Infectious diseases are a major global health burden accounting for approximately 15 million deaths annually, many from drug resistant pathogenic agents, with a significant number of cases occurring in developing countries.[1–7] In particular, the resurgence of tuberculosis (TB) has been accompanied by the rapid spread of multi-drug resistance TB (MDR-TB) resulting from Mycobacterium tuberculosis (Mtb) strains that fail to respond to the first-line drugs, rifampin and isoniazid. Currently, <5% of ~0.5 million MDR-TB cases estimated globally are appropriately diagnosed and treated due in part to the long assay turnaround time associated with conventional culture-based drug susceptibility testing.[8]
Quantitative PCR (qPCR), line probe assays, or home-brewed nucleic acid amplification tests (NAATs), have recently been used to identify MDR-TB. These tests provide shorter assay turnaround times compared to culture, but depend on sophisticated laboratory infrastructure and well-trained personnel to ensure accurate, reliable, and reproducible results. The WHO expert group recently recommended two NAAT line probe assays, INNO-LiPA Rif.TB from Innogenetics and MTBDRplus from Hain Lifescience, both of which employ multiplexed PCR reverse hybridization approaches. Line probe assays, however, are not designed to interrogate a mixed population of drug resistant and susceptible bacterial populations,[9–11] because sequence-specific array hybridization is unable to detect low abundance single base variations due to cross hybridization artifacts, especially in high GC content regions.
Several groups have developed partial or fully integrated microfluidic devices for carrying out NAATs for infectious diseases.[12–17] For example, Cepheid's GeneXpert MDR-TB system performs qPCR using Taqman probes for Mtb and 5 rifampin resistance mutations, providing results in nearly 2 h.[18] However, many of these devices are made from silicon/glass materials and thus require direct photolithographic processing to manufacture the desired structures, which increases the production cost of devices.
Herein, we describe a modular design approach for an assay and hardware to detect/identify MDR-TB. The molecular assay was designed to interrogate single base variations in codons 516, 526, and 531 (using the numbering system of E. coli rpoB) in the rifampin-resistance determining region (RRDR) of the rpoB gene, which was used as surrogate markers for MDR-TB.[19, 20] The multi-step assay was carried out within a modular thermoplastic fluidic cartridge operated by the accompanying support peripherals packaged into a small instrument (1' × 1' × 1'). No operator intervention was required once the clinical sample (sputum) was loaded into the fluidic cartridge. This system offered advantages compared to existing NAAT systems. For example, to accommodate low-resource setting scenarios where operator expertise is limited, the hardware provided full process automation. The fluidic system consisted of a cartridge made from thermoplastics with the desired structures fabricated in a high-production format using molding to keep chip cost low for one-time use applications (in vitro diagnostics) appropriate for resource limited settings.
Additional design concepts employed in the fluidic cartridge included: (1) A hybrid modular architecture, which combined several task-specific modules interconnected to a fluidic motherboard with the material selected to optimize performance. (2) Most of the active elements were poised off-chip to keep chip cost low. (3) The embossing step was used to not only create the fluidic network, but other necessary device components as well, such as the DNA extraction bed, thermal isolation grooves, valve seats and waveguides. The fluidic cartridge could easily be reconfigured without requiring re-engineering to accommodate alternative assays. For example, a universal array module could be replaced with a colorimetric readout module to simplify the readout hardware. Compared to monolithic[12–16] or LEGO approaches,[21, 22] the hybrid modular architecture balanced the efficiency and flexibility in fluidic cartridge design and material selection.
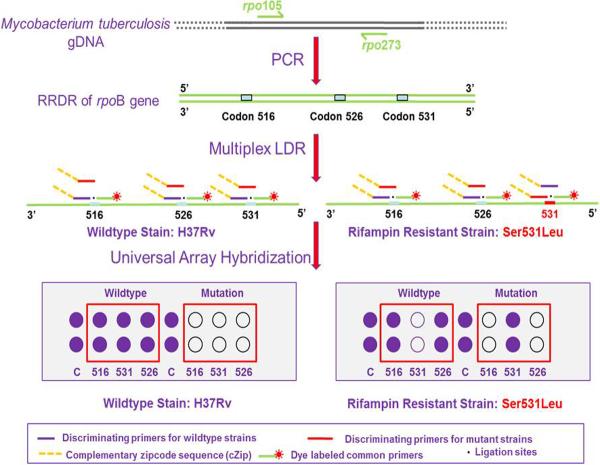
Collectively, more than 95% of resistance to rifampin is associated with missense, insertion, and deletion mutations in the RRDR of the rpoB gene. Single base variations in codons 516, 526 or 531 are most frequently found in rifampin-resistant Mtb strains world-wide. We therefore developed a PCR/LDR/universal array assay to detect sequence variations harbored within these codons (see Figure 1).[23]
Figure 1.
Schematic representation of the molecular assay. Following a primary PCR to generate a 193 bp rpoB amplicon spanning the RRDR, 3 sets of ligase detection reaction (LDR) primers were used for interrogating missense mutations in codons 516, 526 and 531 of the rpoB gene. The discriminating primer contained a base complementary to either the wild-type or mutation sequence at its 3'-end. The common primer was phosphorylated at its 5'-end and contained a fluorescent dye, Cy5, at its 3'-end. A thermally stable ligase covalently links the two primers hybridized to the target if there was a perfect match at the locus being interrogated. The discriminating primer also carried a unique complementary “zipcode” sequence (cZip) at its 5'-end to direct the LDR product to a specific location on the “zipcode” universal array.
The PCR/LDR/universal array approach employed a high fidelity Taq ligase and decoupled the mutation discrimination step from the amplification and hybridization steps.[23–27] The assay demostrated the following important attributes: (1) The closely clustered drug resistance mutations of MDR-TB could be interrogated using a uniplex PCR. Therefore, careful design of primers with similar Tm's and problems with uneven amplification produced by different targets was negated. (2) Only the specific drug resistant sequence variations generate positive results. Silent mutations, which do not confer drug resistance, did not generate false positive results. (3) Hetero-resistant Mtb containing a low abundance of drug resistant strains (~1%) in patients with emerging drug resistance could be identified due to the high discriminatory capability of the ligase enzyme. (4) Two amplification steps were used, an exponential amplification associated with the PCR and a linear amplification resulting from the LDR, which improved the limit-of-detection of the assay.
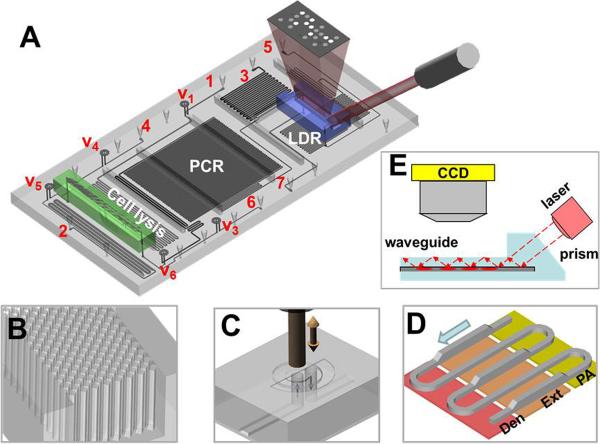
The fluidic cartridge shown in Figures 2, S1 and S2 (Supporting Information) was designed to include five different processing steps; cell lysis, solid phase DNA extraction (SPE), PCR, LDR and universal array hybridization. The thermal steps (cell lysis, PCR and LDR) were situated on a fluidic motherboard while the SPE and universal array were placed on two different modules interconnected to the motherboard. A detailed description of how each module and the motherboard were fabricated is included in the Supporting Information. The modules and motherboard were hot embossed from a particular thermoplastic, selected to optimize performance of each specific processing step. To minimize post-assembly steps, embossing was used not only to generate fluidic networks, but also specific components required for proper operation of the module or motherboard. For example, the SPE module required the use of polycarbonate, which was suitable for the selective capture of nucleic acids to its surface following photoactivation.[28] Further, to increase the available surface area required to increase the DNA load, high aspect ratio micropillars were manufactured into the SPE bed during embossing of the fluidic network, thus obviating the need for packing the SPE bed with externally added beads (see Supporting Information, Figure S1).
Figure 2.
Integrated, modular fluidic cartridge for TB analysis. (A) 3D rendering of the cartridge and the array. 1–7 - fluidic inlets and outlets: 1 - sample inlet, 2 – PCR cocktail inlet, 3 – LDR cocktail inlet, 4 – ethanol and air inlet, 5 – array wash inlet, 6 – vacuum connection, 7 – waste. V1–V6 – on-chip membrane valves (note that V2 is positioned next to SPE module on the cell lysis microchannel and is not visible in the current view). (B) Close-up of the SPE bed showing DNA capture bed filled with an array of high-aspect ratio pillars. (C) Schematic operation of the on-chip membrane valve with mechanical actuation – electrically actuated solenoid presses on the polymer membrane closing the passage of fluid from the bottom layer through the valve and back to the bottom layer. (D) Geometry of the continuous flow PCR reactor with dual-depth microchannels for extended residence time in the extension-zone; Den – denaturation, Ext – extension, PA – primer annealing. (E) Schematic representation of the detection mode. Laser excitation is coupled to a waveguide through an integrated prism. Light travelling through the waveguide excites the labeled LDR products hybridized to the zip code array spotted on the waveguide surface.
Polycarbonate has a relatively high glass transition temperature, which makes it a suitable material for thermal reactions and thus, the ideal material for the fluidic motherboard, which performed thermal reactions requiring 65°C to 95°C operation. Polycarbonate also has a relatively large elongation at break threshold such that it can be used as a microfluidic valve membrane. Using polycarbonate as the cover plate for the motherboard obviated the need for an additional post-molding assembly step as the valve membranes were attached to the fluidic network in the same lamination step used to enclose the fluidic network with the cover plate. However, polycarbonate is not compatible with ultra-sensitive fluorescence detection because of its relatively high background.
Poly(methyl methacrylate), PMMA, demonstrates good optical clarity and minimal non-specific adsorption artifacts, making it an ideal material for construction of the universal array module.[29] The universal array was poised within a microchannel resident on this module to reduce the time for probe addressing by minimizing diffusional constraints.[30–32] Waveguides were fabricated using double-sided embossing with simple plasma activation of the PMMA surface to allow for covalent attachment of the zipcode probes to the waveguide (Supporting Information, Figure S3).[33] Finally, printing of the zipcode probes could be performed using conventional DNA spotting equipment prior to enclosure of the fluidic network because of the low temperature required for bonding the cover plate to the substrate containing the probes.[34]
Figures 2B and S1B provide schematic drawings and SEM images of the SPE module. Design specifics on the SPE module can be found in the Supporting Information. Figure 2C shows a schematic of the operation of the polycarbonate membrane valves with solenoid actuation. These micro-valves could withstand head pressures up to 105 psi without leakage, which is more than an order of magnitude higher pressure load than PDMS valves.[35]
The thermal reactors situated on the fluidic motherboard (Figures 2A, S1A, and S1C) incorporated a continuous flow format, which provided ultrafast PCR amplification because extension times are limited by the kinetics of the polymerase for properly designed thermal reactors.[36] Several thermal management structures were also included to further improve amplification efficiency (Figure S4), such as backside thermal isolation grooves, a thin substrate and copper plates to give a uniform temperature distribution throughout a particular thermal zone.[37] Further, a dual-depth channel (200 μm and 100 μm) was employed to provide sufficient residence time within the polymerase extension zone to generate full length PCR products during each cycle while operating at a fixed linear flow velocity (Figures 2D and S1C). For a detailed discussion on the operation of the fluidic cartridge, see the Supporting Information.
We performed a series of assays using different positive and negative controls that were processed using the modular fluidic cartridge. The results concurred with those expected, as shown in Figure S5. For example, supplying an input of E. coli cells did not result in postive fluorescence signals on the universal array. However, input of a wild-type Mtb strain generated fluorescence signatures at the appropriate spots of the array (Figure S5).
We were interested in evaluating the minimum number of Mtb cells required to provide positive results using our system and whether results could meet the needed level for clinical diagnosis. To determine the limit-of-detection, assays were conducted using a serial dilution of cultured H37Rv cells as the input. As few as 50 Mtb cells could be successfully detected, representing a 100-fold improvement in sensitivity compared to current clinical smear tests, which require 5,000–10,000 bacilli in 1 mL of sputum.
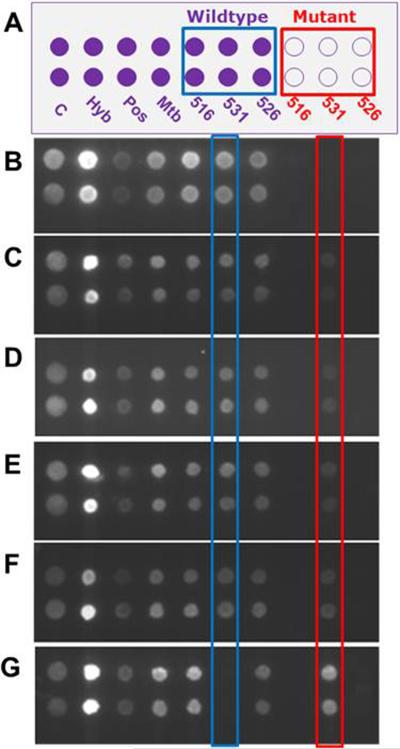
Hetero-resistant Mtb is defined as the coexistence of mixed populations of drug susceptible and resistant Mtb strains in the same patient, which is considered as a preliminary stage to full resistance. These cases result either from super-infection or from selection pressure during antibiotic treatment. The occurrence of hetero-resistance can be as high as 20%.[10, 11] To evaluate the ability of our system to identify a minority population of drug resistant Mtb strains, a series of control hetero-resistance samples containing increasing percentages of a rifampin resistant strain (S531L) – 0%, 1%, 2%, 5%, 10%, 100% – in a drug susceptible strain of H37Rv were prepared and analyzed (Figure 3). The fluorescence intensity from the 531 mutant spot increased with an increased percentage of the rifampin resistant strain present in the sample while the spot intensity from the 531Wt decreased (Figure 3C). The fluorescence from the 531 mutant spots run with samples containing as little as 1% of rifampin resistant strain was calculated to be ~10-fold higher than fluorescence from those run with samples containing 0% rifampin resistant strains (Figure 3C). Therefore, less than 1% of the drug resistant strain could still be discriminated based on a 3-fold standard-deviation higher signal than non-specific signal. Very slight levels of fluorescence were detected from the 531Wt spots run with 100% of the drug resistant strain, which could be attributed to mis-ligation and/or nonspecific hybridization.[38]
Figure 3.
Identification of a mixed-population of drug susceptible (H37Rv) and drug resistant (S531L) Mtb strains. (A) Layout of the universal array. Spots labeled C were printed with 5'-Cy5 oligonucleotides, and used as quality controls (C) of the printing process. Spots labeled as Hyb were hybridization controls. Spots labeled as Pos were positive controls (spiked plasmids). Spots labeled Mtb were Mtb-specific probes targeting an IS6110 insertion fragment. (B–G) Universal array images for different amounts of the drug resistant and drug susceptible strains. The percentage of the sample containing the drug resistant strain was 0% (B), 1% (C), 2% (D), 5% (E), 10% (F) and 100% (G).
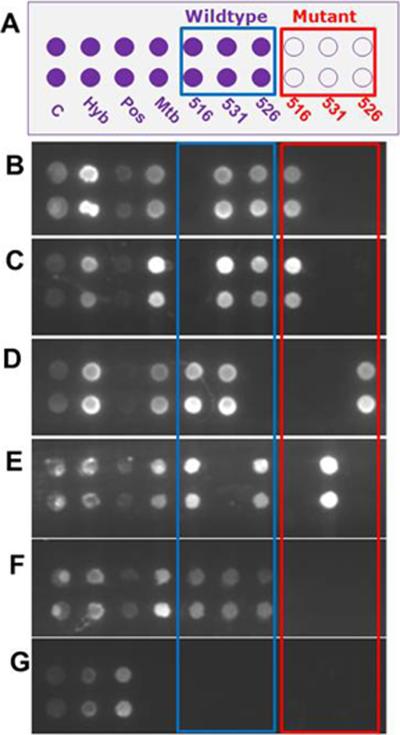
The modular system was challenged with four clinical samples containing either rifampin-resistant or susceptible Mtb strains. Figure 4 represents results that were consistent with the phenotype and the genotype of those clinical samples. The analysis of samples YE69 (A516V), 3976-83 (A516V), YE68 (H526Y), 3908-83 (S531L) and 94-2219 (Wt), Figure 4B–G, identified the correct mutation for each drug-resistant strain as determined by sequence analysis of the rpoB gene of these clinical isolates.
Figure 4.
Universal array hybridization results from clinical samples (sputum). (A) Layout of the universal array with the same designations as that given in Figure 3A. (B) Mtb clinical isolate (YE69) harbors a mutation in codon 516 of rpoB. (C) Mtb(+) sputum sediment (3976-83) harbors a mutation in codon 516 of rpoB. (D) Mtb clinical isolate (YE68) possesses a mutation in codon 526 of rpoB. (E) Mtb(+) sputum sediment (3908-83) harbors a mutation in codon 531 of rpoB. (F) Mtb-(+) sputum sediment (94-2219) contained no mutations in rpoB RRDR (Wt). (G) Mtb(−) sputum sediment. The sample identifier is given in parentheses.
A colorimetric zipcode array module was developed as detailed in the Supporting Information in order to simplify the readout by eliminating the need for laser-induced fluorescence (Figure S6). PMMA wafers containing immobilized zipcode probes were prepared using a PDMS stencil (Figure S6B). Also, 1.4 nm nanogold-labeled common primers were used instead of fluorescent dye-labeled primers as shown in Figure S6A. After LDR and hybridization, the nanogold labels acted as a catalytic site for silver deposition, which was visible to the naked eye and could be recorded by a digital camera. A drug susceptible Mtb stain (H37Rv) and a rifampin-resistant strain (S531L) were tested and the results are shown in Figure S6C. Using this colorimetric test for detection, the Mtb cell limit-of-detection was found to be 50 cells, similar to that observed using laser-induced fluorescence detection.
We have reported the development of a modular design approach for both the software (molecular assay) and hardware (fluidic cartridge and support peripherals) to determine sequence variations in reporter sequences to detect and identify MDR-TB directly from sputum samples. The system provided full process automation minimizing operator expertise requirements and also, generated results in <30 min, which will be a key operating metric by providing rapid results to allow proper treatment of patients, potentially minimizing the generation of additional drug resistant strains due to implementation of a full treatment regimen.
Recently, extremely drug resistant TB (XDR-TB) has been reported. XDR-TB strains contain mutations responsible for MDR-TB as well as sequence variations resulting in resistance to second-line drugs. By incorporating a multiplexed PCR and additional LDR primers, the system presented here can easily be configured to detect sequence variations responsible for XDR-TB without hardware redesign. The system, both software and hardware, holds the potential to be a universal platform for identifying genetic sequence signatures in a variety of applications, including cancer diagnostics,[23, 26, 27] forensic testing and biothreat pathogen detection and identification in both developing and developed countries[24] due to the modular design.
Acknowledgements
The authors would like to thank the NIH (EB010087) for partial financial support of this work as well as the World Class Univeristy (WCU) program of South Korea. Finally, the authors would like to thank Don Patterson, Tana Pittman, Weislaw Stryjewski and Brad Ellison for this technical assistance.
References
- [1].World Health Organization Report on infectious diseases: Removing Obstacles to Healthy Development. 1999 can be found under http://www.who.int/infectious-disease-report/index-rpt99.html.
- [2].World Health Organization The world health report 2002 - Reducing Risks, Promoting Healthy Life. 2002 doi: 10.1080/1357628031000116808. can be found under http://www.who.int/whr/2002/en/ [DOI] [PubMed]
- [3].World Health Organization Scaling up the response to infectious diseases: A way out of poverty. 2002 can be found under http://www.who.int/infectious-disease-report/2002/
- [4].Perkins MD, Roscigno G, Zumla A. Lancet. 2006;367:942–943. doi: 10.1016/S0140-6736(06)68386-4. [DOI] [PubMed] [Google Scholar]
- [5].Morens DM, Folkers GK, Fauci AS. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Okeke IN, Klugman KP, Bhutta ZA, Duse AG, Jenkins P, O'Brien TF, Pablos-Mendez A, Laxminarayan R. Lancet Infectious Diseases. 2005;5:568–580. doi: 10.1016/S1473-3099(05)70217-6. [DOI] [PubMed] [Google Scholar]
- [7].Okeke IN, Laxminarayan R, Bhutta ZA, Duse AG, Jenkins P, O'Brien TF, Pablos-Mendez A, Klugman KP. Lancet Infectious Diseases. 2005;5:481–493. doi: 10.1016/S1473-3099(05)70189-4. [DOI] [PubMed] [Google Scholar]
- [8].World Health Organization Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. 2008 can be found at http://www.who.int/tb/features_archive/xdr_mdr_policy_guidance/en/index.html. [PubMed]
- [9].World Health Organization Expert Group Report. Molecular Line Probe Assays for Rapid Screening of Patients at Risk of Multi-Drug Resistant Tuberculosis (MDR-TB) 2008 can be found at http://www.who.int/tb/features_archive/expert_group_report_june08.p df.
- [10].Hofmann-Thiel S, van Ingen J, Feldmann K, Turaev L, Uzakova GT, Murmusaeva G, van Soolingen D, Hoffmann H. European Respiratory Journal. 2009;33:368–374. doi: 10.1183/09031936.00089808. [DOI] [PubMed] [Google Scholar]
- [11].Rinder H, Mieskes KT, Loscher T. Int. J. Tuberc. Lung Dis. 2001;5:339–345. [PubMed] [Google Scholar]
- [12].Sauer-Budge AF, Mirer P, Chatterjee A, Klapperich CM, Chargin D, Sharon A. Lab on a Chip. 2009;9:2803–2810. doi: 10.1039/b904854e. [DOI] [PubMed] [Google Scholar]
- [13].Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers JP. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Analytical Chemistry. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- [15].Pal R, Yang M, Lin R, Johnson BN, Srivastava N, Razzacki SZ, Chomistek KJ, Heldsinger DC, Haque RM, Ugaz VM, Thwar PK, Chen Z, Alfano K, Yim MB, Krishnan M, Fuller AO, Larson RG, Burke DT, Burns MA. Lab on a Chip. 2005;5:1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- [16].Blazej RG, Kumaresan P, Mathies RA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- [18].Ioannidis P, Papaventsis D, Karabela S, Nikolaou S, Panagi M, Raftopoulou E, Konstantinidou E, Marinou I, K. S. J. Clinical Microbiol. 2011;49:3068–3070. doi: 10.1128/JCM.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Williams DL, Spring L, Collins L, Miller LP, Heifets LB, Gangadharam PRJ, Gillis TP. Antimicrobial Agents and Chemotherapy. 1998;42:1853–1857. doi: 10.1128/aac.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Williams DL, Waguespack C, Eisenach K, Crawford JT, Portaels F, Salfinger M, Nolan CM, Abe C, Stichtgroh V, Gillis TP. Antimicrobial Agents and Chemotherapy. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rhee M, Burns MA. Lab on a Chip. 2008;8:1365–1373. doi: 10.1039/b805137b. [DOI] [PubMed] [Google Scholar]
- [22].Shaikh KA, Ryu KS, Goluch ED, Nam JM, Liu JW, Thaxton S, Chiesl TN, Barron AE, Lu Y, Mirkin CA, Liu C. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9745–9750. doi: 10.1073/pnas.0504082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F. Journal of Molecular Biology. 1999;292:251–262. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- [24].Pingle M, Rundell M, Das S, Golightly LM, Barany F. Methods in Molecular Biology. 2010;632:147–157. doi: 10.1007/978-1-60761-663-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Das S, Pingle MR, Muñoz-Jordán J, Rundell MS, Rondini S, Granger K, Chang GJ, Kelly E, Spier EG, Larone D, Spitzer E, Barany F, Golightly LM. Journal of Clinical Microbiology. 2008;46:3276–3278. doi: 10.1128/JCM.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng YW, Shawber C, Notterman D, Paty P, Barany F. Genome Research. 2006;16:289–292. doi: 10.1101/gr.4181406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Favis R, Day JP, Gerry NP, Phelan C, Narod S, Barany F. Nature Biotechnology. 2000;18:561–564. doi: 10.1038/75452. [DOI] [PubMed] [Google Scholar]
- [28].Witek MA, Llopis SD, Wheatley A, McCarley RL, Soper SA. Nucleic Acids Research. 2006;34 doi: 10.1093/nar/gkl146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu F, Datta P, Wang H, Gurung S, Hashimoto M, Wei S, Goettert J, McCarley RL, Soper SA. Analytical Chemistry. 2007;79:9007–9013. doi: 10.1021/ac7016597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y, Vaidya B, Farquar HD, Stryjewski W, Hammer RP, McCarley RL, Soper SA, Cheng YW, Barany F. Anal. Chem. 2003;75:1130–1140. doi: 10.1021/ac020683w. [DOI] [PubMed] [Google Scholar]
- [31].Situma C, Wang Y, Hupert M, Soper SA. Abstr. Pap. Am. Chem. Soc. 2004;228:U118–U118. [Google Scholar]
- [32].Situma C, Wang Y, Hupert M, Barany F, McCarley RL, Soper SA. Anal. Biochem. 2005;340:123–135. doi: 10.1016/j.ab.2005.01.044. [DOI] [PubMed] [Google Scholar]
- [33].McCarley RL, Vaidya B, Wei SY, Smith AF, Patel AB, Feng J, Murphy MC, Soper SA. J. Am. Chem. Soc. 2006;127:842–843. doi: 10.1021/ja0454135. [DOI] [PubMed] [Google Scholar]
- [34].Ford SM, Davies J, Kar B, Qi SD, McWhorter S, Soper SA, Malek CK. J. Biomech. Eng.-Trans. ASME. 1999;121:13–21. doi: 10.1115/1.2798035. [DOI] [PubMed] [Google Scholar]
- [35].Zhang W, Lin S, Wang C, Hu J, Li C, Zhuang Z, Zhou Y, Mathies RA, Yang CJ. Lab Chip. 2009;9:3088–3094. doi: 10.1039/b907254c. [DOI] [PubMed] [Google Scholar]
- [36].Hashimoto M, Chen PC, Mitchell MW, Nikitopoulos DE, Soper SA, Murphy MC. Lab on a Chip. 2004;4:638–645. doi: 10.1039/b406860b. [DOI] [PubMed] [Google Scholar]
- [37].Chen PC, Nikitopoulos DE, Soper SA, Murphy MC. Biomedical Microdevices. 2008;10:141–152. doi: 10.1007/s10544-007-9119-6. [DOI] [PubMed] [Google Scholar]
- [38].Luo JY, Bergstrom DE, Barany F. Nucleic Acids Research. 1996;24:3071–3078. doi: 10.1093/nar/24.15.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]