Abstract
Sample transport and electrokinetic injection bias are well-characterized in capillary electrophoresis and simple microchips, but a thorough understanding of sample transport on devices combining electroosmosis, electrophoresis, and pressure-driven flow is lacking. In this work, we evaluate the effects of electric fields from 0–300 V/cm, electrophoretic mobilities from 10−4–10−6 cm2/Vs, and pressure-driven fluid velocities from 50–250 µm/s on sample injection in a microfluidic chemical cytometry device. By studying a continuous sample stream, we find that increasing electric field strength and electrophoretic mobility result in improved injection and that COMSOL simulations accurately predict sample transport. The effects of pressure-driven fluid velocity on injection are complex, and relative concentration values lie on a surface defined by pressure-driven flow rates. For high mobility analytes, this surface is flat, and sample injection is robust despite fluctuations in flow rate. For lower mobility analytes, the surface becomes steeper, and injection depends strongly on pressure-driven flow. These results indicate generally that device design must account for analyte characteristics and specifically that this device is suited to high mobility analytes. We demonstrate that for a suitable pair of peptides fluctuations in injection volume are correlated; electrokinetic injection bias is minimized; and electrophoretic separation achieved.
Keywords: chemical cytometry, electrokinetic injection bias, microfluidics, simulation
1 Introduction
Chemical cytometry is an analytical technique in which a single cell is isolated and lysed and its contents separated and detected [1]. The separation step permits quantitative information about multiple analytes to be readily determined. Multi-analyte detection is critical to measuring enzyme activity, determining how biochemical species co-vary, and elucidating regulatory pathways at the level of individual cells (see, e.g., [2–3]). Each of these analytical problems requires accurate comparisons of the abundance of many species in a single cell, and acquisition and validation of these data by chemical cytometry demand a thorough understanding of sample transport on the analytical platform used.
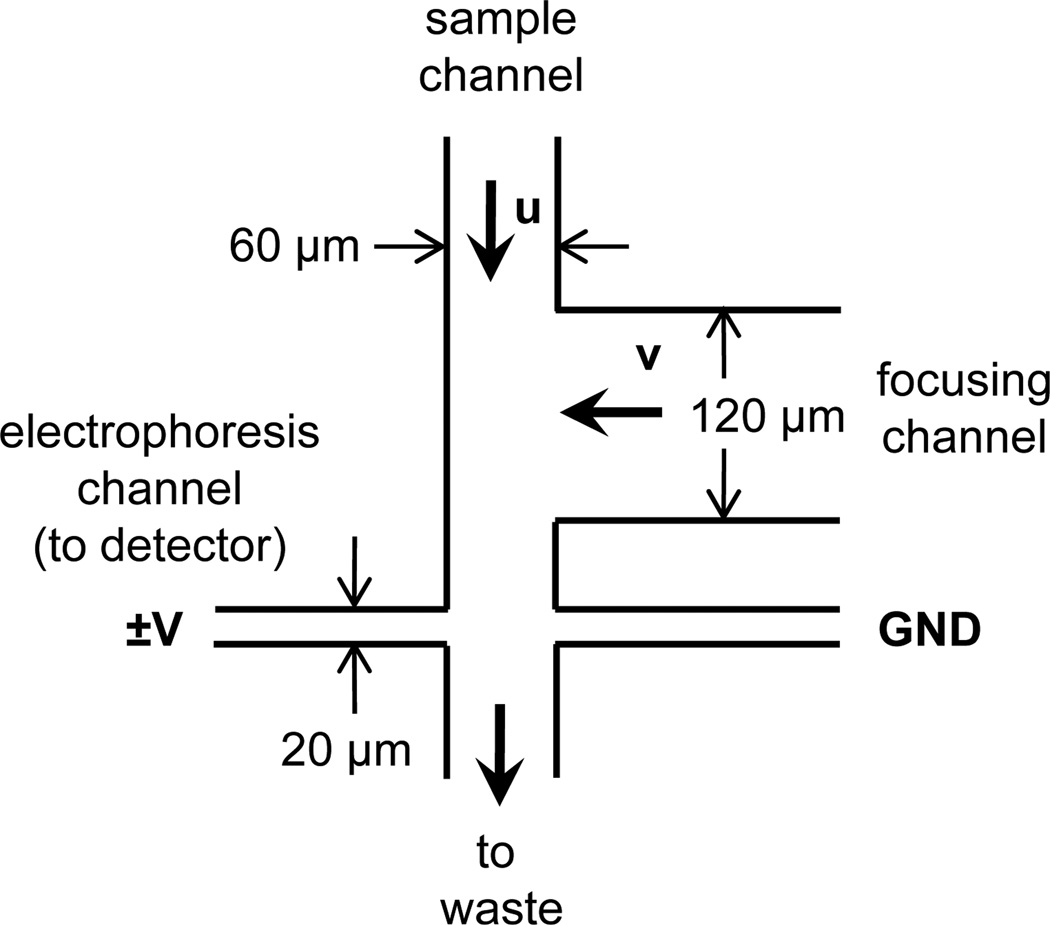
Several microfluidic devices have been developed for single-cell analysis because miniaturized systems facilitate low-volume sample handling and increase throughput by automation [4–5]. Sample injection in chemical cytometry devices depends on the location of cell lysis. In some designs, the cell is loaded into the electrophoresis channel [6–8] or fixed at the channel entrance prior to lysis [9–11]. Analyte movement into the electrophoresis channel occurs via application of an electric field, and all applied forces are along the axis of the electrophoresis channel. In an alternative device design (Figure 1), the cell is lysed while moving perpendicular to the electrophoresis channel [12–14]. This design is favorable because low mobility cellular debris does not enter the electrophoresis channel; however, the same physical principle that excludes low mobility debris from the electrophoresis channel may also induce electrokinetic injection bias.
Figure 1.
Schematic of the device design with channel widths and physical parameters. The arrows u and v indicate the direction, but not necessarily the magnitude, of pressure-driven flow in the sample and focusing channels, respectively. The gap between the focusing channel and the electrophoresis channel was 50 µm. All channels were 30 µm deep. Sample concentration was evaluated in the three potential exit channels for the sample stream: the electrophoresis channel at either side of its intersection with the sample channel and in the waste side of the sample channel.
Electrokinetic injection bias occurs when one analyte is preferentially injected into the electrophoresis channel. The magnitude of electrokinetic injection bias can be described by a bias factor, b: the ratio of the amount of substance 1 injected, Q(1), to the amount of substance 2 injected, Q(2).
| (1) |
This phenomenon has been well-characterized in both capillaries [15–16] and simple microchips [17–19], but no detailed study of electrokinetic injection bias on chemical cytometry microdevices has been performed to date. In capillary electrophoresis, the bias factor can be readily calculated using the electrophoretic mobilities of the analytes (μep,1 and μep,2) and the electroosmotic mobility (μeo) [16]. However, in microfluidic devices, the origins of electrokinetic injection bias can be more complex and depend on the injection scheme used. For example, the orthogonal orientation of pinched injections reduces bias compared to electrokinetic injections on capillaries, but both the loading and injection steps contribute to some bias [19]. Similarly, gated injections are subject to multiple bias effects. In addition to the first-order bias that occurs in capillary injections, gated injections also result in a second-order, transradial bias because analytes of different mobility experience different turning radii [17–18]. In microdevices designed for chemical cytometry, sample transport is typically quite sophisticated because of the small sample volume and the need to solubilize or remove debris after cell lysis. Consequently, sample injection and bias may be influenced by several factors, including pressure-driven fluid velocities and variable buffer compositions, in addition to electrokinetic effects. These factors necessitate empirical studies and computer simulations of sample injection on these devices.
To more thoroughly understand electrokinetic injections in chemical cytometry microchips, we have investigated sample transport using simulations and experiments with a commonly-used device design (Figure 1) [12–14]. In this device, cells are carried through the device by pressure-driven flow, focused to the lysis location using pressure-driven flow from a focusing channel, and lysed. During successful operation, electrokinetic and hydrostatic forces cause analytes of interest from the lysed cell to turn 90° and be injected into the electrophoresis channel, while low mobility cellular debris continues down the sample channel to waste. Using this device, we examined sample injection for three reporter peptides used to assay Abl kinase activity (Table 1). In single-cell enzyme assays, the amounts of unreacted and enzymatically-modified reporter peptide are quantified to determine the activity of the enzyme of interest; however, before the relative amount of each species can be determined, any electrokinetic injection bias introduced by the device design and differing peptide mobilities must be known. Consequently, reporter peptides are attractive analytes for these studies because their application to enzyme assays demands accurate quantitation. The purpose of this study is to evaluate the effects of electric field strength, electrophoretic mobility, and pressure-driven flow on sample injection in a commonly used microfluidic device for chemical cytometry.
Table 1.
Reporter peptides for Abl kinase.
| Name | Sequencea) | μep (× 10−4 cm2/Vs) (unmodified form) |
μep (× 10−4 cm2/Vs) (phosphorylated form) |
|---|---|---|---|
| abl-sub | 5-FAM-EAIYAAPFAKKK-NH2 | −0.01 | −0.36 |
| QZ-V-48B | 5-FAM-GGIYAAP(NMe)FKKKA-NH2 | +0.37 | −0.93 |
| NCMK-1 | 5-FAM-GGIYAAP(NMe)F-NH2 | −1.48 | −2.83 |
(NMe)F = N-methylphenylalanine
2 Materials and Methods
2.1 Materials
Egg phosphatidylcholine (PC) and egg phosphatidylglycerol (PG) were obtained from Avanti. Tris buffer for vesicle formation (Tris-ves) was composed of 10 mM tris(hydroxymethyl)aminomethane (Tris) and 150 mM NaCl (pH 7.4). Small unilamellar vesicles were prepared in Tris-ves as described previously [20]. Extracellular buffer (ECB) was 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2 and 10 mM glucose (pH 7.4). Tris buffer for separations (Tris-sep) was 25 mM Tris (pH 8.4). PDMS prepolymer and curing agent were obtained from Dow-Corning. Silicone tubing (3.2 mm i.d. and 6.4 mm o.d.) for reservoirs was obtained from Cole-Parmer, and coverglass was obtained from Fisher. Silver-silver chloride (Ag/AgCl) wire electrodes were prepared by soaking 1 mm diameter silver wire (Alfa Aesar) in an aqueous solution of 1 M iron (III) chloride and 0.1 M hydrochloric acid overnight.
Three reporter peptides for Abl kinase were synthesized by Anaspec, Inc. in both their unmodified and phosphorylated forms. The peptide sequences are shown in Table 1; the modified forms are phosphorylated at the tyrosine (Y) residue. The electrophoretic mobility of each peptide in ECB was determined based on migration times for gated injections [21] of each peptide on a simple cross chip. For high mobility peptides, gated injections were performed in a PC-coated device; for low mobility peptides, the electrophoretic mobilities were determined in the presence of higher electroosmotic flow using a device coated with 70% PC and 30% PG. To monitor pressure-driven flow in the device, Ba/F3 (mouse leukemic) cells were loaded with Calcein red-orange acetoxymethyl ester (Invitrogen) and fixed with 4% formaldehyde (Thermo Scientific) in phosphate buffered saline. Immediately prior to use as flow tracers, the fixed cells were pelleted by 1 min centrifugation at 1400× g, washed once, and resuspended in either 1 µM analyte in ECB or Tris-sep buffer.
2.2 Device fabrication and preparation
PDMS devices were prepared by soft lithography as described previously [22]. Briefly, a 10:1 mixture of PDMS prepolymer and curing agent were mixed together, degassed, cast against a 30 µm high SU-8 master, and cured at 100 °C for 10 min on a hotplate. The dimensions of the resulting channels are shown in Figure 1. The PDMS channels were exposed to an oxygen plasma (Harrick, PDC-00) and irreversibly sealed to a coverglass. Silicone tubing reservoirs were then plasma-sealed over each inlet and outlet hole [13]. Immediately after assembly, each device was filled by capillary action with small unilamellar phosphatidylcholine vesicles, which spontaneously fused with the hydrophilic channel walls to form a supported bilayer membrane coating [20]. Coated channels were incubated at 4 °C for at least 1 h and up to 1 day before use. Immediately before use, the device was flushed for 10 min with ECB to remove remaining vesicles and Tris ves buffer.
2.3 Injection experiments
At the start of each experiment, the sample reservoir was filled with 1 µM analyte in ECB; the focusing reservoir with Tris-sep, and all other reservoirs with ECB. The sample and focusing channel solutions also contained Calcein red-orange stained, formaldehyde-fixed Ba/F3 cells for use as flow tracers. Gentle (50–250 µm/s) pressure-driven flow was established in the cell and focusing channels by adjusting the fluid levels in the reservoirs. Voltages of 0 V, ±340 V, ±680 V, and ±1020 V were applied to the electrophoresis channel using Ag/AgCl wire electrodes, a LabSmith HVS 448 high voltage sequencer, and Sequencer 1.131 software. Sample transport was monitored using a 20× Plan Fluor objective (Nikon Inc.) and a Nikon TE-300 microscope equipped for epifluorescence with FITC (Semrock) and TRITC (G-2EC, Nikon) filters, a xenon arc lamp, and a CCD camera (CoolSnapfx, Photometrics). Data were collected using Metafluor software (v5.0r1, Molecular Devices, Inc.).
2.4 Image analysis
To quantify sample transport, fluorescence micrographs of the intersection were analyzed using ImageJ [23]. The micrographs were background-subtracted using an image of the device filled with ECB and normalized by dividing the data image by an image of the device filled with 1 µM analyte. This normalization step corrected for variations in excitation intensity across the field of view. Quantitative measures of fluorescence intensity were obtained from the average counts for regions of interest in each of the three potential outlet channels. To determine the fluid velocities in the cell and focusing channels, images of the fixed cells were obtained using long (500 ms) exposure times. As a result, each cell appeared in images as an extended streak. The distance traveled by a cell was equal to the streak length minus the cell diameter, and the velocity of each cell was equal to the distance traveled divided by the exposure time. We used cell concentrations of 106 cells/mL as an acceptable compromise between adequately sampling the radially varying fluid velocity and avoiding channel clogging at higher cell densities. We routinely measured 5 or more cells per channel per field strength, so average fluid velocities in the cell and focusing channels were determined by averaging the velocities of n ≥ 5 cells in each channel.
2.5 COMSOL simulations
COMSOL Multiphysics 3.5 was used to model sample transport in the device. Specifically, electrophoretic transport, pressure-driven transport, and the electric field lines were modeled using the Electrokinetic Flow, Incompressible Navier-Stokes, and Conductive Media DC application modes, respectively. Electroosmotic flow was modeled using a predefined coupling of the Incompressible Navier-Stokes and Conductive Media DC applications modes in the MEMS module. For each simulation, the average peptide concentration (mol/m3) and mass transfer rate (mol/s) were determined for each of three potential outlets: the electrophoresis channel on either side of its intersection with the sample channel and the waste outlet of the sample channel. The Supporting Information includes further details and the physical parameters used for the simulations (Table S1).
3 Results and Discussion
To investigate the effects of electric field strength, electrophoretic mobility, and pressure-driven fluid velocities on sample injection, we chose to load a continuous sample stream into the device rather than lysing individual cells and injecting their contents. This simplified system had several advantages: (i) effects of cell-to-cell variability in peptide loading were eliminated, (ii) enough material was injected to image sample transport with a CCD camera, (iii) the effects of varying field strength and fluid velocity were readily observed in real time (Movie, Supporting Information), and (iv) variability due to the location and extent of cell lysis was eliminated. The voltages and pressures applied to the device yielded a sample stream with the same shape and velocity used for single cell experiments. Thus the behavior of the sample stream should reproduce the trajectory of the sample plug produced upon cell lysis. In this work, the effects of electric field strength and fluid velocity on sample injection were examined empirically and used to determine the ability of COMSOL simulations to accurately model transport on this PDMS/glass hybrid device. The effects of these parameters were evaluated for three peptide substrates for Abl kinase with electrophoretic mobilities spanning three orders of magnitude from −1×10−6 to −2.83×10−4 cm2/Vs.
3.1 Effect of electric field strength
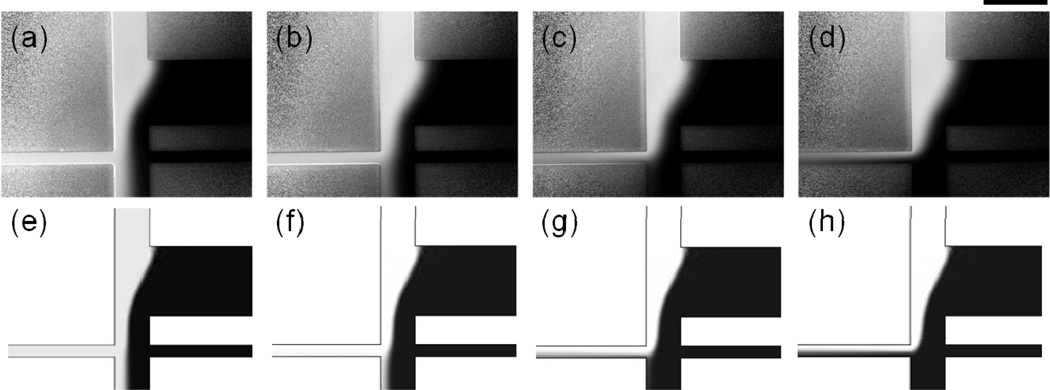
As expected, sample injection improved with increasing electric field strength for all peptides, and the shape of the sample stream and field strength required to turn the stream were well-modeled in COMSOL for the entire range of field strengths tested (0–300 V/cm, Figure 2). For example, the COMSOL simulations accurately predicted the shape of the sample stream as it turned into the electrophoresis channel (Figure 2c,g) and the flow of buffer into the electrophoresis channel at higher field strength (Figure 2d,h). The only consistent qualitative difference between the COMSOL simulations and the experimental results was that the right side of the sample stream, which was not bounded by a channel wall, was broader and more diffuse in the experimental results than in simulations. For phosphorylated abl-sub, a second qualitative difference between the experimental and simulated results was observed: the peptide migrated toward the anode in experiments and toward the cathode in simulations. The unmodified and phosphorylated forms of abl-sub have low magnitude electrophoretic mobilities that were overwhelmed by the higher magnitude cathodic electroosmotic flow in simulations. The electrophoretic mobility of p-abl-sub (−0.36 × 10−4 cm2/Vs) was close in magnitude and opposite in sign compared to the electroosmotic mobility of the PC-coated channels (+0.52 ± 0.19 × 10−4 cm2/Vs). The large standard deviation of the measured electroosmotic mobility suggested that the relative velocities of these phenomena could not determined, and the miscalculation of migration direction likely represented a limitation of the data rather than a failure of the simulation. In general , we expect accurate simulations for values of μep at least one standard deviation larger or smaller in magnitude than μeo. More precisely measured electroosmotic mobilities, such as those obtained for glass devices, should therefore lend themselves to even more accurate simulations.
Figure 2.
(a–d) Experimental and (e–h) simulated results showed good qualitative agreement for electric field strengths of 0–300 V/cm, as evidence by p-NCMK-1 peptide transport at (a, e) 0 V/cm, (b, f) +100 V/cm, (c, g) +200 V/cm, and (d, h) +300 V/cm. For the experimental data, the pressure-driven fluid velocities in the sample and focusing channels were ~100 µm/s and ~225 µm/s, respectively. For the simulations, the fluid velocity in both channels was 150 µm/s. The micrographs in (a–d) were background subtracted, normalized, and set to the same brightness/contrast scale. The scale bar is 200 µm.
Overall, qualitative agreement between experiments and simulations was obtained despite differences in buffer composition. This device design has often been operated with different buffer solutions in the cell and focusing channels [12–14]. For example, filling the focusing channel with lower salt Tris-sep buffer reduced fouling of the waste and electrophoresis channels compared to a device filled entirely with ECB [14], so these conditions were used in the experiments reported here. The COMSOL models, however, were run for devices filled entirely with ECB to simplify the simulations. To further validate our comparisons of the experimental and simulated data, we compared experimental results for sample transport in devices filled entirely with ECB to those obtained with Tris-sep in the focusing channel and found minimal difference between the two conditions (Figure S2). This suggested that Tris-sep buffer from the focusing channel did not enter the electrophoresis channel in significant amounts; indeed, separations on this device have yielded similar migration times and separation metrics to those performed in a simple cross chip filled with ECB-glucose [14].
Quantitative evaluation of electrokinetic injection bias requires knowledge of the amount of each substance injected, Q(i), or in the case of the continuous injections used here, the amount of sample injected as a function of time, i.e., the mass transfer rate, Q(i)/t. While this mass transfer information was readily extracted from simulations, the experimental data consisted of fluorescence counts, which were linearly proportional to sample concentration (R2 = 0.998). Consequently, the relative concentration of sample in the detector side of the electrophoresis channel was calculated with Equation (2),
| (2) |
where C is analyte concentration in each outlet channel. Simulated data indicated that this value was a suitable stand-in for mass transfer data (Figure S1), so relative concentration was used to compare experimental and simulated results.
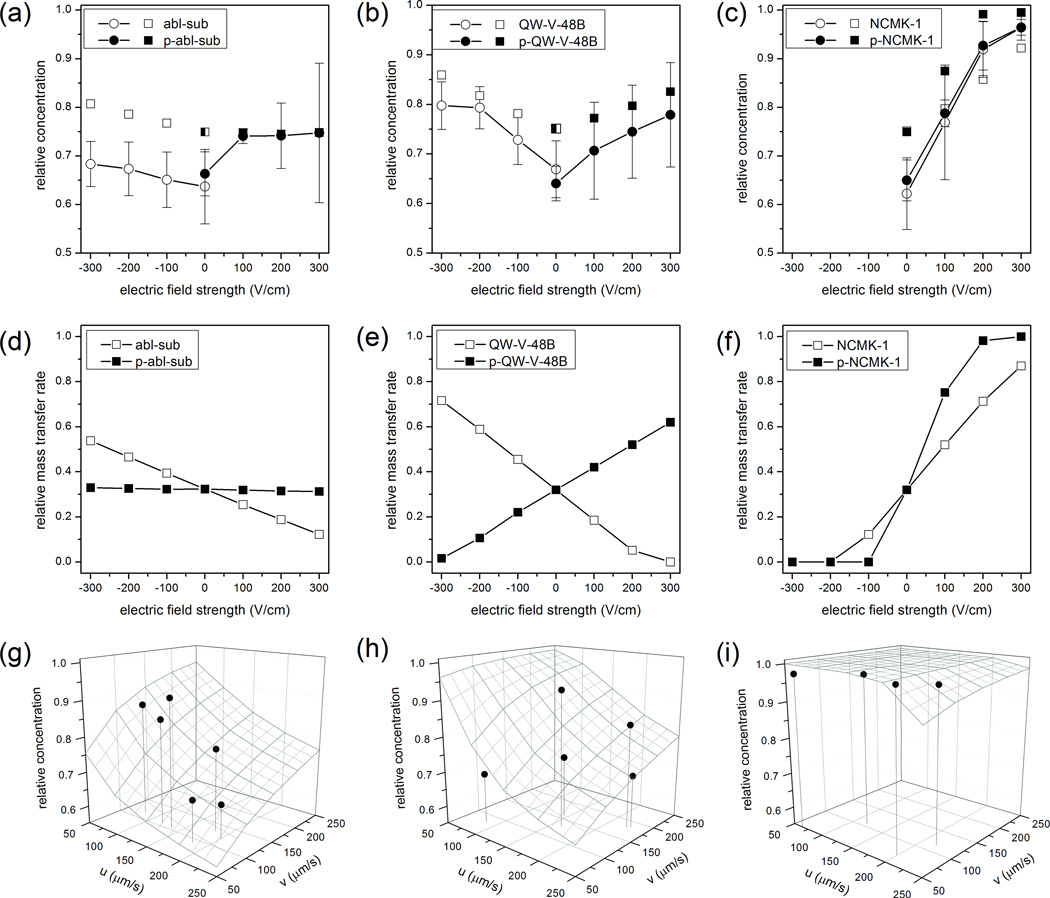
Overall, quantitative agreement between simulations and experiments was good (Figure 3a–c). Specifically, for moderate to high mobility analytes in the presence of an electric field, the COMSOL models showed excellent agreement with the experimental results. For these data, the difference between experiments and simulations ranged from 0.1% to 10%. In contrast, COMSOL simulations systematically overestimated the amount of material injected in the absence of an electric field (0 V/cm, Figure 3a–c) and for the low mobility peptide abl-sub at all field strengths (Figure 3a). In the absence of an applied field, or for a peptide with an extremely low electrophoretic mobility (μep,abl-sub = −1×10−6 cm2/Vs), transport was dominated by pressure-driven flow and diffusion. We suspect that the discrepancy between the simulated and experimental results at low values of E and μep resulted from underestimation of the sample stream broadening in COMSOL (Figure 2 and Supporting Information). Notably, COMSOL accurately predicted the relative effect of increasing electric field strength on abl-sub injection: although the simulated results were offset from the experimental data, the two plots possessed the same slope (Figure 3a).
Figure 3.
Sample injection as a function of electric field strength (a–f) and fluid velocities (g–i). In all plots, circles represent experimental data; squares are simulated data; and unmodified and phosphorylated (p-) peptides are shown using open and filled symbols, respectively. (a–c) Overall, simulated and experimental results showed good agreement on the effect of electric field strength on sample injection. Error bars show the standard deviation for n = 4–6 runs total on at least three different devices. (d–f) The simulated mass transfer rates demonstrate the profound effect of electrophoretic mobility on sample transport and were used to determine electrokinetic injection bias. (g–i) The effects of sample (v) and focusing channel (u) velocities on the relative concentrations of the phosphorylated peptides, (g) p-abl-sub (νek = 50 µm/s), (h) p-QW-V-48B (νek = 120 µm/s), and (i) p-NCMK-1 (νek = 690 µm/s), are shown for a fixed field strength of +300 V/cm. The wire frame surfaces represent data simulated in COMSOL, and the solid black circles are individual experimental runs. Drop lines indicate the fluid velocities for each experimental point, which were determined using fixed, stained cells as flow tracers.
3.2 Effect of analyte electrophoretic mobility: comparing reporter peptides
The Abl kinase reporter peptides studied here – abl-sub [24], QW-V-48B (manuscript in preparation), and NCMK-1 – represent a range of electrophoretic mobilities spanning three orders of magnitude. Consequently, we were able to evaluate the effect of analyte mobility on sample injection. All three peptides and their phosphorylated forms were roughly the same size (MW = 1166 – 1774 Da) and were calculated to have similar diffusion coefficients (Supporting Information). As a result, in the absence of an electric field, sample injection into the electrophoresis channel was statistically the same for all peptides (p = 0.353, one-way ANOVA). When voltage was applied, the peptides’ different electrophoretic mobilities resulted in different transport profiles.
Although the relative concentration data in Figure 3a–c was useful for comparing experimental and simulated results, the mass transfer rate in each channel, Q(i)/t, was necessary to determine the expected electrokinetic injection bias for the three reporter peptides. Relative mass transfer rates from the simulations were defined as the rate of sample transfer toward the detector divided by the total sample transfer rate toward all three potential outlets (Figure 3d–f). Notably, the unmodified forms of abl-sub and QW-V-48B migrated toward the cathode, while the phosphorylated forms migrated toward the anode. As a result, favorable electric field conditions for injecting one form of either peptide were unfavorable for the other form, resulting in a high degree of electrokinetic injection bias (Figure 3d, e). For example, at a field strength of +300 V/cm and fluid velocities of 150 µm/s, the bias factors, b, for abl-sub and QW-V-48B and their phosphorylated forms would be 2.55 and 109, respectively. The electrokinetic injection bias for QW-V-48B is excessive at +300 V/cm because the phosphorylated form loads well while the unmodified form of the peptide effectively does not enter the electrophoresis channel. At lower field strengths (±100 V/cm), small amounts (20–50%) of both peptides enter the electrophoresis channel, but in a mass-limited sample, such as a single-cell lysate, this degree of sample loss is unacceptable. Consequently, both abl-sub and QW-V-48B would make poor reporters for Abl kinase on the device used here.
Results for the peptide NCMK-1 (Figure 3c, f) were much more promising. The unmodified and phosphorylated forms of NCMK-1 both migrated toward the anode; the relative concentration of peptide in the electrophoresis channel was more reproducible; and both forms of the peptide were efficiently directed toward the detector at moderate field strength. Additionally, despite the fact that the unmodified and phosphorylated forms had different electrophoretic mobilities, both peptides were sufficiently mobile to be almost completely injected into the electrophoresis channel, and electrokinetic injection bias was minimal (Figure 3f). For example, at 300 V/cm, the experimental ratio of the relative concentration of NCMK-1 to p-NCMK-1 was 0.9995. In simulations, the same conditions corresponded to 87% of NCMK-1 and 100% of the p-NCMK-1 being transferred into the electrophoresis channel (Figure 3f, b = 0.87).
3.3 Effects of pressure-driven fluid velocities
To investigate the effects of hydrodynamic fluid velocities on sample injection, we compared experimental and simulated results for the phosphorylated forms of each peptide at fluid velocities from 50–250 µm/s in the sample and focusing channels and a constant electric field strength of +300 V/cm. A previous report on a similar device suggested that efficient sample injection required that the analyte’s electrokinetic velocity (νek) be greater than its pressure-driven velocity (u) [12]. Indeed, the relative magnitudes of the sample’s hydrodynamic and electrokinetic velocities contributed to injection efficiency; however, the relationship between these parameters was complex.
For a fixed electric field strength of +300 V/cm, the sample and focusing channel velocities determined the analyte’s location on a relative concentration surface (Figure 3g–i). For lower mobility analytes, including p-abl-sub (Figure 3g) and p-QW-V-48B (Figure 3h), this surface was fairly steep. As expected from previous work, increasing pressure-driven fluid velocity in the sample channel negatively affected sample injection. In fact, this was true even when the sample’s hydrodynamic velocity (u) was less than its electrokinetic velocity (Figure 3h). In contrast, higher focusing channel velocities (v) corresponding to improved sample injection. Presumably this occurred because flow from the focusing channel diverted the sample channel stream, bringing it closer to the entrance of the electrophoresis channel. Similar trends were observed for a higher mobility analyte, p-NCMK-1 (Figure 3i), but the relative concentration surface was fairly flat. Relative concentration only dropped off at very high sample channel velocities (~250 µm/s) and very low focusing channel velocities (~50 µm/s).
Because the relative concentration surface was steeper for low mobility analytes, their transport was more susceptible to fluctuations in pressure-driven flow. Indeed, these results (Figure 3g,h) suggested that variation in pressure-driven fluid velocities contributed to variation in relative concentration and explained in part the large error bars for some data in Figure 3a,b. For example, if we assume fluid velocities in the sample and focusing channels of 135–165 µm/s (i.e., 20% variation around an average velocity of 150 µm/s), p-abl-sub transport would vary by 11%. The same variation in fluid velocities would result in only 0.02% variation in p-NCMK-1 transport. We conclude that this device design is best suited to analytes with high electrophoretic mobilities; these molecules were efficiently injected into the electrophoresis channel with minimal bias and were less susceptible to variations in pressure-driven fluid flow.
3.4. Simultaneous injection of two peptides
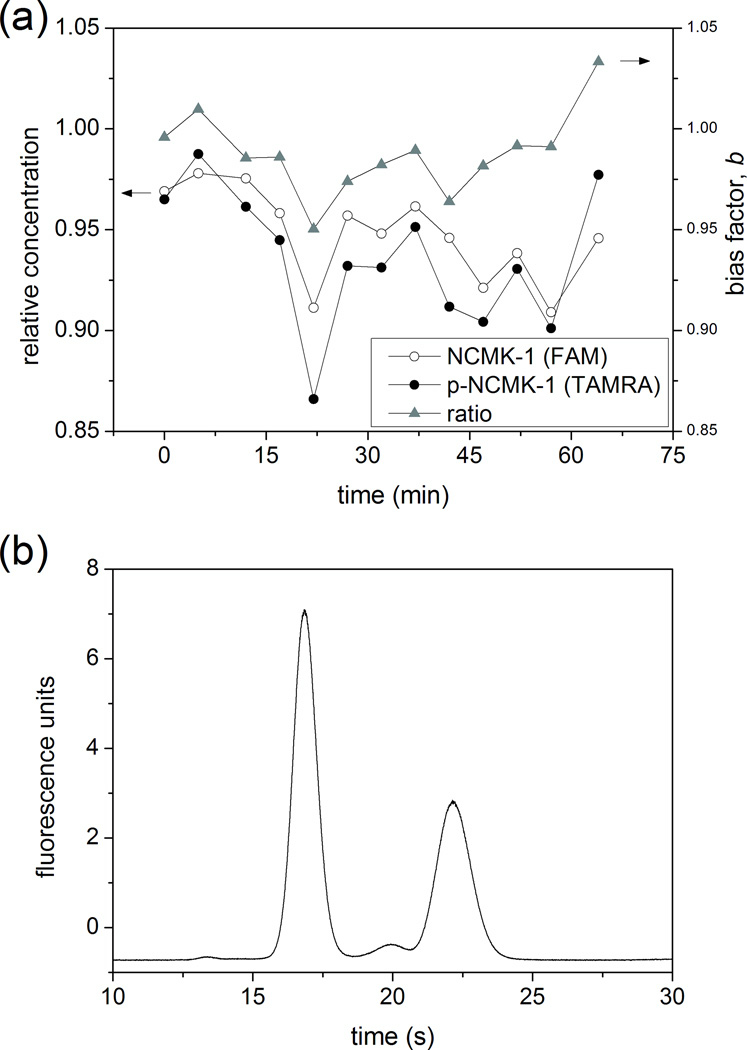
In single-cell enzyme assays, the unmodified and phosphorylated forms of a reporter peptide are released from the lysed cell, injected into the electrophoresis channel, separated, and detected. The ratio of the peak areas of the unmodified and phosphorylated forms is then used to determine enzyme activity. As a result, peptides must be injected with minimal bias with respect to each other (b ≈ 1), and the relative amount of each peptide injected must be stable in time (i.e., b must be stable) so that ratiometric signals from numerous cells can be compared. We therefore evaluated the bias factor, b, as a function of time for two related peptides: 5-FAM-labeled NCMK-1 (NCMK-1) and 5-TAMRA-labeled p-NCMK-1 (p-T-NCMK-1) (Figure 4a). The spectrally-resolved fluorescent labels allowed us to measure sample transport for two peptides on the same device at the same time. The bulky, neutral TAMRA label slightly decreased the mobility of p-T-NCMK-1 relative to that of FAM-labeled p-NCMK-1 (μep, p-T-NCMK-1 = −2.43 × 10−4 cm2/Vs, μep, p-NCMK-1 = −2.83 × 10−4 cm2/Vs); however, the comparison of NCMK-1 and p-T-NCMK-1 still provided valuable information about how transport of two related peptides co-varied on the device. The results indicated minimal bias for injections of these two peptides (b = 0.99 ± 0.02). Additionally, while the relative concentrations of both peptides fluctuated with time (RSDNCMK-1 = 2.4%, RSDp-T-NCMK-1 = 3.6%), the fluctuations were similar in magnitude and direction for both peptides. As a result, variation in the bias factor, b, was alleviated by correlation between the fluctuations in the two peptides’ transport (RSD = 2.0%).
Figure 4.
(a) Stability of peptide transport (circles) and injection ratio (triangles) as a function of time for unmodified and phosphorylated (p-)NCMK-1 at +300 V/cm. The unmodified peptide (open) was labeled with 5-carboxyfluorescein (FAM), and the phosphorylated peptide (filled) was labeled with 5-carboxytetramethylrhodamine (TAMRA). The relative concentration of each peptide in the electrophoresis channel is reported on the left axis, and the ratio of the two signals is given on the right axis. (b) Electropherogram of 1 µM NCMK-1 and 1 µM p-NCMK-1 in ECB on a PC-coated simple cross chip. The electric field strength was 110 V/cm, and the detection point was 5 mm from the injection cross.
While electrokinetic injection bias is eliminated by minimizing difference in analyte electrophoretic mobility, subsequent electrophoretic separation depends on this difference to resolve analytes. Both NCMK-1 and p-NCMK-1 were sufficiently mobile to be almost completely injected into the electrophoresis channel at a field strength of +300 V/cm (Figure 3c, f). To confirm that the difference in their electrophoretic mobilities was sufficient for electrophoretic separations on-chip, we separated unmodified and phosphorylated NCMK-1 in ECB on a PC-coated simple cross chip (Figure 4b). For an electric field strength of 110 V/cm, the two peptides were baseline resolved at a detection point 5 mm from the injection intersection.
4 Concluding Remarks
Although a few multi-purpose microfluidic devices have been demonstrated (e.g., [24]), most microdevices are still designed for specific assays of specific analytes. In some cases, researchers optimize a device using convenient model analytes, such as fluorescent dyes, with the goal of eventually applying the optimized design to biologically-relevant analytes [12,14]. This strategy is only effective, however, if the analytes-of-interest are sufficiently similar to the model analytes. The results presented here demonstrate the utility of COMSOL modeling in answering this question. Additionally, this work demonstrates that this device, optimized for separation of small, highly charged fluorescent dyes, is best-suited to analysis of high electrophoretic mobility analytes (μep ≥ ± 1.5 × 10−4 cm2/Vs) at moderate to high field strengths (≥ 300 V/cm). Lower mobility analytes are not effectively injected into the electrophoresis channel, and these analytes are more susceptible to electrokinetic injection bias and fluctuations in pressure-driven flow. This effect could potentially be offset by an increase in electroosmotic flow at the expense of discrimination between analytes and cellular debris. In general, these results demonstrate that thoughtful pairing of device design with analyte characteristics is necessary as more microanalysis systems graduate from model analytes to real-world applications.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health: Minority Opportunities in Research division of the National Institute of General Medical Sciences (NIGMS) grant #K12GM000678 (MLK) and EB11763 and CA139599 (NLA).
Abbreviations
- ECB
extracellular buffer
- p-
phosphorylated
- PC
phosphatidylcholine
Footnotes
The authors disclose no financial interest.
References
- 1.Cohen D, Dickerson JA, Whitmore CD, Turner EH, Palcic MM, Hindsgaul O, Dovichi NJ. Annu. Rev. Anal. Chem. 2008;1:165–190. doi: 10.1146/annurev.anchem.1.031207.113104. [DOI] [PubMed] [Google Scholar]
- 2.Krutzik PO, Crane JM, Clutter MR, Nolan GP. Nat. Chem. Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 3.Jiang D, Sims CE, Allbritton NL. Faraday Discuss. 2011;149:187–200. doi: 10.1039/C005362G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 5.Zare RN, Kim S. Annu. Rev. Biomed. Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Wheeler A, Zare RN. Proc. Nat. Acad. Sci. 2004;101:12809–12813. doi: 10.1073/pnas.0405299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munce NR, Li J, Herman PR, Lilge L. Anal. Chem. 2004;76:4983–4989. doi: 10.1021/ac0496906. [DOI] [PubMed] [Google Scholar]
- 8.Xia F, Jin W, Yin X, Fang Z. J. Chromatogr. A. 2005;1063:227–233. doi: 10.1016/j.chroma.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Kleparnik K, Horky M. Electrophoresis. 2003;24:3778–3783. doi: 10.1002/elps.200305667. [DOI] [PubMed] [Google Scholar]
- 10.Gao J, Yin X-F, Fang Z-L. Lab Chip. 2004;4:47–52. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]
- 11.Hellmich W, Pelargus C, Leffhalm K, Ros A, Anselmetti D. Electrophoresis. 2005;26:3689–3696. doi: 10.1002/elps.200500185. [DOI] [PubMed] [Google Scholar]
- 12.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Anal. Chem. 2003;75:5646–5655. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 13.Phillips KS, Kang KM, Licata L, Allbritton NL. Lab Chip. 2010;10:864–870. doi: 10.1039/b921817c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips KS, Lai H-H, Johnson E, Sims CE, Allbritton NL. Lab Chip. 2011;11:1333–1341. doi: 10.1039/c0lc00370k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jorgenson JW, Lukacs KD. Anal. Chem. 1981;53:1298–1302. [Google Scholar]
- 16.Huang XH, Gordon MJ, Zare RN. Anal. Chem. 1988;60:375–377. [Google Scholar]
- 17.Slentz BE, Penner NA, Regnier F. Anal. Chem. 2002;74:4835–4840. doi: 10.1021/ac020301m. [DOI] [PubMed] [Google Scholar]
- 18.Blas M, Delauney N, Ferrigno R, Rocca J-L. Electrophoresis. 2007;28:2961–2970. doi: 10.1002/elps.200600692. [DOI] [PubMed] [Google Scholar]
- 19.Alarie JP, Jacobson SC, Ramsey JM. Electrophoresis. 2001;22:312–317. doi: 10.1002/1522-2683(200101)22:2<312::AID-ELPS312>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Phillips KS, Kottegoda S, Kang KM, Sims CE, Allbritton NL. Anal. Chem. 2008;80:9756–9762. doi: 10.1021/ac801850z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson SC, Koutny LB, Hergenroder R, Moore AWJ, Ramsey JM. Anal. Chem. 1994;66:3472–3476. [Google Scholar]
- 22.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 23.Rasband WS. ImageJ. < http://rsb.info.nih.gov/ij/> (1997–2009).
- 24.Songyang Z, Carraway KLI, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BJA, Mayer BJ, Cantley LC. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.