed] [Google Scholar]
- 13.Bosco EE, Mulloy JC, Zheng Y. Rac1 GTPase: a "Rac" of all trades. Cell Mol Life Sci. 2009;66:370–374. doi: 10.1007/s00018-008-8552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seol HJ, Smith CA, Salhia B, Rutka JT. The guanine nucleotide exchange factor SWAP-70 modulates the migration and invasiveness of human malignant glioma cells. Transl Oncol. 2009;2:300–309. doi: 10.1593/tlo.09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, et al. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 18.Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, et al. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J Biol Chem. 2004;279:25226–25233. doi: 10.1074/jbc.M313792200. [DOI] [PubMed] [Google Scholar]
- 19.Berens ME, Rief MD, Loo MA, Giese A. The role of extracellular matrix in human astrocytoma migration and proliferation studied in a microliter scale assay. Clin Exp Metastasis. 1994;12:405–415. doi: 10.1007/BF01755884. [DOI] [PubMed] [Google Scholar]
- 20.Nakada M, Niska JA, Miyamori H, McDonough WS, Wu J, Sato H, et al. The phosphorylation of EphB2 receptor regulates migration and invasion of human glioma cells. Cancer Res. 2004;64:3179–3185. doi: 10.1158/0008-5472.can-03-3667. [DOI] [PubMed] [Google Scholar]
- 21.Petropoulos CJ, Payne W, Salter DW, Hughes SH. Appropriate in vivo expression of a muscle-specific promoter by using avian retroviral vectors for gene transfer [corrected] J Virol. 1992;66:3391–3397. doi: 10.1128/jvi.66.6.3391-3397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci U S A. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 24.Berzat A, Hall A. Cellular responses to extracellular guidance cues. EMBO J. 2010;29:2734–2745. doi: 10.1038/emboj.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/Mphases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]
- 27.Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J Cell Biol. 1998;141:1147–1157. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz AA, Govek EE, Bottner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261:1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 29.Federspiel MJ, Bates P, Young JA, Varmus HE, Hughes SH. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc Natl Acad Sci U S A. 1994;91:11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner M, Kisseberth WC, Su Y, Besnard F, Messing A. GFAP promoter directs astrocyte-specific expression in transgenic mice. J Neurosci. 1994;14:1030–1037. doi: 10.1523/JNEUROSCI.14-03-01030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 32.Holland EC, Hively WP, Gallo V, Varmus HE. Modeling mutations in the G1 arrest pathway in human gliomas: overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev. 1998;12:3644–3649. doi: 10.1101/gad.12.23.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casper KB, McCarthy KD. GFAP-positive progenitor cells produce neurons and oligodendrocytes throughout the CNS. Mol Cell Neurosci. 2006;31:676–684. doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, et al. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature. 2002;416:759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]
- 35.Kiyokawa E, Hashimoto Y, Kobayashi S, Sugimura H, Kurata T, Matsuda M. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–3336. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etienne-Manneville S. Cdc42–the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, et al. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Oh E, Thurmond DC. Glucose-stimulated Cdc42 signaling is essential for the second phase of insulin secretion. J Biol Chem. 2007;282:9536–9546. doi: 10.1074/jbc.M610553200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baird D, Feng Q, Cerione RA. The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr Biol. 2005;15:1–10. doi: 10.1016/j.cub.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 40.Bellanger JM, Lazaro JB, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 41.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oceguera-Yanez F, Kimura K, Yasuda S, Higashida C, Kitamura T, Hiraoka Y, et al. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J Cell Biol. 2005;168:221–232. doi: 10.1083/jcb.200408085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu XF, Ishida H, Raziuddin R, Miki T. Nucleotide exchange factor ECT2 interacts with the polarity protein complex Par6/Par3/protein kinase Czeta (PKCzeta) and regulates PKCzeta activity. Mol Cell Biol. 2004;24:6665–6675. doi: 10.1128/MCB.24.15.6665-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, et al. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–1098. [PubMed] [Google Scholar]
- 46.Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- 47.Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50:190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H, Marks JW, Hittelman WN, Yagita H, Cheung LH, Rosenblum MG, et al. Development and characterization of a potent immunoconjugate targeting the Fn14 receptor on solid tumor Cells. Mol Cancer Ther. 2011;10:1276–1288. doi: 10.1158/1535-7163.MCT-11-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]