Abstract
Genetic variations in POR, encoding NADPH-cytochrome P450 oxidoreductase (CYPOR), can diminish the function of numerous cytochromes P450, and also have the potential to block degradation of heme by heme oxygenase-I (HO-1). Purified full-length human CYPOR, HO-1, and biliverdin reductase were reconstituted in lipid vesicles and assayed for NADPH-dependent conversion of heme to bilirubin. Naturally-occurring human CYPOR variants queried were: WT, A115V, Y181D, P228L, M263V, A287P, R457H, Y459H, and V492E. All CYPOR variants exhibited decreased bilirubin production relative to WT, with a lower apparent affinity of the CYPOR∙HO-1 complex than WT. Addition of FMN or FAD partially restored the activities of Y181D, Y459H, and V492E. When mixed with WT CYPOR, only the Y181D CYPOR variant inhibited heme degradation by sequestering HO-1, whereas Y459H and V492E were unable to inhibit HO-1 activity suggesting that CYPOR variants might have differential binding affinities with redox partners. Titrating the CYPOR-HO-1 complex revealed that the optimal CYPOR:HO-1 ratio for activity was 1:2, lending evidence in support of productive HO-1 oligomerization, with higher ratios of CYPOR:HO-1 showing decreased activity. In conclusion, human POR mutations, shown to impact P450 activities, also result in varying degrees of diminished HO-1 activity, which may further complicate CYPOR deficiency.
Keywords: CYPOR, HO-1, P450, reductase, heme, oxygenase, POR deficiency
1. Introduction
A number of disease states arise from metabolic deficiencies linked to variation in POR [1–13], the human gene encoding NADPH-cytochrome P450 oxidoreductase (CYPOR). Many microsomal enzymes, including more than four dozen type II cytochrome P450 monooxygenases (P450s) [14], squalene monooxygenase [15], cytochrome b5 [16], fatty acid desaturase [17], and 7-dehydrocholesterol reductase [18] and heme oxygenase [19], form productive redox complexes with CYPOR, in which electron transfer is required for catalysis. Loss of CYPOR function, therefore, results in a complex mixture of deficiencies in endobiotic and xenobiotic metabolism and can lead to severe developmental malformations including midface hypoplasia, humeroradial synostosis, bowing and fracture of femora, and sexual dimorphisms [20]. The formation of biliverdin IXα by heme oxygenase-1 requires three moles of oxygen for the three monooxygenation steps and electrons from NADPH supplied by CYPOR [21], thereby emphasizing the impact of CYPOR deficiencies on this system.
Regulated heme degradation is critical due to the toxic nature of free heme [22, 23]. Heme oxygenase-1 (HO-1) is the CYPOR-dependent, stress-inducible, membrane-bound enzyme that breaks heme down to carbon monoxide (CO), ferrous iron (Fe2+), and biliverdin. Biliverdin is further degraded to bilirubin by the cytosolic enzyme biliverdin reductase (BVR) [24, 25], in human spleen, liver, and kidney [26, 27]. The C-terminal transmembrane segment of HO-1 was previously shown to drive high-affinity functional complexation with CYPOR [28] and was recently shown to be crucial for HO-1 oligomerization, thereby contributing to its stability and function [29]. The byproducts of heme degradation by HO-1 also have important physiological effects, such as the anti-oxidant/anti-inflammatory activity of bilirubin [30–34], as well as the vasodilatory and anti-inflammatory activity of CO [35–38]. The cumulative antioxidant effects of HO-1 activity are best demonstrated by the increased susceptibility to oxidative injury and shortened lifespan observed in a patient with HO-1 gene deficiency [39]. Regulated heme catabolism also ensures efficient sequestration, via ferritin induction, and availability of free iron for incorporation by other critical metalloproteins [40].
Targeted gene deletion in mice has been used to study the roles of both CYPOR [41–45] and HO-1 (reviewed [46]) in a multitude of physiological processes ranging from growth and development to disease progression. In general, CYPOR-knock-out mice (CPR−/−) exhibit total embryonic lethality due to severe developmental defects, whereas HO-1-deleted mice exhibit partial embryonic lethality and a high incidence of postnatal morbidity. It was previously hypothesized that embryonic lethality observed in the CPR−/− mouse was at least partially the result of diminished/absent HO-1 activity and subsequent heme toxicity [43], although this assertion remains unsubstantiated.
In order to assess the extent to which HO-1 enzymatic activity is influenced by POR variation, we first developed and optimized a coupled spectrophotometric assay for the conversion of heme to bilirubin [47], relying on purified recombinant human CYPOR and HO-1, reconstituted in lipid vesicles, as well as soluble BVR, and including catalase to preclude oxidative interference. Using that system, the following naturally-occurring full-length CYPOR variants, representing a spectrum of potential phenotypic outcomes, were assayed for the capacity to support heme degradation by full-length HO-1: WT, A115V, Y181D, P228L, M263V, A287P, R457H, Y459H, and V492E. Although a similar study was recently published by Pandey et al. [48], numerous and substantial differences in experimental design and methodology, as well as outcomes described herein, give new mechanistic insights into the nature of the CYPOR-HO-1 functional interaction. Furthermore, this is the first attempt to examine the relative affinities among the CYPOR mutants and HO-1, as a redox partner. Using kinetic analysis in reconstituted systems, differential affinities among the various human CYPOR mutants with HO-1 are demonstrated.
2. Materials and Methods
2.1 Materials
Heme, NADPH, EDTA, glycerol, dilauroylphosphatidylcholine (DLPC), FAD, FMN, catalase from bovine liver, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). 2’5’ ADP Sepharose 4B was obtained from GE Healthcare (Piscataway, NJ). All spectrophotometric analyses of HO-1 activity were performed on a SpectraMax M5 plate reader from Molecular Devices (Sunnyvale, CA). HEK293T cells (CRL-11268) were purchased from ATCC (Manassas, VA). Dulbecco’s Modified Eagle Medium, gentamycin, phosphate-buffered saline (PBS), and lipofectamine were purchased from Invitrogen (Carlsbad, CA). Coelenterazine 400a and coelenterazine h were purchased from Gold Biotechnology (St. Louis, MO) and Promega (Madison, WI), respectively. The BRET vectors were obtained from Perkin Elmer (Waltham, MA). BRET was measured using a TriStar LB 941 plate reader (Berthold Technologies, Bad Wildbad, Germany).
2.2 CYPOR Expression and Purification
Full-length human CYPOR was recombinantly expressed as an OmpA3 fusion protein, extracted and purified from Escherichia coli membranes by 2’5’-ADP-Sepharose 4B affinity according to previously described methods [47, 49, 50]. Site-directed mutagenesis of the pPORh plasmid was used to generate expression constructs for A115V, P228L, M263V, A287P, and R457H variants, as described previously for preparation of V492E and Y459H variants [49], and the Y181D variant [51]. These expression systems [47] varied somewhat from those originally described by Flück et al. [1], which deleted 27 N-terminal residues, in that the full-length proteins were cloned and expressed. In addition to the full-length CYPOR, a soluble variant lacking the N-terminal 66 residues (Δ66) determined from structural considerations [52] was expressed as a poly-histidine fusion protein, and purified from E. coli by immobilized metal affinity chromatography (IMAC) as described previously [49].
2.3 HO-1 Expression and Purification
Full-length, human HO-1, containing an R254K mutation to prevent proteolytic degradation of the C-terminal trans-membrane segment, was recombinantly expressed as a glutathione-S-transferase fusion protein, extracted and purified from E. coli membranes by glutathione affinity chromatography, and quantified as described previously [53, 54].
2.4 BVR Expression and Purification
Recombinant human BVR was also expressed as a poly-histidine fusion protein in E. coli and purified by IMAC as described previously [47].
2.5 Reconstitution of HO-1 and CYPOR in DLPC Liposomes
Reconstituted systems (RCSs) were prepared as described previously [47], in which 350:1 was demonstrated to be the optimal DLPC:HO-1 ratio, and incubation of the lipid-protein mixture for 2 hrs at room temperature resulted in the highest levels of protein incorporation and productive complex formation within the liposomes. The final concentration of HO-1 in all reconstituted systems was 0.05 µM, unless otherwise stated. CYPOR levels were varied over a range from 0–0.5 µM as indicated for each individual experiment. Such titrations were found to be absolutely necessary to produce optimal activities in these systems.
2.6 HO-1 Activity Measurements
Bilirubin formation was monitored as the difference in absorbance at 464 and 530 nm, and the enzyme activity was calculated using the delta extinction coefficient of 40 mM−1cm−1 [55, 56]. Reconstituted systems were prepared and pre-incubated for 2 h at room temperature before the addition of other assay components. The final volume for all assays was 0.1 ml, contained in 100 mM potassium phosphate (pH 7.4). All activity assays were allowed to pre-incubate at 37°C for 2 min before the addition of 0.5 mM NADPH to initiate the reaction. Each reaction was performed in triplicate, and the turnover number was expressed as nanomoles of bilirubin formed per minute per nanomole of HO-1 (nmol/min/nmol). The substrate, heme, was added at 15 µM. BVR was included at 0.05 µM, equivalent to HO-1 in most assays. Fatty acid-free BSA was included at 0.25 mg/ml. Catalase was added to a concentration of 0.25 U/ml. The effect of H2O2 on the linearity of the reaction was previously determined in the reconstituted systems and the necessity for including catalase was demonstrated [47].
2.7 GST Pulldown of HO-1:CYPOR Complexes from Reconstituted Systems
RCS containing HO-1, mutant/WT CYPOR, and DLPC were prepared at a final concentration of 0.2 µM of each protein (as indicated in Figure 6) in a volume of 0.25 ml in binding buffer containing 10 mM sodium phosphate, 1.8 mM potassium phosphate, 2.7 mM potassium chloride, 5 mM EDTA, 5 mM EGTA, and 100 mM sodium chloride (pH 7.3). High performance glutathione Sepharose beads (GE Healthcare, Piscatawy, NJ) were washed 5 times in the binding buffer, and 0.02 ml of a 1:1 suspension was added to the RCS described above. The samples were rocked at room temperature for 2 hr before being washed 6 times with 0.25 ml binding buffer by centrifugation (3500 × g for 15 sec) and resuspension. After the last wash, the beads were resuspended in 0.05 ml loading buffer (10 % SDS, 0.02 % bromophenol blue, 5 % β-mercaptoethanol, and 100 mM Tris-HCl (pH 6.8)) and were boiled for 15 min before loading 0.02 ml on a 10% SDS PAGE gel and running for 1 hr at 200 V. The amounts of CYPOR and HO-1 recovered were determined by densitometry using three concentrations of the two standard proteins (Lanes 8–10). Densitometry results are reported as the mean ratio ± standard error.
2.8 Bioluminescence Resonance Energy Transfer (BRET) to Detect HO-1∙HO-1 Complexes
2.8.1 BRET Vectors
BRET was used to determine if HO-1 exists as a homomeric complex in HEK293T cells, and if this complex is disrupted in the presence of CYPOR. Plasmids for the transient expression of Renilla luciferase-HO1 and GFP (green fluorescence protein)-HO-1 fusion proteins were prepared using the pRluc-C1 and pGFP2-C1 parent vectors, respectively, with the tags fused at the N-terminal portion of the rat HO-1 molecule. The CYPOR-null transcript was prepared by subcloning full-length rabbit CYPOR DNA into the pRluc-N2 vector with a stop codon inserted between the CYPOR and Rluc genes by site-directed mutagenesis.
2.8.2 Cell Culture and Transfection
HEK 293T/17 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum under a humidified atmosphere of 5% CO2. Cells were distributed onto 6-well plates at 7.5 × 105 cells/well and were then allowed to grow for 24 hours prior to transfection. Lipofectamine 2000 was used to transiently transfect the cells with up to 500 ng DNA per well according to the manufacturer’s protocol. In the absence of CYPOR, the total amount of HO-1 DNA transfected was 300 ng per well, and in the presence of CYPOR, 200 ng of unlabeled-CYPOR DNA, and 300 ng of labeled-HO-1 DNA were transfected. Total DNA in a given experiment was kept constant for each well by co-transfecting a complementary amount of pUC19. Four hours after transfection, DMEM was replaced with fresh DMEM supplemented with both Antibiotic-Antimycotic and Gentamicin solutions.
2.8.3 BRET Determination
Twenty-four hours after transfection, media was removed and cells were suspended in 1 ml PBS and pelleted at 300 × g for 10 minutes. The supernatant was removed and cells were resuspended in 750 µl fresh PBS. Cells were then plated in quadruplicate onto a white 96-well Optiplate at 100 µl per well. BRET measurements were performed on a TriStar LB 941 plate reader with automatic injectors. Coelenterazine 400a was injected to a final concentration of 5 µM into three of the four wells for each sample. The plate was then shaken for 1 second, and the luciferase signal (at 400 nm) and GFP signal (at 515 nm) were measured sequentially for 3 seconds each.
The BRET ratio was calculated by dividing the GFP signal by the luciferase signal after using untransfected cells as a blank. Since a small amount of the Rluc signal is detectable through the GFP filter, the baseline BRET ratio, which was measured using cells expressing only Rluc-HO-1, was subtracted from each point.
2.8.4 Relative Protein Quantification
GFP fluorescence and Rluc luminescence were used to determine the relative amounts of GFP- and Rluc-tagged proteins. The cells used for protein quantification came from the same samples as those used for BRET measurement to avoid variation in cell density. Cells were plated at 100 µL per well into a black, clear-bottom 96-well plate, and GFP fluorescence was measured by reading the emission at 510 nm (excitation at 395 nm) using a SpectraMax M5 plate reader. Luminescence was measured on the TriStar LB 941 plate reader using 100 µL cell aliquots on a white 96-well Optiplate. Coelenterazine h was manually added to a final concentration of 5 µM and luminescence was measured at 480 nm for 1 second. Coelenterazine h was used here as a luciferase substrate to avoid the quenching of the signal that occurs due to energy transfer when coelenterazine 400a is used.
The relative amounts of GFP-HO-1 and Rluc-HO-1 were estimated based on the GFP fluorescence and Rluc luminescence when compared to a GFP-Rluc fusion protein, which was assumed to have a GFP-Rluc ratio of 1:1. This method allowed us to estimate the relative amounts of the fusion proteins, without relying on the transfected DNA levels.
3. Results
3.1 HO-1 Activity of Reconstituted Systems
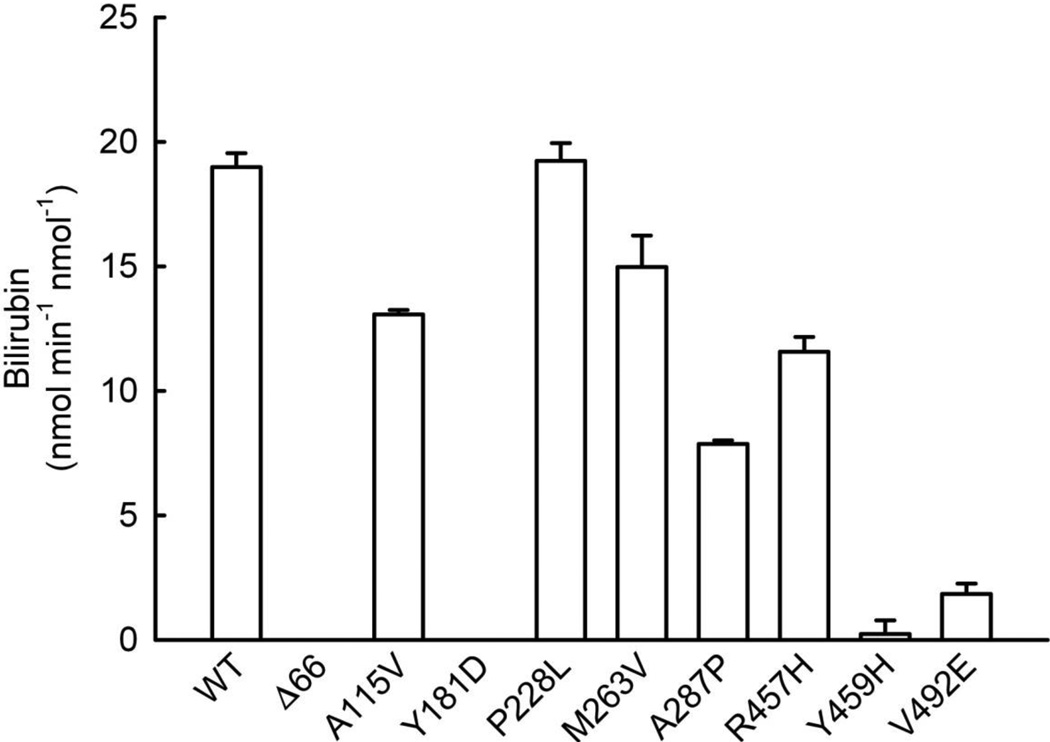
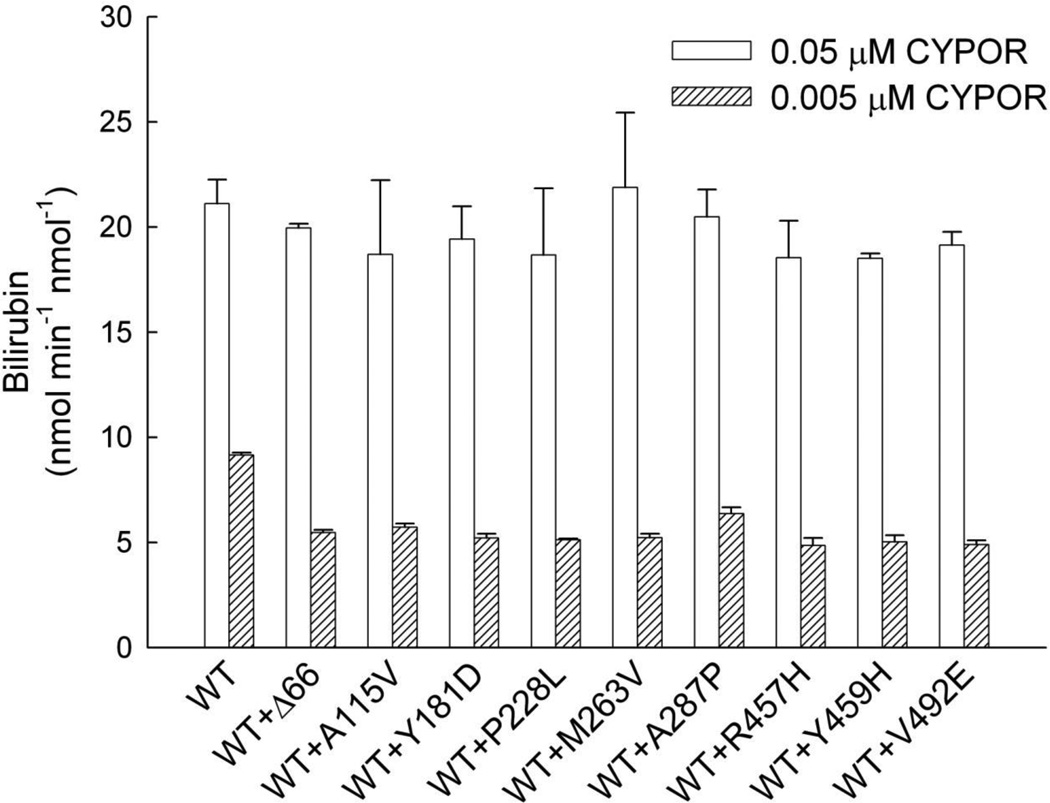
Initially, DLPC reconstituted systems were prepared with HO-1 and each of the CYPOR variants at 1:1 molar ratio ([HO-1] = [CYPOR] = 0.05 µM). The following CYPOR variants were included: WT, WTΔ66, A115V, Y181D, P228L, M263V, A287P, R457H, Y459H, and V492E. The HO-1 activity of each of the reconstituted systems was then assayed for the NADPH-dependent conversion of heme to bilirubin, supported by BVR. The results, Figure 1, showed that WT CYPOR supported the highest level of HO-1 activity with a rate of ~ 20 nmol min−1 nmol−1. While the HO-1 activity of the P228L CYPOR-reconstituted system was equal to that of WT CYPOR, each of the other variants compromised the reconstituted activity to varying extents. A115V, R457H, and M263V CYPOR variants each retained > 60% of WT CYPOR activity, while the A287P, V492E, and Y459H variants retained 41%, 10%, and 1% of WT activity, respectively. The V492E and Y459H variants have been shown to affect FAD binding [49], while A287P and R457H also can affect FAD binding due to their proximity to the FAD-binding pocket in the CYPOR molecule, but less dramatically. On the other hand, the P228L variant residue is located on the surface of the FMN-binding domain of CYPOR where it could affect interactions with redox partners. A115V is located in the FMN-binding domain on the interior of the molecule where a disruption of structure could result in some compromise of activity, although the effect on HO-1 activity was not dramatic. Even with the optimized assay system, HO-1 activity was completely undetectable with either the Y181D variant, exhibiting compromised FMN binding [51], or the Δ66 variant, which due to the lack of the hydrophobic N-terminus was unable to reconstitute interactions with its membrane-bound or hydrophobic redox partners [49, 57].
Figure 1. Survey of CYPOR Variant Effects on HO-1 Activity.
Reconstituted systems were prepared containing each of the CYPOR variants shown ([CYPOR] = [HO-1] = 0.05 µM) and assayed for HO-1 catalyzed conversion of free heme (15 µM initial) to biliverdin, as indicated by the rate of the bilirubin formation by BVR in the coupled assay. Each bar represents the mean ± standard deviation of triplicate assays. Δ66 refers to WT CYPOR with N-terminal 66 residues deleted.
3.2 Effects of Flavin Addition on HO-1 Activity of Reconstituted Systems
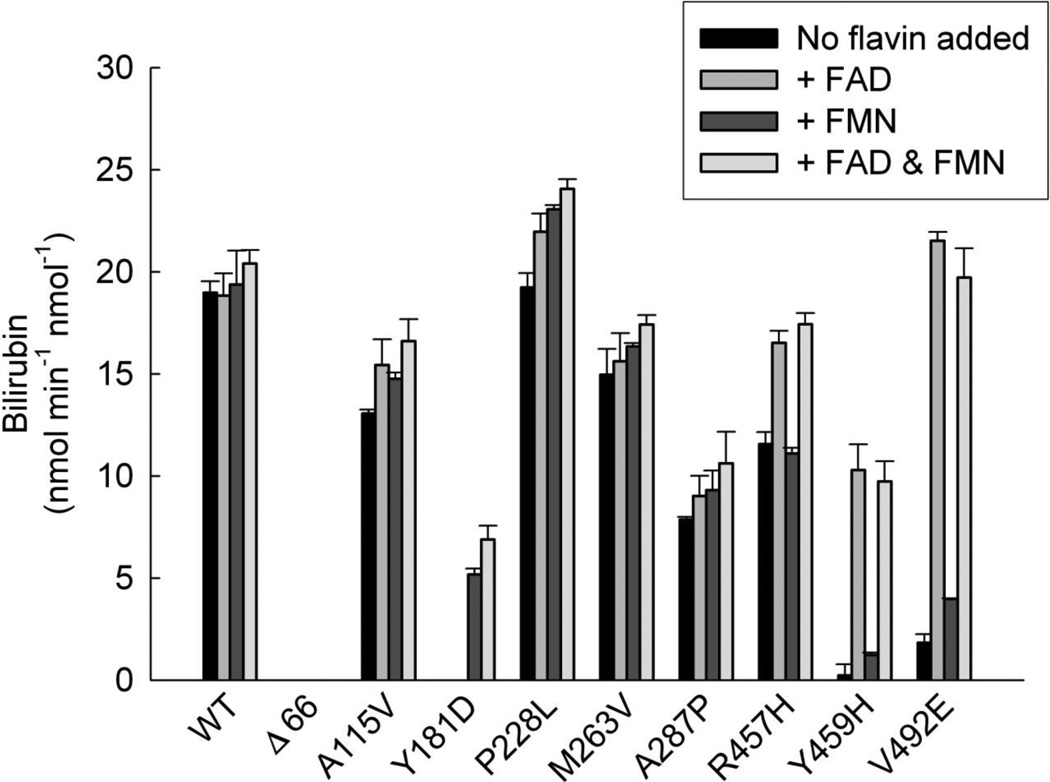
Since we demonstrated previously that addition of exogenous flavins partially restored electron transfer from the purified CYPOR variants, Y459H and V492E (lacking FAD) [49, 51] to cytochrome c and P4504A4 and to cytochrome c and P4501A2 from Y181D (lacking FMN) [51], we wanted to determine the effects of flavin supplementation on heme degradation by the reconstituted HO-1 systems. The rates of bilirubin formation were determined in the presence/absence of 20 µM FAD, FMN, or both as shown in Figure 2. WT CYPOR activity was not significantly enhanced upon addition of flavins to the reaction, as expected. The same was true of the A287P and M263V variants. Flavin supplementation also had no effect on the soluble Δ66 variant which remained devoid of measurable activity in the system. Low-level activation (< 20%) was observed for A115V and P228L variants upon flavin addition. FAD strongly activated the R457H, Y459H, and V492E variants restoring 87%, 54%, and 113% of WT activity, respectively. Likewise, FMN restored the activity of the Y181D variant to 27% of WT. These findings were in good agreement with our previous studies demonstrating rescue of electron transport function to various degrees of the Y459H, V492E, and Y181D variants by flavin supplementation in NADPH-cytochrome c reduction [49, 51, 58], P450 4A4-mediated ω-hydroxylase [49], and P450 1A2-mediated Odealkylase assays [51, 58]. Flavin supplementation has subsequently been verified to increase the ability of CYPOR mutants to support the reduction of cytochrome b5 and monooxygenation by P450 3A4 [59]. We therefore decided to perform flavin titrations for each of these variants in order to estimate flavin affinities. The resulting curves (Supplemental Figure 1) suggested FAD affinities of 0.9 and 0.6 µM for the Y459H and V492E variants, respectively, whereas the FMN affinity of Y181D was 3 µM. These values, derived kinetically, are orders of magnitude higher than the affinities of WT CYPOR for its flavin cofactors. For example, it has been experimentally difficult to remove FAD from CYPOR, without unfolding the protein [60] and the binding affinity for FMN is quite high as well (~10 nM).
Figure 2. Flavin Activation of CYPOR Variants.
Reconstituted systems were prepared containing each of the CYPOR variants shown ([CYPOR] = [HO-1] = 0.05 µM) and assayed for HO-1 activity in the presence/absence of 20 µM FAD, FMN, or both. Each bar represents the mean ± standard deviation of triplicate assays.
3.3 CYPOR Titration of Reconstituted Systems
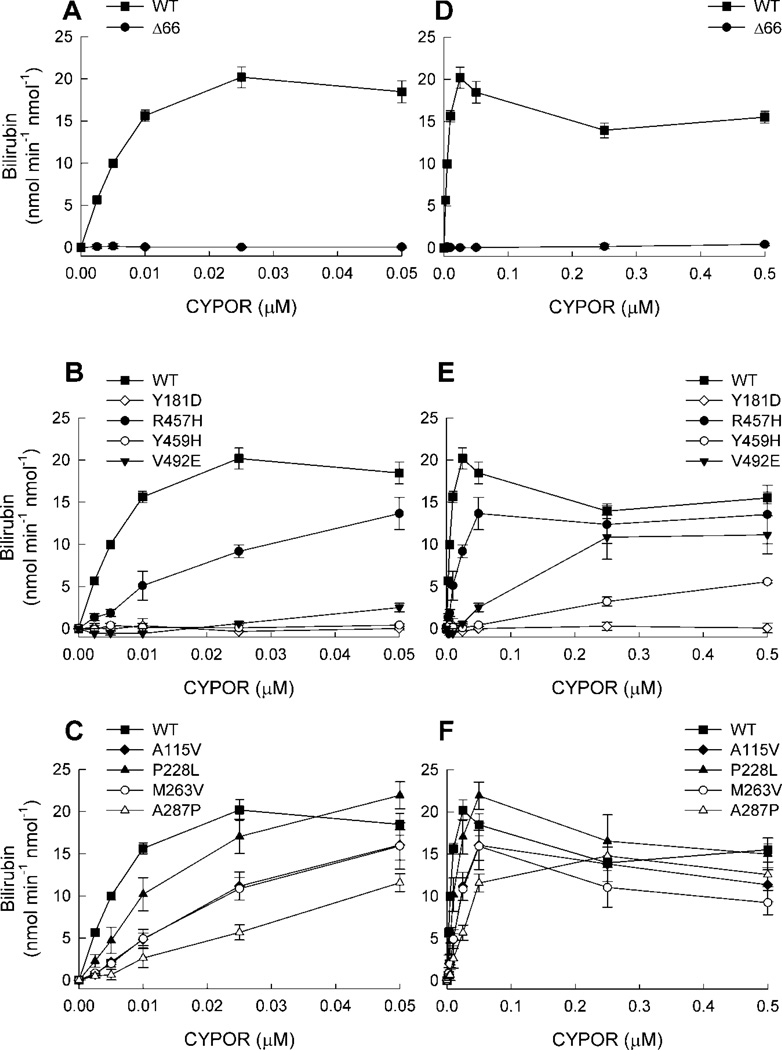
Having demonstrated a wide range of capacities among the naturally occurring CYPOR variants to support HO-1 activity when reconstituted at 1:1 CYPOR:HO-1, we wanted to estimate the CYPOR:HO-1 affinity of each of the complexes by varying [CYPOR], while holding [HO-1] constant at 0.05 µM. The resulting titration curves are shown in Figure 3. In panel (A), WT CYPOR and the WT Δ66 variant were titrated from 0–0.05 µM. WT produced the highest observed activity at [CYPOR] = 0.025 µM, corresponding to a CYPOR:HO-1 ratio of 1:2. Fitting the data to a single binding site saturation curve (Supplemental Figure 2) yielded an apparent Km = 8.6 ± 0.9 nM for the functional CYPOR:HO-1 complex. Regardless of concentration, WT Δ66 failed to support a measurable rate of reaction, verifying the critical importance of the N-terminal trans-membrane helix in CYPOR-HO-1 complex formation. These data cast considerable doubt on the physiological significance of studies utilizing the truncated forms of CYPOR and/or HO-1 for reconstitution and affinity studies, despite the fact that bilirubin formation has been demonstrated with these preparations.
Figure 3. Titration of HO-1 with CYPOR Variants.
Reconstituted systems were prepared with varying concentrations of CYPOR, [HO-1] = 0.05 µM and assayed for HO-1 activity. In each plot, each point represents the mean ± standard deviation of triplicate assays. For comparison, WT CYPOR is shown in each panel. Panels (A), (B), and (C) represent the low range of [CYPOR] ≤ 0.05 µM, and panels (D), (E), and (F) represent the broad range of [CYPOR] ≤ 0.5 µM. See insets for symbol legends.
Identical titrations were repeated with each of the naturally occurring CYPOR variants. The results for the flavin-deficient variants (Y181D, R457H, Y459H, and V492E) are shown in Figure 3B. R457H supported just less than half of the activity seen with WT CYPOR, while the other flavin-deficient variants were totally inactive. The results for the remaining variants (A115V, P228L, M263V, and A287P) are shown in Figure 3C. Each of these variants supported increased HO-1 activity in a concentration-dependent manner, although each was less active than WT at [CYPOR] ≤ 0.025 µM. None of the curves for the variants appeared to have neared saturation over the range of [CYPOR] titrated, so the range was expanded 10-fold, up to 0.5 µM. Figure 3D shows the results for WT and WT Δ66, where again, no HO-1 activity was detectable for WT Δ66. Surprisingly, the data for WT deviated from the simple (single binding site) saturation that was observed at low [CYPOR], with higher [CYPOR] resulting in partial, but significant, inhibition of HO-1. The results of the corresponding studies of the CYPOR variants are given in Figure 3E for the flavin-deficient variants, and Figure 3F for the other variants. In almost all cases, the same phenomenon of HO-1 inhibition by high concentrations of CYPOR was observed. Collectively, these data suggested a CYPOR-dependent transition in HO-1 function from optimal at low [CYPOR] to suboptimal at high [CYPOR]. That the transition point appeared to occur at different [CYPOR], depending upon the variant in question, suggests differential HO-1 binding affinities among them. In an attempt to estimate the affinity of the optimally active complexes, curve-fitting was repeated for each of the variants using only the activity values corresponding to sub-saturating [CYPOR] (see Supplemental Figure 2). The extrapolated Vmax and apparent Km values are reported in Table I, along with the aggregate Vmax/Km values, representing the overall catalytic efficiency of each CYPOR:HO-1 complex. It should be noted that the turnover numbers for bilirubin formation are 27.5 min−1 in the current studies compared to 1.16 min−1 in the Pandey et al. report, more than a twenty fold difference [48], suggesting that none of their reported activities were at Vmax. Kinetic comparisons between wild type and mutants under sub-optimal Vmax conditions could lead to erroneous conclusions. Vmax values obtained from data fitting for each of the variants was within ± 50% of that calculated for WT CYPOR, but all of the CYPOR variants tested had significantly higher apparent Km values than WT (i.e., lower apparent affinity of CYPOR for HO-1) ranging from an approximate 3-fold increase for P228L to >200-fold increase for Y459H. Two of the flavin-deficient variants, namely Y459H and V492E, failed to inhibit HO-1 activity at high [CYPOR], instead exhibiting simple saturation kinetics over the range from 0–0.5 µM CYPOR (10:1 CYPOR:HO-1, Figure 3E). Because all of the variants were titrated without flavin supplementation, it is hypothesized that the binding-dissociation equilibria were reflective of the mixed populations of apo- and holo-CYPOR shown previously to exist amongst the Y181D, Y459H, and V492E variants [49, 51], and likely to exist for R457H based on the FAD activation observed herein (Figure 2).
Table 1.
Kinetic parameters extrapolated from curve-fitting of HO-1 activity versus [CYPOR]. Raw data and curve-fitting is shown in Supplemental Figure 2.
| CYPOR variant | Vmax
* (nmol/min/nmol) |
KmCYPOR
§ (nM) |
Vmax/Km (min−1nM−1) |
Relative Vmax/Km (% WT) |
|---|---|---|---|---|
| WT | 27.5 ± 1.2 | 8.6 ± 0.9 | 3.2 ± 1.3 | - |
| A115V | 37.3 ± 7.0 | 64.5 ± 18.9 | 0.6 ± 0.4 | 18 |
| Y181D | ND# | ND# | ND# | - |
| P228L | 33.3 ± 3.0 | 25.1 ± 4.6 | 1.3 ± 0.6 | 42 |
| M263V | 37.7 ± 8.7 | 66.6 ± 23.8 | 0.6 ± 0.4 | 18 |
| A287P | 18.2 ± 1.2 | 44.7 ± 7.9 | 0.4 ± 0.2 | 13 |
| R457H | 26.8 ± 4.5 | 47.8 ± 13.8 | 0.6 ± 0.3 | 18 |
| Y459H | 27.8 ± 14.8 | 1990 ± 1300 | 0.01 ± 0.01 | 0 |
| V492E | 17.2 ± 3.5 | 220 ± 110 | 0.08 ± 0.03 | 3 |
Vmax is a mathematical value extrapolated from a Michaelis-Menten plot (Supplemental Fig. 2) and represents the maximal reaction rate were CYPOR not inhibitory of HO-1 activity at higher concentrations.
Km refers to the [CYPOR] at which half maximal HO-1 activity would be predicted.
ND = not determined because activity below the limit of detection
3.4 Inhibition of Functional HO-1:WT CYPOR Complexation by Inactive CYPOR Variants
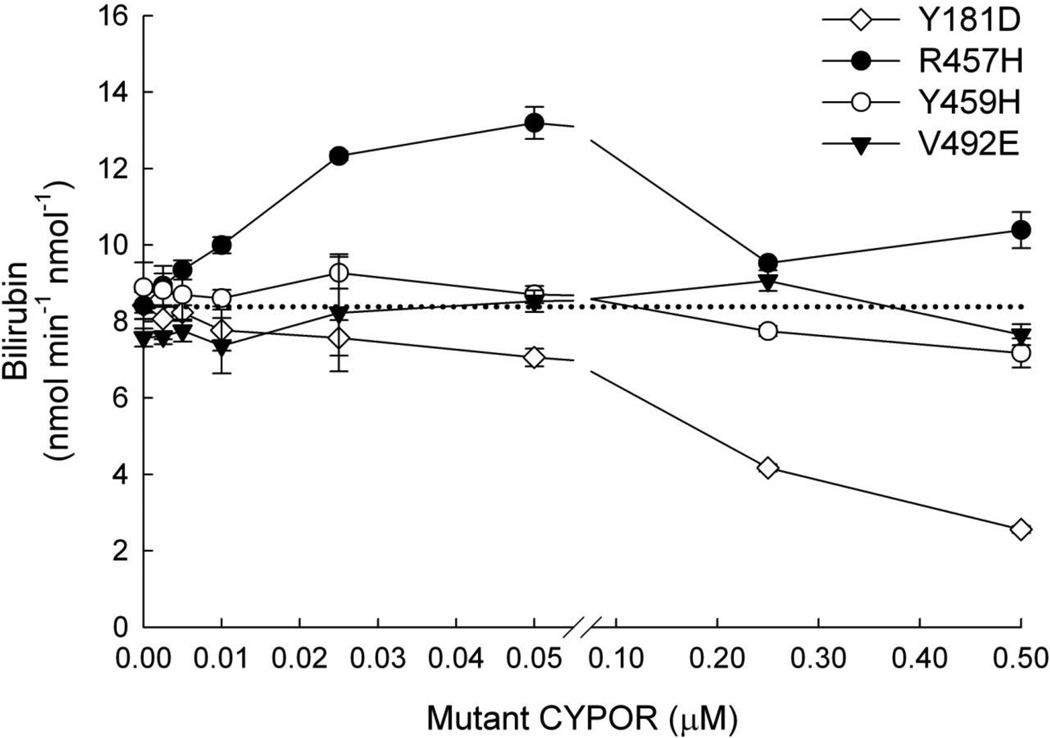
Since the lowest levels of activity were observed among the flavin-deficient variants, we decided to test whether HO-1 could be functionally sequestered from WT CYPOR by any of these variants. The starting point for each titration was a reconstituted system with 0.05 µM HO-1 and 0.005 µM WT CYPOR that exhibited HO-1 activity at ~ 8.3 nmol min−1 nmol−1. The resulting data are presented in Figure 4. Since the R457H variant had higher activity than the other flavin deficient variants, it was hypothesized that titration with this variant would have similar effects to titrating with WT CYPOR, namely increasing activity at low [CYPOR] with inhibition of HO-1 activity at higher CYPOR;HO-1 ratios. This was the case as titration with R457H up to 0.05 µM increased HO-1 activity (~60%), but higher concentrations, up to 0.5 µM, neutralized the gain in activity, as was shown previously (Figure 3). Y459H and V492E did not compete effectively with WT but Y181D titration resulted in significant inhibition of the system with ~30% activity remaining at [Y181D] = 0.5 µM. While this demonstrated the capacity of a naturally occurring CYPOR variant to inhibit the interaction of WT CYPOR with HO-1, the fact that a 100:1 ratio of Y181D:WT was required to achieve ~50% HO-1 inhibition suggested that the phenomenon would be unlikely to occur physiologically, since HO-1 is so highly inducible relative to CYPOR, and [HO-1], under induced conditions, greatly exceeds [CYPOR] [61].
Figure 4. Titration of WT CYPOR-HO-1 Complex with CYPOR Variants.
Reconstituted systems were prepared with [WT CYPOR] = 0.005 µM, [HO-1] = 0.05 µM, and varying concentrations of Y181D, R457H, Y459H, or V492E variants of CYPOR. Each system was assayed for HO-1 activity. Each point on the plot represents the mean ± standard deviation of triplicate assays. The dashed line represents the baseline rate of reaction of the system without addition of the variant CYPOR.
3.5 HO-1 Activity of Reconstituted Systems with Mixed CYPOR Variants
To estimate the effects of POR variation on the HO-1 system in heterozygous individuals (one mutated and one WT copy of POR), we prepared mixed reconstituted systems with equal concentrations of both WT and variant CYPOR at [CYPOR]total = 0.05 µM or 0.005 µM and at [HO-1] = 0.05 µM. Rates of bilirubin production from these mixed systems, representing 1:1 and 1:10 CYPOR:HO-1 ratios, were determined as plotted in Figure 5. All of the systems had the same level of activity, within the margin of error of the assay, at 1:1 CYPOR:HO-1, but at 1:10, each of the mixed systems was less active than that containing WT CYPOR alone. This was not unexpected, considering that each of the variants had less activity than WT at low [CYPOR]. Since HO-1 expression is so highly inducible, whereas CYPOR expression is much less so, the mixed systems at 1:10 CYPOR:HO-1 are more likely to be representative of those in tissues/cells exposed to the types of physiological/environmental stress that induce HO-1. These data suggest that under those conditions, POR heterozygous individuals would have lower HO-1 function than those without POR variations.
Figure 5. Assay of HO-1 Activity Using Mixtures of WT and CYPOR Variants.
Reconstituted systems were prepared with [HO-1] = 0.05 µM and [CYPOR]total = 0.05 µM (open bars) or 0.005 µM (shaded bars), where WT and variant CYPORs were mixed at 1:1. Each system was assayed for HO-1 activity. Each bar represents the mean ± standard deviation of triplicate assays.
3.6 GST Capture of HO-1:CYPOR Complexes from Reconstituted Systems
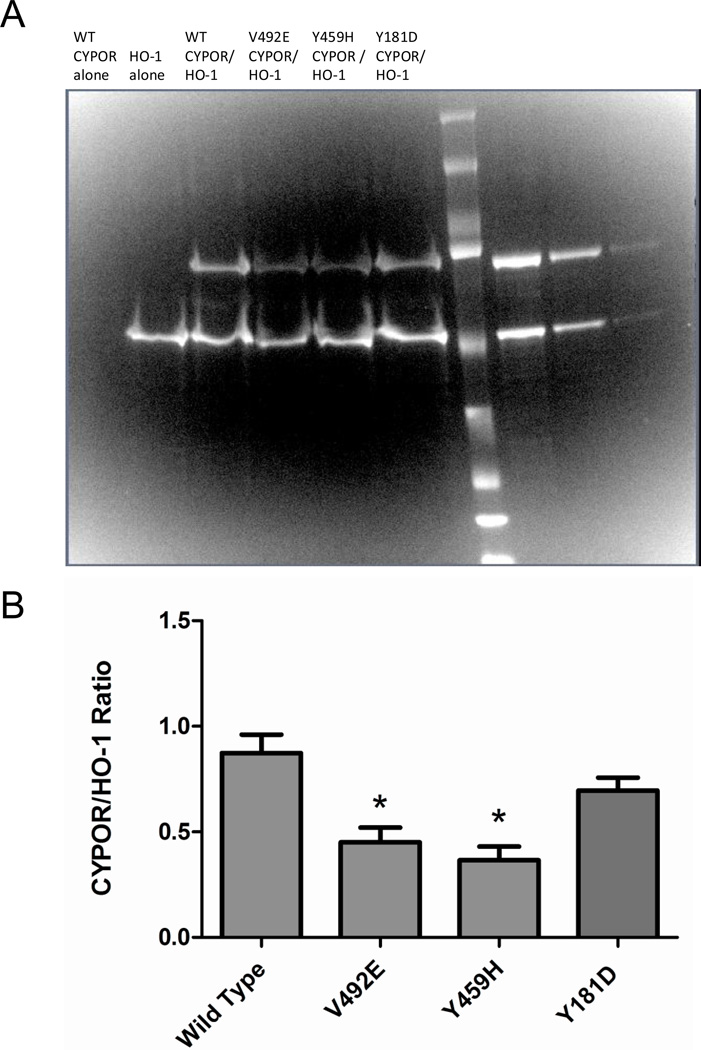
In order to estimate relative binding capacities of the CYPOR variants with HO-1, pull down experiments were performed. Since the HO-1 protein used in these studies contained an N-terminal GST fusion tag, glutathione Sepharose beads were used to capture HO-1-containing complexes from reconstituted systems. Following GST capture, samples were analyzed by SDS-PAGE as shown in Figure 6A. The negative control showed that no wild type CYPOR was pulled down in the absence of HO-1. Densitometry analysis of the HO-1 and CYPOR bands on the gel, Figure 6B, revealed differences in the ratio of HO-1:CYPOR contained in the captured complexes from the reconstituted systems. The data in Figure 4 are consistent with that in Figure 6 with respect to the inability of Y459H and V492E to inhibit HO-1 activity. However, Y181D, which was able to compete with WT in the enzymatic assay, also showed a tighter interaction in the GST-pull down experiment.
Figure 6. GST Pulldown of HO-1:CYPOR Complexes from Reconstituted Systems.
Glutathione sepharose beads were used to capture HO-1 containing complexes from reconstituted systems. SDS-PAGE (panel A) analysis of captured proteins in lanes 1–6 compared to CYPOR and HO-1 standards in lanes 8–10 and densitometry was used to calculate the CYPOR:HO-1 ratios (panel B) where bars represent the mean ratio ± standard error of 4 experimental replicates. Significantly less V492E and Y459H than WT CYPOR variants were recovered, suggesting lower relative affinity for HO-1 among these variants.
3.7 BRET Detection of Homomeric HO-1 Complexes in HEK293 Cells
The non-Michaelis-Menten response observed with the kinetic data in Figure 3 (where HO-1 activity was increased with CYPOR concentrations up to a 1:2 CYPOR:HO-1 ratio, followed by the decline observed at higher CYPOR levels) was suggestive of more complex interactions between CYPOR and HO-1. Such interactions could include either allosteric effects resulting from the binding of CYPOR to HO-1 or potential effects of CYPOR on the organization of these proteins. In a recent report by Hwang et al. [29], HO-1 was shown to form homomeric complexes when transfected into HEK293 cells. Therefore, we wanted to confirm that HO-1∙HO-1 complexes could be detected using the BRET technique in HEK293 cells transiently expressing Rluc-HO-1 and GFP-HO-1 fusion proteins. As a first step, the subcellular localization of transfected HO-1 was confirmed. The data in Supplemental Figure 3 show that transfected GFP-HO-1 co-localized with the ER marker DsRed-ER.
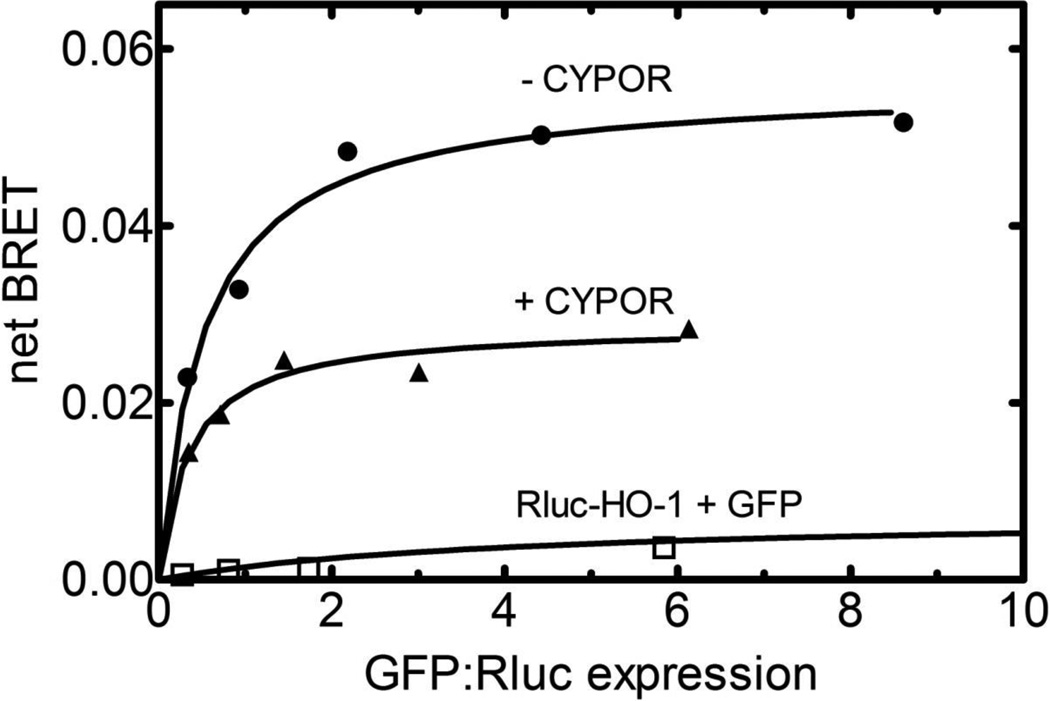
Next, HO-1∙HO-1 complex formation was verified using BRET, where Rluc-HO-1 and GFP-HO-1 constructs were co-transfected into HEK293T cells. Addition of the luciferase substrate, coelenterazine 400a, leads to the generation of luminescence at 410 nm. In the event that a GFP is within 75 Å of the Rluc tag, there is a greater than 50% probability of energy transfer to the GFP, leading to fluorescence emission at 510 nm. In these experiments (Figure 7), the ratio of the Rluc- and GFP-containing DNA constructs was varied. At low GFP-HO-1:Rluc-HO-1 ratios the BRET signal (taken as the ratio of 510 nm fluorescence/410 nm luminescence) was low, due to the low levels of GFP-construct to accept the energy transfer. However, as the ratio of the GFP- and Rluc-containing constructs was increased, a substantial increase in the BRET ratio was observed, saturating at high GFP/Rluc ratios. The data in Figure 7 (upper curve) demonstrate that a specific interaction between the two HO-1 fusion proteins occurred in this cell line, as transfection of Rluc-HO-1 with the naked GFP vector exhibited a decreased BRET signal that did not increase with increasing GFP:Rluc ratio (lower curve).
Figure 7. Effect of CYPOR on HO-1∙HO-1 Complex Formation Using BRET.
Homomeric complex formation between HO-1 molecules was demonstrated by co-transfection of Rluc-HO-1 and GFP-HO-1 into HEK293T cells at varying ratios of the GFP- and Rluc-containing constructs (upper curve). The total amount of transfected HO-1 DNA was held constant at 300 ng. The effect of CYPOR on the HO-1∙HO-1 complex was examined by co-transfection with unlabeled CYPOR (middle curve). In these experiments, the quantities of CYPOR and HO-1 constructs were 200 ng and 300 ng, respectively. As in the absence of CYPOR, the relative amounts of the Rluc-HO-1 and GFP-HO-1 constructs were varied, maintaining a constant total amount of HO-1 DNA transfected. As a control experiment, Rluc-HO-1 and naked GFP were cotransfected at increasing GFP to Rluc-HO-1 ratios (lower curve). Details are described in Materials and Methods.
Co-transfection of CYPOR had a significant effect on the HO-1∙HO-1 complex. The presence of untagged CYPOR caused an inhibition of the HO-1∙HO-1 BRET response, consistent with the ability of CYPOR to influence HO-1∙HO-1 complex formation either by disrupting or otherwise influencing the conformation/configuration of the HO-1 multimers.
4. Discussion
The defensive role of HO-1 against oxidative stress, mediating a number of physiological and pathological processes, has been well established (reviewed [62]). Since CYPOR activity is essential to HO-1 catalytic function [19], it was important to evaluate the impact of genetic variations in POR. To that end, wild type and 8 variant CYPOR proteins were tested for the capacity to support HO-1 activity in a lipid vesicle-reconstituted system.
A survey of heme breakdown by reconstituted systems containing equimolar CYPOR and HO-1 revealed a wide range of activities (Figure 1). With the exception of Y181D, all CYPOR variants exhibited measurable activity, contrary to the report of Pandey et al. (47), which reported total loss of activity for R457H, Y459H, and V492E. This could be attributable to the very low activities obtained even with WT CYPOR in those studies. A115V, P228L, M263V, and R457H each had >60% of WT activity. A287P, V492E, and Y459H variants retained 41%, 10%, and 1% of WT activity, respectively. Deletion of the CYPOR N-terminal 66 residues, comprising a trans-membrane anchoring helix, completely obliterated the capacity of the enzyme to interact productively with full-length lipid reconstituted HO-1.
We demonstrated previously that addition of free flavin(s) to some CYPOR-mediated reactions can enhance the catalytic efficiency of certain CYPOR variants, namely by adding FAD to R457H, V492E, or Y459H [49, 58], and by adding FMN to Y181D [51]. HO-1 activity was also affected by flavin addition (Figure 2), the best example of which was the >1000-fold activation of Y181D by FMN. These studies provide further impetus to investigate the potential of riboflavin therapy to rescue certain CYPOR-dependent activities in individuals with certain POR mutations, as was first suggested by this laboratory in 2006 [49].
Measurement of HO-1 activity as a function of [CYPOR] showed some surprising results. At low CYPOR concentrations, a typical saturation phenomenon was observed; however, this response appeared to saturate at a 1:2 CYPOR:HO-1 ratio (Figure 3), rather than the expected equimolar ratio. As the CYPOR:HO-1 ratio was further increased, there was a generalized decrease in HO-1 activities. These results suggested a more complex interaction between these proteins. Recent studies showing that HO-1 exists in HEK293 cells as an oligomer support this suggestion [29]. Interestingly, our results confirm the existence of HO-1 as an oligomer, and suggest that CYPOR is capable of modulating complex formation (Figure 8). Although the exact mechanistic details and stoichiometries need to be resolved, the data clearly demonstrate that HO-1 exists in the membrane as an aggregate that is disrupted or structurally modified in the presence of CYPOR, explaining both the optimal catalytic activities observed at subsaturating CYPOR levels, and the observed inhibition of HO-1 function at higher [CYPOR]. The dynamic organization of the microsomal electron transport chain was highlighted by a recent study, in which both P450-mediated monooxygenase activities and generation of reactive oxygen species were dramatically diminished in liver microsomes prepared from rats treated with cadmium, a potent HO-1 inducer [61].
Rough estimates of apparent Km for the functional CYPOR-HO-1 complex, derived from fitting only those data points where [CYPOR] ≤ optimal to a single binding site model (Supplemental Figure 2), are given in Table I. The results, despite the large statistical error ranges, suggest massive variation in HO-1 affinities among the CYPOR variants tested with WT and Y459H defining the range with Km values of ~9 nM and ~2 µM, respectively. Vmax/Km values further indicate the vast differences in catalytic efficiency of the HO-1 system that could result from POR variation with WT exhibiting ~ 200-fold higher efficiency than Y459H. Y181D activities were not sufficiently high for Km estimation and further highlight the differences in catalytic efficiency among CYPOR variants. The current studies also strongly suggest that experiments in which the components are not titrated to determine optimal conditions can lead to misleading results. It has been well-established that the ratio of cytochromes P450 to CYPOR in endoplasmic reticulum is extremely high, varying upon induction conditions between ~5–20:1[63] Routinely, most reconstitution assays are performed under conditions of excess CYPOR.
In these studies, it has been shown that titration of WT CYPOR with an inactive CYPOR variant can functionally sequester HO-1 away from WT (Figure 4). A variant:WT ratio of 100:1 was required for the Y181D variant to inhibit the HO-1 system by 50%, suggesting the relatively low affinity of Y181D for HO-1. Mixing each of the variants with WT CYPOR at 1:1 CYPOR:HO-1 in reconstituted systems (Figure 5) failed to distinguish any of the variants from WT, but at 1:10 CYPOR:HO-1 ratio, WT CYPOR was significantly more active than any of the variants. This suggests that a single mutated POR allele would be well-tolerated in terms of basal HO-1 function but normal individuals would be predicted to have higher HO-1 function than heterozygotes when under HO-1-inducing stress. Consideration of the inducibility of HO-1, as well as many of the P450s, and preliminary evidence of widely ranging affinities among CYPOR and its microsomal partners, suggests that a careful analysis of the stoichiometry of these critical proteins will be required to fully appreciate the physiological implications of POR variation. Stoichiometry is an important parameter to be determined in estimating these interactions and their affinities and should be examined as carefully as possible in cellular systems, as well as in purified, reconstituted systems. This consideration has been taken into account in using the engineered E. coli system of Kranendonk et al. [58] in which the ratio of CYPOR to CYPs is maintained at 1:5–10.
Although the corresponding data in this study are in general agreement with those of Pandey et al. [48], there are several important differences that need to be addressed. First, the HO-1 activities were about 20-fold larger than obtained by Pandey et al. [48]. Part of this discrepancy is likely due to differences in experimental conditions, such as, the absence of catalase in their assay mixture, the presence of which would lead to higher activities, and their measurement of activities at supersaturating CYPOR:HO-1 ratios. As noted by the current study, use of such high CYPOR:HO-1 ratios would be expected to lead to a decrease in the reported HO-1 catalytic activity. A second discrepancy is that 3 CYPOR variants (R457H, Y459H, and V492E) are completely devoid of activity according to Pandey et al. [48], whereas in the present study these same variants support clearly measurable rates of HO-1 function (up to ~60% of WT for R457H variant). Our results, demonstrating partial restoration of HO-1 activities by flavin supplementation, suggest that this difference may be due to significantly lower flavin levels in their system. Reed et al observed a range of CYPOR:HO-1 ratios from 4:1 in untreated rat liver microsomes to 1:10 after induction by cadmium treatment [61], the latter representing the physiological condition under which HO-1 activity is most critical. Furthermore, the data herein suggest that at high CYPOR:HO-1 ratios, one would expect CYPOR to disrupt HO-1 oligomerization, a phenomenon that can greatly affect HO-1 function, as first demonstrated by Hwang et al. [29], and one that has been confirmed herein.
In conclusion, the data in this manuscript demonstrate that HO-1 activity is decreased by several of the naturally occurring CYPOR mutations, with the greatest inhibition being observed by those mutants in the FAD- and FMN-binding regions. These inhibited activities from mutations in this region are due to flavin depletion, and the activities can be restored by flavin replacement, suggesting a major reversible conformational disruption. Interestingly, titrations of HO-1 activity as a function of CYPOR concentration showed a bimodal response, reaching its maximal activity at a 1:2 CYPOR:HO-1 ratio, with higher CYPOR levels leading to inhibition of heme degradation. This unusual kinetic response is possibly due to the ability of CYPOR to modulate the aggregation state of HO-1 (Figure 7). HO-1 is shown to exist in a homomeric complex within cells, with CYPOR being capable of disrupting the aggregate. Although the mechanistic details remain to be identified, it is clear that there is interplay between CYPOR and HO-1, with CYPOR not only providing the electrons necessary for catalysis, but also serving as a modulator of HO-1 function, potentially through an alteration in the HO-1 aggregation state.
Supplementary Material
Acknowledgements
This work was supported in part by NIH Grant GM081568 (to BSSM. who is The Robert A. Welch Distinguished Professor in Chemistry, AQ-0012), NIH Grant ES004344 (to WLB), and the Granting Agency of Czech Republic grant GACR P301/10/1426 (to PM). The UTHSCSA Nucleic Acids Technology Core Facility was responsible for primer synthesis and DNA sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Preliminary presentations of this work were given in poster format at the following meetings: 17th International Symposium of Microsomes and Drug Oxidations, Saratoga, NY, 2008; 34th FEBS Congress, Prague, Czech Republic, 2009; 6th International Congress on Heme Oxygenases, Miami Beach, Florida, 2009.
References
- 1.Fluck CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, Jabs EW, Mendonca BB, Fujieda K, Miller WL. Nat Genet. 2004;36:228–230. doi: 10.1038/ng1300. [DOI] [PubMed] [Google Scholar]
- 2.Arlt W, Walker EA, Draper N, Ivison HE, Ride JP, Hammer F, Chalder SM, Borucka-Mankiewicz M, Hauffa BP, Malunowicz EM, Stewart PM, Shackleton CH. Lancet. 2004;363:2128–2135. doi: 10.1016/S0140-6736(04)16503-3. [DOI] [PubMed] [Google Scholar]
- 3.Adachi M, Asakura Y, Tachibana K, Shackleton C. Pediatr Int. 2004;46:583–589. doi: 10.1111/j.1442-200x.2004.01960.x. [DOI] [PubMed] [Google Scholar]
- 4.Adachi M, Tachibana K, Asakura Y, Yamamoto T, Hanaki K, Oka A. Am J Med Genet A. 2004;128:333–339. doi: 10.1002/ajmg.a.30169. [DOI] [PubMed] [Google Scholar]
- 5.Fukami M, Horikawa R, Nagai T, Tanaka T, Naiki Y, Sato N, Okuyama T, Nakai H, Soneda S, Tachibana K, Matsuo N, Sato S, Homma K, Nishimura G, Hasegawa T, Ogata T. J Clin Endocrinol Metab. 2005;90:414–426. doi: 10.1210/jc.2004-0810. [DOI] [PubMed] [Google Scholar]
- 6.Huang N, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, Jabs EW, Van Vliet G, Sack J, Fluck CE, Miller WL. Am J Hum Genet. 2005;76:729–749. doi: 10.1086/429417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homma K, Hasegawa T, Nagai T, Adachi M, Horikawa R, Fujiwara I, Tajima T, Takeda R, Fukami M, Ogata T. J Clin Endocrinol Metab. 2006;91:2643–2649. doi: 10.1210/jc.2005-2460. [DOI] [PubMed] [Google Scholar]
- 8.Dhir V, Ivison HE, Krone N, Shackleton CHL, Doherty AJ, Stewart PM, Arlt W. Mol Endocrinol. 2007;21:1958–1968. doi: 10.1210/me.2007-0066. [DOI] [PubMed] [Google Scholar]
- 9.Hart SN, Li Y, Nakamoto K, Wesselman C, Zhong XB. Drug Metab Pharmacokinet. 2007;22:322–326. doi: 10.2133/dmpk.22.322. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal V, Huang N, Miller WL. Pharmacogenet Genomics. 2008;18:569–576. doi: 10.1097/FPC.0b013e32830054ac. [DOI] [PubMed] [Google Scholar]
- 11.Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. Pharmacogenet Genomics. 2008;18:11–24. doi: 10.1097/FPC.0b013e3282f2f121. [DOI] [PubMed] [Google Scholar]
- 12.Hart SN, Zhong XB. Expert Opin Drug Metab Toxicol. 2008;4:439–452. doi: 10.1517/17425255.4.4.439. [DOI] [PubMed] [Google Scholar]
- 13.Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM. Pharmacogenomics. 2009;10:579–599. doi: 10.2217/pgs.09.7. [DOI] [PubMed] [Google Scholar]
- 14.Guengerich FP. Mol Interv. 2003;3:194–204. doi: 10.1124/mi.3.4.194. [DOI] [PubMed] [Google Scholar]
- 15.Ono T, Bloch K. J Biol Chem. 1975;250:1571–1579. [PubMed] [Google Scholar]
- 16.Schenkman JB, Jansson I. Pharmacol Ther. 2003;97:139–152. doi: 10.1016/s0163-7258(02)00327-3. [DOI] [PubMed] [Google Scholar]
- 17.Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, Redline R. Proc Natl Acad Sci U S A. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishino H, Ishibashi T. Arch Biochem Biophys. 2000;374:293–298. doi: 10.1006/abbi.1999.1602. [DOI] [PubMed] [Google Scholar]
- 19.Schacter BA, Nelson EB, Marver HS, Masters BS. J Biol Chem. 1972;247:3601–3607. [PubMed] [Google Scholar]
- 20.Miller WL, Huang N, Agrawal V, Giacomini KM. Molecular and Cellular Endocrinology. 2009;300:180–184. doi: 10.1016/j.mce.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Ryter SW, Alam J, Choi AM. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 22.Abraham NG, Kappas A. Pharmacol Rev. 2008;60:79–127. doi: 10.1124/pr.107.07104. [DOI] [PubMed] [Google Scholar]
- 23.Alam J. Antioxid Redox Signal. 2002;4:559–562. doi: 10.1089/15230860260220049. [DOI] [PubMed] [Google Scholar]
- 24.Maines MD, Trakshel GM, Kutty RK. J Biol Chem. 1986;261:411–419. [PubMed] [Google Scholar]
- 25.Tenhunen R, Marver HS, Schmid R. J Biol Chem. 1969;244:6388–6394. [PubMed] [Google Scholar]
- 26.Martasek P, Solangi K, Goodman AI, Levere RD, Chernick RJ, Abraham NG. Biochem Biophys Res Commun. 1988;157:480–487. doi: 10.1016/s0006-291x(88)80274-2. [DOI] [PubMed] [Google Scholar]
- 27.Tenhunen R, Marver HS, Schmid R. Proc Natl Acad Sci U S A. 1968;61:748–755. doi: 10.1073/pnas.61.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber WJ, III, Scruggs BA, Backes WL. Biochemistry. 2009;48:190–197. doi: 10.1021/bi801912z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang H-W, Lee J-R, Chou K-Y, Suen C-S, Hwang M-J, Chen C, Shieh R-C, Chau L-Y. Journal of Biological Chemistry. 2009;284:22672–22679. doi: 10.1074/jbc.M109.028001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dore S, Takahashi M, Ferris CD, Zakhary R, Hester LD, Guastella D, Snyder SH. Proc Natl Acad Sci U S A. 1999;96:2445–2450. doi: 10.1073/pnas.96.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keyse SM, Tyrrell RM. Proc Natl Acad Sci U S A. 1989;86:99–103. doi: 10.1073/pnas.86.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TS, Chau LY. Nat Med. 2002;8:240–246. doi: 10.1038/nm0302-240. [DOI] [PubMed] [Google Scholar]
- 33.Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 34.Willis D, Moore AR, Frederick R, Willoughby DA. Nat Med. 1996;2:87–90. doi: 10.1038/nm0196-87. [DOI] [PubMed] [Google Scholar]
- 35.Otterbein LE, Bach FH, Alam J, Soares M, Tao Lu H, Wysk M, Davis RJ, Flavell RA, Choi AM. Nat Med. 2000;6:422–428. doi: 10.1038/74680. [DOI] [PubMed] [Google Scholar]
- 36.Ryter SW, Otterbein LE, Morse D, Choi AM. Mol Cell Biochem. 2002;234–235:249–263. doi: 10.1023/A:1015957026924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens CF, Wang Y. Nature. 1993;364:147–149. doi: 10.1038/364147a0. [DOI] [PubMed] [Google Scholar]
- 38.Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 39.Yachie A, Niida Y, Wada T, Igarashi N, Kaneda H, Toma T, Ohta K, Kasahara Y, Koizumi S. J Clin Invest. 1999;103:129–135. doi: 10.1172/JCI4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maines MD. Annu Rev Pharmacol Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 41.Masters BS, Marohnic CC. Drug Metab Rev. 2006;38:209–225. doi: 10.1080/03602530600570065. [DOI] [PubMed] [Google Scholar]
- 42.Otto DM, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, Adams RH, Tickle C, Wolf CR. Mol Cell Biol. 2003;23:6103–6116. doi: 10.1128/MCB.23.17.6103-6116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen AL, O'Leary KA, Kasper CB. J Biol Chem. 2002;277:6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Gu J, Cui H, Zhang QY, Behr M, Fang C, Weng Y, Kluetzman K, Swiatek PJ, Yang W, Kaminsky L, Ding X. J Pharmacol Exp Ther. 2005;312:35–43. doi: 10.1124/jpet.104.073353. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, Zhang QY, Zhuo X, Xie Q, Ding X. Genesis. 2003;36:177–181. doi: 10.1002/gene.10214. [DOI] [PubMed] [Google Scholar]
- 46.Soares MP, Bach FH. Trends Mol Med. 2009;15:50–58. doi: 10.1016/j.molmed.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Huber WJ, III, Marohnic CC, Peters M, Alam J, Reed JR, Masters BSS, Backes WL. Drug Metab Dispos. 2009;37:857–864. doi: 10.1124/dmd.108.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pandey AV, Fluck CE, Mullis PE. Biochem Biophys Res Commun. 2010;400:374–378. doi: 10.1016/j.bbrc.2010.08.072. [DOI] [PubMed] [Google Scholar]
- 49.Marohnic CC, Panda SP, Martasek P, Masters BS. J Biol Chem. 2006;281:35975–35982. doi: 10.1074/jbc.M607095200. [DOI] [PubMed] [Google Scholar]
- 50.Yasukochi Y, Masters BS. J Biol Chem. 1976;251:5337–5344. [PubMed] [Google Scholar]
- 51.Marohnic CC, Panda SP, McCammon K, Rueff J, Masters BS, Kranendonk M. Drug Metab Dispos. 2010;38:332–340. doi: 10.1124/dmd.109.030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Proc Natl Acad Sci U S A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber WJ, 3rd, Backes WL. Biochemistry. 2007;46:12212–12219. doi: 10.1021/bi701496z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber WJ, 3rd, Backes WL. Anal Biochem. 2008;373:167–169. doi: 10.1016/j.ab.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maines M. Methods Enzymol. 1996;268:473–488. doi: 10.1016/s0076-6879(96)68049-5. [DOI] [PubMed] [Google Scholar]
- 56.Maines MD, Kappas A. Proc Natl Acad Sci U S A. 1974;71:4293–4297. doi: 10.1073/pnas.71.11.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Black SD, French JS, Williams CH, Jr, Coon MJ. Biochem Biophys Res Commun. 1979;91:1528–1535. doi: 10.1016/0006-291x(79)91238-5. [DOI] [PubMed] [Google Scholar]
- 58.Kranendonk M, Marohnic CC, Panda SP, Duarte MP, Oliveira JS, Masters BS, Rueff J. Arch Biochem Biophys. 2008;475:93–99. doi: 10.1016/j.abb.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicolo C, Fluck CE, Mullis PE, Pandey AV. Mol Cell Endocrinol. 2010;321:245–252. doi: 10.1016/j.mce.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 60.Narayanasami R, Horowitz PM, Masters BS. Arch Biochem Biophys. 1995;316:267–274. doi: 10.1006/abbi.1995.1037. [DOI] [PubMed] [Google Scholar]
- 61.Reed JR, Cawley GF, Backes WL. Drug Metab Lett. 2011;5:6–16. doi: 10.2174/187231211794455253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gozzelino R, Jeney V, Soares MP. Annu Rev Pharmacol Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 63.Estabrook RW, Franklin MR, Cohen B, Shigamatzu A, Hildebrandt AG. Metabolism. 1971;20:187–199. doi: 10.1016/0026-0495(71)90091-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.