Although fluorine is not a naturally occurring component of genetic material, fluorine-containing drugs play roles in many medical applications.[1] Fluorine is highly electronegative and 2′-deoxy-2′-fluoro analogs of nucleosides adopt C3′-endo conformations characteristic of the sugars in RNA helices. The 2′-F substitution was first introduced in ribozymes[2] and then evaluated in 1993 in the context of antisense oligonucleotides.[3] Macugen (pegaptanib), an oligonucleotide therapeutic agent substituted with 2′-F pyrimidines, has been approved by the US FDA. It is an RNA aptamer that selectively antagonizes vascular endothelial growth factor (VEGF) and is used clinically in treatment of exudative (wet) age-related macular degeneration.
RNA interference (RNAi) is a mechanism for regulation of gene expression that operates in organisms ranging from plants to flies to human. Synthetic duplexes, called short interfering RNAs (siRNAs), enter the RNAi pathway and can be used to silence expression from virtually any gene. As part of a protein complex called RISC, siRNA recognizes and cleaves mRNA strands complementary to the “antisense” or “guide” strand of the siRNA. The mRNA is cleaved between the nucleotides paired to bases 10 and 11 of the siRNA guide strand.[4] With molecular weights in the range 13 kDa and with about 40 negatively charged phosphates, siRNAs are not typical “drug-like” molecules. Native oligonucleotides are rapidly degraded in serum and are not readily taken up by cells. Chemical modification can impart “drug-like” properties to siRNA,[5] but these modifications may also inhibit various steps in the process that leads to gene silencing.[6]
In cell culture, siRNAs substituted with 2′-F have activity similar or superior to unsubstituted controls independent of position or strand indicating that the modification is well tolerated by RISC.[7–9] Although siRNAs modified with 2′-F clearly have greater stability toward nucleases and a prolonged half-life in human plasma compared to 2′-OH containing siRNAs[7,10,11] and are less immunostimulatory than unmodified siRNA,[12,13] conflicting results have been obtained when these modified siRNAs were evaluated in vivo. The increase in stability did not translate into enhanced or prolonged reduction of target gene expression in mice in two separate studies.[10,14] In contrast, in a study in which 2′-F-modified siRNA and target HBV DNA were co-injected in a liposome particle, the modified siRNA was significantly more potent than unmodified siRNA.[11] To resolve these discrepancies, we set out to evaluate the properties of the 2′-F-modified siRNA in vitro and in vivo in an established model system targeting the endogenously expressed coagulation factor VII.
The siRNAs A and B were designed to target Factor VII (FVII) mRNA; this siRNA sequence was previously shown to effectively inhibit Factor VII expression in rodent and primate animal models.[15,16] Factor VII is a blood clotting factor and is ideal for evaluating the efficacy of siRNAs as it is a secreted protein readily measured in serum. In addition, the protein has a short half-life, so silencing of translation can be measured with minimal lag. An siRNA duplex with 2′-F at all pyrimidines (Table 1, siRNA B) was synthesized using previously reported methods (see the supporting information file). The 2′-F modification imparted significant thermal stability to the siRNA duplex. The melting temperature (Tm) of siRNA B was almost 15°C higher than that of the unmodified duplex (Table 1). siRNA B was significantly more stable when incubated in serum than unmodified siRNA A (Suppl. Figure S1). The unmodified siRNA was completely degraded within 4 hours, whereas the 2′-F-modified siRNA had a t1/2 greater than 24 h. The 2′-F-modified siRNA was not immunostimulatory in an assay in human peripheral blood mononuclear cells: The unmodified siRNA stimulated production of INFα and TNFα; the 2′-F siRNA did not (Suppl. Figure S2).
Table 1.
siRNA sequences and chemical modifications used in the in vivo FVII silencing study.a
| siRNA | Sense strand (5′ to 3′) Antisense strand (5′ to 3′) |
Modification | Tm [°C] |
|---|---|---|---|
| A | Unmodified | 71.8 | |
| B | 86.2 | ||
| C | 80.0 | ||
| D | 87.1 | ||
| E | >100.0 | ||
| F | 83.0 | ||
| G | 91.0 | ||
| H | ~94.0 |
The modifications in each strand are indicated by color coding corresponding to that in the “Modification” column".
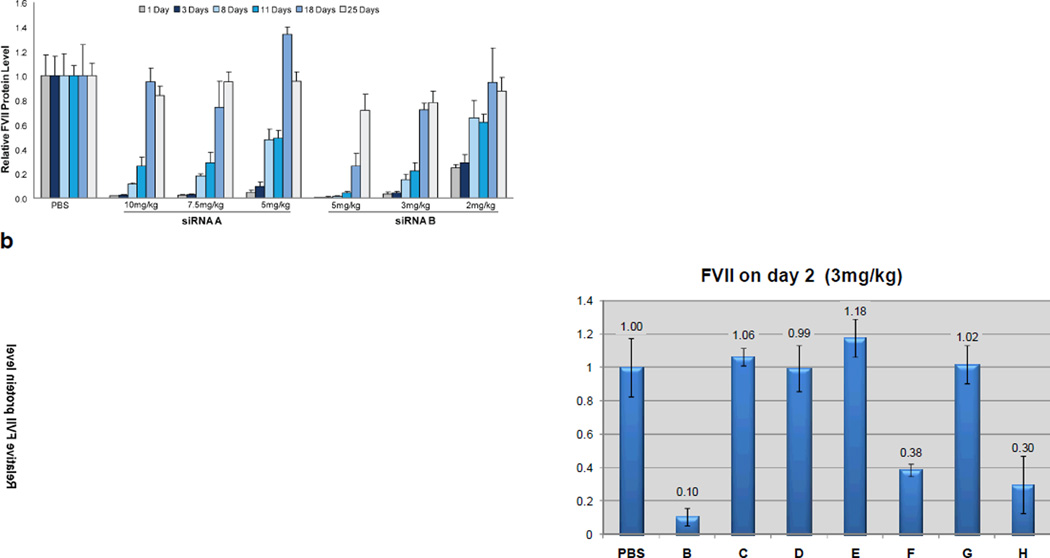
When siRNAs were transfected into HeLa cells that stably express mouse FVII using Lipofectamine 2000, siRNA B proved roughly 2-fold more potent than the unmodified control: The IC50 for siRNA B was 0.50 nM and that of siRNA A was 0.95 nM (data not shown). To evaluate activity of the 2′-F-modified siRNA in vivo, siRNAs were formulated with LNP01, a liver-specific liposome formulation previously described.[16] Silencing was evaluated in mice given a single intravenous injection of formulated siRNA. As shown in Figure 1a, siRNA B was ca. twice as potent as siRNA A.
Figure 1.
In vivo activities of 2′-F-modified siRNA and comparison relative to native RNA and other modifications. a) 2′-F-modified siRNA is approximately twice as potent as the unmodified siRNA in vivo. Mice (n = 5) received a single i.v. dose of LNP01-formulated siRNA A or siRNA B or PBS. FVII protein levels were measured from serum collected at the indicated time points post administration using a chromogenic assay (Coaset Factor VII, DiaPharma Group or Biophen FVII, Aniara Corp.). b) In vivo FVII gene silencing as a function of chemistries using siRNA sequences and chemical modifications described in Table 1.
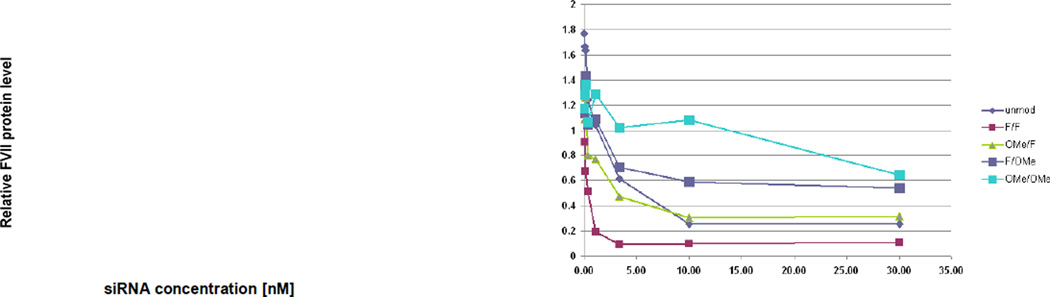
Many examples exist of potent siRNAs containing several types of chemical modifications.[13,17] In some cases stability toward nuclease degradation or potency for these siRNA with several modifications were higher than for the same sequence of siRNA with individual modifications.[11,18] Individual siRNA strands were synthesized with 2′-F, 2′-O-methyl (2′-O-Me), or 2′-O-methoxyethyl (2′-O-MOE) at all pyrimidines or locked nucleic acid (LNA) at uridines as shown in Table 1. Not all pyrimidines were modified in the LNA siRNA as even with only all uridines modified, the duplex had a Tm above 100°C (Table 1). As strand separation is required for RISC activity, exceptionally high duplex stability may be detrimental to siRNA activity. The siRNAs shown in Table 1 were evaluated in the mouse model for silencing of FVII. Mice were given a single intravenous injection of LNP01-formulated siRNA (3 mg/kg) or PBS. As shown in Figure 1b, siRNA B with 2′-F pyrimidines on both sense and antisense strand was the most active. Neither 2′-O-Me nor LNA were tolerated on the antisense strand; 2′-O-MOE was not tolerated on either strand. Results of an in vitro dose-response evaluation of the 2′-F and 2′-O-Me substituted duplexes are shown in Figure 2.
Figure 2.
In vitro activity of 2′-F- and 2′-OMe-modified and unmodified siRNAs in the FVII assay.
To shed light on the increased siRNA activity in vitro and in vivo of 2′-F-modified RNA relative to RNA and other 2′-modifications, we first carried out detailed calorimetric and UV melting experiments with the octamers r(CGAAUUCG) and f(CGAAUUCG) (Table 2). These data revealed, surprisingly, that the higher stability of 2′-F-RNA was primarily the result of a favorable enthalpy. Therefore, the common assumption that conformational preorganization for the target strand and a resultant entropic benefit is the main cause of the superior RNA affinity of the 2′-F modification may not be correct. That the entropic contributions to the pairing stability of RNA and 2′-F-RNA were virtually the same was particularly unexpected given fluorine’s reputation as a poor hydrogen bond acceptor.[19]
Table 2.
Thermodynamic stability and osmotic stressa analysis (Δnw) for RNA and 2′-F-RNA octamers
|
Sequence (r = RNA and f = 2′-F-RNA) |
Tm [°C] (UV mltg.) |
ΔH [kcalmol−1] (DSC) |
ΔH [kcalmol−1] (UV concentration |
ΔS [eu] depend.) |
Δnw (ethylene glycol) |
Δnw (glycerol) |
Δnw (acetamide) |
|---|---|---|---|---|---|---|---|
| 5′-r(CGAAUUCG)-3′ | 34.1 ± 0.6 | 39.2 ± 1.4 | 58.0 ± 9.4 | 189.7 ± 30.6 | 18.8 ± 4.9 | 22.5 ± 6.1 | 37.9 ± 5.8 |
| 5′-f(CGAAUUCG)-3′ | 53.3 ± 0.3 | 53.5 ± 0.7 | 62.3 ± 9.8 | 192.1 ± 30.2 | 1.2 ± 3.8 | 3.0 ± 6.9 | 14.8 ± 3.3 |
Δnw = number of water molecules released upon melting of the duplex (for a discussion of osmotic stress see ref. 24).
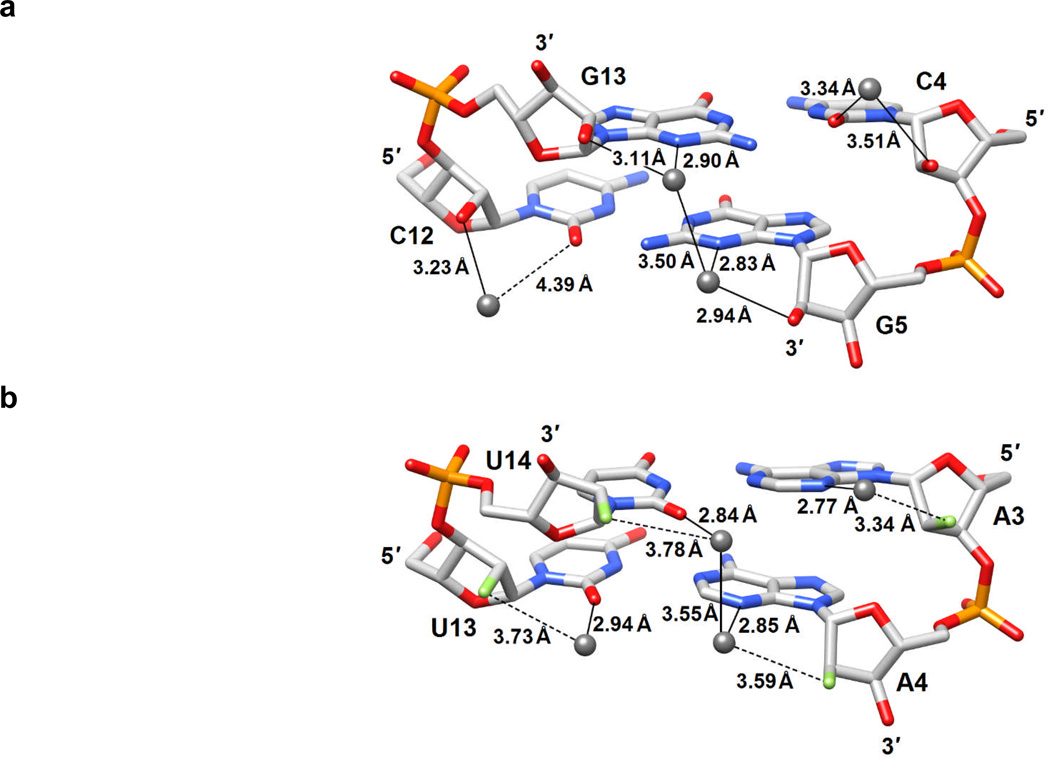
RNA 2′-hydroxyl groups stabilize an intricate water structure in the minor groove, as visualized first in the crystal structure of the duplex [r(CCCCGGGG)]2 (Figure 3a).[20] In principle one would expect this feature to be associated with an entropic penalty. The 2′-O-Me,[21] 2′-O-MOE,[22] and LNA[23] modifications also affect hydration favorably. To analyze the effects of the 2′-F-modification on the minor groove water structure, we determined the crystal structure of [f(CGAAUUCG)]2, the first for any 2′-F-modified nucleic acid, at 1.2 Å resolution. The structure revealed that the fluorine atom at the 2′-position of the sugar moiety does not participate in hydrogen bonds to water molecules associated with either the minor groove base edges or phosphates (Figure 3b). Thus, fluorine was unable to act as a bridgehead in the stabilization of water bridges across the minor groove.
Figure 3.
Differences in the minor groove hydration of a) RNA and b) 2′-F-RNA illustrated by the water structure at individual base-pair steps in the crystal structures of r(CCCCGGGG)]2[20] and [f(CGAAUUCG)]2 (this work). 2′-F atoms are colored in green, water molecules are gray spheres, and hydrogen bonds are thin solid lines.
The structural data were consistent with the outcomes of osmotic stressing experiments[24] that indicated poor hydration of the 2′-F-RNA duplex (Table 2). Considerable amounts of water were released upon melting of the duplex [r(CGAAUUCG)]2, however, in line with the wet minor groove in the crystal structure.[20] Changes in hydration are likely not the only consequence of the 2′-F modification. It is possible that the fluorine substituent polarizes the nucleobase, thereby strengthening Watson-Crick hydrogen bonds and enhancing base stacking. Such effects would reconcile the structural and osmotic stressing data with those from the thermodynamic studies, and could explain the chiefly enthalpic origin of the stability gains afforded by 2′-F-RNA relative to RNA. The data presented here provide evidence that the unique activity displayed by 2′-F-modified siRNAs is mirrored in unique physical attributes such as an unusually “dry” minor groove and a highly favorable enthalpic contribution to duplex formation. A more in-depth analysis of the role of the 2′-substituents in nucleic acid stability will be required to fully explain the in vivo data.
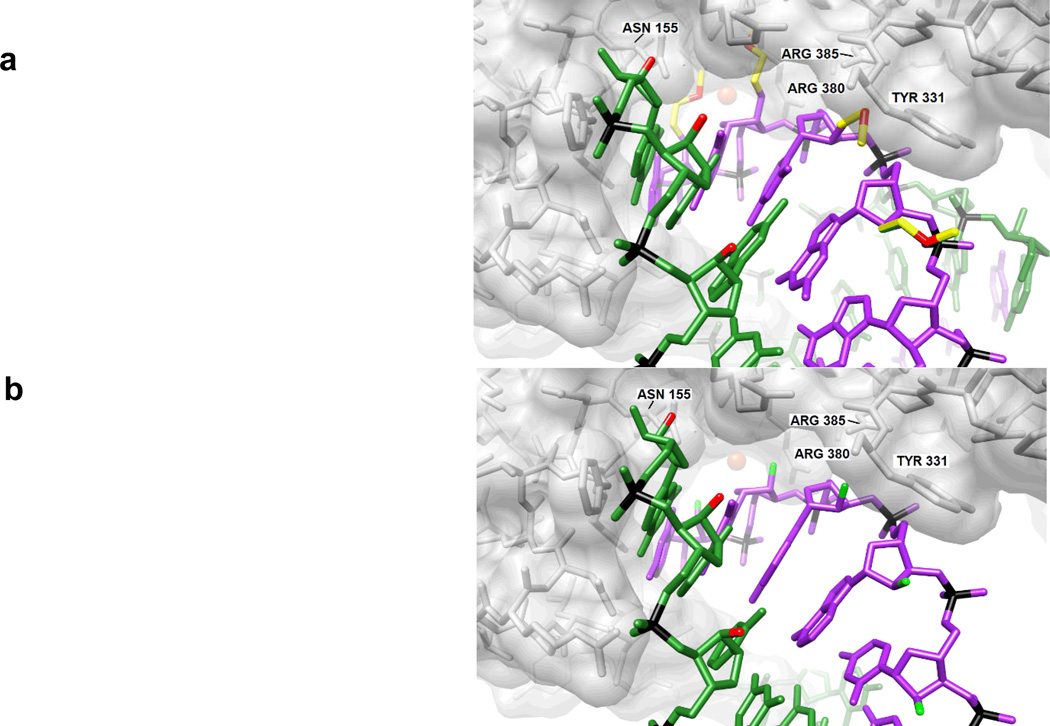
In order to silence gene expression, siRNAs must be loaded into the RISC, the sense strand must be dissociated from the complex, and finally the mRNA target must be recognized and cleaved. The modifications that disrupt siRNA activity may inhibit any of these steps. For example, when designing siRNAs, one walks a thermodynamic affinity tightrope: High affinity is desirable so that target recognition is effective but if the stability is too high, the siRNA duplex may not be dissociated by RISC. It is clear that large groups are not tolerated on the sense strand even though duplex affinity is not compromised.[9] The relatively bulky 2′-O-MOE modification likely compromises binding by proteins involved in the RNAi pathway because of steric clashes (i.e., the PIWI domain; Figure 4a). By comparison, certain unique properties of the 2′-F modification, such as the small size (Figure 4b), high electronegativity paired with hydrophobicity, and the resulting dry minor groove that precludes the need for dehydration upon binding, appear to have beneficial effects in regard to interactions with proteins.
Figure 4.
Interactions between antisense RNA (magenta):mRNA (green) duplexes and Piwi protein. The models of the Piwi complexes with a) 2′-O-MOE RNA:RNA and b) 2′-F-RNA:RNA are based on the crystal structure of the protein from Archaeoglobus fulgidus (AfPiwi) bound to an siRNA-like duplex that mimics the 5′-end of a guide RNA strand bound to an overhanging target mRNA (PDB ID 2bgg).[25] The models were built by superimposing crystal structures of either 2′-O-MOE-RNA[22] or 2′-F-RNA (this work) onto the RNA duplex in the experimental crystal structure. Carbon atoms of 2′-O-MOE substituents are yellow, 2′-F atoms are green, 2′-oxygen atoms are red, and selected Piwi residues are labeled. A metal ion that anchors the 5′-nucleotide of the guide RNA is depicted as a red sphere. There are clashes between 2′-O-MOE substituents and protein atoms.
Overall, the characteristics of the 2′-F modification make it particularly suitable for the design of highly effective siRNAs. Its small size combined with high electronegativity allows for position-independent incorporation into both strands and locks the sugar in the RNA-compatible C3′-endo conformation, respectively. Detailed calorimetric and UV melting experiments revealed that the higher thermal stability of 2′-F-modified duplexes was predominantly due to increased enthalpy rather than entropic effects. We also found that siRNAs modified with 2′-F exhibited increased nuclease stability, significantly reduced immune stimulation in an in vitro model, and, in some cases, improved in vitro and in vivo activity relative to the unmodified control RNA. Fully modified RNAs with purine and pyrimidine 2'-F modifications are being evaluated now and their siRNA activities will be reported in future communications.
Experimental Section
See supporting information for further details.
Footnotes
We are grateful to the US National Institutes of Health for continuous support (R01 GM055237 to M.E. and R01 GM071461 to E.R.). Vanderbilt University is a member institution of LS-CAT at the APS, Argonne, IL. Use of the APS at Argonne National Lab was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.2010
Contributor Information
Muthiah Manoharan, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA), mmanoharan@alnylam.com.
Akin Akinc, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Rajendra K. Pandey, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA)
June Qin, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Philipp Hadwiger, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Matthias John, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Kathy Mills, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Klaus Charisse, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Martin A. Maier, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA)
Lubomir Nechev, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA).
Emily M. Greene, Department of Chemistry, State University of New York at Binghamton, Binghamton, NY 13902 (USA)
Pradeep S. Pallan, Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232-0146 (USA)
Eriks Rozners, Department of Chemistry, State University of New York at Binghamton, Binghamton, NY 13902 (USA).
Kallanthottathil G. Rajeev, Alnylam Pharmaceuticals, 300 Third Street, Cambridge, MA 02142 (USA)
Martin Egli, Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232-0146 (USA), Fax: (+1) (615) 322-7122, martin.egli@vanderbilt.edu, Homepage: http://structbio.vanderbilt.edu/~eglim/.
References
- 1.a) Strunecká A, Patočka J, Connett P. J. Appl. Biomed. 2004;2:141–150. [Google Scholar]; b) Hagmann WK. J. Med. Chem. 2008;51:4359. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- 2.Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Science. 1991;253:314. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Dan Cook P. J. Med. Chem. 1993;36:831. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 4.Pratt AJ, MacRae IJ. J. Biol. Chem. 2009;284:17897. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Manoharan M. Curr. Opin. Chem. Biol. 2004;8:570. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]; b) Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. Nat. Chem. Biol. 2006;2:711. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deleavey GF, Watts JK, Damha MJ. Curr. Prot. Nucleic Acid Chem. 2009;39:16.3.1–16.3.22. doi: 10.1002/0471142700.nc1603s39. [DOI] [PubMed] [Google Scholar]
- 7.a) Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. Biochemistry. 2003;42:7967. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]; b) Chiu YL, Rana TM. RNA. 2009;9:1034. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Antisense Nucleic Acid Drug Dev. 2003;13:83. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]; d) Muhonen P, Tennila T, Azhayeva E, Parthasarathy RN, Janckila AJ, Vaananen HK, Azhayev A, Laitala-Leinonen T. Chem. Biodivers. 2007;4:858. doi: 10.1002/cbdv.200790073. [DOI] [PubMed] [Google Scholar]
- 8.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. J. Med. Chem. 2005;48:901. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 9.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. J. Med. Chem. 2005;48:4247. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 10.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. RNA. 2004;10:766. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, Polisky BA, Zinnen S. Hepatology. 2005;41:1349. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 12.a) Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B. Nat. Biotechnol. 2005;23:1002. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]; b) Sioud M. Eur. J. Immunol. 2006;36:1222. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]; c) Cekaite L, Furset G, Hovig E, Sioud M. J. Mol. Biol. 2007;365:90. doi: 10.1016/j.jmb.2006.09.034. [DOI] [PubMed] [Google Scholar]; d) Sioud M. New Biotechnol. 2010;27:236. doi: 10.1016/j.nbt.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartman G. Nat. Med. 2005;11:263. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 14.Viel T, Boisgard R, Kuhnast B, Jego B, Siquier-Pernet K, Hinnen F, Dolle F, Tavitian B. Oligonucleotides. 2008;18:201. doi: 10.1089/oli.2008.0133. [DOI] [PubMed] [Google Scholar]
- 15.a) Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Bushini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Jayaprakash KN, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher H-P, Zimmerman TS, Langer R, Anderson DG. Nature Biotechnol. 2008;26:561. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, Soutschek J, Vornlocher HP, Manoharan M, Stoffel M, Langer R, Anderson DG, Horton JD, Koteliansky V, Bumcrot D. Nature. 2007;449:745. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, Jayaprakash KN, Jayaraman M, Rajeev KG, Manoharan M, Koteliansky V, Rohl I, Leshchiner ES, Langer R, Anderson DG. Molecular Therapy, 2009;17:872. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koller E, Propp S, Murray H, Lima W, Bhat B, Prakash TP, Allerson CR, Swayze EE, Marcusson EG, Dean NM. Nucleic Acids Res. 2006;34:4467. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dande P, Prakash TP, Sioufi N, Gaus H, Jarres R, Berdeja A, Swayze EE, Griffey RH, Bhat B. J. Med. Chem. 2006;49:1624. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 19.Dunitz JD. ChemBioChem. 2004;5:614. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 20.Egli M, Portmann S, Usman N. Biochemistry. 1996;35:8489. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 21.Auffinger P, Westhof E. Angew. Chem. Int. Ed. 2001;40:4648. doi: 10.1002/1521-3773(20011217)40:24<4648::aid-anie4648>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 22.Teplova M, Minasov G, Tereshko V, Inamati G, Cook PD, Manoharan M, Egli M. Nat. Struct. Biol. 1999;6:535. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 23.Egli M, Minasov G, Teplova M, Kumar R, Wengel J. Chem. Comm. 2001:651. [Google Scholar]
- 24.Rozners E, Moulder J. Nucleic Acids Res. 2004;32:248. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker JS, Roe SM, Barford D. Nature. 2005;434:663. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]