Abstract
Circadian clocks enable organisms to adapt to a 24 h diurnal cycle and anticipate rhythmic changes in the environment. The Arabidopsis central oscillator contains three genes encoding core clock components. CIRCADIAN CLOCK ASSOCIATED 1 (CCA1)/LATE ELONGATED HYPOCOTYL (LHY) and TIMING OF CAB EXPRESSION 1 (TOC1) reciprocally repress genes encoding each other and are critical for the generation of circadian rhythms controlling many clock outputs. A precise regulation of transcriptional events is, therefore, essential for proper circadian function. Here, we investigated histone 3 (H3) tail modifications of CCA1, LHY and TOC1 under various conditions. We found specific association of only H3K4Me3 and H3K9/14Ac with the translational start site of these three genes. These H3 marks were enriched at circadian time points of their increased transcription at different photoperiods and under free-running conditions, suggesting circadian regulation of H3 modifications. Analysis of clock-compromised CCA1-overexpressing lines provided evidence that light/dark photoperiods signal the establishment of these chromatin changes which are gated by the clock.
Keywords: Arabidopsis thaliana, CCA1, Chromatin modifications, Circadian clock, LHY, TOC1
Introduction
Many organisms possess an internal oscillator that allows them to detect the time of the day and adjust several of their biological processes to a 24 h cycle. This circadian clock is able to anticipate certain rhythmic changes, such as the end/beginning of the day, and execute the appropriate physiological responses. There is evidence that a functional clock is required for increased fitness of an organism (Ni et al. 2009). Circadian mutants that are unable to match their internal free-running periods to the appropriate duration of the day show reduced fitness when compared with wild-type (WT) plants (Green et al. 2002, Dodd et al. 2005). Entrained by environmental stimuli, such as photoperiod and temperature, the circadian clock is comprised of several interconnected feedback loops that eventually control the rhythms of expression of output genes (Imaizumi 2010, Pruneda-Paz and Kay 2010).
In Arabidopsis, the central oscillator relies on multiple interconnected loops that generate robust rhythms. At the heart of the central oscillator is a negative feedback loop between the morning expressed CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL) and the evening expressed TOC1 (TIMING OF CAB EXPRESSION 1 or PSEUDO RESPONSE REGULATOR 1). Recent reports have shown that similar to the other PRR family members (Nakamichi et al. 2010), TOC1 also functions as a repressor, and in this capacity TOC1 binds to the promoters of CCA1/LHY to repress expression of the latter two genes (Gendron et al. 2012, Huang et al. 2012, Pokhilko et al. 2012). CCA1 and LHY mRNAs accumulate at dawn, and their encoded proteins inhibit TOC1 transcription by binding to the evening element on the TOC1 promoter (Schaffer et al. 1998, Wang and Tobin 1998, Green and Tobin 1999, Alabadi et al. 2001, Alabadí et al. 2002, Green and Tobin 2002). Initial reports indicated that TOC1 mRNA accumulates at dusk and the protein indirectly promotes CCA1 and LHY transcription (Alabadi et al. 2001, Alabadí et al. 2002, Pruneda-Paz et al. 2009). However, the 12 h delay between the accumulation of TOC1 and the expression of CCA1/LHY was difficult to reconcile with this mechanism. Recently, TOC1 was shown to associate directly with the CCA1/LHY promoters and act as a repressor in early night (Gendron et al. 2012, Huang et al. 2012), whereas the other PRRs (PRR9, 7 and 5) maintain that repression throughout the day (Nakamichi et al. 2010). In this new model, the Arabidopsis clock structure includes a repressilator, a three-component negative feedback ring in which each member will repress the next one in the loop (Pokhilko et al. 2012). Members of the repressilator which connects the morning and evening loops were shown to repress each other sequentially as follows: (i) CCA1/LHY repress genes (LUX, ELF3 and ELF4) from the evening complex (EC) (Nusinow et al. 2011); (ii) the EC proteins will repress PRR genes (Dixon et al. 2011, Helfer et al. 2011); and (iii) PRRs (PRR9, 7, 5 and TOC1) will repress CCA1/LHY, thus closing the ring (Nakamichi 2011, Pokhilko et al. 2012). Although described initially in synthetic biology, the repressilator has also been proposed to function in animal clocks, where it accounts for a part of the whole central oscillator (Ukai-Tadenuma et al. 2011). In summary, the interplay between these different components largely accounts for the generation of output rhythms. Proper circadian function therefore depends on accurate transcriptional regulation of these three core clock genes: CCA1, LHY and TOC1.
Work in the last two decades has highlighted the importance of chromatin structure in transcriptional regulation. Modifications of histone N-terminal tails have been shown to determine the degree of compactness of nucleosomes, thereby modulating the transcriptional status of a certain gene locus (Benhamed et al. 2006, Feng et al. 2010). Acetylation and methylation of specific lysines within histone 3 (H3) and H4 tails are some of the major processes involved in the generation of activating or repressive chromatin marks. Recently, such dynamic chromatin modifications have been associated with circadian clock function in animals (Etchegaray et al. 2003, Naruse et al. 2004). CLOCK, a positive regulator within the mouse central oscillator, has been reported to possess histone acetyltransferase (HAT) activity (Doi et al. 2006, Hardin and Yu 2006, Nakahata et al. 2007). Moreover, this regulator is able to associate with MLL1, a mammalian homolog of Drosophila Trithorax with H3K4Me3 activity (Katada and Sassone-Corsi 2010, Masri and Sassone-Corsi 2010). Together, these findings suggest that circadian clock function comprises different layers of regulation, and dynamic changes in chromatin structure can be associated with the generation of circadian rhythmicity. Indeed, in Arabidopsis, TOC1 expression was shown to correlate with the accumulation of activating histone marks, such as acetylation, at the TOC1 promoter, an event that was inhibited by CCA1 binding (Perales and Más 2007, Stratmann and Más 2008). However, the acetylated residues on the H3 tails were not mentioned, and contributions from other H3 activating or repressive marks to TOC1 expression were not investigated. Recently, H3 acetylation and H3K4Me3 were shown to associate with the transcriptional rhythms of CCA1, LHY and TOC1, whereas H3K36Me2 levels displayed a negative correlation with the expression of these three circadian genes (Song and Noh 2012). Taken together, these reports provided the first insights into how dynamic histone modifications associate with the circadian waving patterns of genes constituting the central oscillator.
The final acetylation status of histone tails in nucleosomes is determined by the relative activities of HATs and histone deacetylases (HDACs). The transcriptional regulation of several light-responsive genes is controlled by TAF1 (HAT) and HD1 (HDAC) (Bertrand et al. 2005, Benhamed et al. 2006, Jang et al. 2011). However, whether these acetylating/deacetylating enzymes also play a role in clock gene regulation has not been explored. On the other hand, recent reports have implicated Jumonji domain-containing protein JMJ30/JMJD5, with putative histone demethylation activity, as well as protein arginine methyltransferase 5, in circadian regulation (Hong et al. 2010, Jones et al. 2010, Lu et al. 2011, Sanchez et al. 2010). These results suggest a role for histone modifications in clock function, but the molecular mechanisms underlying this relationship remain poorly understood.
Here we investigated H3 modifications typical of repressive (K9Me2, K9Me3 and K27Me3) or activating states (K4Me3, K9Ac, K14Ac and K9/14Ac) associated with the three central oscillator genes CCA1, LHY and TOC1. We found enrichment of only H3K4Me3 and H3K9/14Ac in sequences surrounding the translational start site (TSS) of these three genes. Moreover, dynamic changes of these activating H3 marks tracked closely the transcriptional waving patterns in different photoperiods and under free-running conditions, suggesting circadian modulation of chromatin modifications. To evaluate further the role of the circadian clock in chromatin modification, we performed a similar analysis in CCA1-overexpressing (OX) plants which are compromised in clock function (Wang and Tobin 1998). Under light/dark cycles, changes in activating chromatin marks associated with CCA1/LHY and TOC1 in the CCA1-OX plants mimicked the transcript profile of each gene, although with dampened amplitude. Under free-running conditions, however, there was hardly any enrichment of activating H3 marks associated with the three core clock genes in the CCA1-OX plants, and this paralleled the loss of oscillation of the corresponding transcripts. Our results suggest a mechanism in which photoperiod initiates the establishment of activating H3 marks within the TSS of the three core clock genes but these dynamic chromatin changes are gated and maintained by the circadian clock to restrict gene transcription to the appropriate times of the day/night cycles.
Results
H3 dynamics under short-day photoperiods
The central oscillator components CCA1, LHY and TOC1 display antiphasic transcription patterns. CCA1 and LHY transcripts peak in the early morning, whereas TOC1 accumulates at dusk (Stratmann and Más 2008). We first used WT plants grown under short-day (SD; 8 h light/16 h dark) conditions to study changes in H3 tail modifications accompanying transcriptional changes of these three genes. Chromatin immunoprecipitation (ChIP) experiments were performed at 4 h intervals over one light/dark cycle using antibodies specific for seven different H3 modifications (K4Me3, K9Ac, K14Ac, K9/14Ac, K9Me2, K9Me3 and K27Me3). Four primer pairs were used for each gene, each of which interrogated a specific genomic region, as follows: primer pair 1 was specific for a region upstream of the TSS; 2 for the sequence around the TSS; 3 for the gene body; and 4 for the 3′ untranslated region (3′ UTR).
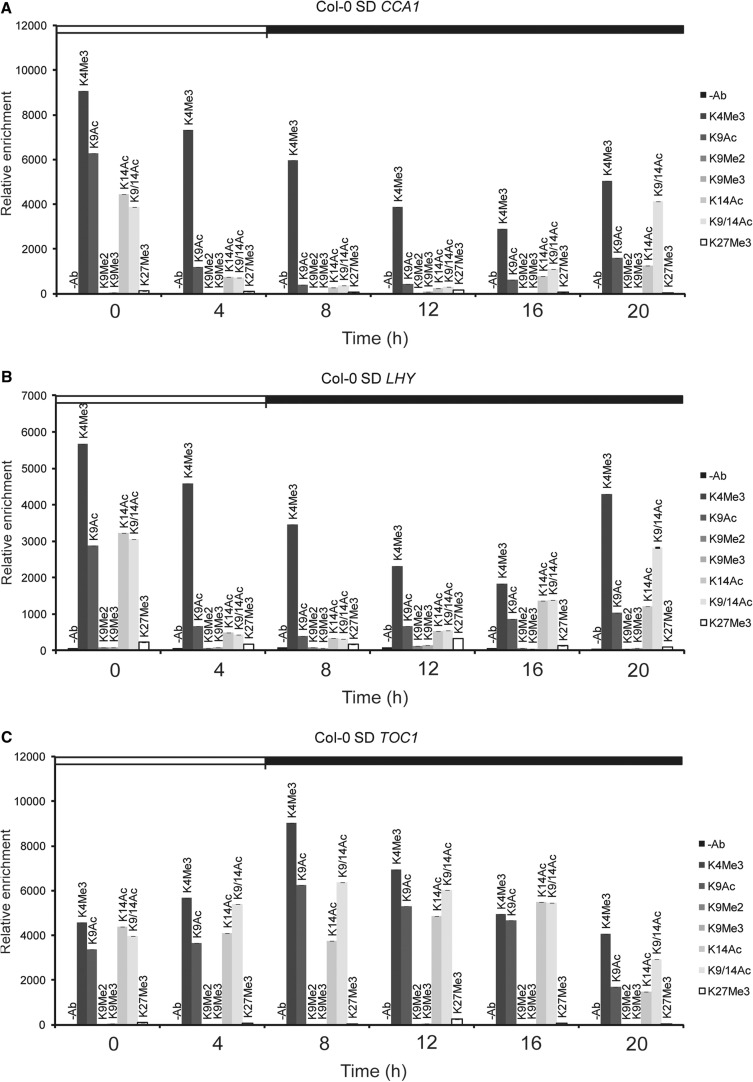
Changes in H3 tail modifications at different times of the day were seen only with primer pair 2 which amplified the TSS regions of CCA1, LHY or TOC1 (data not shown). Fig. 1 shows that the most prominent changes were found with H3K4Me3, H3K9Ac, H3K14Ac and H3K9/14Ac antibodies, all of which detect marks associated with active transcription. No significant changes were detected with the three repressive marks, H3K9Me2, H3K9Me3 and H3K27Me3 (Fig. 1), in agreement with earlier reports (Ni et al. 2009). More important, activating H3 marks associated with the CCA1 TSS followed a similar pattern to CCA1 mRNA accumulation under the same circadian conditions (Fig. 1A; Supplementary Fig. S1A). Similar results were obtained with LHY and TOC1 (Fig. 1B, C; Supplementary Fig. S1A). These results suggest that under SD conditions, activating H3 modifications that signal activated states are enriched at the TSS of central oscillator genes at times of active transcription.
Fig. 1.
Activating chromatin marks are associated with core clock genes, CCA1, LHY and TOC1 at times of their active transcription. (A–C) Sequences surrounding the translational start site region of CCA1 (A), LHY (B) and TOC1 (C) were interrogated in a 24 h period starting at dawn (ZT0). White and black boxes indicate day and night, respectively. ChIP was performed using antibodies against H3K4Me3, H3K9Ac, H3K9Me2, K3K9Me3, H3K14Ac, H3K9/14Ac or H3K27Me3. Control ChIP was performed without antibodies. Relative enrichment of the amplified region was plotted. Error bars represent the standard deviation (n = 3).
Histone 3 activating marks associated with the TSSs of CCA1/LHY and TOC1 are circadian regulated
Supplementary Fig. S1 shows that changes in CCA1, LHY and TOC1 transcript levels under different day lengths and in free-running conditions were basically similar to those published previously (Schaffer et al. 1998, Wang and Tobin 1998, Green and Tobin 1999, Alabadi et al. 2001). The oscillation pattern of CCA1/LHY transcript levels in seedlings grown under SDs peaked at the end of the night [Zeitgeber time (ZT) 20] and at dawn (ZT0), and was minimal at ZT12. Under long-day (LD) conditions (16 h light/8 h dark), the trough of CCA1/LHY expression was maintained at ZT12 but the peak was shifted at ZT0, showing a slight phase delay. In free-running experiments, SD-entrained seedlings were released into continuous dark (DD) for 48 h, whereas LD-grown seedlings were released into continuous light (LL) for the same period of time. As expected, CCA1 and LHY transcript levels oscillated with a phase similar to that seen under the original entrainment conditions but with decreased amplitude, which became more dampened under DD conditions (Supplementary Fig. S1). On the other hand, TOC1 transcript levels showed a phase delay in seedlings grown under LD as compared with SD, similar to that reported previously and to PRR5 expression (Kiba et al. 2007, Perales and Más 2007). Under free-running conditions, we found a decrease in the amplitude of TOC1 waving pattern, which was more pronounced under DD conditions (Supplementary Fig. S1), especially on the second subjective day.
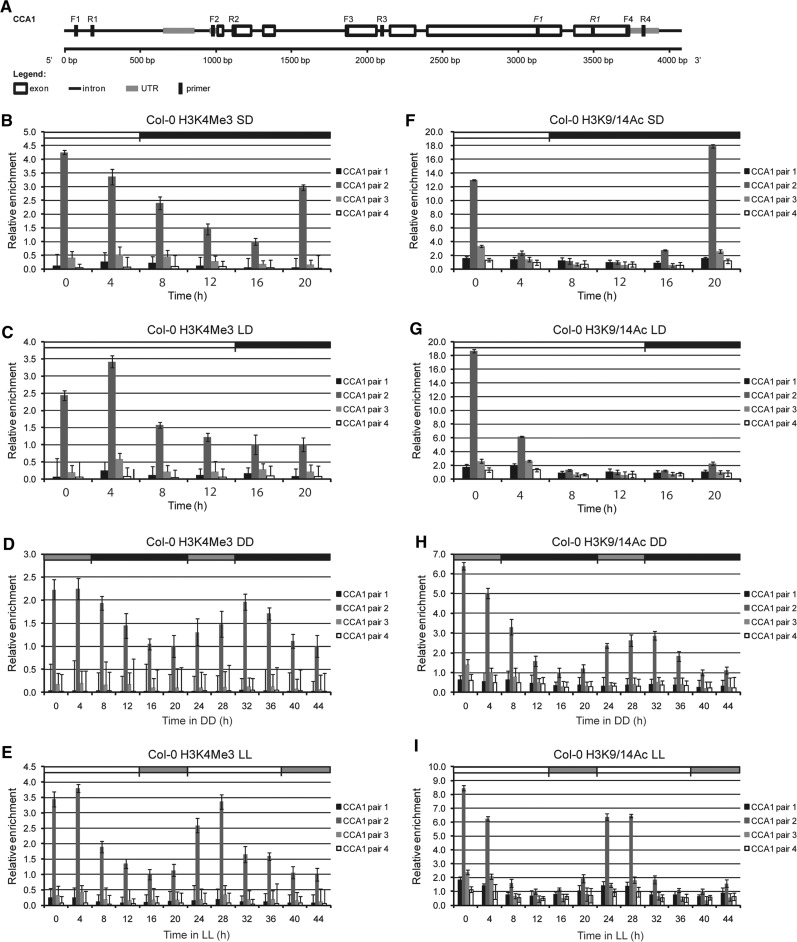
Next we investigated whether H3 modifications previously found in CCA1/LHY and TOC1 TSSs were under circadian control. Because the results obtained with the H3K9/K14Ac-specific antibody were similar to those obtained with anti-H3K9Ac and anti-H3K14Ac, we performed subsequent analysis with anti-H3K9/K14Ac. Initially, we determined changes in activating marks (H3K4Me3 and H3K9/K14Ac) surrounding the CCA1 TSS, and examined the three other regions of the CCA1 locus as well (Fig. 2). Fig. 2 shows that changes in H3K4Me3 and H3K9/K14Ac enrichment levels at the CCA1 TSS paralleled changes in CCA1 transcript levels in SD or LD and mirrored the slight delayed phase of transcript levels found in longer photoperiods. These modifications were specific of the TSS as they were absent in the other three tested regions (5′ upstream region, gene body and 3′ UTR). A similar enrichment in H3K4Me3 and H3Ac around the TSS of CCA1 and LHY has been recently reported even though seedlings were grown under 12 h light/12 h dark conditions and only two time points were investigated, ZT0 and ZT12 (Song and Noh 2012). Our analysis also showed that the peak of H3K9/14Ac enrichment preceded that of H3K4Me3 at times when the CCA1 transcript level was maximal (compare Fig. 2B with F and C with G).
Fig. 2.
Chromatin modifications of Histone 3 associated with CCA1. (A) Schematic representation of CCA1. The four primers used for quantitative PCR analysis of ChIP fragments are indicated. Primers used for quantitative RT–PCR analysis of transcripts are in italics. (B–I) ChIP analysis of CCA1 using antibodies against H3K4Me3 (B–E) or H3K9/14Ac (F–I) by quantitative PCR analysis (ACTIN2/7 as internal control). Error bars represent standard deviation values from two biological replicates, each sample being analyzed in triplicate (n = 6). (B, F) Short-day conditions [SD (8 h light : 16 h dark)]. (C, G) Long-day conditions [LD (16 h light : 8 h dark)]. (D, H) SD conditions followed by 48 h dark (DD). (E, I) LD conditions followed by 48 h light (LL). White and black boxes indicate day and night, respectively. Gray boxes represent subjective day (in DD) or subjective night (in LL).
Under free-running conditions (LL and DD), these activating H3 marks overlapped with the time points of CCA1 active transcription, suggesting circadian regulation of these modifications, in agreement with recent results (Song and Noh 2012) (Fig. 2; Supplementary Fig. S1). Moreover, under LL and DD, the waving patterns of H3K4Me3 and H3K9/14Ac modifications showed reduced amplitude with increased dampening under DD, similarly to the CCA1 transcript waving pattern (Fig. 2; Supplementary Fig. S1). Overall comparison of H3 modification oscillation patterns revealed a greater enrichment of H3K9/K14Ac (12- to 18-fold) compared with H3K4Me3 (4.0- to 6.0-fold), resulting in higher amplitude of the H3K9/14Ac waveform. Similar results were also obtained with activating H3 marks associated with LHY TSS (Supplementary Fig. S2). However, H3K4Me3 associated with CCA1/LHY TSSs for an extended time when compared with H3K9/K14Ac. This effect was also reported for the TSS of LHY when H3K4Me3 and H3Ac marks were investigated by Song and Noh (2012).
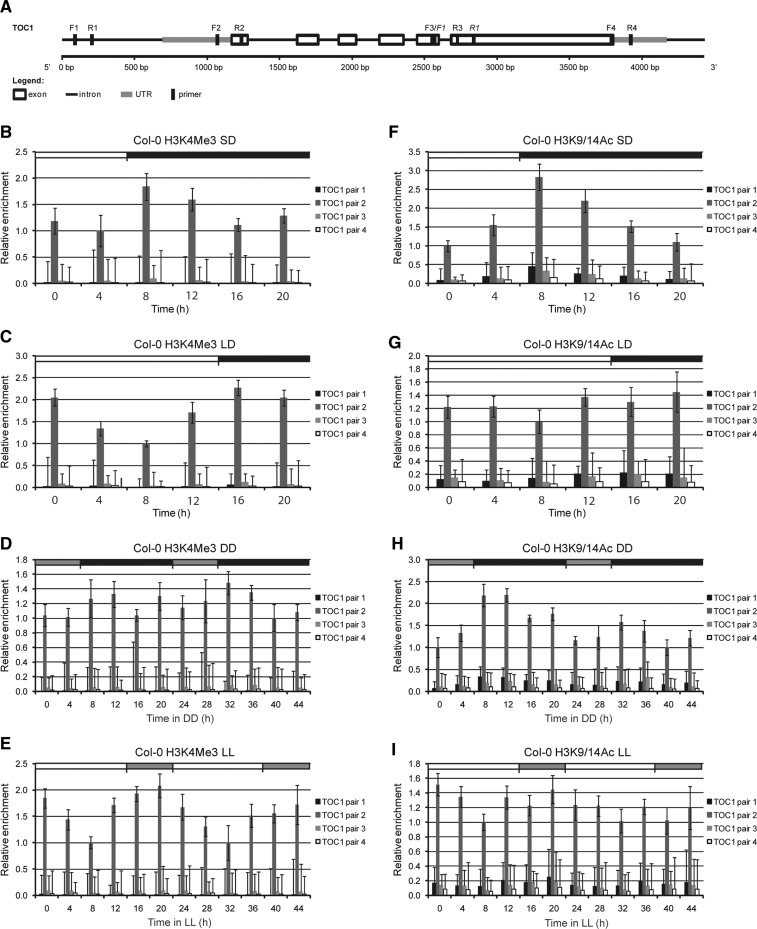
Because TOC1 expression is antiphasic to CCA1 and LHY, we also investigated changes in H3 modifications associated with the four different regions of the TOC1 locus under different circadian conditions. Similarly to CCA1/LHY, significant changes in H3 marks were found only in sequences surrounding the TSS (Fig. 3). Notwithstanding its antiphasic expression, under SD and LD, changes in H3K4Me3 and H3K9/K14Ac enrichment closely followed TOC1 rhythmic expression, similar to previous results (Song and Noh 2012). Moreover, the shift in the TOC1 transcription peak found in LD conditions when compared with SD conditions correlated with a delay in the peaks of H3K4Me3 and H3K9/14Ac marks associated with the TOC1 TSS (Fig. 3B, C, F, G; Supplementary Fig. S1A, B).
Fig. 3.
Chromatin modifications of Histone 3 associated with TOC1. (A) Schematic representation of TOC1. The four primers used for quantitative PCR analysis of ChIP fragments are indicated. Primers used for quantitative RT–PCR analysis of transcripts are in italics. (B–I) ChIP analysis of TOC1 using antibodies against H3K4Me3 (B–E) or H3K9/14Ac (F–I) by quantitative real-time PCR analysis (ACTIN2/7 as internal control). Error bars represent standard deviation values from two biological replicates, each sample being analyzed in triplicate (n = 6). (B, F) Short-day conditions [SD (8 h light : 16 h dark)]. (C, G) Long-day conditions [LD (16 h light : 8 h dark)]. (D, H) SD conditions followed by 48 h dark (DD). (E, I) LD conditions followed by 48 h light (LL). White and black boxes indicate day and night, respectively. Gray boxes represent subjective day (in DD) or subjective night (in LL).
Under free-running conditions, however, the changes in H3K4Me3 and H3K9/14Ac modifications associated with the TOC1 TSS did not completely overlap with the TOC1 transcript oscillation, in contrast to CCA1 and LHY (Fig. 3D, E, H, I; Supplementary Fig. S1C, D). This was especially the case when plants were released into LL and analyzed for H3K9/14Ac modifications (Fig. 3I; Supplementary Fig. S1D). These results differ from those of previous reports (Perales and Más 2007, Song and Noh 2012), although the H3Ac antibodies used in those studies were not specified but probably differ from those used here and the conditions of entrainment preceding the release into LL were not the same.
Taken together, our results show that H3K4Me3 and H3K9/K14Ac marks associated with the CCA1, LHY and TOC1 TSSs displayed an enrichment waving pattern that mostly matched the timing of expression of those genes under the same circadian conditions. Moreover, the patterns of H3K4Me3 and H3K9/14Ac enrichment were still maintained under free-running conditions, albeit with some dampening of the amplitude, indicating that these H3 tail modifications are regulated by the circadian clock.
The circadian clock and photoperiod mediate H3K4 tri-methylation and H3K9/14 acetylation
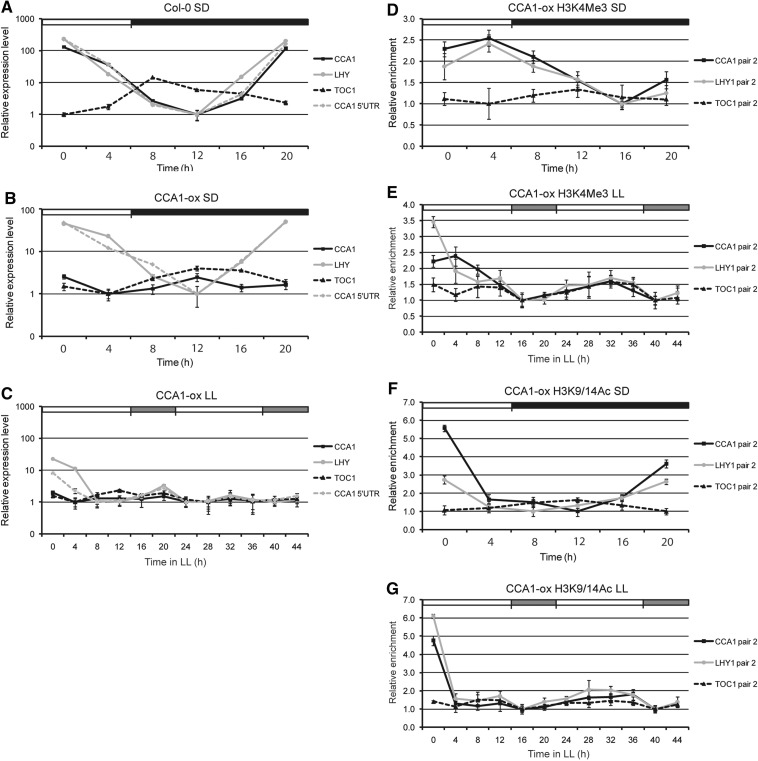
To obtain additional evidence of circadian regulation of H3K4Me3 and H3K9/14Ac marks associated with the three core clock genes and to distinguish the relative contribution of photoperiod and circadian clock function, we examined changes in these activating H3 marks in CCA1-OX transgenic plants. Previous work has shown that overexpression of CCA1 from a 35S promoter compromises clock function in CCA1-OX transgenic plants (Wang and Tobin 1998). To determine the endogenous CCA1 transcript levels, we designed a specific primer pair to amplify the CCA1 5′ UTR. We confirmed its specificity since in CCA1-OX plants grown under SD conditions the endogenous CCA1 and LHY transcript displayed a similar waving pattern to WT plants (peak at ZT0 and trough at ZT12) although with a reduced amplitude, and this rhythmic behavior was lost when a non-discriminating primer pair was used (Fig. 4A, B). The WT results that are shown in Supplementary Fig. S1A were used again in Fig. 4A to allow comparison with the CCA1-OX results.
Fig. 4.
Circadian rhythms of CCA1, LHY and TOC1 transcription and H3 activating marks in CCA1-OX plants. (A–C) Quantitative RT–PCR analysis (ACTIN2 as control) of CCA1, LHY and TOC in Col-0 (A) and CCA1-OX plants (B, C) under SD conditions (A, B) or free-running conditions (C). In CCA1-OX plants, both the endogenous and transgenic CCA1 transcripts were amplified using CCA1 primers, whereas the endogenous CCA1 transcript was determined using the CCA1 5′ UTR primers. Relative mRNA levels are represented on a logarithmic scale. (D–G) ChIP analysis of translation start site regions of CCA1, LHY or TOC1 using antibodies against H3K4Me3 (D, E) or H3K9/14Ac (F, G) by quantitative real-time PCR analysis (ACTIN 2/7 as control) under SD (D, F) or free-running conditions (E, G). Error bars represent standard deviation values from two biological replicates, each sample being assayed in triplicate (n = 6). White, black and gray boxes indicate day, night and subjective night, respectively.
Confirming earlier reports, the circadian pattern of TOC1 transcript in CCA1-OX plants grown under SD showed a reduction in amplitude and a delay in the appearance of its peak (at ZT8 in Col-0 and ZT12 in CCA1-OX) (Perales and Más 2007). Under LL, however, CCA1, LHY and TOC1 transcription was repressed and their typical circadian rhythms were lost (Fig. 4C) (Wang and Tobin 1998, Perales and Más 2007).
We monitored changes in H3K4Me3 and H3K9/K14Ac marks in CCA1-OX plants grown under SD and LL (Fig. 4D–G). Similar to previous experiments, we tested the four different primer pairs for each gene, and the detailed results are presented in Supplementary Figs. S3 and S4. H3 enrichment was seen in the region related to the TSS that is detected by primer pair 2. These results are shown in Fig. 4. In SD, the H3K4Me3 pattern associated with CCA1 and LHY TSS showed a delay of 4 h in the peak and trough when compared with the corresponding transcript levels. Although we detected a moderate oscillation in this pattern, there was some dampening of the wave when compared with the WT (Fig. 4D; Supplementary Fig. S3A, B). Consistent with the inhibition of TOC1 transcription in CCA1-OX plants (Perales and Más 2007), the H3K4Me3 mark associated with the TOC1 TSS showed reduced amplitude and loss of rhythmicity (Fig. 4D; Supplementary Fig. S3C). Under LL, we detected a stronger dampening of the oscillation pattern of H3K4Me3 associated with CCA1 and LHY, and loss of rhythmicity of this activating mark associated with the TOC1 TSS (Fig. 4E; Supplementary Fig. S4A–C).
Next, we analyzed the circadian pattern of H3K9/14Ac marks in CCA1-OX plants under the same conditions. In SD, H3K9/14Ac associated with the CCA1 and LHY TSSs was highly enriched at times of active transcription but decreased rapidly (Fig. 4F; Supplementary Fig. S3D, E), generating a waving pattern with reduced amplitude when compared with the WT. In contrast, H3K9/14Ac enrichment in the TOC1 TSS was very much reduced, with loss of oscillation (Fig. 4F; Supplementary Fig. S3F). Similar to H3K4Me3 marks, the decrease in amplitude with loss of waving patterns was even more pronounced under LL conditions. We found that H3K9/14Ac marks associated with CCA1, LHY and TOC1 TSSs showed reduced or even loss of rhythmic oscillation (Fig. 4G; Supplementary Fig. S4D–F). Moreover, similar to the WT, the overall enrichment in H3K9/14Ac marks was higher when compared with H3K4Me3 modifications.
In summary, both H3K4Me3 and H3K9/14Ac marks associated with CCA1, LHY and TOC1 TSSs showed a reduced enrichment in CCA1-OX plants, in agreement with the reduced oscillation patterns of their corresponding transcripts, and this effect was even more pronounced when light/dark entrainment was removed, e.g. under LL conditions. These results suggest that changes in activating H3 marks associated with the core clock genes are affected when circadian function is compromised, and this effect is accentuated in the absence of light/dark cycles.
Acetylation status of H3K9/14 in core clock genes is differently affected by TAF1 and HD1
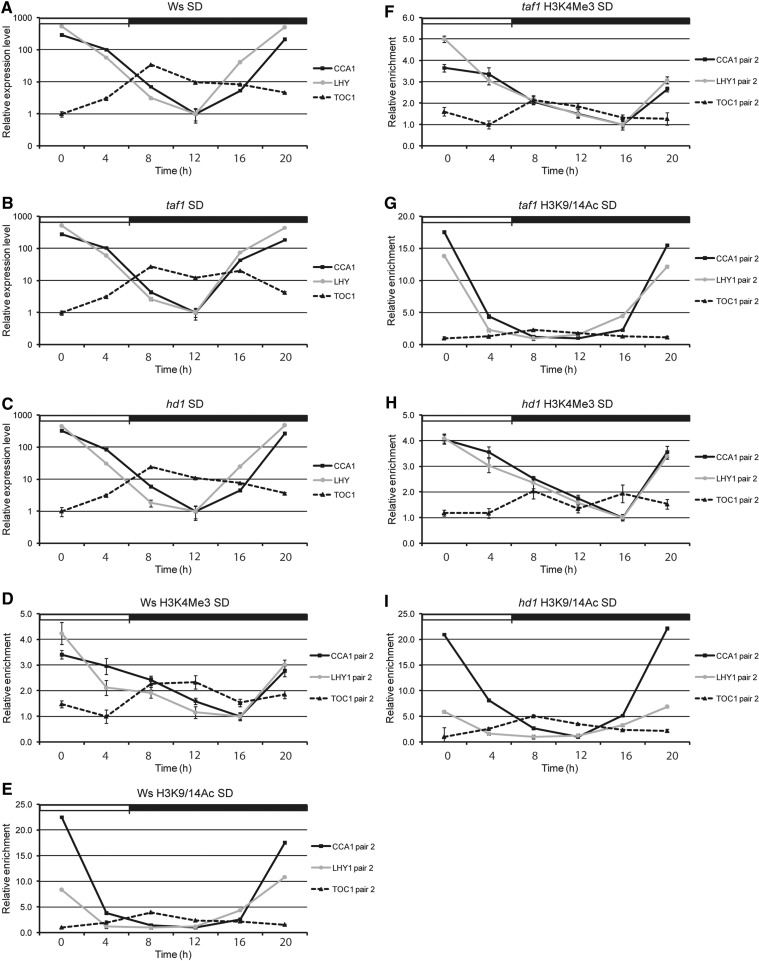
So far, our results suggest that the interplay between light/dark cycles and circadian clock function results in changes in H3K9/14Ac enrichment that are important for the regulation of CCA1, LHY and TOC1 transcription. Therefore, it is reasonable to assume the existence of a dynamic mechanism mediating H3 acetylation/deacetylation under photoperiod and circadian control. Previous reports have uncovered a role for TAF1, a HAT, and HD1, a HDAC, in the regulation of several light-responsive genes (Bertrand et al. 2005, Benhamed et al. 2006). To see whether these two histone modification enzymes are involved in the regulation of the three clock genes, we entrained wild type [Wassilewskija (Ws)], taf1 and hd1 mutant plants in SD and determined CCA1, LHY and TOC1 transcript waving patterns. In comparison with the WT (Ws), taf1 and hd1 mutants showed a very similar oscillation pattern for the three genes, with CCA1/LHY increasing in the early morning (ZT0) and minimal at ZT12. TOC1 waving showed a peak at dusk (ZT8) and a trough at ZT0, although in taf1 mutants there was a small increase in TOC1 transcript at ZT16 (Fig. 5A–C). Taken together these results suggest that taf1 and hd1 mutants possess a running clock.
Fig. 5.
Transcriptional and H3 oscillation patterns of CCA1, LHY and TOC1 in hd1 and taf1 mutants. (A–C) Quantitative RT–PCR analysis (ACTIN2 as internal control) of CCA1, LHY and TOC1 in Arabidopsis thaliana Ws (A), taf1 (B) and hd1 (C) plants over a 24 h period under SD conditions. Relative mRNA levels are represented on a logarithmic scale. (D–I) ChIP results showing H3K4Me3 (D, F, H) or H3K9/14Ac (E, G, I) enrichment in CCA1, LHY and TOC1 TSS in Ws (D, E), taf1 (F, G) and hd1 (H, I) plants. The immunoprecipitated CCA1, LHY or TOC1 fragments were detected by quantitative PCR analysis. As internal control, ACTIN2/7, was used for normalization, and error bars represent standard deviation values from two independent biological replicates, each sample being analyzed in triplicate (n = 6). White and black boxes indicate day and night, respectively.
Next, we investigated how H3K4Me3 and H3K9/14Ac marks associated with CCA1, LHY and TOC1 were affected in these mutants. As before, we failed to obtain enrichment in any genomic regions outside the TSS (Supplementary Figs. S5–S7). In both taf1 and hd1 mutants, H3K4Me3 modifications associated with the CCA1 TSS displayed a waving pattern similar to the WT (Ws) (Fig. 5D, F, H; Supplementary Figs. S5A, S6A, S7A). However, in the case of H3K9/14Ac, there was a delay of 4 h in the rate of loss of CCA1 TSS acetylation in hd1 when compared with the WT (Fig. 5E, I; Supplementary Figs. S5D, S7D), although the overall circadian oscillation of CCA1 H3K9/14Ac marks remained unaffected. In contrast, changes in H3K9/14Ac modifications in taf1 showed a circadian pattern similar to the WT, with a peak at ZT0 and a trough at ZT16 (Fig. 5E, G; Supplementary Figs. S5D, S6D). We also determined the circadian oscillation pattern of these H3 marks associated with the LHY TSS. Similarly to CCA1, we failed to detect a significant change in the waving of H3K4Me3 marks in both taf1 and hd1 mutants (Fig. 5D, F, H; Supplementary Figs. S5B, S6B, S7B). However, in comparison with the WT, we did not find any delay in H3K9/14Ac associated with the LHY TSS but rather a slight dampening of the wave in hd1 and an opposite small increase in amplitude in taf1 (Fig. 5G, I; Supplementary Figs. S5E, S6E, S7E).
As mentioned above, TOC1 transcription is antiphasic to CCA1 and LHY, and this waving pattern was still maintained in both taf1 and hd1 (Fig. 5A–C). We examined the H3K4Me3 mark at the TOC1 TSS and found no changes in taf1, but a small decrease in these marks at ZT12 in hd1 plants when compared with the WT (Fig. 5D, F, H; Supplementary Figs. S5C, S6C, S7C). In the case of H3K9/14Ac enrichment at the TOC1 TSS, the oscillation pattern of hd1 was similar to that of the WT. However, in taf1 plants, we detected dampening of the wave (approximately 2-fold reduction), suggesting a moderate loss in acetylation marks in these mutants (Fig. 5E, G, I; Supplementary Figs. S5F, S6F, S7F).
Together, our results indicate that TAF1 and HD1 may contribute, to some extent, to TOC1 acetylation and CCA1 deacetylation, respectively. Considering that 12 HAT and 18 HDAC genes have been identified in Arabidopsis (Benhamed et al. 2006), it is likely that, because of gene function redundancy, prominent changes in H3K9/14 acetylation/deacetylation can be seen only when double/triple mutants are analyzed.
Discussion
Circadian clock function relies on the coordinate action of different molecular events that restrict transcription of circadian-regulated genes to specific times of the day. Components of the central oscillator are interlocked in multiple feedback loops in which different regulators affect each other and their own transcription, generating the necessary rhythms underpinning the physiological responses (Bell-Pedersen et al. 2005, Pruneda-Paz and Kay, 2010). Within the Arabidopsis central oscillator, three major components have been identified, CCA1 and LHY, two Myb-domain transcription factors (Schaffer et al. 1998, Wang and Tobin, 1998), and TOC1, a PRR (Strayer et al. 2000, Alabadi et al. 2001). A perfect time-keeping mechanism critically depends on the transcription of these components at the appropriate circadian times. Recent reports have described findings similar to those described here. Song and Noh (2012) identified H3 activating marks (H3K4Me3 and H3Ac) that associate with CCA1, LHY and TOC1 TSSs at the times of their active transcription under light and dark cycles as well as free-running conditions. They also provided the first evidence of a H3 negative mark (H3K36Me2) that accumulated in the promoter regions of CCA1, LHY and TOC1 at the times of their lowest expression. Here, we have confirmed their findings but also provided further evidence on how dynamic histone modifications affect the expression of these core clock genes. We showed that light and dark maintain the oscillation in chromatin modifications when the clock function is impaired, such as CCA1 overexpression, but the clock gates these modifications to the appropriate times of the day. We also investigated in further detail the acetyltransferases and deacetylases that could account for these events. Taken together, our results considerably extend the existing knowledge and provide a more complete overview of how photoperiod, clock and chromatin remodeling control the daily oscillations of CCA1, LHY and TOC1 transcripts. These findings are discussed in detail below.
Activating H3 marks associated with the TSS of core clock genes
Transcriptional regulation is associated with chromatin modifications including methylation, acetylation and phosphorylation of histone N-terminal tails (Feng et al. 2010). We investigated changes in H3 N-terminal tail modifications associated with the genomic loci for the three core components of the central oscillator. Besides their fundamental role in clock function, CCA1, LHY and TOC1 transcript levels oscillate 100- (CCA1 and LHY) to 10-fold (TOC1) between peak and trough. These large fluctuations in transcript levels allowed detection of clear changes in H3 modifications (2- to 18-fold enrichment). For each of these three genes, we interrogated four separate genomic regions, comprising the 5′ upstream region of the TSS, the TSS, the gene body and the 3′ UTR. Changes in enrichment in seven different H3 modifications, K4Me3, K9Ac, K14Ac, K9/14Ac, K9Me2, K9Me3 and K27Me3, were monitored. We found only activating H3 marks, namely H3K4Me3 and H3K9/14Ac, to be specifically associated with the TSS of CCA1, LHY and TOC1. These results suggest that circadian clock function relies on dynamic chromatin modifications that are hallmarks of actively transcribed sites (Pfluger and Wagner 2007, Cazzonelli et al. 2009, Ni et al. 2009).
Photoperiod regulates dynamic rhythms of activating H3 marks
Irrespective of the photoperiod used, an increase in transcript levels was accompanied by an enrichment of both H3K4Me3 and H3K9/14Ac, in agreement with previous findings (Song and Noh 2012). However, we failed to detect any phase advance for histone marks in relation to transcription initiation as previously described (Perales and Más 2007). Correspondingly, a decrease in transcript level was accompanied by a decrease in H3K4Me3 and H3K9/14Ac marks. However, the accumulation of H3K4Me3 and H3K9/14Ac marks at each TSS followed different oscillatory patterns. H3K9/14Ac marks at CCA1 and LHY TSSs peaked at times of highest transcription but quickly declined, whereas H3K4Me3 accumulation/disappearance greatly overlapped with transcript accumulation. Moreover, we found that under longer photoperiods there was a phase delay in terms of both CCA1/LHY transcription and associated H3 activating marks. This was also the case for TOC1, confirming previous results, although we failed to see any increase in amplitude (Perales and Más 2007). When compared with CCA1/LHY, H3 modifications associated with the TOC1 TSS displayed a highly similar profile of daily changes that mimicked the oscillation of TOC1 transcripts. These results suggest that proper accumulation of these three core clock transcripts depends on the establishment of activating H3 marks at their TSS, and erasure of these modifications correlates with down-regulation of their transcription. The correlation found between circadian-regulated transcription and these H3 modifications has also been described in animals, where rhythmic H3K9/14 acetylation and H3K4 tri-methylation have been shown to overlap and regulate transcription in the clock (Etchegaray et al. 2003, Naruse et al. 2004, Katada and Sassone-Corsi 2010). In fact one of the core components of the mouse central oscillator, CLOCK, is a H3K9/14 HAT whose function is essential for circadian regulation (Doi et al. 2006, Hardin and Yu, 2006).
We have recently reported that phyA active transcription in the dark was marked by enrichment of activating H3K4Me3, H3K9/14Ac and H3K27Ac at the TSS (Jang et al. 2011). On the other hand, light-mediated transcriptional repression was accompanied not only by a decrease in these activating marks but also by an increase in the repressive H3K27Me3 mark. The simultaneous implementation of activating and repressive histone marks appears to provide a tight mechanism for a rapid turn on and switch-off of phyA transcription in response to dark and light. In contrast, our results with CCA1, LHY and TOC1 indicate an association between the loss of activating histone marks and transcriptional repression of these genes, since we failed to see any enrichment in the repressive marks tested. However, we did not investigate the enrichment in H3K36Me2 on the promoters of the three core clock genes, which was shown to accumulate at the times of their lowest transcription (Song and Noh 2012), suggesting a repressive role for this H3 modification.
The circadian clock controls the oscillation pattern of H3K4Me3 and H3K9/14Ac at the TSS of core clock genes
In agreement with a recent report (Song and Noh 2012), under free-running conditions (LL or DD) the association between H3K9/14Ac and H3K4Me3 and CCA1 and LHY TSSs still displayed a clear waving pattern. For these two genes, changes in the enrichment of these activating H3 marks generally followed the transcript oscillation pattern, but there was a clear wave dampening especially in the second cycle of DD. In the case of TOC1 under DD conditions, the enrichment of H3K9/14Ac and H3K4Me3 showed a similar pattern and mimicked the transcript accumulation. However, under LL conditions, the H3K4Me3 mark oscillated but there was a partial loss of H3K9/14Ac rhythmicity, and we failed to see a complete association between this mark and TOC1 transcript oscillation as has been reported by Song and Noh (2012). Nevertheless, because oscillation of H3K4Me3 and H3K9/14Ac was still preserved under DD/LL in most cases, our results on the three core clock genes provide evidence of circadian control of changes in these activating H3 marks. Moreover, the dampening of the H3 modification wave form suggests that light/dark cycles may contribute to the relative enrichment level of the activating H3 marks.
To distinguish the relative contribution of the circadian clock and photoperiod to the regulation of these activating histone modifications, we analyzed H3K4Me3 and H3K9/14Ac in CCA1-OX plants grown under SD or LL conditions (Wang and Tobin 1998). H3 modifications associated with CCA1/LHY still oscillated under SD, similar to their transcripts, albeit with the same different dynamics that we found in the WT. However, the H3 modification waving pattern was lost under free-running conditions, mimicking the repression in CCA1/LHY transcription (Wang and Tobin 1998). Consistent with TOC1 repression in CCA1-OX plants (Perales and Más, 2007), H3 modifications displayed a significant wave dampening under SD and a loss of rhythmicity under LL. These results suggest that the coordinate action of photoperiod and circadian clock function accounts for H3K4Me3 and H3K9/14Ac dynamics at the TSSs of the three core clock genes. A reasonable hypothesis is to assume that photoperiod might set in motion these dynamic changes but the circadian clock gates them to the appropriate times of the day when each gene is transcribed.
Dynamic acetylation/deacetylation of H3K9/14 accounts for proper expression of core circadian genes
Our analysis of the three core clock genes uncovered two major acetylation patterns at these loci: (i) for the morning regulators CCA1/LHY, H3K9/14Ac peaks quickly but transiently at times of highest transcription and (ii) for the evening gene TOC1, H3K9/14Ac accumulates steadily until reaching the time of highest transcription and then slowly disappears from this locus. These patterns suggest the existence of opposing action of H3 acetylase or deacetylase regulated by the circadian clock. In animals these opposite processes lay quite close to the central oscillator, with CLOCK promoting acetylation of H3K9/14 and SIRT1 (a HDAC) deacetylating these H3 marks (Doi et al. 2006, Nakahata et al. 2008). Moreover, the SIRT1/CLOCK complex is required for proper acetylation rhythms and circadian regulation of chromatin remodeling (Nakahata et al. 2008). In Arabidopsis, TAF1 (a HAT) and HD1 (HDAC1) were shown to regulate expression of light-responsive genes by promoting H3K9 acetylation/deacetylation, respectively (Bertrand et al. 2005, Benhamed et al. 2006). Recently, we showed that HD1 is one of the deacetylases involved in the transcriptional repression of the phyA locus in the light (Jang et al. 2011). Therefore, we investigated whether the circadian rhythms of H3K9/14Ac at TSSs of the three clock genes were TAF1 or HD1 dependent. We found that CCA1, but not LHY, deacetylation was slowed down in hd1 mutants, suggesting differential regulation of the two genes by different HDACs. In the case of TOC1, there was some dampening of the H3K9/14Ac oscillation in taf1 plants, indicating a modest impairment of these activating H3 marks. The small effect of these mutants is not surprising since Arabidopsis encodes about 18 HDACs and at least 12 HATs (Benhamed et al. 2006) which are likely to be functionally redundant.
Circadian regulation and histone cross-talk
Our results show a similar circadian profile between H3K4Me3 and H3K9/14Ac at the TSSs of the three clock genes under different photoperiods and free-running conditions. Moreover, disruption of the circadian clock and loss of photoperiod regulation equally affected these waving patterns, suggesting the existence of a cross-talk between H3K4Me3 and H3K9/14Ac modifications, as has been previously reported in plants, yeast and animal cells (Lee et al. 2010, Jang et al. 2011). Recently, the mouse histone methyltransferase MLL1 was shown to interact with CLOCK. The CLOCK/MLL1 complex promotes cyclic and sequential H3K4 methylation and H3K9/14 acetylation critical for proper transcriptional activity of CLOCK and circadian gene expression (Katada and Sassone-Corsi 2010). This and our results provide the first line of evidence of histone cross-talk and clock function. Although our work here provides an overview on the circadian regulation of these activating H3 marks, future identification of the H3-modifying enzymes and associated factors is needed to deepen our mechanistic understanding of circadian regulation and their effect on Arabidopsis clock function.
Materials and Methods
Plant material and growth conditions
All analyses were carried out using wild-type Arabidopsis thaliana ecotype Columbia (Col-0) or Ws. The following mutant alleles were used: haf2-1 (Bertrand et al. 2005) and athd1-t (Tian et al. 2003) in the Ws background. CCA1-OX (Wang and Tobin, 1998) was in the Col-0 background. Seeds stratified at 4°C for 72 h were germinated on Murashige and Skoog (MS) medium supplemented with sucrose (1%). Seedlings were grown for 18–22 d in a growth chamber under SD (8 h light : 16 h dark) or LD (16 h light : 8 h dark) conditions at 21°C under white light (120 µmol m−2 s−1). Samples were harvested at the indicated ZTs for a period of 24 h. Each experiment was repeated at least twice with independent biological material.
RNA extraction and quantitative RT–PCR analysis
Total RNA was extracted from about 0.1 g of seedlings using the RNeasy Plant Mini Kit (Qiagen). RNA was treated with DNase I (Qiagen) on the column, and 1.5–2.0 µg of RNA was used for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen) with oligo(dT) primers. cDNA was added to 20 µl of PCR buffer containing SYBR Premix Ex Taq (TAKARA) and fluorescence detected using a CFX96 Real-Time System (Biorad) starting with 30 s at 95°C, followed by 40 cycles of 95°C (5 s), 60°C (30 s). A 10 s 95°C step preceded the melt curve step (from 65 to 95°C, at increments of 0.5°C for 5 s). Primer pairs used for CCA1, LHY and TOC1 were according to Ding et al. (2007). The primer pair for ACTIN2 has been described by Ni et al. (2009). Each RNA sample was assayed in triplicate. Expression levels were calculated relative to ACTIN2 using a comparative threshold cycle (Ct) method with ΔΔCt = (Ct,sample – Ct,actin2)ZT x – (Ct,sample – Ct,actin2)ZT y, where ZT x is any ZT and ZT y represents the 1x expression of the target gene normalized to ACTIN2.
Chromatin immunoprecipitation
About 2.5 g of seedlings was collected at the indicated ZTs and immediately cross-linked with 37 ml of 1% (v/v) formaldehyde under vacuum for 10 min. ChIP analysis was performed as described (Jang et al. 2011). Antibodies used were anti-H3K4Me3 (Active Motif, 39159), anti-H3K9Me2 (Millipore, 07-441), anti-H3K9Me3 (Active Motif, 39161), anti-H3K27Me3 (Active Motif, 39155), anti-H3K9Ac (Millipore, 07-352), anti-H3K14Ac (Millipore, 07-353), anti-H3K9/14Ac (Millipore, 06-599) and anti-H3S10p (Diagenode, CS-116-100).
ChIP quantitative real-time PCR analysis
Quantitative PCR was performed to determine the amounts of genomic DNA immunoprecipitated in the ChIP experiments. A 1 µl aliquot of recovered DNA was added to 20 µl of PCR buffer containing SYBR Premix Ex Taq (TAKARA) and fluorescence detected using a CFX96 Real-Time System (Biorad) starting with 30 s at 95°C, followed by 40 cycles of 95°C (5 s), 55°C (30 s). A 10 s 95°C step preceded the melt curve step (from 65 to 95°C, at increments of 0.5°C for 5 s). Each sample was assayed in triplicate, and enrichment levels were calculated relative to ACTIN2/7 using a comparative Ct method with ΔΔCt = (Ct,sample – Ct,actin2/7)ZT x – (Ct,sample – Ct,actin2/7)ZT y, where ZT x is any ZT and ZT y represents the 1x enrichment of the TSS fragment (amplified by primer pair 2) normalized to ACTIN2/7. Primers pairs used were: ACTIN2/7 as described by Johnson et al. (2002) and the primers listed in Supplementary Table S1 for CCA1, LHY and TOC1.
Genotyping of mutant lines
Genotyping of the haf2-1 and athd1-t lines was performed using previously published primer pairs and PCR programs (Tian et al. 2003, Bertrand et al. 2005).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Institutes of Health [grant No. GM44640 to N-H.C.].
Supplementary Material
Acknowledgments
We thank members of the Chua lab for discussion.
Glossary
Abbreviations
- CCA1
CIRCADIAN CLOCK ASSOCIATED 1
- ChIP
chromatin immunoprecipitation
- DD
continuous dark
- H3
histone 3
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- LD
long day
- LHY
LATE ELONGATED HYPOCOTYL
- LL
continuous light
- PRR
PSEUDO RESPONSE REGULATOR
- RT–PCR
reverse transcription–PCR: SD, short day
- TOC1
TIMING OF CAB EXPRESSION 1
- TSS
translation start site
- UTR
untranslated region
- WT
wild type
- ZT
Zeitgeber time.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou D-X. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C, Benhamed M, Li YF, Ayadi M, Lemonnier G, Renou JP, et al. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 2005;280:1465–1473. doi: 10.1074/jbc.M409000200. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C, Millar T, Finnegan E, Pogson B. Promoting gene expression in plants by permissive histone lysine methylation. Plant Signal. Behav. 2009;4:484–488. doi: 10.4161/psb.4.6.8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ. TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell. 2007;19:1522–1536. doi: 10.1105/tpc.106.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Etchegaray J-P, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl Acad. Sci. USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang Z-Y, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl Acad. Sci. USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. The role of CCA1 and LHY in the plant circadian clock. Dev. Cell. 2002;2:516–518. doi: 10.1016/s1534-5807(02)00184-3. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Yu W. Circadian transcription: passing the HAT to CLOCK. Cell. 2006;125:424–426. doi: 10.1016/j.cell.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Song H-R, Lutz K, Kerstetter RA, Michael TP, McClung CR. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I-C, Chung PJ, Hemmes H, Jung C, Chua N-H. Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell. 2011;23:459–470. doi: 10.1105/tpc.110.080481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl Acad. Sci. USA. 2010;107:21623–21628. doi: 10.1073/pnas.1014204108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua N. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, et al. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat. Neurosci. 2010;13:1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Grimaldi B, Sahar S, Hirayama J, Sassone-Corsi P. Signaling to the circadian clock: plasticity by chromatin remodeling. Curr. Opin. Cell Biol. 2007;19:230–237. doi: 10.1016/j.ceb.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N. Molecular mechanisms underlying the Arabidopsis circadian clock. Plant Cell Physiol. 2011;52:1709–1718. doi: 10.1093/pcp/pcr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N, Sakakibara H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse Y, Oh-hashi K, Iijima N, Naruse M, Yoshioka H, Tanaka M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004;24:6278–6287. doi: 10.1128/MCB.24.14.6278-6287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 2007;10:645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernandez AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15:259–265. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Song H-R, Noh Y-S. Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells. 2012;34:279–287. doi: 10.1007/s10059-012-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann T, Más P. Chromatin, photoperiod and the Arabidopsis circadian clock: a question of time. Semin. Cell Dev. Biol. 2008;19:554–559. doi: 10.1016/j.semcdb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, et al. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics. 2003;165:399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.