Abstract
Developing short peptides into useful probes and therapeutic leads remains a difficult challenge. Structural rigidification is a proven method for improving the properties of short peptides. In this work, we report a strategy for stabilizing peptide macrocycles by introducing side-chain-to-side-chain staples, producing peptide bicycles with higher affinity, selectivity, and resistance to degradation. We have applied this strategy to G1, an 11-residue peptide macrocycle that binds the Src homology 2 (SH2) domain of growth-factor-bound protein 2 (Grb2). Several homodetic peptide bicycles were synthesized entirely on-resin with high yields. Two rounds of iterative design produced peptide bicycle BC1, which is 60-fold more potent than G1 and 200-fold more selective. Also, BC1 is completely intact after 24 hours in buffered human serum, conditions under which G1 is completely degraded. Our peptide bicycle approach holds promise for the development of selective inhibitors of SH2 domains and other pTyr-binding proteins, as well as inhibitors of many other protein-protein interactions.
Keywords: peptides, cyclization, signal transduction, protein-protein interactions, drug design
Introduction
Inhibition of protein-protein interactions remains a difficult challenge for chemical biology, and is still a relatively new frontier for drug design.[1] Structure-based design, phage display, and other powerful methods have allowed the development of peptides that inhibit a large variety of protein-protein interactions. However, peptides are difficult starting points for the development of intracellular tools and novel therapeutics.[2] Peptides have high desolvation energies that prevent them from passing through membranes, they are susceptible to proteolytic degradation, and their conformational flexibility can limit interaction affinity and selectivity. Even so, peptides have many of the advantages of large protein biologics, yet are small enough to be prepared and modified synthetically. These traits have fueled intense interest in strategies for peptide drug development.[3] In this work, we present a simple and direct approach to improving cyclic peptides via intramolecular cross-linking. This strategy produces peptide bicycles with enhanced target affinity, selectivity, and resistance to degradation.
There is a long history of understanding and improving peptide function via conformational restriction.[4] While peptide macrocyclization does not automatically confer desirable properties, judicious application of macrocyclization has produced peptides with increased affinity, selectivity, proteolytic stability, and cell penetration.[5] Intramolecular side-chain-to-side-chain cross-linking, or “stapling,” is a related modification that has also produced peptides with improved properties.[6] Recent results have demonstrated the extraordinary effects of stapling α-helices using α-methylated, olefin-containing amino acids in (i,i+4) positions.[6a–c] In these cases, on-resin ring-closing metathesis was used to produce stable α-helices with an all-hydrocarbon staple.[7] This strategy has resulted in stapled helices of up to 36 residues, many of which have dramatically enhanced affinity, selectivity, protease resistance and cell penetration.[6a–c, 8] These results highlight how the structural and physicochemical effects of intramolecular cross-linking can be highly cooperative within short peptides.
While α-helices are a prominent secondary structure with substantial representation at protein-protein interfaces,[9] the diversity of interaction surfaces necessitates the development of diverse platforms for inhibitor development. To this end, we sought to explore how side-chain-to-side-chain stapling would affect peptide function in the context of head-to-tail peptide macrocycles. Peptide macrocycles share many appealing features of short α-helices, including nascent structure, capacity for internal backbone-to-backbone hydrogen bonding, and presentation of a sizeable interaction surface within a small polypeptide. However, because macrocycles adopt a much wider range of three-dimensional structures compared to α-helices, there will not be a single optimal staple for cyclic peptides in general. Application of this strategy to any given cyclic peptide will thus require systematic exploration of cross-link positions and geometries to generate peptide bicycles with enhanced functional properties.
There are many cyclic peptides that inhibit protein-protein interactions that could be used as starting points for engineering peptide bicycles. Most of these are disulfide-bridged macrocycles of 7–12 amino acids, derived from phage display experiments.[10] Compounds in this class are useful in vitro tools but are difficult to translate into cellular probes and therapeutics. Peptide G1 (Fig. 1A) is an 11-residue disulfide-bridged macrocycle that binds the Src-homology 2 (SH2) domain of growth-factor-bound-protein 2 (Grb2).[11] We chose G1 as our starting point for testing the peptide bicycle strategy for several reasons. First, inhibiting Grb2 inside living cells prevents activation of the Ras pathway by growth factor receptors.[12] This pathway is a well-established target for breast and other cancers, making Grb2 inhibitors of high clinical interest.[13] Second, Grb2-SH2 has been the subject of extensive peptidomimetic design and screening efforts, as well as biophysical analyses of protein-ligand binding.[12, 14] This provides an ample framework for understanding the determinants of ligand binding to SH2 domains. Third, G1 itself has been the subject of many rounds of optimization using traditional medicinal chemistry approaches.[14c, 15] Thus, we could take advantage of G1’s known structure-activity relationships to help design our peptide bicycles. For example, while an alanine scan of G1 seemed to indicate that nearly all residues were required to target Grb2-SH2,[11] later structure-activity relationships of a thioether-bridged analog (G1TE) demonstrated that the binding epitope is largely restricted to the side chains of Glu2, Tyr4 and Asn6 (Fig. 1A).[15b, 15d] Glu2 and Tyr4 form an epitope that binds the SH2 domain in place of its natural ligand, phosphotyrosine (pTyr), while Asn6 occupies an adjacent specificity pocket.[14c] These structure-activity relationships would assist in the design of bicyclic G1 analogs. Finally, G1 was an optimal starting point because reduction of the disulfide bond abolished all binding activity.[11, 15c] These data indicated that G1 activity is highly conformation-dependent, making it plausible that further conformational restriction within the macrocycle would lead to further gains in affinity and selectivity.
Figure 1.
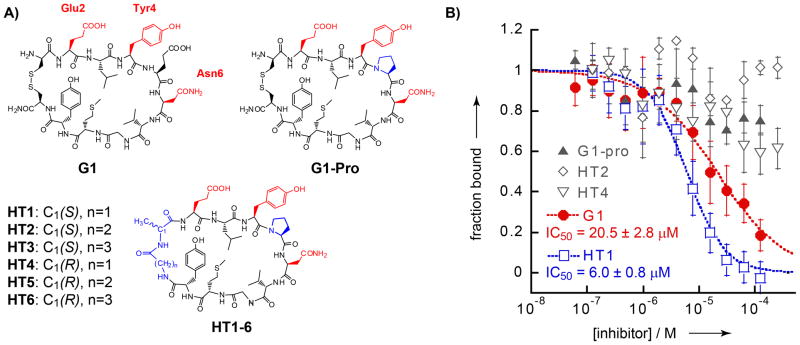
A head-to-tail macrocycle based on G1. A) Structures of G1, G1-Pro, and HT1-6. Side-chains shown in red are implicated in direct SH2 domain binding in the known structure-activity relationships of previous G1 analogs.[15b, 15d] Amino acids that differ from the parent peptide G1 are shown in blue. B) Competition assay data for G1, G1-Pro, HT1, HT2, and HT4. Data for HT3, HT5 and HT6 are similar to those for HT2 and HT4 and are shown in Figure S2. IC50 values reflect the curve fits shown.
Results and Discussion
Head-to-tail peptide macrocycles based on G1
First, we sought to replace the disulfide bond of G1 with a head-to-tail macrocycle linker.[16] Six head-to-tail G1 analogs (HT1-6, Fig. 1A) were synthesized with different linker lengths (glycine, β-alanine, or γ-aminoisobutyric acid in one linker position) and stereochemistries (L-alanine or D-alanine in the other linker position). These peptides also differ from G1 by substitution of Glu5 with a proline, which was desirable to promote cyclic structure and eliminate unnecessary charge. Previous structure-activity relationships indicated that, while substitution of Glu5 decreased G1 activity, a Glu5-to-Pro substitution was well-tolerated within the thioether analog G1TE.[11, 15a] Cyclic peptides were synthesized through side-chain attachment of Fmoc-Glu-OAll to Wang resin, followed by Fmoc solid-phase synthesis of the peptide chain, deprotection of the allylated C-terminus, and on-resin head-to-tail macrocyclization (Scheme S1).
After HPLC purification, peptides were tested for inhibitory potency using a fluorescence polarization competition assay.[17] Each G1 variant was incubated at various concentrations with Grb2-SH2 and a fluorescein-labelled probe that contained a pTyr-containing Grb2 ligand (pTyr-Val-Asn-Val).[18] This assay was designed to avoid dye-labelling the macrocycles, as well as to ensure that binding was occurring at the pTyr-binding site of the SH2 domain. Of the six macrocycles tested, only HT1 showed any inhibition of Grb2-SH2 (Fig. 1B). HT1 had an IC50 of 6.0 ± 0.8μM, roughly threefold better than the parent compound G1 (IC50 = 20.5 ± 2.8 μM). G1 and HT1 have two major differences, the linker region and the Glu5-to-Pro substitution. To deconvolute the roles of these differences, we synthesized G1-Pro, which is identical to G1 but contains the Glu5-to-Pro substitution. G1-Pro shows little inhibitory activity (Fig. 1B), demonstrating that the Glu-5-to-Pro substitution actually diminishes activity in the disulfide-bonded macrocycle. However, replacement of the disulfide in G1-Pro with the amide bond in HT1 apparently has an overriding effect on the overall peptide macrocycle, more readily accommodating the proline residue. We also note that switching a single stereocenter (HT2) or adding a single methylene unit (HT4) within the linker region of HT1 alters the overall conformation so much that the macrocycle no longer inhibits Grb2-SH2 within detectable limits. All the above data indicate that HT1 activity is extremely dependent on a specific macrocycle conformation. This provided additional motivation for testing rigidified peptide bicycles based on HT1.
Peptide bicycles that inhibit Grb2-SH2
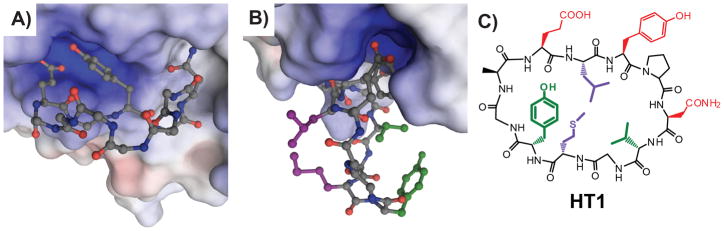
To produce peptide bicycles based on HT1, we first needed to predict which side chains to cross-link. To this end, an energy-minimized model of HT1 binding to Grb2-SH2 was constructed (Fig. 2A,B). An initial conformation of HT1 was based on the canonical structure of Grb2-SH2 bound to a peptide hairpin,[18] and assumed that HT1 binds the SH2 domain using its Glu2, Tyr4 and Asn6 residues in a similar binding mode as G1 and G1TE (Fig. 2A,B).[14c, 15e] This model was then energy-minimized using AMBER99 or OPLS-AA force fields with similar results. Within this model, two pairs of side chains were positioned in close proximity (Fig. 2B,C). While the model is not intended to be a rigorous docking exercise, it was sufficient to provide reasonable starting points for executing the peptide bicycle strategy.
Figure 2.
Models help identify side-chain pairs for intramolecular cross-linking. A) Energy-minimized model of HT1 (ball-and-stick, nitrogens shown in blue, oxygens in red, carbons in gray, and hydrogens omitted) bound to Grb2-SH2 (surface colored to indicate electrostatics). Side-chains of HT1 not involved in direct Grb2-SH2 binding are omitted for clarity. This energy-minimized model is based on the structure of a native peptide ligand and established SAR data on G1 and its analogs.[14c, 18] Modeling and energy minimization was performed using Molecular Operating Environment (Chemical Computing Group) using AMBER99 and OLPS-AA force fields (negligible differences seen between the two). Selected local water molecules from the crystal structure were included explicitly (not shown). Soft constraints were used on the protein and the peptide was allowed to move freely within the pocket. B) The identical model as in A), but rotated to show relative positions of Leu3 and Met9 (purple), and Val7 and Tyr10 (green). C) Structure of HT1 with side-chains color-coded as in B).
Guided by this model, we synthesized four bicyclic variants of HT1 (BC1-4, Fig. 3A) using lactam staples that incorporate commercially available amino acids.[19] Several related classes of peptide bicycles have been previously reported,[20] but there are very few reports of head-to-tail cyclic, lactam-stapled peptides.[21] These few reports have noted that the order in which macrocycles are closed has a large impact on overall yield, and that solution-phase cyclization was often needed to successfully close the second macrocycle. Furthermore, the consensus among these prior works was that synthetic efficiency was highest when head-to-tail cyclization was performed prior to side-chain-mediated cyclization. In synthesizing BC1-4, we chose to vary the order in which the two macrocyclization reactions were performed in order to optimize the yields of these particular scaffolds. The optimal order for macrocyclization reactions was highly dependent on the positions of the side-chain-to-side-chain crosslinks: BC1 and BC2 were optimally synthesized by closing the side-chain macrocycle first, while BC3 and BC4 were optimally synthesized by closing the head-to-tail macrocycle first. This procedure led to efficient syntheses of BC1-4 entirely on-resin, with crude purities ranging from 61 to 75%. These synthetic yields demonstrate that high-yielding, on-resin synthesis of peptide bicycles can be readily achieved.
Figure 3.
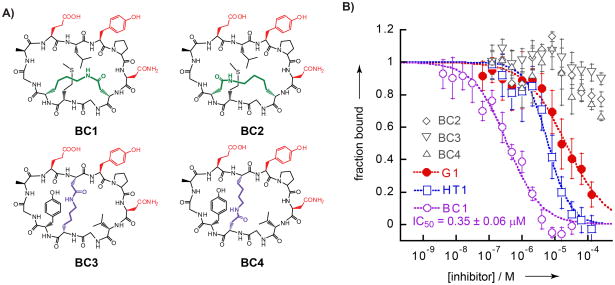
Peptide bicycles based on HT1. A) Chemical structures of BC1-4. Side-chains shown in red are implicated in direct SH2 domain binding in the known structure-activity relationships of G1. Purple and green side-chains comprise staples in positions modelled to be in close proximity, as shown in Fig. 2. B) Competition assay data for G1, HT1, and BC1-4. IC50 values reflect the curve fits shown.
Next, we tested BC1-4 in the competition assay (Fig. 3B). Of these, only BC1 inhibited the binding of the pTyr-containing probe to Grb2-SH2. BC1 had an IC50 of 0.35 ± 0.06 μM, 17-fold more potent than HT1 and nearly 60-fold more potent than the original G1 peptide. This result demonstrates that peptide bicyclization can result in large increases in inhibitory potency, and that simple structure-activity data and modelling can inform correct placement of the side-chain-to-side-chain staple.
Stringent requirements for the staple
To understand which aspects of the staple contributed to the higher potency of BC1, we tested analogs lacking a covalently linked staple. HT7 differs from HT1 in that an Asn is substituted for Val7, and HT8 differs from HT1 in that an acetylated Lys is substituted for Tyr10 (Fig. S2). Each of these possesses part of the staple of BC1, including the amide group, but is still monocyclic. HT7 and HT8 show little inhibitory activity (Fig. S3). These results show that the conformational constraint of the staple, and not the simple addition of a specific amide group, was responsible for BC1’s increased potency. They also provide further evidence of the exquisite conformational sensitivity of the macrocycle scaffold, since HT7 and HT8 have effectively lost the inhibitory potency of HT1. We then tested two additional variants of BC1 which maintained the staple length but altered the position and orientation of the amide bond within the staple (BC5 and BC6, Fig. S2). All of these were also inactive in the competition assay (Fig. S3). Small changes in the staple thus led to large losses in inhibitory potency, implying that the conformation of BC1 is precisely controlled by the staple’s composition and geometry.
Bicyclization improves serum stability
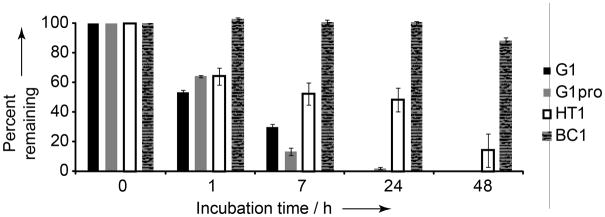
Natural peptide bicycles such as α-amanitin and sunflower trypsin inhibitor I are uniquely resistant to proteolytic degradation.[22] To test whether our designed peptide bicycles were similarly stable, we incubated 0.9 mg/mL of G1, G1-Pro, HT1, and BC1 in human serum and monitored their degradation by HPLC (Fig. 4). G1 and G1-Pro showed steady degradation, with less than 30% of the peptide remaining after seven hours and no detectable amounts of peptide remaining after 24 hours. HT1 was only 50% degraded after 24 hours. BC1, by contrast, remained completely intact after 24 hours and was less than 15% degraded after 48 hours in human serum.
Figure 4.
Peptide bicycles are resistant to degradation in human serum. Degradation of peptide macrocycles and bicycles was monitored after zero, 1, 7, 24 and 48 hours in buffered human serum. Percent peptide remaining was calculated by integrating HPLC peaks and normalizing to the zero time point.
Bicyclization improves selectivity for Grb2-SH2 over another SH2 domain
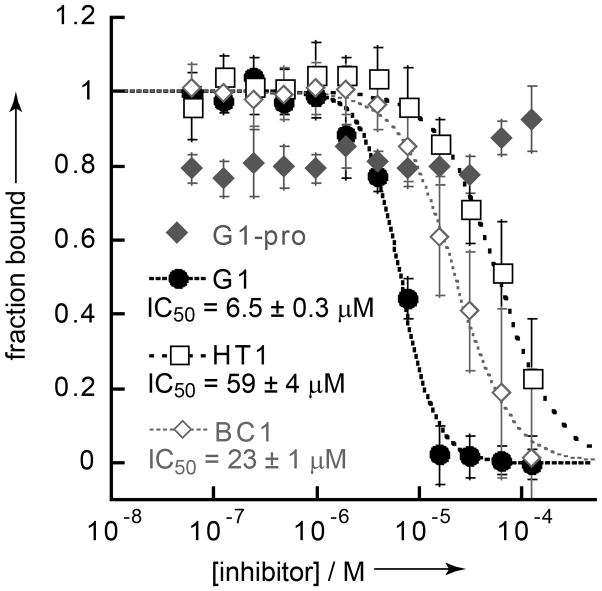
A great deal of structure-activity data on G1, G1TE, and now BC1 has demonstrated that the ability to bind Grb2-SH2 is highly conformation-dependent.[14c, 15] The exquisite sensitivity of BC1 to any conformational perturbation led us to ask whether it might be highly selective for Grb2-SH2 over other SH2 domains. Since binding requires not only the pTyr-mimicking epitope formed by Glu2 and Tyr4, but also the correct conformation of the entire molecule, we reasoned that BC1 might discriminate between variant loops outside the pTyr-binding pocket. To test the specificity of our compounds, we set up a second competition fluorescence polarization assay using the SH2 domain from tensin-1 (tensin-SH2). We found that tensin-SH2 binds the same pTyr-Val-Asn-Val probe that binds Grb2-SH2, with a similar dissociation constant (see Fig. S1). This finding is consistent with values from other assays, and also with prior work establishing similar consensus binding sequences for these SH2 domains.[23] Surprisingly, G1 inhibits tensin-SH2 more potently than it does Grb2-SH2 (IC50 for tensin-SH2 of 6.4 ± 0.3μM), while HT1 and BC1 inhibit tensin-SH2 with much less potency (IC50’s of 59 ± 4 and 23 ± 1μM, respectively). Thus, BC1 has 66-fold selectivity for Grb2-SH2 over tensin-SH2, compared to 0.31-fold for G1. This represents a 200-fold increase in selectivity for Grb2-SH2.
Bicyclization pre-organizes the phosphotyrosine-mimicking epitope
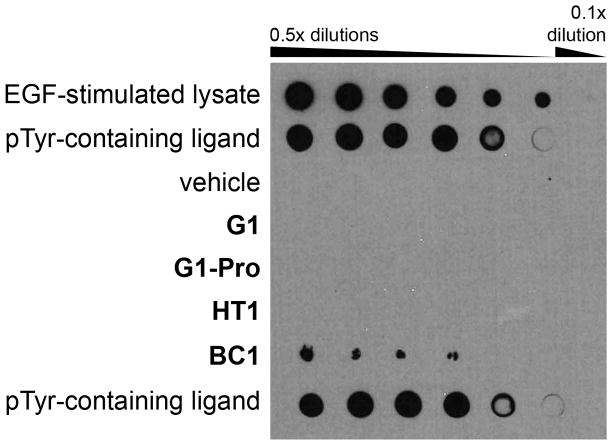
The peptide bicycle strategy is based on the idea that structural stabilization of the binding epitope will promote greater inhibitory potency and selectivity. For BC1, this would mean that the bicyclic structure rigidly pre-organizes the pTyr-mimicking epitope. To test this hypothesis, we spotted serial dilutions of G1, G1-Pro, HT1, and BC1 onto nitrocellulose membranes and probed them with the commonly used anti-pTyr antibodies 4G10 and PY20 (Figs. 6 and S4). EGF-stimulated cell lysate and our own pTyr-containing probe peptide were used as controls to validate the peptide spotting and blotting procedures. BC1 consistently captured both anti-pTyr antibodies, while G1, G1-Pro, and HT1 were unable to capture anti-pTyr antibodies. This demonstrated that BC1 can maintain a conformation that mimics pTyr even when adsorbed to nitrocellulose. This provides direct evidence that the bicyclic structure of BC1 successfully pre-organizes the molecule into its protein-binding structure.
Figure 6.
Anti-pTyr dot blot assay. Peptides were normalized to 750 μM, then serially diluted as shown and spotted onto nitrocellulose. The membranes were blocked with bovine serum albumin, probed with anti-pY antibodies 4G10 (above) or PY20 (see Fig. S4), probed with secondary antibody, and then images were developed using chemoluminescent detection. Controls included EGF-stimulated cell lysate (highest concentration is 1 mg/mL total protein), the dye-labeled, pTyr-containing probe peptide used in the fluorescence polarization assays (pTyr-containing ligand), and vehicle (50% DMSO).
Conclusion
Peptide bicycles are a largely overlooked class of molecules with great potential as chemical probes and therapeutics. Most prior work on these rigidified scaffolds has focused on engineering natural bicyclic scaffolds such as conotoxins, cyclotides, defensins and trypsin inhibitors.[24] In this work, we describe a complementary approach for engineering peptide bicycles by introducing successive conformational constraints within peptide loops or macrocycles. Applying this strategy to G1 resulted in a 60-fold improvement in inhibitory potency and a 200-fold improvement in selectivity after only two rounds of iterative design. In addition, the SH2-domain-binding peptide bicycle is only one-half to one-third the size of stabilized α-helices and other bicyclic peptidomimetic scaffolds.[6, 25] The lactam staples used in this work result in high synthetic yields and take advantage of existing orthogonal protection strategies and commercially available amino acids.
Stapled α-helices have been described for decades.[6e, 19, 26] However, recent work has shown that helices bearing an all-hydrocarbon staple have particularly favourable properties, including considerable cell penetration and activity in whole-animal cancer models.[6a–c] We are currently testing whether olefin staples will produce similar benefits for small peptide bicycles.
BC1 represents a unique, conformation-dependent pTyr mimic. The structure-activity relationships of BC1, its selectivity for Grb2-SH2, and its ability to capture anti-pTyr antibodies all provide evidence that it presents a pTyr-mimicking epitope without requiring any phosphate groups. More detailed structural analysis of BC1 and its complex with Grb2-SH2 will help fill in molecular-level details regarding how BC1 mimics pTyr and how it binds Grb2-SH2 so selectively. These structural data will also help explain the observed requirements for staple geometry. For now, the peptide bicycle strategy requires iterative testing to determine optimal staple positioning, length and geometry. Additional structural information that explains the structure-activity relationships of BC1 will also be useful for streamlining this process, leading towards a more structure-based design approach. This would unlock the ability to design bicycles that selectively target other SH2 domains and other pTyr-binding proteins. We are also exploring the cell-penetrating capabilities of these and other peptide bicycles, to determine whether they represent useful probes for cancer biology. Such probes would be useful to explore cancer therapies that synergize with existing treatments targeting EGFR and Ras-dependent pathways.[27]
Experimental Section
Protein preparation
Constructs for expression of the SH2 domains of Grb2 and Tensin-1 in the pMAL C2x plasmid vector were generous gifts from Dehua Pei.[23b] Grb2-SH2 and tensin-SH2 were expressed as maltose-binding protein fusions essentially as described.[23b] BL21(DE3) E. coli (New England Biolabs) were transformed with the expression plasmids and single colonies were grown in 1L 2XYT with 20% glucose at 37 deg. C. In log phase, cultures were induced with 0.35 mM IPTG and incubated at 30 deg. C for 2.5 hours. Bacteria were pelleted, re-suspended in buffer containing 20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol and protease inhibitor cocktail (Sigma-Aldrich). Cells were lysed by sonication and cell debris was pelleted. Cleared lysate was diluted 1:2 in buffer and purified by amylose column chromatography (New England Biolabs). Fractions were analyzed by SDS-PAGE, pooled and concentrated using centrifugal 9K MWCO protein concentrators (Thermo Scientific).
Peptide synthesis and purification
All solid-phase resins, Fmoc-amino acids, and coupling reagents were purchased from EMD Biosciences unless otherwise noted. Linear, cyclic and bicyclic peptides were synthesized using established solid-phase peptide synthesis procedures (see schemes S1-S3). Peptides were purified to ≥ 95% purity by reverse-phase HPLC using water/acetonitrile in 0.1% TFA using a preparatory-scale C8 column. Lyophilized peptides were dissolved in DMSO, and concentrations of DMSO in subsequent assays were normalized to ≤ 1%.
Fluorescence polarization competition assay
Fluorescence polarization assays were conducted in flat-bottom, black 96-well plates (Corning). Direct-binding experiments (Fig. S1) were performed by incubating 5 nM fluoresceinated phosphotyrosine-containing ligand (Fluorescein-βAla-βAla-pTyr-Val-Asn-Val) with different concentrations of Grb2-SH2 or tensin-SH2 in phosphate-buffered saline (pH 7.2). This allowed the selection of optimum SH2 domain concentrations for competitive binding assays. Competition assays were carried out by incubating 5 nM pYVNV ligand with 0.91 μM Grb2-SH2 (or 1.4 μM tensin-SH2) with different concentrations of inhibitor. Data was normalized to values obtained for fluorescent ligand alone (0% bound) and ligand with protein and no inhibitor (100% bound). IC50 curve fits were obtained using non-linear regression analysis (Kaleidagraph, Synergy Software). Error bars represent three or more independent trials.
Human serum stability assays
The human serum stability assay was performed essentially as described.[28] Human male type-AB serum (Sigma-Aldrich) was diluted 9:1 in phosphate-buffered saline. Peptides were incubated in buffered serum at 37 deg. C at a final concentration of 0.9 mg/mL. At the indicated times, 50 μL aliquots were removed and quenched by adding 200 μL ice-cold methanol. Serum proteins were pelleted, and the supernatant was analyzed by HPLC. Values were normalized to the zero time point. Error bars represent three independent trials.
Peptide dot blots
Peptides were diluted to 500 μM in Tris-buffered saline, pH 7.4, with 0.1% Tween-20. These stocks were serially diluted and 1 μL drops were spotted directly onto a nitrocellulose membrane. Spots were allowed to completely dry. The membrane was blocked with 0.2% bovine-serum albumin for 1 h. The membrane was then incubated with anti-pTyr antibody (4G10 or PY20, Millipore), followed by HRP-conjugated secondary antibody (Sigma-Aldrich). The membrane was then washed and incubated with SuperSignal Chemiluminescent Substrate (Pierce) and imaged on film.
Figure 5.
Inhibition of tensin-SH2 by peptide macrocycles and bicycles. This fluorescence polarization competition assay used the same dye-labeled ligand as the Grb2-SH2 competition assay. IC50 values reflect the curve fits shown.
Acknowledgments
This work was supported by a Smith Family Award for Excellence in Biomedical Research and an NIH New Innovator Grant (DP-2OD007303) to J.A.K.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
References
- 1.a) Berg T. Curr Op Drug Disc Dev. 2008;11:666–674. [PubMed] [Google Scholar]; b) Wells JA, McClendon CL. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 2.Kritzer JA. Nat Chem Biol. 2010;6:868–870. doi: 10.1038/nchembio.469. [DOI] [PubMed] [Google Scholar]
- 3.a) Bauer RA, Wurst JM, Tan DS. Curr Opin Chem Biol. 2010;14:308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Driggers EM, Hale SP, Lee J, Terrett NK. Nat Rev Drug Disc. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]; c) Hruby VJ. Nat Rev Drug Discov. 2002;1:847–858. doi: 10.1038/nrd939. [DOI] [PubMed] [Google Scholar]; d) Kohli RM, Walsh CT, Burkart MD. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 4.a) Hruby VJ, Balse PM. Curr Med Chem. 2000;7:945–970. doi: 10.2174/0929867003374499. [DOI] [PubMed] [Google Scholar]; b) Hruby VJ, Alobeidi F, Kazmierski W. Biochem J. 1990;268:249–262. doi: 10.1042/bj2680249. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pelton JT, Kazmierski W, Gulya K, Yamamura HI, Hruby VJ. J Med Chem. 1986;29:2370–2375. doi: 10.1021/jm00161a037. [DOI] [PubMed] [Google Scholar]; d) Ovadia O, Greenberg S, Laufer B, Gilon C, Hoffman A, Kessler H. Exp Op Drug Disc. 2010;5:655–671. doi: 10.1517/17460441.2010.493935. [DOI] [PubMed] [Google Scholar]; e) Kessler H. Angew Chem Int Ed Engl. 1982;21:512–523. [Google Scholar]
- 5.a) White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SSF, Kalgutkar AS, Bauman JN, Zhang YZ, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. Nat Chem Biol. 2011;7:810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J Am Chem Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]; c) Fairlie DP, Abbenante G, March DR. Curr Med Chem. 1995;2:654–686. [Google Scholar]; d) Kwon YU, Kodadek T. Chem Biol. 2007;14:671–677. doi: 10.1016/j.chembiol.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 6.a) Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007;129:2456. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]; d) Blackwell HE, Grubbs RH. Angew Chem Int Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; e) Harrison RS, Shepherd NE, Hoang HN, Ruiz-Gómez G, Hill TA, Driver RW, Desai VS, Young PR, Abbenante G, Fairlie DP. Proc Natl Acad Sci USA. 2010;107:11686–11691. doi: 10.1073/pnas.1002498107. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Pelton JT, Gulya K, Hruby VJ, Duckles SP, Yamamura HI. Proc Natl Acad Sci USA. 1985;82:236–239. doi: 10.1073/pnas.82.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YW, Grossmann TN, Verdine GL. Nat Protocols. 2011;6:761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 8.a) Bird GH, Madani N, Perry AF, Princiotto AM, Supko JG, He XY, Gavathiotis E, Sodroski JG, Walensky LD. Proc Natl Acad Sci USA. 2010;107:14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stewart ML, Fire E, Keating AE, Walensky LD. Nat Chem Biol. 2010:6. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullock BN, Jochim AL, Arora PS. J Am Chem Soc. 2011;133:14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]; b) Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, Kantor C, Gahmberg CG, Salo T, Konttinen YT, Sorsa T, Ruoslahti E, Pasqualini R. Nat Biotechnol. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]; c) Koivunen E, Gay DA, Ruoslahti E. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 11.Oligino L, Lung FDT, Sastry L, Bigelow J, Cao T, Curran M, Burke TR, Wang SM, Krag D, Roller PP, King CR. J Biol Chem. 1997;272:29046–29052. doi: 10.1074/jbc.272.46.29046. [DOI] [PubMed] [Google Scholar]
- 12.a) Song YL, Peach ML, Roller PP, Qiu S, Wang SM, Long YQ. J Med Chem. 2006;49:1585–1596. doi: 10.1021/jm050910x. [DOI] [PubMed] [Google Scholar]; b) Machida K, Mayer BJ. Biochim Biophys Acta Proteins Proteomics. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]; c) Wei CQ, Gao Y, Lee K, Guo R, Li BH, Zhang MC, Yang DJ, Burke TR. J Med Chem. 2003;46:244–254. doi: 10.1021/jm0203635. [DOI] [PubMed] [Google Scholar]
- 13.a) Burke T. Int J Pept Res Ther. 2006;12:33–48. doi: 10.1007/s10989-006-9014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Garcia-Echeverria C. Curr Med Chem. 2001;8:1589–1604. doi: 10.2174/0929867013371905. [DOI] [PubMed] [Google Scholar]; c) Fretz H, Furet P, Garcia-Echeverria C, Rahuel J, Schoepfer J. Curr Pharm Des. 2000;6:1777–1796. doi: 10.2174/1381612003398546. [DOI] [PubMed] [Google Scholar]
- 14.a) DeLorbe JE, Clements JH, Whiddon BB, Martin SF. ACS Med Chem Lett. 2010;1:448–452. doi: 10.1021/ml100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) DeLorbe JE, Clements JH, Teresk MG, Benfield AP, Plake HR, Millspaugh LE, Martin SF. J Am Chem Soc. 2009;131:16758–16770. doi: 10.1021/ja904698q. [DOI] [PubMed] [Google Scholar]; c) Jiang S, Li P, Peach ML, Bindu L, Worthy KW, Fisher RJ, Burke TR, Nicklaus M, Roller PP. Biochem Biophys Res Commun. 2006;349:497–503. doi: 10.1016/j.bbrc.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 15.a) Li P, Zhang MC, Long YQ, Peach ML, Liu HP, Yang DJ, Nicklaus M, Roller PP. Bioorg Med Chem Lett. 2003;13:2173–2177. doi: 10.1016/s0960-894x(03)00385-8. [DOI] [PubMed] [Google Scholar]; b) Lung FDT, Long YQ, King CR, Varady J, Wu XW, Wang S, Roller PP. J Pept Res. 2001;57:447–454. doi: 10.1034/j.1399-3011.2001.00833.x. [DOI] [PubMed] [Google Scholar]; c) Lung FDT, King CR, Roller PP. Lett Pept Sci. 1999;6:45–49. [Google Scholar]; d) Long YQ, Voigt JH, Lung FDT, King CR, Roller PP. Bioorg Med Chem Lett. 1999;9:2267–2272. doi: 10.1016/s0960-894x(99)00379-0. [DOI] [PubMed] [Google Scholar]; e) Li P, Zhang MC, Peach ML, Zhang XD, Liu HP, Nicklaus M, Yang DJ, Roller PP. Biochem Biophys Res Commun. 2003;307:1038–1044. doi: 10.1016/s0006-291x(03)01291-9. [DOI] [PubMed] [Google Scholar]
- 16.Foister S, Taylor LL, Feng JJ, Chen WL, Lin A, Cheng FC, Smith AB, Hirschmann R. Org Lett. 2006;8:1799–1802. doi: 10.1021/ol060278h. [DOI] [PubMed] [Google Scholar]
- 17.a) Nam NH, Ye G, Sun G, Parang K. J Med Chem. 2004;47:3131–3141. doi: 10.1021/jm040008+. [DOI] [PubMed] [Google Scholar]; b) Nachman J, Gish G, Virag C, Pawson T, Pomes R, Pai E. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0011215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, Caravatti G, Fretz H, Schoepfer J, Grutter MG. Nat Struct Biol. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 19.Taylor JW. Pept Sci. 2002;66:49–75. [Google Scholar]
- 20.a) Goncalves V, Gautier B, Garbay C, Vidal M, Inguimbert N. J Pept Sci. 2008;14:767–772. doi: 10.1002/psc.965. [DOI] [PubMed] [Google Scholar]; b) Karskela T, Virta P, Lonnberg H. Curr Org Synth. 2006;3:283–311. [Google Scholar]; c) Falb E, Salitra Y, Yechezkel T, Bracha M, Litman P, Olender R, Rosenfeld R, Senderowitz H, Jiang S, Goodman M. Bioorg Med Chem. 2001;9:3255–3264. doi: 10.1016/s0968-0896(01)00234-6. [DOI] [PubMed] [Google Scholar]; d) Smith DD, Slaninova J, Hruby VJ. J Med Chem. 1992;35:1558–1563. doi: 10.1021/jm00087a009. [DOI] [PubMed] [Google Scholar]; e) Robinson AJ, van Lierop BJ, Garland RD, Teoh E, Elaridi J, Illesinghe JP, Jackson WR. Chem Comm. 2009:4293–4295. doi: 10.1039/b909056h. [DOI] [PubMed] [Google Scholar]
- 21.a) Veber DF, Holly FW, Nutt RF, Bergstrand SJ, Brady SF, Hirschmann R, Glitzer MS, Saperstein R. Nature. 1979;280:512–514. doi: 10.1038/280512a0. [DOI] [PubMed] [Google Scholar]; b) Bartoloni M, Kadam RU, Schwartz J, Furrer J, Darbre T, Reymond JL. Chem Comm. 2011;47:12634–12636. doi: 10.1039/c1cc15704c. [DOI] [PubMed] [Google Scholar]; c) Schiller PW, Berezowska I, Nguyen TMD, Schmidt R, Lemieux C, Chung NN, Falcone-Hindley ML, Yao WQ, Liu J, Iwama S, Smith AB, Hirschmann R. J Med Chem. 2000;43:551–559. doi: 10.1021/jm990461z. [DOI] [PubMed] [Google Scholar]; d) Teixido M, Altamura M, Quartara L, Giolitti A, Maggi CA, Giralt E, Albericio F. J Comb Chem. 2003;5:760–768. doi: 10.1021/cc0301059. [DOI] [PubMed] [Google Scholar]
- 22.a) Korsinczky MLJ, Schirra HJ, Craik DJ. Curr Prot Pept Sci. 2004;5:351–364. doi: 10.2174/1389203043379594. [DOI] [PubMed] [Google Scholar]; b) Wieland T, Faulstich H. Experientia. 1991;47:1186–1193. doi: 10.1007/BF01918382. [DOI] [PubMed] [Google Scholar]
- 23.a) Wavreille AS, Pei D. ACS Chem Biol. 2007;2:109–118. doi: 10.1021/cb600433g. [DOI] [PubMed] [Google Scholar]; b) Zhang YY, Zhou SG, Wavreille AS, DeWille J, Pei D. J Comb Chem. 2008;10:247–255. doi: 10.1021/cc700185g. [DOI] [PubMed] [Google Scholar]
- 24.a) Garcia AE, Tai KP, Puttamadappa SS, Shekhtman A, Ouellette AJ, Camarero JA. Biochemistry. 2011;50:10508–10519. doi: 10.1021/bi201430f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sancheti H, Camarero JA. Adv Drug Deliv Rev. 2009;61:908–917. doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Clark RJ, Jensen J, Nevin ST, Callaghan BP, Adams DJ, Craik DJ. Angew Chem Int Ed. 2010;49 doi: 10.1002/anie.201000620. online in advance of print. [DOI] [PubMed] [Google Scholar]; d) Clark RJ, Fischer H, Dempster L, Daly NL, Rosengren KJ, Nevin ST, Meunier FA, Adams DJ, Craik DJ. Proc Natl Acad Sci USA. 2005;102:13767–13772. doi: 10.1073/pnas.0504613102. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Craik DJ, Cemazar M, Daly NL. Curr Op Drug Disc Dev. 2006;9:251–260. [PubMed] [Google Scholar]; f) Korsinczky MLJ, Clark RJ, Craik DJ. Biochemistry. 2005;44:1145–1153. doi: 10.1021/bi048297r. [DOI] [PubMed] [Google Scholar]; g) Lesner A, Legowska A, Wysocka M, Rolka K. Curr Pharm Des. 2011;17:4308–4317. doi: 10.2174/138161211798999393. [DOI] [PubMed] [Google Scholar]
- 25.a) Heinis C, Rutherford T, Freund S, Winter G. Nat Chem Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]; b) Angelini A, Cendron L, Chen S, Touati J, Winter G, Zanotti G, Heinis C. ACS Chem Biol. 2012 doi: 10.1021/cb200478t. [DOI] [PubMed] [Google Scholar]
- 26.a) Phelan JC, Skelton NJ, Braisted AC, McDowell RS. J Am Chem Soc. 1997;119:455–460. [Google Scholar]; b) Bracken C, Gulyas J, Taylor JW, Baum J. J Am Chem Soc. 1994;116:6431–6432. [Google Scholar]
- 27.a) Downward J. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]; b) Adjei AA. J Natl Cancer Inst. 2001;93:1062–1074. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 28.Getz JA, Rice JJ, Daugherty PS. ACS Chem Biol. 2011;6:837–844. doi: 10.1021/cb200039s. [DOI] [PMC free article] [PubMed] [Google Scholar]