Abstract
Purpose
The aim of this study was to use on- tissue reduction followed by MALDI-IMS to identify an m/z 5812.85 peak which is over-expressed in healthy human pancreatic tissue compared to Type one Diabetes (T1D) tissue.
Experimental design
A major constraint of MALDI-IMS is identification of compounds with m/z ≥ 4000. On-tissue reduction using tris (2-carboxyethyl) phosphine (TCEP) breaks the inter-domain disulphide bonds generating low molecular weight peptides amenable to direct MS/MS analysis. Pancreatic tissues from healthy (n=1) and diabetic subjects (n=1) were profiled by MALDI-IMS with/without reduction.
Results
On-tissue reduction resulted in the loss of the over expressed 5812.85 m/z peak and the simultaneous appearance of a 3430.664 m/z peak in healthy tissue. The latter peak presumably derived from the 5812.85 m/z peak was identified as the insulin B chain by MS/MS. MALDI-IMS images show that both the 5812.85 insulin peak before reduction and the 3430.664 peak after reduction co-localized with the healthy pancreatic islets.
Conclusion and clinical relevance
On-tissue reduction followed by MALDI-IMS resulted in the identification of insulin and localization of pancreatic islets of langerhans. The approach will be useful in the future identification of novel therapeutic molecular targets to beta-cells lost during T1D.
Keywords: MALDI-IMS, Insulin, Βeta-cell, Disulphide bridge, Reduction
Mass Spectrometry Imaging (MSI) has become a universal technique which allows the combination of the chemical specificity and analogous detection of mass spectrometry with microscopic imaging capabilities and comprehensive visualization of the spatial distribution of biomarker candidates within a tissue type[1]. The ability to concurrently obtain images from all analytes detected, from atomic to macromolecular ions, allows for the exploration of the chemical organization of a sample and to correlate this with physical features. MALDI-MSI greatly enhances the ability to identify potential candidates for new biomarkers. It has the capability to directly analyze numerous proteins from a tissue section, and the combination of their spatial coordinates allows for their molecular localization while maintaining tissue integrity [2, 3].
Though in-situ MS-MS is extremely useful in the identification of molecules directly from tissues, it is often difficult to characterize and identify biomolecules of interest with molecular masses ≥ 4000 Da. In the linear mode of MALDI instruments the resolution significantly reduces, with an increase in the molecular mass. This results in the formation of broader peaks where natural isotope envelopes are distributed the different populations of molecules across multiple species. In addition, a larger amount of energy is required to ionize larger molecules compared to their smaller counterparts hence, the ability to detect these molecules are decreased [4]. Due to the constraints of MALDI-TOF mass spectrometers to resolve [5] and detect higher molecular weight compounds most signals detected are under m/z 30,000 [6]. The difficulty in localizing higher molecular weight compounds can be overcome by the in situ digestion of proteins in a tissue section either by trypsin or by the treatment with reducing agents such as tris (2-carboxyethyl) phosphine (TCEP) to break disulphide bridges. In-situ digestion generates smaller peptide fragments that not only represent the abundance of the parent protein but allow for efficient detection by MALDI-IMS. Information on spatial positions of the parent protein can be inferred from the localization of the fragments in reflectron mode [7], hence no data is lost.
Here we demonstrate that using in-situ reduction followed by alkylation, allows for the direct identification of insulin in a pancreatic tissue section of a healthy subject. The experimental strategy in which the known inter-disulphide bridges of insulin are broken allows us to identify the protein with a molecular mass of 5812 Da which is beyond the mass range (usually 600–4000 Da) for protein identification by standard MS/MS by conventional MALDI-TOF mass spectrometry. After reduction, the individual smaller domains of the protein are amenable to MS/MS fragmentation, sequencing and identification by MALDI-TOF. Upon reduction with TCEP, the inter-disulphide bonds within insulin are broken with the formation of two fragments the A chain with m/z 2383 and B chain with m/z 3429. These molecules will still represent the overall abundance of the mature multi domain protein and within the range for detection, identification and quantitation by MALDI-TOF MS. It should be noted that the advantages of using reduction and alkylation as the key step to identifying proteins with inter-disulphide bridges will not be the same for all globular or tightly folded proteins with a significantly higher number of intra-disulphide bonds.
Reduction and alkylation was done in the ImagePrep™ workstation. The fixed and dried tissue sample mounted on the MALDI-MSI slide was sprayed homogeneously with 10 mM TCEP dissolved in water for 30 minutes. This was followed by alkylation (55 mM iodoacetamide) for 30 minutes in darkness. The tissue was washed in 0.1%TFA/95% ethanol for 30 seconds to remove the reduction and alkylation reagents. Finally, the tissue section was sprayed with α-CHCA matrix, and subjected to MS analysis (Supplementary File 1).
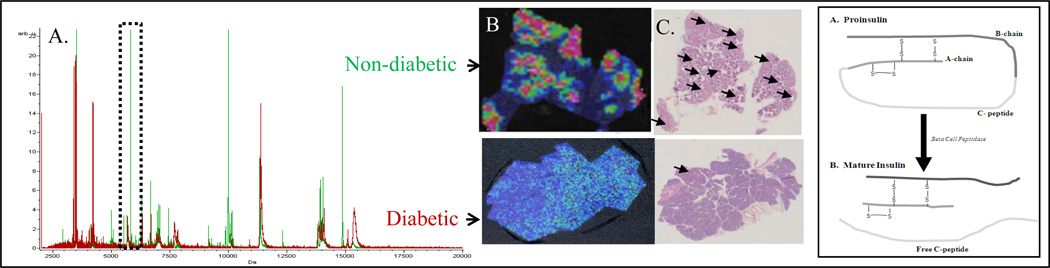
As a means to compare the overall spectral differences between diabetic and non diabetic tissues, multiple regions of interest (ROI) were randomly collected from the nondiabetic and diabetic samples (Supplementary File 1). Using light microscopy, islet morphology and areas that once contained healthy islets (as in the case of the diabetic tissue) were identified and the areas surrounding these islets were used as ROIs as those are the major areas of mononuclear cell invasion leading to islet destruction in T1D. The composite spectra were imported and compared in ClinProt Tools 2.0™ (Bruker Daltonics). Although there are a lot more islets in a healthy pancreas, the pancreas of a diabetic may also contain islets. The major difference being that these islets have a poor morphology and do not produce insulin, hence they are not visible at mass 5812.85 in MSI. Figure 1A shows the composite spectra of the two classes represented in red and green. These correspond to the diabetic and non-diabetic tissues respectively. In Fig. 1B pancreatic tissues of non-diabetic patients visualized via MALDI-MSI display clusters with a high expression of a peak with m/z 5812.85 compared to that of diabetic patients. These clusters seen within the non-diabetic patient’s pancreas corresponded to the location of pancreatic islet of langerhans in hematoxylin and eosin stained slides indicated by black arrows (Figure 1C and supplementary Fig 1). Comparative analysis of the data show 3 prominent peaks in the range of 5000–16000 m/z that are significantly over expressed in the non-diabetic tissue compared to the diabetic. The peak at ~ 5– 6 kDa was in the range of the theoretical mass of mature insulin. The direct identification of this molecule from the tissues by standard MS/MS using conventional MALDI-TOF mass spectrometry would be difficult due to the aforementioned instrumentation limitations.
Figure 1.
Panel A shows representative mass spectrum comparing human pancreatic tissue of diabetic vs. non diabetic patients. The spectra were obtained using MSI from the ROI surrounding the islets using MSI. The presence of insulin is shown at m/z 5812.85. Panel B depicts the corresponding MSI of the tissues. C shows the hematoxylin and eosin staining corresponding to the MALDI-MSI images in B. Islets are indicated by black arrows. The box to the far right shows a schematic of the tertiary structure of Insulin depicting its disulphide bridges.
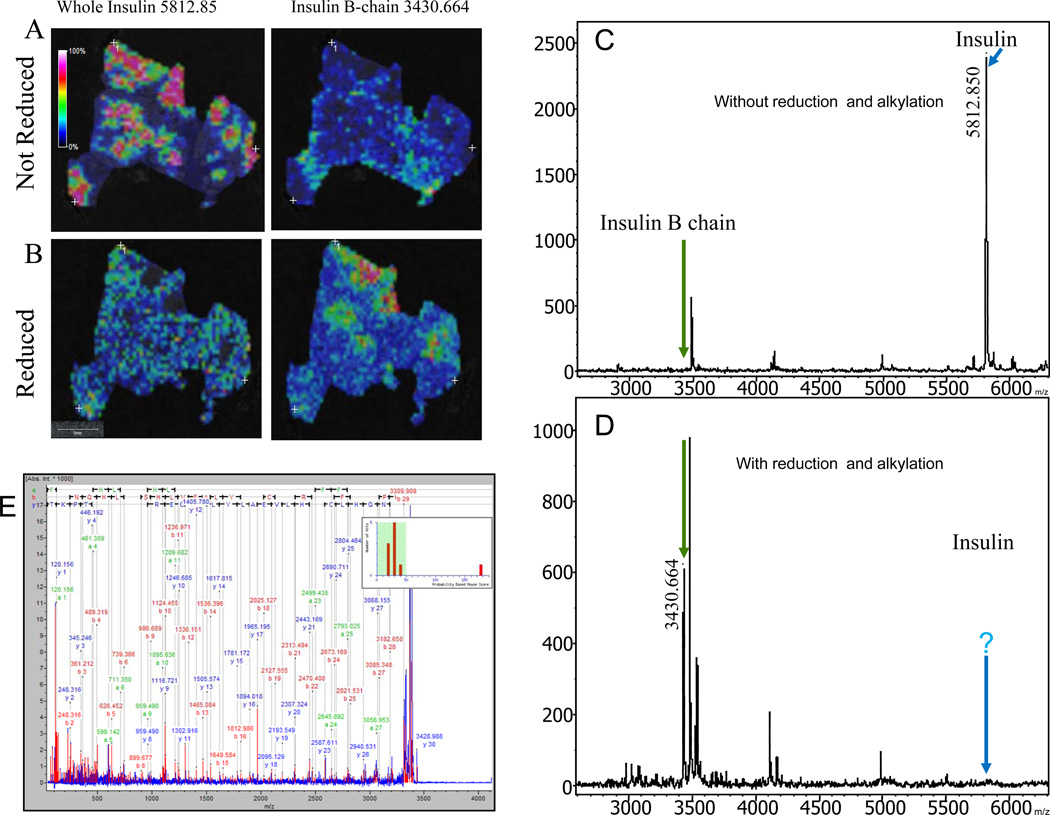
A peak with m/z 5812.85 was highly expressed in the human pancreatic tissues from non-diabetic patients compared to that of diabetic patients when visualized via MALDI-MSI. It was reasonable to assume that the high intensity peak at m/z 5812.85 was insulin since the peak was in the range of the theoretical mass of insulin. Since exact masses and the use of protein identification programs such as TagIdent are inaccurate, further analysis was necessary to characterize and identify the peak. Based on the knowledge of the tertiary structure of insulin (Figure 1D), reduction and alkylation was used to generate fragments that are amenable to MS/MS analysis for the identification and characterization of the molecule. To verify and validate that the molecule with m/z 5812.85 expressed within the islet clusters was in fact insulin a control experiment was performed where two consecutive slices of tissue from the same specimen were processed for MSI. One was processed with and the other without the reduction and alkylation steps. The dried tissue sample was sprayed homogeneously with a matrix solution of α-CHCA for the reduced tissue and sinapinic acid (10 mg/ml) containing 0.13% TFA and 75% ACN for the non-reduced and data was collected as described in Supplementary File 1. Figure 2 shows MALDI images of the insulin chains before and after reduction and alkylation within the islet themselves in the pancreatic head section of a healthy 10 year old patient.
Figure 2.
MSI of the pancreatic head portion of a healthy human pancreas. A, the left image shows the distribution of healthy islet cells with high expressions of insulin at mass 5812.85. The right image shows the absence of the B chain at mass 3430.664 prior to reduction. B, the left image shows the disappearance of insulin at m/z 5812.85 after reduction. In the bottom right image the B chain at mass 3430.664 is shown after reduction and cleavage of the cysteine disulfide bridges. 2C shows the MS spectrum with intact insulin at m/z 5812.85 before reduction (blue arrow) and the absence of the B chain as depicted by the green arrow. 2D shows the MS spectrum without insulin at m/z 5812.85 (blue arrow) and the emergence of the B chain at m/z 3430.664 (green arrow) after reduction. 2E annotated MS/MS spectrum identifying insulin from healthy human tissue homogenates.
The top left image shows the distribution of whole intact insulin molecule with m/z 5812.85 before reduction. The highlighted areas correspond to the insulin producing β-cells within the islets of langerhans (Fig. 2A left panel). This mass directly corresponded to the mass of intact insulin, comprising both the A and B chains connected via their inter-disulphide bridges. The right panel (Fig. 2A), shows the distribution of insulin B chain at m/z 3430.664 prior to reduction. Since the data acquisition and images were processed prior to reduction, the disulphide bond connecting the A and B chains are intact hence, it is not surprising that there is no peptide expression or detection at m/z 3430.664. Before reduction, the only molecule that is visualized by MSI is that of intact insulin at m/z 5812.85. Figure 2B, bottom left panel, shows the expression of m/z 5812.85 after in-situ reduction by TCEP. The reduced tissue does not have the molecule with m/z 5812.85 since reduction results in the cleavage of the disulphide bonds that bind the A and B insulin chains resulting in the formation of the individual chains with m/z of 2383 and 3430.664 respectively (Fig. 2B left panel). The expression of the insulin B chain at m/z 3430.664 is apparent after reduction as seen in the bottom right panel of Fig. 2B. The corresponding MALDI-MSI spectra of the healthy human pancreatic tissue before (Fig. 2C) and after (Fig. 2D) reduction and alkylation shows the presence of whole insulin at m/z 5812.85 (Fig. 2C blue arrow) and the absence of the B chain at m/z 3430.664 before reduction (Fig. 2C green arrow). The appearance of the B chain at m/z 3430.664 and simultaneous loss of the whole insulin at m/z 5812.85 post reduction are highlighted by the green and blue arrows respectively (Fig. 2D).
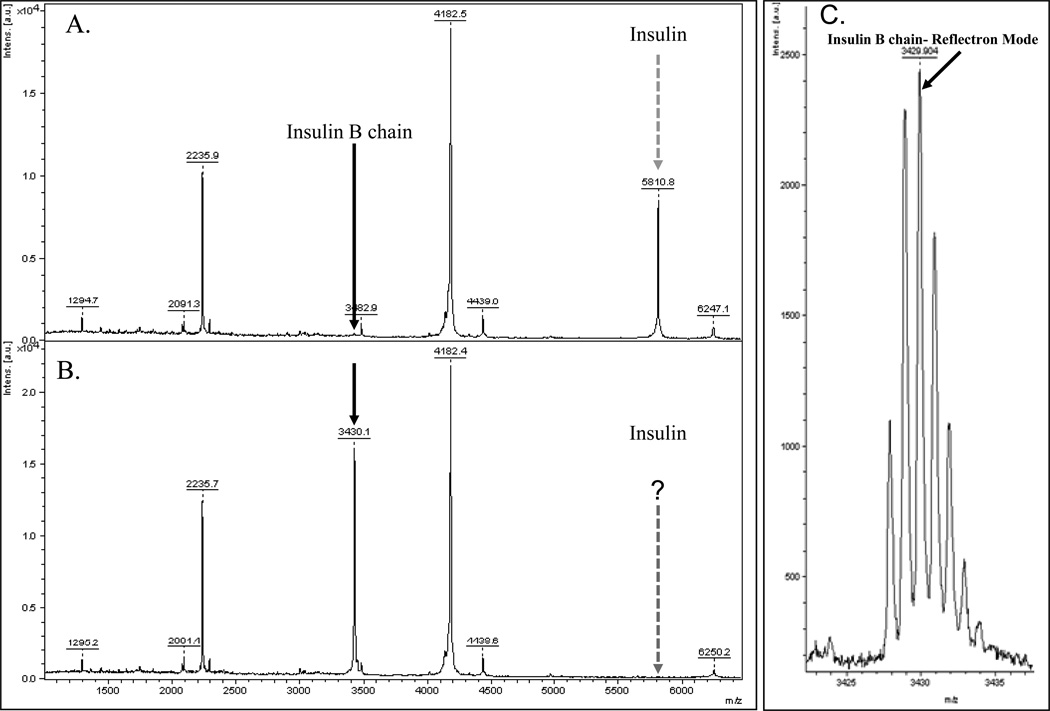
To validate the results seen within the whole tissue sections using MSI, tissue lysates were made as described in supplementary file 1 from the remaining block of the healthy nondiabetic sample and the peak of interest was identified via MS/MS (Figure 2E). The experimental sample and a control sample (without reduction and alkylation) were spotted on a plate in a ratio of 1:1 with matrix (50mg/mL CHCA with 0.1% TFA in 50% ACN) and analyzed by MS and tandem MS [8] on a Ultraflex II MALDI-TOF/TOF instrument (Bruker Daltonics) in positive ion mode in both linear and reflectron modes (Fig 3C) with a total of 2000 laser shots). MS/MS data was used for protein identification in MASCOT [9] after spectral processing by FlexAnalysis™ and BioTools™ (Supplementary File 1). Protein scores greater than 77 were considered to be significant (p < 0.05) [10]. Figure 3A, shows a spectrum from a healthy pancreatic lysate before reduction and alkylation. Figure 3B shows the spectrum after reduction and alkylation. There is a high abundant the peak at m/z 5810.8 prior to reduction. The peak is completely obliterated from the spectrum upon reduction by TCEP, indicating that the breaking of inter-disulfide (S-S) bonds within a protein with multiple domains. Instead, a new peak appears prominently in the spectrum with m/z 3430.1. MS/MS analysis identifies the peptide as the reduced B chain of insulin with the sequence FVNQHLCGSHLVEALYVCGERGFFYTPKY with a significant Mascot score and complete y- and b- fragment ion series (Fig. 2E). The theoretical monoisotopic MH+ of the peptide is 3430.939. Figure 3C depicts the reflectron mode spectrum of the same healthy pancreatic lysate with a peak at m/z 3429.004. The difference in masses ~319 ppm can be accounted for by the different modes of acquisition. In the linear mode of the MS instrument the m/z of the insulin B chain is 3430.1, in the high resolution reflectron mode the m/z was 3429.004. The identification of the peptide at m/z 3430.1 as the B chain of insulin directly confirms that the highly abundant peak at m/z 5810.8 (prior to reduction) is in fact the full length double domain protein comprising of the B and A chains of insulin covalently bonded through a disulfide bridge. However, after the reduction step, we would expect to generate the A and B chains with m/z 2383 and 3430.1 in the MS spectrum respectively. In our analysis, we see the expected B chain peak at m/z 3430.1 but the 2383 peak corresponding to the A chain of the protein is absent. A closer examination of the primary amino acid sequence of the peptide (GIVEQCCTSICSLYQLENYCN) reveals that there are no basic amino acids lysine (K) and arginine (R) in the peptide. In addition, the other two amino acid residues that can capture and retain protons during the peptide ionization process in mass spectrometer, histidine (H) and proline (P) are also absent in the sequence.
Figure 3.
Representative spectrum from healthy human pancreatic tissue lysate. Tissue lysate before (3A) and after (3B) reduction. The expected m/z of insulin is 5810.8 (shown with dashed arrows). The Insulin B chain appears at m/z 3430.1 after reduction with TCEP (black arrows). (3C) The MS spectrum in reflectron mode showing the isotopic cluster with m/z 3429.004 which was used in the MS/MS analysis to identify insulin.
These data show that MSI is an innovative tool that along with in-situ chemical modifications such as reduction is a valuable technique for confident and accurate identification of proteins directly from human pancreatic tissue sections. We have shown that identification of insulin a relatively large polypeptide, within healthy pancreatic sections was feasible after the in-situ reduction to form chains with molecular masses that are amenable to MS/MS sequencing. Recent availability of human pancreatic tissue from people with or at risk of diabetes or normal controls could help elucidate the cause and new treatments for the disease. Identification of insulin will be a key biomarker for β-cell regions of the pancreas. The identification of insulin by MS after in-situ reduction enabled us to localize the β-cells within the islets of langerhans. Insulin is a marker for the beta-cell region because only the beta-cells secrete insulin and hence in MSI, islets containing functioning beta-cells over-express the insulin mass of 5812. These experiments have been done multiple times (n= 4) reproducibly on tissue from different patients and have also been extended to tissues from three individual NOD mice (data not shown). This study provides proof-of concept on the use of MALDI-MSI in the in situ identification of proteins within pancreatic sections. These experiments are a first step for future experiments that will be used to identify other novel biomarkers in the milieu surrounding the islets of langerhans. Having a way of always identifying the islets during MSI experimentation will greatly facilitate the identification of major differentially expressed inflammatory proteins surrounding the islets. This will help identify potentially useful targets for therapy to preserve beta-cell mass in Type one or Type 2 diabetes.
Many proteins, especially those that are secreted from cells, or adhere to the cell surface [11] contain cysteine residues that can be oxidized to form these covalent bond disulphide bridges [12]. These bonds act as thermodynamic stabilizer of the native structure of the protein by decreasing the unfolded conformations. The cysteines involved in the formation of the disulphide bridge can be implicated in either an intra or inter bond, depending on whether they belong to the same polypeptide chain or link two different chains, respectively[13]. MALDI-MSI complemented with chemical modifications such as reduction, is a highly specific and targeted technique and tool for molecular biomarker discovery in diabetes.
Supplementary Material
Statement of clinical relevance.
Type one diabetes (T1D) is a chronic debilitating disease characterized by insulitis and the subsequent loss of the insulin producing beta cells. In T1D the body is unable to produce insulin leading to hyperglycemia, a condition which causes the major complications of the disease. Current T1D research focuses on the mechanisms involved in islet autoimmunity, and how members of the immune system modulate the autoimmune response. Presently, there is no known cure for T1D and diabetics endure the invasive conventional treatment of insulin replacement therapy. Here, we apply MALDI-IMS after on- tissue reduction to mine the proteome of the pancreatic islet of langerhans as a means of identifying key players involved in insulitis. This study showed that insulin, a relatively large molecule can be easily identified on human pancreatic sections via MALDI-IMS. This approach will be used in future studies to determine differential protein expression between non-diabetic and diabetic pancreas. We expect to identify candidates to be used in the development of novel cell based therapies that specifically target beta-cell growth and regeneration.
Acknowledgments
This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by the Juvenile Diabetes Research Foundation International (JDRF) and by funds from NIH/NIDDK ROI DK55240 to Jerry L. Nadler and NIH/NCI U01 CA085067 to O. John Semmes.
Abbreviations
- β-cell
Beta Cell
- LIFT
Laser Induced Fractionation Technique
- nPOD
Network for Pancreatic Organ donors of Diabetes
- OCT
Optimal Cutting Temperature
- TCEP
tris (2-carboxyethyl) phosphine
- T1D
Type One Diabetes
- T2D
Type Two Diabetes
Footnotes
All authors have declared no conflict of interest
References
- 1.McDonnell LA, MAR H. Imaging Mass Spectromety. Mass Spectrometry Reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 2.Chaurand P, Schwartz SA, Caprioli RM. Assessing protein patterns in disease using imaging mass spectrometry. J. Proteome Res. 2004b;3:245–252. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- 3.Caprioli RM, Farmer TB, Gile J. Molecular Imaging of Biological Samples: Localization of Peptides and Proteins Using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 4.Guerrera IC, Kleiner Application of Mass Spectrometry in Proteomics. Biosci. Rep. 2005;25:71–93. doi: 10.1007/s10540-005-2849-x. [DOI] [PubMed] [Google Scholar]
- 5.Bahr U, et al. Delayed extraction time-of-flight MALDI mass spectrometry of proteins above 25,000 Da. J. Mass Spectrom. 1997;32:1111–1116. doi: 10.1002/(SICI)1096-9888(199711)32:10<1111::AID-JMS567>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Westmancott G, et al. Investigating ion-surface collisions with a niobium superconducting tunnel junction detector in a time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2000;14:600–607. doi: 10.1002/(SICI)1097-0231(20000415)14:7<600::AID-RCM915>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Groseclose MR, et al. Identification of proteins directly from tissue: in situ triptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 8.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- 9.[ http://www.matrixscience.com], Matrix Science [Google Scholar]
- 10.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Deponte M, Hell K. Disulphide bind formation in the intermembrane space of mitochondria. Journal of Biochemistry. 2009;146:599–608. doi: 10.1093/jb/mvp133. [DOI] [PubMed] [Google Scholar]
- 12.Matsumara M, Signor G, Matthews BW. Substantial increase in protein stability by multiple disulphide-bonds. Nature. 1989;342:291–342. doi: 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- 13.Mormann M, Eble J, Christian S, Mesters RM, et al. Fragmentation of intra-peptide and inter-peptide disulfide bonds of proteolytic peptides by nanoESI collision-induced dissociation. Anal. Bioanal. Chem. 2008;392:831–838. doi: 10.1007/s00216-008-2258-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.