Abstract
Aggregation of amyloid-β peptides (Aβ) into fibrils is the key pathological feature of many neurodegenerative disorders. Typical drugs inhibit Aβ fibrillation by binding to monomers in 1:1 ratio and display low efficacy. Here, we report that model CdTe nanoparticles (NPs) can efficiently prevent fibrillation of Aβ associating with 100–330 monomers at once. The inhibition is based on the binding multiple Aβ oligomers rather than individual monomers. The oligomer route of inhibition is associated with strong van der Waals interactions characteristic for NPs and presents substantial advantages in the mitigation of toxicity of the misfolded peptides. Molar efficiency and the inhibition mechanism revealed by NPs are analogous to those found for proteins responsible for prevention of amyloid fibrillation in human body. Besides providing a stimulus for finding biocompatible NPs with similar capabilities, these data suggest that inorganic NPs can mimic some sophisticated biological functionalities of proteins.
Keywords: amyloid beta-peptides, fibril, nanoparticles, inhibition, nanoscale assemblies
Aβ fibrils (or nanowires, NWs) have been implicated in many neurodegenerative disorders such as Alzheimer's disease.[1] Both soluble oligomers and mature fibrils from Aβ are neurotoxic and cause death of brain cells.[1–3] The inhibition of Aβ assembly has been considered as the primary therapeutic strategy for the neurodegenerative diseases. Short peptides, in particular KLVFF residues, have been designed to interfere with β-structured aggregation via hydrophobic interactions.[4,5] Proteins capable of binding to Aβ also have an inhibitory effect on Aβ fibrillation.[4,6] Recently, antibodies against Aβ as well as surface-modified proteins with Aβ-residues have been applied to prevent Aβ aggregation and reduce the toxicity.[4,7] Although peptide or protein analogs with specific binding sites might have the primary importance for fundamental studies and diagnostics of the neurodegenerative disorders, therapeutic agents have not been developed within this strategy. Some of the problems are the blood-brain barrier permeability,[4,8] complexity of their synthesis, low in vivo stability, and low efficacy which can be attributed in part to the fact that these agents are designed bind to peptide monomers in 1:1 ratio.[1d,4,5,9]
Considering the fact that many NPs are capable of self-organization into similar structures as Aβ peptides,[10] it is intriguing to investigate the nexus of self-organization processes between NPs and peptides especially because assembly behavior of NPs reveal similarities with those of biological species. Such studies have mostly fundamental importance but may also reveal new aspects of NPs toxicology and provide alternative methodology for preventing the agglomeration of Aβ peptides.[11] Although not all NPs are biocompatible, they might worth some consideration as therapeutic agents because they are easy to synthesize and have great in-vivo stability. In this respect, the non-biodegradable nature of inorganic NPs can be of potential advantage and can help to fully utilize their activity over long period of time.
The existing data on the effects of both organic and inorganic NPs on peptide assembly are controversial. Overall, the presence of NPs has typically promoted aggregation of Aβ, which was explained in terms of condensation-ordering mechanism.[12] Since the fibrillation occurs by nucleation-dependent kinetics, the increased local concentration of peptides in the vicinity of NPs due to electrostatic attraction greatly accelerates the fibril formation. For example, 70 nm and 200 nm polymeric NPs of N-isopropylacrylamide and N-tert-butylacrylamide, 16 nm cerium oxide NPs, 16 nm polymer-coated quantum dots, and carbon nanotubes catalyzed the fibrillation of amyloid protein of β2-microglobulin.[12b] Similarly, the strong absorption capacity of 20 nm TiO2 NPs to the amyloid peptide promoted fibril formation.[12c] Also, it has been further reported that biological molecules,[13a,b] large colloidal particles,[13c] liquid-air, and liquid-solid interfaces[13d,e] promote the fibril formation as well. At the same time, some 40 nm polymeric NPs have been found to slow down the rate of Aβ fibrillation by depleting the amount of free monomeric peptides, although the fibril formation still could not be prevented.[14a] Surface-modified 14 nm nanogels,[14b] micelles[14c] and fullerene[14d] were able to alter the conformation of the peptides and reduce the toxic activity. 3–5 nm N-acetyl-L-cysteine-capped quantum dots also displayed inhibitory activity towards fibrillation which was attributed to the formation of hydrogen bonds,[14e] albeit the same bonds are also involved in peptide-NP systems which promote assembly of amyloid peptides. The formation of dense NP coatings on fibrils was observed and the computer model of a kinetic model based on nucleation and growth inhibition was discussed.[14e]
Focusing now on the finding an efficient fibrillation inhibitor as the most challenging task, one needs to admit that there is very little clarity what forces to engage and what nano- or molecular scale species would be preferential for this purpose. As a new step in this direction, we report here the strong inhibition of Aβ fibrillation by TGA (thioglycolic acid)-stabilized CdTe NPs. These NPs were originally selected because they have general resemblance in terms of the size (Figure 1A), charge, and association behavior to some proteins.[15] We reasoned that it is possible that a minor fraction of NPs would have proper local stereochemistry to specifically self-assemble with (mis)folded peptides that can either accelerate or frustrate the fibrillation. The eventual mechanism of NP-peptide interaction was found to be markedly different from the one we envisioned and from those considered before for NPs[12,14] but very similar to that found for proteins in the past to inhibit peptide fibrillation.[6]
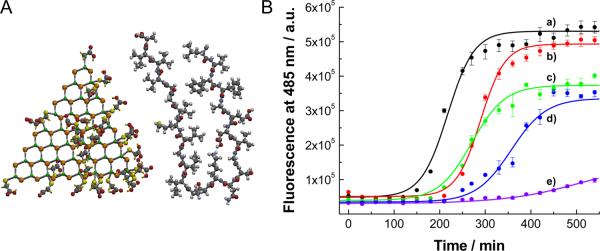
Figure 1.
(A) Molecular structure of CdTe NPs and Aβ1-42 peptide. Some of the molecules of TGA adsorbed to the surface of CdTe are removed for clarity. Peptide is folded in the configuration characteristic for fibrils. Molecular modeling was carried out by SPARTAN. (B) Kinetics of Aβ1-40 fibrillation with and without CdTe NPs. Time-dependent ThT fluorescence was monitored at 485 nm with excitation wavelength of 450 nm. The molar ratio of [CdTe]/[Aβ1-40] was varied from 0 to 0.005: (a) 0, (b) 0.001, (c), 0.005, (d), 0.01 and (e) 0.05. Fluorescence intensity of ThT at 485 nm is proportional to the amount of fibrils.
To examine the influence of CdTe NPs on the fibril formation of amyloid-β peptides consisting of 40 residues (denoted as Aβ1-40 hereafter), incubation solutions of the Aβ1-40 with and without NPs were prepared. The kinetics of fibrillation can be monitored by a dye-binding assay with thioflavin T (ThT), the fluorescent spectrum of which can be altered with the growth of fibrils (Figure 1B).[12b,c,14a] As for the neat peptide (Figure 1B trace a), the fibrillation follows a conventional nucleated growth mechanism, that can be described by a lag phase, a rapid exponential growth, and a final equilibrium state. Accordingly, the experimental data were fit by a sigmoidal equation (solid lines) to extract kinetic constants.[16]
When the NPs were introduced (Figure 1B traces b–e), the fluorescent intensity at saturation gradually decreased with the increase of the [CdTe]/[Aβ1-40] molar ratio, indicating consistent inhibition of fibrillation in a dose-dependent manner by CdTe NPs. At the same time, the lag phase was extended from 159.5 min for pure peptide to 236.3 min, 197.5 min and 279.9 min for [CdTe]/[Aβ1-40] of 0.001, 0.005, and 0.01, respectively. The lag time for the case of [CdTe]/[Aβ1-40] = 0.05 (Figure 1B trace e) could not be determined because fibrillation was completely inhibited. The slight increase in the fluorescence intensity from [CdTe]/[Aβ1-40] of 0.05 can be most likely ascribed to the formation of NP-peptide agglomeration, the structure of which will be presented later. The slope of the sigmoidal curve is proportional to the elongation rate and it also rapidly declines with the amount of NPs. These findings show that both nucleation and the growth of fibrils were considerably inhibited with CdTe NPs. Overall, the amount of CdTe NPs 2–3 orders of magnitude smaller than that of peptide was sufficient to inhibit Aβ1-40 fibrillation. Comparatively, in the case of small molecules and short-peptide inhibitors, equivalent or excess amount of agents is generally required to prevent the fibrillation.[1d,4,5,9]
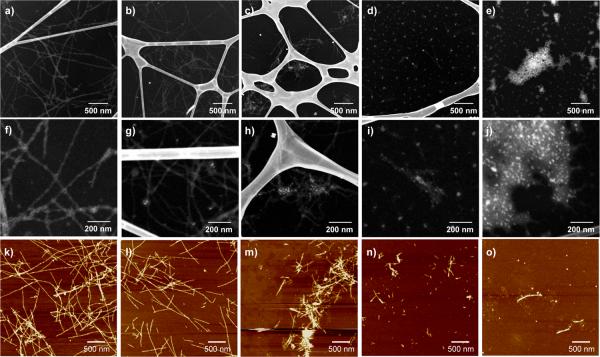
The inhibited fibrillation by CdTe NPs was further confirmed by transmission electron microscopy using the high angle annular dark field (HAADF) technique (Figure 2). The images show the gradually shortening and disappearance of fibrils as the [CdTe]/[Aβ1-40] ratio increases, confirming the fluorescence assay data. All the samples were prepared after the ThT fluorescence had reached a plateau to ensure the visualization of fibrils in equilibrium. In the case of neat Aβ1-40 without CdTe NPs (Figure 2a, 2f), Aβ1-40 self-organized into well-defined fibril structures with lengths more than several micrometers. The diameter of fibrils varied from around 10 nm to 40 nm, which can be ascribed to the lateral association of several fibrils.[1] No short fibrils could be found on the entire TEM grid, which is indicative of full fibrillation.
Figure 2.
TEM (a – j) and AFM (k – o) images of Aβ1-40 incubated with and without CdTe NPs. The molar ratios of [CdTe]/[Aβ1-40] were (a, f, k) 0, (b, g, l) 0.001, (c, h, m) 0.005, (d, i, n) 0.01, and (e, j, o) 0.05. In TEM images, NPs appeared as bright spots that can be easily distinguished from the fibrils even for small concentrations of NPs. The Z-contrast nature of the images enables the direct visualization of the peptide structures without a staining process.
For [CdTe]/[Aβ1-40] = 0.005 (Figure 2c, 2h), the fibrils became considerably shorter with an average length of about 400 nm; the bright spots of NPs, can be seen inside the fibrils. Further increases in the molar ratio resulted in the additional inhibition on the fibril formation. For [CdTe]/[Aβ1-40] = 0.01 (Figure 2d, 2i), pseudo-spherical aggregations were mainly observed, although we still found short fibril-like aggregates with NPs in them with a length less than 100 nm. The fairly large diameter (~ 18 nm) of the spherical aggregates in the images indicated that individual CdTe NPs (~ 3.5 nm) were covered with thick layer of peptide molecules. No uncoated NPs have been observed and, on many occasions more than one NP is included in the larger spherical agglomerates (see Figure S1). Considering [CdTe]/[Aβ1-40] = 0.01 as a threshold for fibrillation inhibition, one can estimate that one NP binds at least 100 monomers.
When the molar ratio of [CdTe]/[Aβ1-40] reached to 0.05, we could not find any indication of fibril formation (Figure 2e, 2j) and all the particles and peptides formed spherical aggregates. The detailed structures of NP-peptide aggregates were further analyzed by high-resolution (HR) TEM images, which clearly showed the lattice structure of NP inside the agglomerates (see Figure S1). AFM results confirm the NP effect on the self-organization of Aβ1-40. Exactly the same conclusions about drastic shortening of the fibrils and dominant formation of spherical agglomerates after [CdTe]/[Aβ1-40] = 0.01 threshold can be reached by analyzing Figure 2k – 2o. From the diameter of the spheroid obtained from both TEM and AFM data, one can also calculate that ca.330 Aβ1-40 monomers are bound in one spheroid (see Figure S2, S3 for details). Since TEM data indicate that some spheroids can contain 2–3 NPs, this number matches quite well with the estimate obtained above.
Now it is important (1) to rationalize why the NPs that we use here give results different from the studies conducted before with other NPs[12] and (2) to understand better the mechanism behind it which might be different than those expected previously.[12,14] Monomeric Aβ1-40 initially self-associates into disorganized oligomers, which then further assemble into β-structured aggregates termed as protofibrils.[1,4,14a] There is a kinetic equilibrium among monomers, oligomers, and protofibirls.[1,4,14a] Once the protofibrils are formed, they can act like templates for the further growth of fibrils by allowing rapid association of both monomers and protofibrils, resulting in the sigmoidal growth as in the ThT assay (Figure 1B). NPs can potentially interfere with any stage of this process. For instance, concomitant studies with molecular modeling can suggest that association of individual peptides with NPs via a thiol bond or hydrogen bonding can result in scrambling of monomeric peptide (see Figure S2).
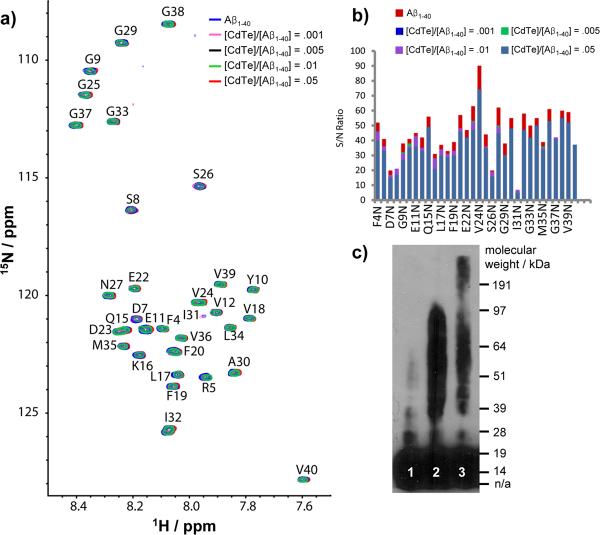
The interactions of NPs and Aβ1-40 and inhibition processes were investigated by several complementary microscopy and spectroscopy techniques. As such, SOFAST-HMQC NMR spectra of 15N-labeled Aβ1-40 were obtained for [CdTe]/[Aβ1-40] mole ratios of 0.0, .001, .005, .01 and .05 (Figure 3a). Conditions when dissolved Aβ1-40 is primarily monomeric and stable were intentionally used (see Experimental section). Intensity of NMR spectra was unchanged for many weeks which is indicative of negligible conversion of the monomer species to larger aggregates.[17] Since the observable peaks in the NMR spectra come exclusively from the Aβ1-40 monomer,[17] perturbations in the NMR spectra upon addition of CdTe NPs provide a sensitive test for the interaction of CdTe NPs with Aβ1-40 monomers. Interestingly, significant changes in chemical shift were not observed for any residues upon the addition of CdTe NPs (Figure 3a). While the long rotational correlation time of the Aβ1-40-CdTe NP complex most likely broadens the signal of the complex beyond detection, the equilibrium between the Aβ1-40-CdTe NP complex and the Aβ1-40 monomer can be monitored by measuring the signal intensity and line-width, which are reflective of the exchange rate between the monomer and the Aβ1-40-CdTe NP complex. Changes in line-width suggestive of chemical exchange on the μs-ms timescale were not observed after the addition of CdTe NPs. The intensities of the signals were practically unaffected by the presence of CdTe NPs, as well. Only a slight and mostly uniform decrease in the signal intensity (~20%) was observed as the CdTe NPs concentration is increased (Figure 3b), setting an upper bound on the degree of the depletion of the monomer concentration by the NPs. The lack of a significant interaction of CdTe NPs with Aβ1-40 monomer indicates that depletion of the monomeric peptide[14a] is not a likely source for the strong inhibition of Aβ1-40 observed in Figure 1 and 2.
Figure 3.
NMR spectra (a, b) and Western blot (c) with and without CdTe NPs. (a) 2D SOFAST-HMQC spectra of freshly dissolved Aβ1-40 with increasing amounts of CdTe NPs at 10° C; (b) The change in the signal-to-noise ratio of Aβ1-40; (c) Western blot of freshly prepared Aβ1-42 (lane 1), 1 day incubated Aβ1-42 in the absence (lane 2) and presence (lane 3) of CdTe NPs. In Western blotting, the molar ratio of [CdTe]/[Aβ1-42] was 0.05. Notations for 1-letter abbreviation of each residue can be found in SI Text.
The NMR, AFM, TEM, and fluorescence spectroscopy data suggest that the formation of the CdTe+Aβ1-40 spheroids (Figure 2) is due to association of oligomers rather than monomers with NPs. This fact can be further confirmed by Western Blot analysis (Figure 3c). The freshly dissolved peptide consists mainly of monomers (lane 1, Figure 3c), which aggregate into Aβ1-42 oligomers with a molecular weight up to ~ 100 kDa (lane 2, Figure 3c) after incubation. When the same process occurred in presence of CdTe NPs, the oligomeric bands became noticeably weaker but considerably elongated above 100 kDa (lane 3, Figure 3c). Since the molecular weight of CdTe NP having a diameter of 3.5 nm can be roughly estimated as 80 kDa, the elongated band indicates the binding of CdTe NPs to oligomers.
The distinction between binding to oligomers and monomers as the mechanism of inhibition is quite significant for several reasons. First of all, the difference between binding modalities give marked difference in efficiency of fibrillation inhibition. Secondly, the oligomers represent the most neurotoxic species among other Aβ1-40 agglomerates and their blocking into NP complexes is expected to have much greater biological effect.[1,3]
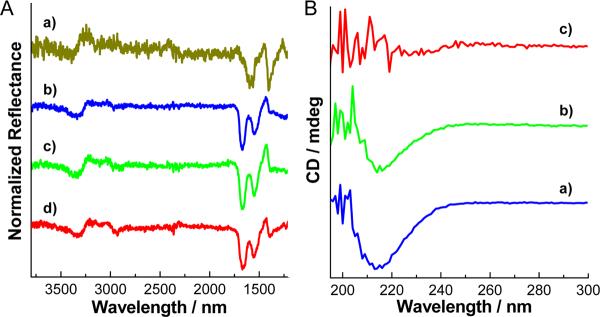
To understand better the molecular reasons for preferential binding of NPs to oligomers and not to monomers, it is instructive to discuss the interactions between them that may include hydrophobic, electrostatic, van der Waals (vdW) interactions, and hydrogen bonding.[15] Hydrophobic interactions between the monomers are known to be the reason for the oligomerization process of the peptide.[1,3a,4] They certainly play a role in stabilization of NP-Aβ1-40 spheroids, however, hydrophobic forces between NPs and oligomers cannot be strong because TGA coating is highly hydrophilic.[18] Interestingly, electrostatic interactions are actually acting against the association of NPs and peptides because they are both negatively charged, with zeta potentials of −31.2 and −16.0 mV, respectively. To investigate the hydrogen bonding that can potentially be the driving force for the assembly,[14e] infrared (IR) spectra were obtained after 1 day incubation (Figure 4A). The vibrational bands of TGA on NPs, such as COO− stretching vibrations at 1585 and 1406 cm−1, and those of peptide, such as amide I (C=O stretching) at 1670 cm−1 and amide II (N-H bending) bands at 1551 cm−1, remain remarkably unchanged in the NP-oligomer complex (in Figure 4A 1670 cm−1 peak slightly moved to 1659 cm−1). The N-H stretching peak attributed to NH2 groups in peptides also maintained at 3323 cm−1 in neat Aβ1-40 and at 3313 cm −1 in [CdTe]/[Aβ1-40] = 0.05. Importantly, no considerable broadening or peak shift typical of any bands that might be responsible for hydrogen bonding interactions between peptide and NPs, in particularly, for COOH groups in TGA on NP surface can be observed.[19] Overall, we do not see any sufficient IR evidence of extensive hydrogen bonding between NPs and peptide molecules. The same conclusion can also be reached based on NMR spectra because hydrogen bonding between monomers and NPs must strongly shift the peak positions and coupling constants for the same groups. Considering that there is already strong H-bonding in the oligomers, the TGA molecules are likely to have difficulties competing with these bonds strengthened by specific conformation of the peptide.
Figure 4.
(A) FT-IR spectra of Aβ1-42 with and without CdTe NPs. (a) CdTe NPs, (b – d) Aβ1-42 with CdTe NPs. The molar ratios of [CdTe]/[Aβ1-42] were (b) 0, (c) 0.01, and (d) 0.05. (B) CD spectra of Aβ1-42 with and without CdTe NPs. The molar ratios of [CdTe]/[Aβ1-42] were (a) 0, (b) 0.01, and (c) 0.05. Both FT-IR and CD spectra were obtained after 1 day incubation. In Figure 4B trace c, CD signal below ~ 215 nm became noisy by the strong absorbance of CdTe NPs.
The conundrum behind the formation of NP-oligomer complexes can be explained by the presence of strong vdW interactions. They are nonspecific short-range, but powerful interactions scaling with molecular weight of the interacting species. The latter feature can explain a number of observations in the context of this and other studies. The preferential binding of NPs to oligomers (Figure 2 and 3), while having negligible association with the monomers (Figure 3a, b) is the direct consequence of the fact that vdW forces are much stronger between NPs and Aβ1-40 oligomers than with monomers. Also, the powerful vdW interactions associated with high electron density on atoms of CdTe NPs differentiate them from organic nanoparticles studied before[12b,14a–d] and, therefore, provide the atomic basis for the dissimilar effect on fibrillation. The energy of vdW interaction of Aβ with CdTe NPs is as much as 3.5 times higher than that with typical organic NPs (see Supporting Information for details), and so, instead of accelerating self-assembly of peptides by condensation-ordering mechanism,[12] they inhibit it by immobilizing oligomers in the spheroids around NPs. Also note that CdTe NPs are tetrahedral and comparable in size to the peptides (Figure 1A). Strong attraction between elongated oligomers/protofibrils and NPs will result in their “wrapping” around the edges of the semiconductor core and concomitant distortion of the amyloid pattern. To verify that, the secondary structure of oligomers after the formation of spheroids was examined by circular dichroism (CD) spectroscopy very sensitive to conformational changes. Aβ1-42 oligomers displayed one negative peak at 215 nm corresponding to β-sheet packing of the peptides.[20] It completely disappeared with the addition of CdTe NPs (Figure 4B), indicating scrambling of the peptide chains.
It is also instructing to compare this inhibitory activity of CdTe NPs to that of proteins known for protective function against AD. Human serum albumin (HSA) is one of the most potent endogenous inhibitors of amyloid fibrillization acting as an “external sink” and does not need to cross the blood-brain barrier. HSA forms complexes with Aβ oligomers but does not interact appreciably with monomers.[6a,b] One HSA associates with ca. 20 monomers and the ratio of [HSA]/[Aβ] ~ 0.056 effectively prevents fibrillation.[6a] This is substantially smaller than what was observed for NPs (Figure 1 and 2). Also note that this is much more efficient than regular small AD drugs that are typically effective in the equivalent or excess ratio, i.e. [small drug]/[Aβ] ≥ 1, and made to bind to peptide monomers. Unlike NPs, however, HSA takes advantage of mostly hydrophobic interactions and binds to the exposed hydrophobic domain of the oligomers, while other interactions play mostly secondary role. The size of HSA is ~ 3.3 nm,[21] which is slightly larger than CdTe NPs used here. Another inhibitory protein, apolipoprotein E3 (apoE3), prevents fibrillation at a ratio of [apoE3]/[Aβ] = 0.001.[6d] The mechanism is associated with binding to disorganized (pre-nucleus) oligomers and not to monomers; immobilization of the monomer in this state prevents their further crystallization, which is analogous to that proposed here for NPs. The nature of interactions between apoE3 and Aβ is not known.
Overall, it is quite clear that CdTe NPs reveal surprisingly many similarities in the mechanism of the inhibition to that of proteins, although originating from different intermolecular interactions. Despite the fact that CdTe NPs are cytotoxic and cannot be used in vivo, this model demonstrates that NPs can reach equal or better efficiency of fibrillation inhibition than the best known proteins. This work also offers a blueprint for nanoscale engineering of NPs from biocompatible materials with similar properties. Biocompatible NP system mimicking the structural characteristics of CdTe NPs, particularly those with a sharp faceted structure, can be suggested to replace toxic CdTe NPs in the process of inhibiting amyloid fibrillation.
Experimental Section
Aβ1-40 was purchased from Invitrogen and used as received. Stock solution of Aβ1-40 was typically prepared by dissolving the lyophilized peptide into dimethyl sulfoxide (DMSO) to a final concentration of 1 mM. For the fibrillation, the stock solution of Aβ1-40 was added to phosphate buffered saline (PBS buffer, pH 7.4) to yield a 25 μM Aβ1-40 solution. Thioglycolic acid (TGA)-stabilized CdTe NPs were prepared as in our previous report.[15] UV-Vis and fluorescence spectra of CdTe NPs are shown in Figure S4, and they are slightly varied with each synthesis. From the UV-Vis absorption, the size of NPs was estimated about 3.5 nm.[22]
To investigate the influence of CdTe NPs on the fibrillation of Aβ1-40, different amount of CdTe NPs was added to the incubation solutions. The molar ratio of CdTe NPs to Aβ1-40 was adjusted as 0, 0.001, 0.005, 0.01, and 0.05. Then the incubation solutions with and without CdTe NPs were stirred at room temperature typically for 1 day. The fibrillation of Aβ1-40 was also monitored by ThT (thioflavin T) assay by adding stock solution of Aβ1-40 and CdTe NPs to PBS buffer containing 20 μM ThT. Photoluminescence spectra for ThT assay were recorded using Fluoromax-3 spectrofluorometer (Jobin Yvon/SPEX Horiba, NJ) with excitation wavelength of 450 nm and monitoring wavelength of 485 nm.
The morphology of Aβ1-40 with and without CdTe NPs were studied by a transmission electron microscope (JEOL 2010F) operated in scanning transmission electron microscopy (STEM) mode. Atomic force microscopy (AFM) imaging was performed with a Nanoscope III (Digital Instruments/Veeco Metrology Group). For the AFM measurement, lyophilized Aβ1-40 was first dissolved into deionized water and then diluted into PBS buffer with and without NPs.
NMR samples were prepared from 15N labeled Aβ1-40 (rPeptide) by first dissolving the peptide in 1% ammonium hydroxide, lyophilizing, and then resuspending in DMSO to 1 mM peptide concentration. The peptide was then diluted to a final concentration of 76.6 μM in 20 mM sodium phosphate buffer, pH 7.5, with 50 mM NaCl. All NMR experiments were performed on a Bruker spectrometer operating at 600.13 MHz (1H frequency), equipped with a cryogenic probe. The interactions of Aβ1-40 with CdTe NPs were followed by performing a series of 2D SOFAST-HMQC experiments[23] at 10°C with increasing concentrations of CdTe NPs as described. Each spectrum was obtained from 128 t1 experiments, 4 scans, and a 100 ms recycle delay. The final 2D data matrix size was 2048 × 2048 after zero-filling in both dimensions. 2D data were processed using TOPSPIN 2.1 (from Bruker). Squared sine-bell function was employed in both the dimension with a shift of pi/4. Resonance assignment and volume fit calculations were performed using SPARKY 3.113.
Acknowledgments
This work was supported by NSF grants 0932823, 0933384, 0938019, NIH grants 1R21CA121841-01A2, 5R01EB007350-02, and in part by DARPA W31P4Q-08-C-0426. S.I.Y. acknowledges the support of the Korea Research Foundation Grant KRF-2008-357-D00078.
References
- [1].a) Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]; b) Lansbury PT, Lashuel HA. Nature. 2006;443:774. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]; c) Ross CA, Poirier MA. Nature Rev. Mol. Cell. Biol. 2005;6:891. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]; d) Hardy J, Selkoe DJ. Science. 2002;297:353. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- [2].a) Petkova AT, Leapman RD, Guo Z, Yau W-M, Mattson MP, Tycko R. Science. 2005;307:262. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]; b) Lorenzo A, Yankner BA. Proc. Natl. Acad. Sci. USA. 1994;91:12243. doi: 10.1073/pnas.91.25.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Haass C, Selkoe DJ. Nature Rev. Mol. Cell. Biol. 2007;8:101. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]; b) Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Nature Neurosci. 2004;8:79. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]; c) Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Proc. Natl. Acad. Sci. USA. 2003;100:330. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Nature. 2002;416:535. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]; e) Klein WL, Krafft GA, Finch CE. Trends Neurosci. 2001;24:219. doi: 10.1016/s0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- [4].Takahashi T, Mihara H. Accounts Chem. Res. 2008;41:1309. doi: 10.1021/ar8000475. [DOI] [PubMed] [Google Scholar]
- [5].a) Gordon DJ, Sciarretta KL, Meredith SC. Biochemistry. 2001;40:8237. doi: 10.1021/bi002416v. [DOI] [PubMed] [Google Scholar]; b) Lowe TL, Strzelec A, Kiessling LL, Murphy RM. Biochemistry. 2001;40:7882. doi: 10.1021/bi002734u. [DOI] [PubMed] [Google Scholar]; c) Findeis MA, Musso GM, Arico-Muendel CC, Benjamin HW, Hundal AM, Lee J-J, Chin J, Kelley M, Wakefield J, Hayward NJ, Molineaux SM. Biochemistry. 1999;38:6791. doi: 10.1021/bi982824n. [DOI] [PubMed] [Google Scholar]; d) Tjernberg LO, Näslund J, Lindqvist F, Johansson J, Karlström AR, Thyberg J, Terenius L, Nordstedt C. J. Biol. Chem. 1996;271:8545. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]; e) Soto C, Kindy MS, Baumann M, Frangione B. Biochem. Biophys. Res. Commun. 1996;226:672. doi: 10.1006/bbrc.1996.1413. [DOI] [PubMed] [Google Scholar]
- [6].a) Milojevic J, Raditsis A, Melacini G. Biophys J. 2009;97:2585. doi: 10.1016/j.bpj.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Milojevic J, Esposito V, Das R, Melacini G. J. Am. Chem. Soc. 2007;129:4282. doi: 10.1021/ja067367+. [DOI] [PubMed] [Google Scholar]; c) Bohrmann B, Tjernberg L, Kuner P, Poli S, Levet-Trafit B, Näslund J, Richards G, Huber W, Döbeli H, Nordstedt C. J. Biol. Chem. 1999;274:15990. doi: 10.1074/jbc.274.23.15990. [DOI] [PubMed] [Google Scholar]; d) Evans KC, Berger EP, Cho C-G, Weisgraber KH. Proc. Natl. Acad. Sci. USA. 1995;92:763. doi: 10.1073/pnas.92.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Klyubin I, Walsh DM, Lemere CA, Cullen WK, Shankar GM, Betts V, Spooner ET, Jiang L, Anwyl R, Selkoe DJ, Rowan MJ. Nature Med. 2005;11:556. doi: 10.1038/nm1234. [DOI] [PubMed] [Google Scholar]; b) Hock C, Konietzko U, Papassotiropoulos A, Wollmer A, Streffer J, von Rotz RC, Davey G, Moritz E, Nitsch RM. Nature Med. 2002;8:1270. doi: 10.1038/nm783. [DOI] [PubMed] [Google Scholar]
- [8].Poduslo JF, Curran GL, Berg CT. Proc. Natl. Acad. Sci. USA. 1994;91:5705. doi: 10.1073/pnas.91.12.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Findeis MA. Biochim. Biophys. Acta. 2000;1502:76. doi: 10.1016/s0925-4439(00)00034-x. [DOI] [PubMed] [Google Scholar]
- [10].a) Bae Y, Kim NH, Kim M, Lee KY, Han SW. J. Am. Chem. Soc. 2008;130:5432. doi: 10.1021/ja800898v. [DOI] [PubMed] [Google Scholar]; b) DeVries GA, Brunnbauer M, Hu Y, Jackson AM, Long B, Neltner BT, Uzun O, Wunsch BH, Stellacci F. Science. 2007;315:358. doi: 10.1126/science.1133162. [DOI] [PubMed] [Google Scholar]; c) Klokkenburg M, Houtepen AJ, Koole R, de Folter JWJ, Erné BH, van Faassen E, Vanmaekelbergh D. Nano Lett. 2007;7:2931. doi: 10.1021/nl0714684. [DOI] [PubMed] [Google Scholar]; d) Yu S-H, Cölfen H, Tauer K, Antonietti M. Nature Mater. 2005;4:51. doi: 10.1038/nmat1268. [DOI] [PubMed] [Google Scholar]; e) Cho K-S, Talapin DV, Gaschler W, Murray CB. J. Am. Chem. Soc. 2005;127:7140. doi: 10.1021/ja050107s. [DOI] [PubMed] [Google Scholar]; f) Lu W, Gao P, Jian WB, Wang ZL, Fang J. J. Am. Chem. Soc. 2004;126:14816. doi: 10.1021/ja046769j. [DOI] [PubMed] [Google Scholar]; g) Tang ZY, Kotov NA, Giersig M. Science. 2002;297:237. doi: 10.1126/science.1072086. [DOI] [PubMed] [Google Scholar]
- [11].a) Xia Y. Nature Mater. 2008;7:758. doi: 10.1038/nmat2277. [DOI] [PubMed] [Google Scholar]; b) Lynch I, Dawson KA. Nanotoday. 2008;3:40. [Google Scholar]; c) Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Nature Mater. 2003;2:577. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- [12].a) Auer S, Trovato A, Vendruscolo MA. Plos Comput. Biol. 2009;5:e1000458. doi: 10.1371/journal.pcbi.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Linse S, Cabaleiro-Lago C, Xue W-F, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Proc. Natl. Acad. Sci. USA. 2007;104:8691. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wu WH, Sun X, Yu YP, Hu J, Zhao L, Liu Q, Zhao YF, Li YM. Biochem. Biophy. Res. Commun. 2008;373:315. doi: 10.1016/j.bbrc.2008.06.035. [DOI] [PubMed] [Google Scholar]
- [13].a) Relini A, Canale C, De Stefano S, Rolandi R, Giorgetti S, Stoppini M, Rossi A, Fogolari F, Corazza A, Esposito G, Gliozzi A, Bellotti V. J. Biol. Chem. 2006;281:16521. doi: 10.1074/jbc.M513827200. [DOI] [PubMed] [Google Scholar]; b) Myers SL, Jones S, Jahn TR, Morten IJ, Tennent GA, Hewitt EW, Radford SE. Biochemistry. 2006;45:2311. doi: 10.1021/bi052434i. [DOI] [PubMed] [Google Scholar]; c) Sluzky V, Tamada JA, Klibanov AM, Langer R. Proc. Natl. Acad. Sci. USA. 1991;88:9377. doi: 10.1073/pnas.88.21.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lu JR, Perumal S, Powers ET, Kelly JW, Webster JRP, Penfold J. J. Am. Chem. Soc. 2003;125:3751. doi: 10.1021/ja0292290. [DOI] [PubMed] [Google Scholar]; e) Powers ET, Kelly JW. J. Am. Chem. Soc. 2001;123:775. doi: 10.1021/ja005528i. [DOI] [PubMed] [Google Scholar]
- [14].a) Cabaleiro-Lago C, Quinlan-Pluck F, Lynch I, Lindman S, Minogue AM, Thulin E, Walsh DM, Dawson KA, Linse S. J. Am. Chem. Soc. 2008;130:15437. doi: 10.1021/ja8041806. [DOI] [PubMed] [Google Scholar]; b) Ikeda K, Okada T, Sawada S-I, Akiyoshi K, Matsuzaki K. FEBS Lett. 2006;580:6587. doi: 10.1016/j.febslet.2006.11.009. [DOI] [PubMed] [Google Scholar]; c) Pai AS, Rubinstein I, Onyüksel H. Peptides. 2006;27:2858. doi: 10.1016/j.peptides.2006.04.022. [DOI] [PubMed] [Google Scholar]; d) Kim JE, Lee M. Biochem. Biophys. Res. Commun. 2003;303:576. doi: 10.1016/s0006-291x(03)00393-0. [DOI] [PubMed] [Google Scholar]; e) Xiao L, Zhao D, Chan W-H, Choi MMF, Li H-W. Biomaterials. 2010;31:91. doi: 10.1016/j.biomaterials.2009.09.014. [DOI] [PubMed] [Google Scholar]
- [15].a) Tang Z, Zhang Z, Wang Y, Glotzer SC, Kotov NA. Science. 2006;314:274. doi: 10.1126/science.1128045. [DOI] [PubMed] [Google Scholar]; b) Zhang Z, Tang Z, Kotov NA, Glotzer SC. Nano Lett. 2007;7:1670. doi: 10.1021/nl0706300. [DOI] [PubMed] [Google Scholar]
- [16].The fitting curves were obtained from an equation in ref. [14a] of y=y0+(ymax − y0)/(1+e−(t−t1/2>)k), where y is the fluorescence intensity at a give time t, y0 and ymax are the initial and maximum fluorescence intensity, respectively, t1/2 is the time required to reach half the maximum fluorescence intensity, and k is the apparent first-order aggregation constant. The lag time can be calculated by t1/2 − 2/k. Since the fibrillation was completely inhibited in the case of [CdTe]/[Aβ1-40] = 0.05, we were not able to include a fitting curve. The solid line in Fig. 1B trace e is a mere guide line.
- [17].Fawzi NL, Ying J, Torchia DA, Clore GM. J. Am. Chem. Soc. 2010;132:9948. doi: 10.1021/ja1048253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yaroslavov AA, Sinani VA, Efimova AA, Yaroslavova EG, Rakhnyanskaya AA, Ermakov YA, Kotov NA. J. Am. Chem. Soc. 2005;127:7322. doi: 10.1021/ja051095d. [DOI] [PubMed] [Google Scholar]
- [19].a) Shang Q, Wang H, Yu H, Shan G, Yan R. Colloids Surf. A Physicochem. Eng. Aspects. 2007;2 94:86. [Google Scholar]; b) Li Z, Du Y, Zhang Z, Pang D. React. Func. Polym. 2003;55:35. [Google Scholar]
- [20].Manavalan P, Johnson WC. Nature. 1983;305:831. [Google Scholar]
- [21].Olivieri JR, Craievich AF. Eur. Biophys. J. 1995;24:77. doi: 10.1007/BF00211402. [DOI] [PubMed] [Google Scholar]
- [22].Yu WW, Qu L, Guo W, Peng X. Chem. Mater. 2003;15:2854. [Google Scholar]
- [23].Schanda P, Brutscher B. J. Am. Chem. Soc. 2005;127:8014. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]