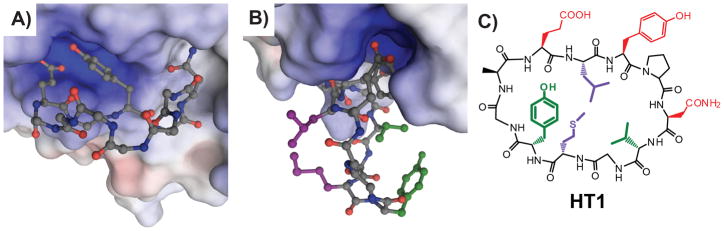
Figure 2.
Models help identify side-chain pairs for intramolecular cross-linking. A) Energy-minimized model of HT1 (ball-and-stick, nitrogens shown in blue, oxygens in red, carbons in gray, and hydrogens omitted) bound to Grb2-SH2 (surface colored to indicate electrostatics). Side-chains of HT1 not involved in direct Grb2-SH2 binding are omitted for clarity. This energy-minimized model is based on the structure of a native peptide ligand and established SAR data on G1 and its analogs.[14c, 18] Modeling and energy minimization was performed using Molecular Operating Environment (Chemical Computing Group) using AMBER99 and OLPS-AA force fields (negligible differences seen between the two). Selected local water molecules from the crystal structure were included explicitly (not shown). Soft constraints were used on the protein and the peptide was allowed to move freely within the pocket. B) The identical model as in A), but rotated to show relative positions of Leu3 and Met9 (purple), and Val7 and Tyr10 (green). C) Structure of HT1 with side-chains color-coded as in B).