The growing prevalence of antibiotic-resistant pathogens continues to drive the need for antibiotics with novel modes of action. Of particular concern are infections caused by multidrug- resistant Mycobacterium tuberculosis, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), penicillin-resistant Streptococcus pneumoniae, fluoroquinolone-resistant Pseudomonas aeruginosa (FQRP), and β-lactam-resistant Gram-negative organisms, since many of these pathogens are intransigent to multiple classes of drugs.[1–3] Among discovery strategies, the empirical screening of chemical entities that are structurally distinct from clinically established agents represents an effective approach to developing novel antibiotics.
Historically, the majority of antimicrobial agents originate from natural products or are semi-synthetic derivatives.[4–7] As part of our ongoing effort to discover novel antitubercular and antibacterial agents and to exploit natural product as scaffolds for chemical diversity, we have been interested in following up emerging and underexplored naturally occurring compounds showing good antimicrobial activities. These promising natural product leads may provide a valuable starting point for the discovery and development of novel antimicrobial chemotypes. In this regard, natural phytochemicals are actively being pursued for their antibacterial properties.[8]
Flavonoids are a large family of naturally occurring polyphenolic phytochemicals, which have been reported to have various desirable pharmacological properties including anticancer[9, 10] and cancer chemopreventive,[11–13] antimalarial,[14] antimicrobial,[15, 16] and antioxidant[17, 18] effects. As such, flavonoids have been the focus of research programs seeking to develop potential chemotherapeutic and chemopreventive agents, traditional Chinese medicine, and dietary supplements with favorable low toxicity profiles.[19–23] The general chemical scaffold of flavonoids consists of a characteristic ring system A-C, chemically known as 2-phenyl-chrome-4-one or 2-phenyl-benzo-γ-pyrone, a regioisomer of coumarin (Figure 1). One such compound, (2S)-naringenin, was recently reported to show good antituberculosis activity with a minimum inhibitory concentration (MIC) of 2.8 µg/mL.[24] To further explore if this novel flavonoid scaffold possesses any tractable antituberculosis and antibacterial activities based on its structure-activity relationship (SAR), we systematically evaluated a small focused compound collection consisting of commercial and in-house synthesized flavonoid analogues, as well as their structurally related resveratrol analogues. In particular, the recently described, abyssinone II, a naturally occurring prenylated flavonoid[25] with aromatase inhibitory activity and breast cancer chemopreventive[26] and antiprostate cancer[27] properties, was included in the screen due to its close structural similarity with naringenin. Moreover, diethylstilbestrol (DES), a known pre-penicillin antibiotic was also included for comparison to further develop the SAR. Briefly, the initial screening library (Figure 2) included four compound series. The first series (compounds 1-7) belongs to flavanone derivatives; the second compound series contains flavone (8-19) or isoflavone (20) analogues; the third compound series (21-24) is 3-hydroxyflavone derivatives; and the final series (compounds 25-34) belongs to resveratrol analogues. Compounds 1-6, 8, 10-24, and 26-34 were purchased from Sigma-Aldrich (purity >97%). The remaining compounds, such as 4'-bromoflavone[11], abyssinone II and analogues[26], and resveratrol[28], were previously synthesized and studied in anticancer and cancer chemopreventive programs. We subsequently screened the collated flavonoid and resveratrol library against M. tuberculosis and a panel of Gram-positive and Gram-negative bacterial pathogens, including Enterococcus faecalis, Staphylococcus aureus, Streptococcus pneumoniae, Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, and Pseudomonas aeruginosa. Their anti-TB and antibacterial activities are summarized in Table 1.
Figure 1.
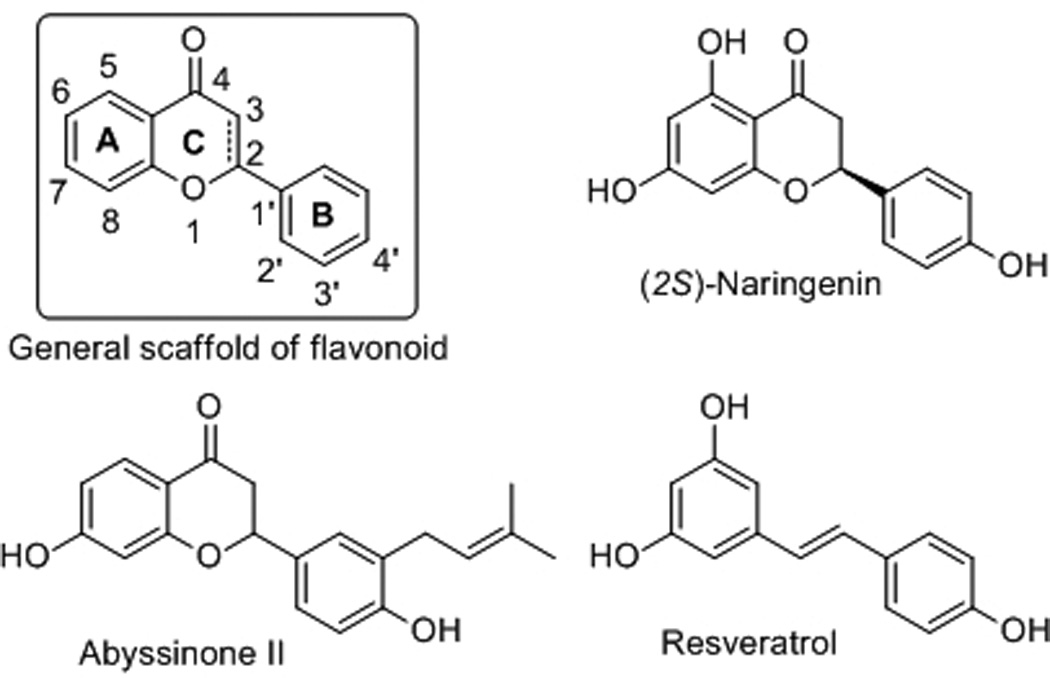
General structural scaffold of flavonoid and chemical structures of naringenin, abyssinone II, and resveratrol.
Figure 2.
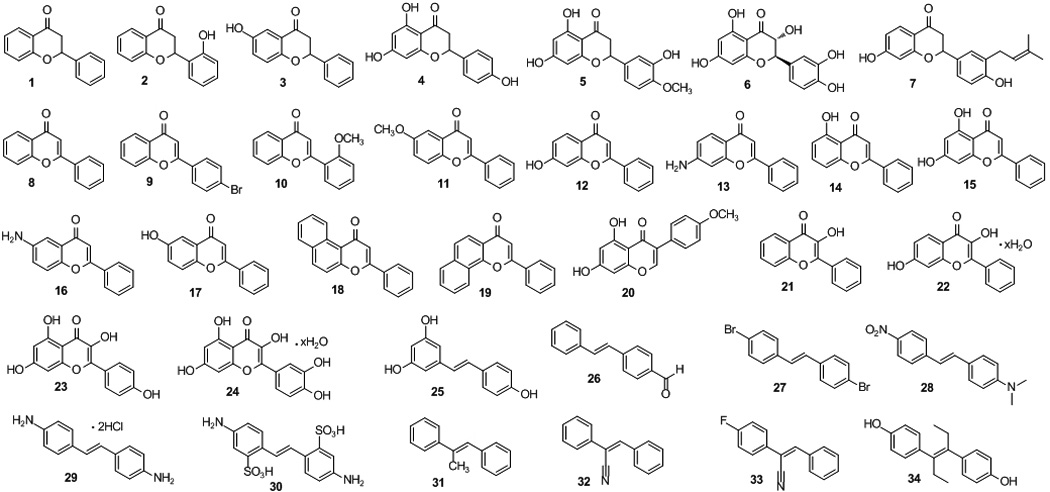
The chemical structures of flavonoid and resveratrol analogues subjected to initial screening.
Table 1.
| Compound |
M. tuberculosis (H37Rv) |
E. faecalis (ATCC29212) |
S. aureus (N315) |
S. pneumoniae (HM145) |
Compound |
M. tuberculosis (H37Rv) |
E. faecalis (ATCC29212) |
S. aureus (N315) |
S. pneumoniae (HM145) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | >100 | >100 | >100 | 18 | >200 | >100 | >100 | >100 |
| 2 | >200 | >100 | >100 | >100 | 19 | >200 | >100 | >100 | >100 |
| 3 | >200 | >100 | >100 | >100 | 20 | >200 | >100 | >100 | >100 |
| 4 | 200 | >100 | >100 | >100 | 21 | >200 | >100 | >100 | >100 |
| 5 | 100 | >100 | >100 | >100 | 22 | >200 | >100 | >100 | >100 |
| 6 | >200 | >100 | >100 | >100 | 23 | >200 | >100 | >100 | >100 |
| 7 | 50 | 25 | 12.5 | 25 | 24 | >200 | 25 | >100 | >100 |
| 8 | 100 | >100 | >100 | >100 | 25 | 100 | 100 | >100 | 100 |
| 9 | >200 | >100 | >100 | >100 | 26 | 200 | >100 | >100 | >100 |
| 10 | 50 | >100 | >100 | >100 | 27 | >200 | >100 | >100 | >100 |
| 11 | >200 | >100 | >100 | >100 | 28 | >200 | >100 | >100 | >100 |
| 12 | >200 | >100 | >100 | >100 | 29 | 100 | >100 | >100 | >100 |
| 13 | 200 | >100 | >100 | >100 | 30 | >200 | >100 | >100 | >100 |
| 14 | >200 | >100 | >100 | >100 | 31 | >200 | >100 | >100 | >100 |
| 15 | >200 | >100 | >100 | >100 | 32 | >200 | >100 | >100 | >100 |
| 16 | >200 | >100 | >100 | >100 | 33 | >200 | >100 | >100 | >100 |
| 17 | >200 | >100 | >100 | >100 | 34 | >200 | 6.25 | 6.25 | 6.25 |
The MICs of control antibiotics were 0.032 µg/mL (INH, M. tuberculosis), 0.4 µg/mL (ciprofloxacin, E. faecalis ), 0.4 µg/mL (ciprofloxacin, S. aureus), 0.8 µg/mL (ciprofloxacin, S. pneumoniae).
No test compounds were active against Gram-negative bacteria including K. pneumoniae (ATCC13883), A. baumannii (ATCC19606), E. coli (K12), and P. aeruginosa (PAO1).
Antitubercular evaluation revealed that flavanone derivatives 1, 4, 5, and 7 exhibited weak to moderate anti-TB activities with MICs ranging from 50–200 µg/mL. Interestingly, abyssinone II (7), bearing the 3'-prenyl group, demonstrated the most potent antituberculosis activity with a MIC value of 50 µg/mL. The prototype flavanone 1 is two-fold less active (MIC = 100 µg/mL) compared with abyssinone. Introduction of a 2'-OH or a 6-OH group to the flavanone nucleus led to the complete loss of anti-TB activity by comparing compounds 1 and 2, 3. Further comparison of the activities of 4 and 7 indicates that the lipophilic prenyl group at the B ring is important. Contrary to the report of Chen et al.,[24] racemic (±)-naringenin 4 only showed marginal anti-TB activity in our assay with a MIC value of 200 µg/mL. This is likely due to differences in the test methods, as the naringenin MIC was previously determined on agar with 2-weeks incubation as opposed to the recommended 3-weeks for growth, which yields results comparable to the broth MIC tests.[29] In the flavone series, only the prototype flavone (8) and 2'-methoxyflavone (10) exhibited moderate anti-TB activity (100 and 50 µg/mL, respectively). Incorporation of a 4'-bromo group abolished the anti-TB activity by comparing 8 and 9. All 3-hydroxyflavone compounds (21-24) bearing a conjugated 2,3-enol functionality with the 4-oxo group were not active against M. tuberculosis, which is likely due to the increased polarity from 3-OH group and poor penetration of the mycobacterial cell wall. Moreover, flavanone 1 showed about the same anti-TB activity as flavone 8 (MIC = 100 µg/mL), while 3-hydroxyflavone 21 was inactive. Notably, resveratrol, a structural relative of flavonoid, completely inhibited the mycobacteria growth at 100 µg/mL.
Further antimicrobial assessment showed that abyssinone II also exhibited relatively good activity against Gram-positive bacteria including E. faecalis (ATCC29212), S. aureus (N315), and S. pneumoniae (HM145), with MIC values of 25, 12.5 and 25 µg/mL, respectively. For comparison, the pre-penicillin compound DES (34) also demonstrated good activity against Gram-positive bacteria, with MIC values of 6.25 µg/mL (Table 1). However, most other compounds were relatively inactive. Amid the flavonoid and resveratrol analogues evaluated, no test compounds were active against Gram-negative bacteria including K. pneumoniae (ATCC13883), A. baumannii (ATCC19606), E. coli (K12), and P. aeruginosa (PAO1). This observation is consistent with prior reports that flavonoids and DES primarily show anti-Gram-positive activities that may result from poor penetration of the Gram-negative outer-membrane by lipophilic molecules.[30,31]
Encouraged by the promising antibacterial activity of abyssinone, a small selected number of abyssinone-based analogues 35-40[26] (Figure 3) were examined against the Gram-positive test panel. Their antibacterial activities are shown in Table 2. Interestingly, chalcones 35 and 36 with the opened central pyran C ring, considered as the synthetic precursors of abyssinone II, showed comparable antibacterial activities against E. faecalis, S. aureus, and S. pneumoniae with MIC values ranging from 25–50 µg/mL. Indeed, synthetic and naturally occurring chalcones have been reported with antiinfective and antiinflammatory properties.[32,33] Unfortunately, removal of the 7-hydroxyl group, or substitution for 7-O-methylated or 7-O-MOM protected analogues (37-42), resulted in a loss of antimicrobial activity. These data suggest the free phenol hydroxyl group at the A-ring is essential for antibacterial activity, while the middle C-ring has relatively minimal impact on activity. This led us to speculate that abyssinone may act as an ionophore, resulting from sum effects of its acidic hydroxyl ion carrier and the lipophilic prenyl group that may interact with the cytoplasmic membrane.
Figure 3.
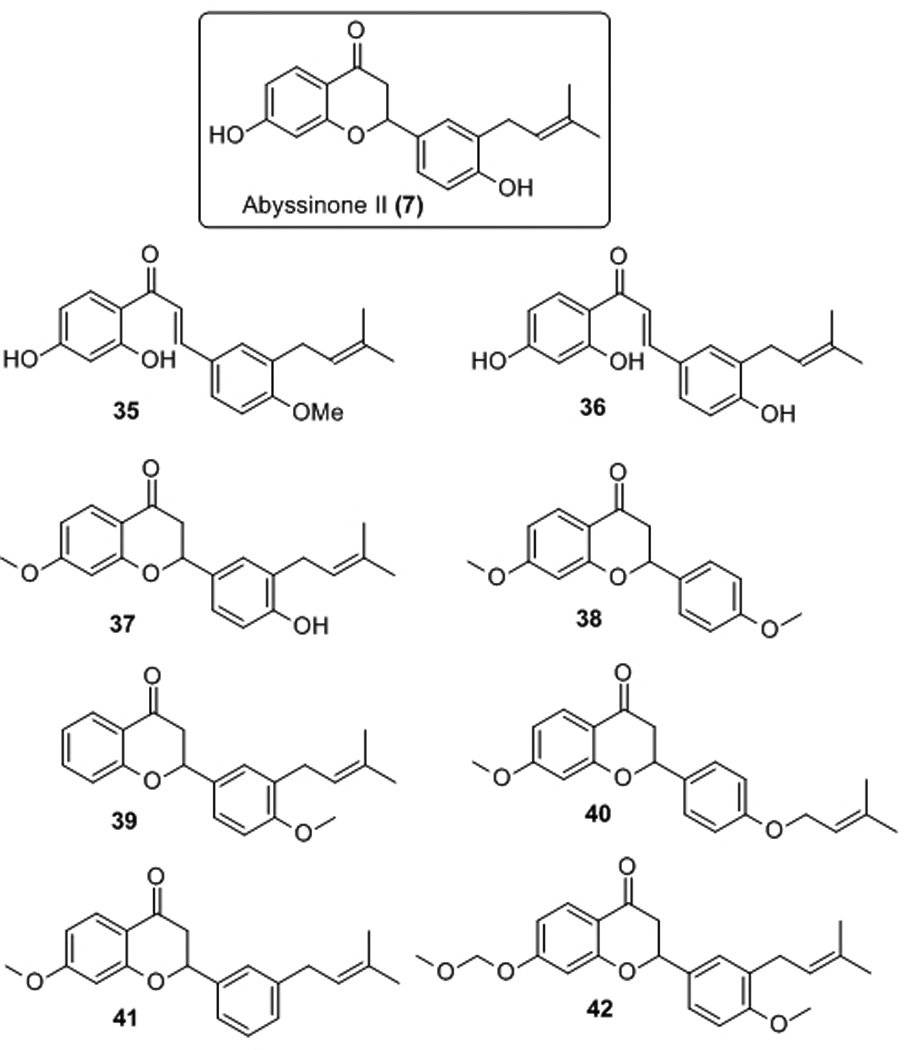
Additional abyssinone II analogues evaluated.
Table 2.
Antibacterial activity (µg/mL) of selected abyssinone analogues.
| Compound |
E. faecalis (ATCC29212) |
S. aureus (N315) |
S. pneumoniae (HM145) |
|---|---|---|---|
| 7 | 25 | 12.5 | 25 |
| 35 | 50 | 25 | 25 |
| 36 | 50 | 50 | >100 |
| 37 | >100 | >100 | >100 |
| 38 | >100 | >100 | >100 |
| 39 | >100 | >100 | >100 |
| 40 | >100 | >100 | >100 |
| 41 | >100 | >100 | >100 |
| 42 | >100 | >100 | >100 |
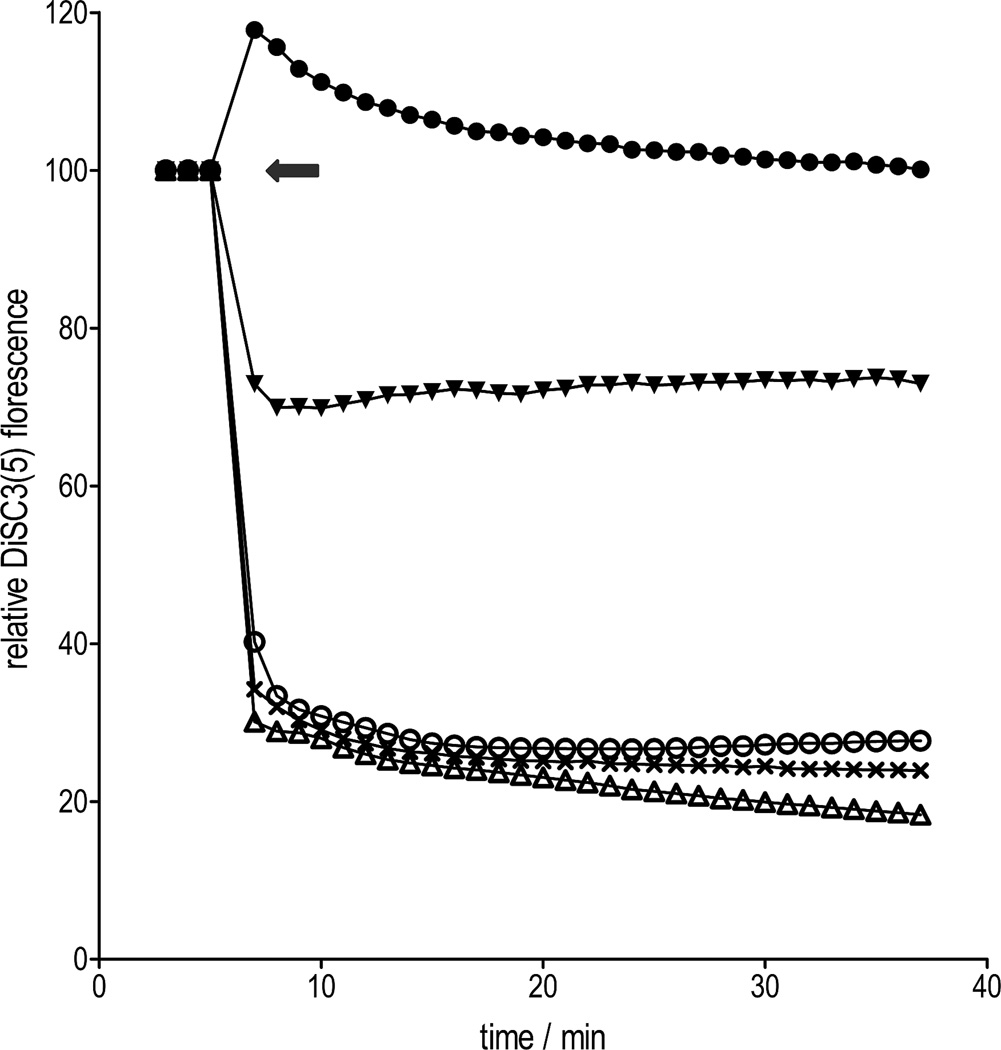
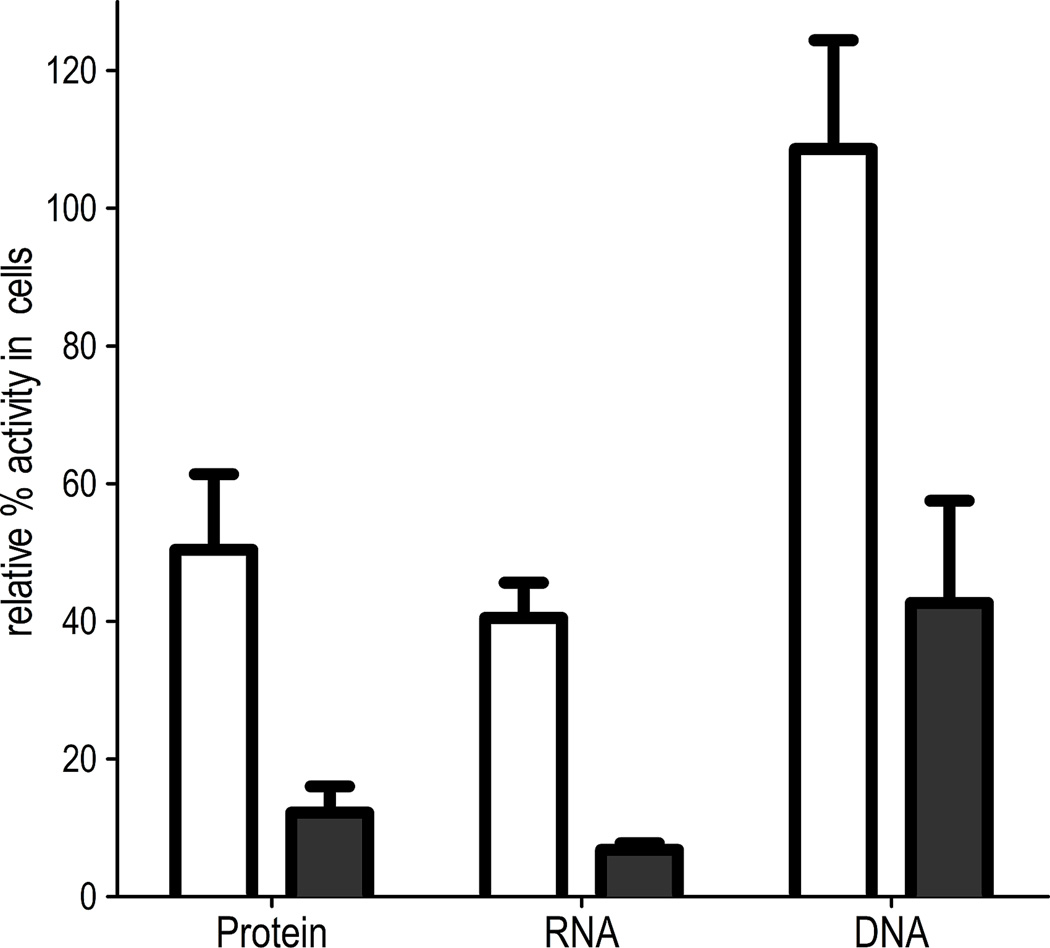
Through membrane potential assays, with the potentiometric dye DiSC3(5), abyssinone (7) was found to rapidly hyperpolarize the staphylococcal membrane, in a manner similar to the proton ionophore CCCP[34] (Figure 4). As anticipated, compound 37, with the 7-hydroxy group replaced by a methoxyl group, failed to hyperpolarize the membrane. Changes in membrane potential status was not associated with structural damage to the membrane integrity, since hyperpolarized cells showed no uptake of propidium iodide, which only penetrates damaged membranes. Rapid hyperpolarization was also induced by the chalcone 36, in contrast to the methoxy-analogue 35 that only had a marginal effect. Overall this suggests that the mode of action of abyssinone and related compounds might result from disruption of the membrane potential that is utilized for cellular energy production. However, this is unlikely to be the sole mechanism, since compound 36 was slightly less active than 35. Further studies revealed that abyssinone inhibited the synthesis of key cellular macromolecules such as protein, RNA and DNA, as part of its global effects (Figure 5), which is consistent with the findings for agents targeting the membrane.[35, 36] Indeed, the membrane-targeted drug daptomycin, which depolarizes Gram-positive bacteria, reduces the biosynthesis of several macromolecules and is one of the most potent agents in the clinic.[35, 36]
Figure 4.
Effects of abyssinone and analogues on bacterial membrane potential at 100 µg/mL. Each data point represents the mean of at least three replicates. The grey arrow indicates the time of addition of compounds; compounds 37 (●), 35 (▼), 7 (○), CCCP (x), 36 (Δ). Compound 37 caused some depolarization but this effect was not sustained.
Figure 5.
Abyssinone affects multiple macromolecular processes in a concentration-dependent manner, 12.5 µg/mL (□), 100 µg/mL (■). The activities of respective macromolecular processes with control antibiotics were as follows: 0.8 µg/mL tetracycline (protein, 46±1.2), 0.06 µg/mL rifampicin (RNA, 18.5±2.6), 0.8 µg/mL ciprofloxacin (DNA, 52±5.2).
In conclusion, the systematic screening of a focused flavonoid and resveratrol library led to the identification of abyssinone II, as an anti-Gram-positive agent that may have a multi-targeted mode of action, resulting from its ability to target the bacterial membrane. Such agents are increasingly becoming attractive therapeutic options owing to their potent actions, likely multi-target effects and limited potential for resistance development. The characterization of abyssinone II as a membrane-targeting molecule therefore presents a promising natural product lead for further medicinal chemistry optimization in an attempt to identify advanced experimental candidates with antimicrobial therapeutic potential.
Experimental Section
MIC determination
MIC values were determined against M. tuberculosis (H37Rv) and other bacteria using the standard microbroth dilution method exactly as previously described,[29] which is based on the methods by the Clinical and Laboratory Standards Institute.[37] The maximum test concentration used against M. tuberculosis was 200 µg/mL and 100 µg/mL against other pathogens.
Membrane potential assays
Dissipation of the transmembrane potential was measured using 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)] from Anaspec (San Jose, CA).[38] S. aureus 8325 was grown to OD600 of 0.4 in Mueller-Hinton broth, washed twice in 50 mM potassium phosphate buffer containing 10 mM glucose. After resuspending cells in the buffer, 5 µM of DiSC3(5) was added and incubated at room temperature for 30 min, before transferring cells to a 96-well plate. When the signal for DiSC3(5) had equilibrated, cells were then exposed to compounds (12.5 and 100 µg/mL) and their fluorescence measured over 30 min in a Synergy 2 plate reader (Biotek). The uptake of propidium iodide by S. aureus 8325 exposed to compounds at 12.5 and 100 µg/mL was assayed as previously described.[39]
Macromolecular synthesis
The effects of abyssinone (at 1 and 8 × MIC = 12.5 µg/mL) on DNA, RNA and protein were determined on S. aureus 8325 (OD600 = 0.4), by measuring the incorporation of the radiolabeled precursors [methyl-3H]thymidine, [5,6-3H]uridine and [4,5-3H]leucine into macromolecular fractions as previously described.[40] The precursors (1 µCi/mL) were added to cultures 10 min before the addition of compounds. Following exposure to compounds for 30 min, cells were lysed with cold 10% TCA and precipitated macromolecules collected on GF/C filters. After washing with 95% ethanol, GF/C filters were analyzed by liquid scintillation counting. The relative % activity in cells exposed to compounds was determined. Ciprofloxacin (DNA), rifampicin (RNA) and tetracycline (protein) were used as controls.
Acknowledgements
This work was gratefully supported by UH Hilo CoP startup fund, NIH INBRE program P20RR016467, R15AI092315 (DS), UT Arlington startup funds, and the American Lebanese Syrian Associated Charities (ALSAC).
References
- 1.Chopra I, Schofield C, Everett M, O'Neill A, Miller K, Wilcox M, Frere JM, Dawson M, Czaplewski L, Urleb U, Courvalin P. Lancet Infect. Dis. 2008;8:133–139. doi: 10.1016/S1473-3099(08)70018-5. [DOI] [PubMed] [Google Scholar]
- 2.Sougakoff W. Clin. Microbiol. Infect. 2011;17:800–805. doi: 10.1111/j.1469-0691.2011.03577.x. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, Livermore DM. J. Infect. 2009;59(Suppl 1):S4–S16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- 4.Arvind SN, Jonnala KK, Suaib L, Dharmendra S, Suman PSK. Med. Res. Rev. 2010;30:603–645. [Google Scholar]
- 5.Copp BR, Pearce AN. Nat. Prod. Rep. 2007;24:278–297. doi: 10.1039/b513520f. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez-Lugo MT, Bewley CA. J. Med. Chem. 2008;51:2606–2612. doi: 10.1021/jm070719i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh C. Nat. Rev. Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 8.Kaur GJ, Arora DS. BMC Complement. Altern. Med. 2009;9:30. doi: 10.1186/1472-6882-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabrera M, Simoens M, Falchi G, Lavaggi ML, Piro OE, Castellano EE, Vidal A, Azqueta A, Monge A, de Cerain AL, Sagrera G, Seoane G, Cerecetto H, Gonzalez M. Bioorg. Med. Chem. 2007;15:3356–3367. doi: 10.1016/j.bmc.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 10.Kosmider B, Osiecka R. Drug Dev. Res. 2004;63:200–211. [Google Scholar]
- 11.Song LL, Kosmeder JW, 2nd, Lee SK, Gerhauser C, Lantvit D, Moon RC, Moriarty RM, Pezzuto JM. Cancer Res. 1999;59:578–585. [PubMed] [Google Scholar]
- 12.Ren W, Qiao Z, Wang H, Zhu L, Zhang L. Med. Res. Rev. 2003;23:519–534. doi: 10.1002/med.10033. [DOI] [PubMed] [Google Scholar]
- 13.Marchand LL. Biomed. Pharmacother. 2002;56:296–301. doi: 10.1016/s0753-3322(02)00186-5. [DOI] [PubMed] [Google Scholar]
- 14.Tasdemir D, Lack G, Brun R, Ruedi P, Scapozza L, Perozzo R. J. Med. Chem. 2006;49:3345–3353. doi: 10.1021/jm0600545. [DOI] [PubMed] [Google Scholar]
- 15.Fowler ZL, Shah K, Panepinto JC, Jacobs A, Koffas MAG. PLoS ONE. 2011;6:e25681. doi: 10.1371/journal.pone.0025681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cushnie TPT, Lamb AJ. Int. J. Antimicrob. Agents. 2005;26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erasto P, Bojase-Moleta G, Majinda RR. Phytochemistry. 2004;65:875–880. doi: 10.1016/j.phytochem.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson DE, Hurst RD. Cell. Mol. Life Sci. 2007;64:2900–2916. doi: 10.1007/s00018-007-7237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Carlo G, Mascolo N, Izzo AA, Capasso F. Life Sci. 1999;65:337–353. doi: 10.1016/s0024-3205(99)00120-4. [DOI] [PubMed] [Google Scholar]
- 20.Beecher GR. J. Nutr. 2003;133:3248S–3254S. doi: 10.1093/jn/133.10.3248S. [DOI] [PubMed] [Google Scholar]
- 21.Marzocchella L, Fantini M, Benvenuto M, Masuelli L, Tresoldi I, Modesti A, Bei R. Recent Pat. Inflammation Allergy Drug Discovery. 2011;5:200–220. doi: 10.2174/187221311797264937. [DOI] [PubMed] [Google Scholar]
- 22.Erlund I. Nutr. Res. 2004;24:851–874. [Google Scholar]
- 23.Horvathova K, Vachalkova A, Novotny L. Neoplasma. 2001;48:435–441. [PubMed] [Google Scholar]
- 24.Chen LW, Cheng MJ, Peng CF, Chen IS. Chem. Biodivers. 2010;7:1814–1821. doi: 10.1002/cbdv.200900227. [DOI] [PubMed] [Google Scholar]
- 25.Kamat VS, Chuo FY, Kubo I, Nakanishi K. Heterocycles. 1981;15:1163–1170. [Google Scholar]
- 26.Maiti A, Cuendet M, Croy VL, Endringer DC, Pezzuto JM, Cushman M. J. Med. Chem. 2007;50:2799–2806. doi: 10.1021/jm070109i. [DOI] [PubMed] [Google Scholar]
- 27.Farmer RL, Biddle MM, Nibbs AE, Huang X, Bergan RC, Scheidt KA. ACS Med. Chem. Lett. 2010;1:400–405. doi: 10.1021/ml100110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CWW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 29.Hurdle JG, Lee RB, Budha NR, Carson EI, Qi J, Scherman MS, Cho SH, McNeil MR, Lenaerts AJ, Franzblau SG, Meibohm B, Lee RE. J. Antimicrob. Chemother. 2008;62:1037–1045. doi: 10.1093/jac/dkn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido H. J. Bioenerg. Biomembr. 1993;25:581–589. doi: 10.1007/BF00770245. [DOI] [PubMed] [Google Scholar]
- 31.Yotis WW, Baman SI. Appl. Microbiol. 1970;19:474–478. doi: 10.1128/am.19.3.474-478.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowakowska Z. Eur. J. Med. Chem. 2007;42:125–137. doi: 10.1016/j.ejmech.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 33.Feldman M, Santos J, Grenier D. J. Nat. Prod. 2011;74:1862–1867. doi: 10.1021/np200174h. [DOI] [PubMed] [Google Scholar]
- 34.Patton TG, Yang SJ, Bayles KW. Mol. Microbiol. 2006;59:1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x. [DOI] [PubMed] [Google Scholar]
- 35.Hobbs JK, Miller K, O'Neill AJ, Chopra I. J. Antimicrob. Chemother. 2008;62:1003–1008. doi: 10.1093/jac/dkn321. [DOI] [PubMed] [Google Scholar]
- 36.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Nat. Rev. Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Seventh Edition: Approved Standard M7-A7. Wayne, PA, USA: CLSI; 2006. [Google Scholar]
- 38.Wu M, Maier E, Benz R, Hancock RE. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 39.O'Neill AJ, Miller K, Oliva B, Chopra I. J. Antimicrob. Chemother. 2004;54:1127–1129. doi: 10.1093/jac/dkh476. [DOI] [PubMed] [Google Scholar]
- 40.Oliva B, Miller K, Caggiano N, O'Neill AJ, Cuny GD, Hoemann MZ, Hauske JR, Chopra I. Antimicrob. Agents Chemother. 2003;47:458–466. doi: 10.1128/AAC.47.2.458-466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]