Abstract
Surface recognition of biosystems is a critical component in the development of novel biosensors, delivery vehicles and for the therapeutic regulation of biological processes. Monolayer-protected nanoparticles present a highly versatile scaffold for selective interaction with biomacromolecules and cells. Through engineering of the monolayer surface, nanoparticles can be tailored for surface recognition of biomolecules and cells. This review highlights recent progress in nanoparticle-biomacromolecule/cellular interactions, emphasizing the effect of the surface monolayer structure on the interactions with proteins, DNA and cell surfaces. The extension of these tailored interactions to hybrid nanomaterials, biosensing platforms and delivery vehicles is also discussed.
Keywords: Nanoparticles, surface monolayer, biomacromolecule recognition, cellular interaction, bionanotechnology
1. Introduction
Effective and selective biomacromolecular interactions rely on large receptor contact areas and structurally and dynamically well-defined architectures for high affinity binding.[1] However, the recognition of biomolecular surfaces is highly challenging owing to their large size and topological/surface complexity.[2] Nanoparticles (NPs) are an attractive alternative to classical small molecule receptors.[3] NPs feature several structural attributes for the creation of biomacromolecular receptors, leading to numerous applications in biosensing, clinical diagnostics and therapeutics.[4] The wide variety of available core materials (metal or semiconductor) featuring inherent optoelectronic and magnetic properties provide useful physical attributes.[5] Additionally, the tunable core-size of NPs (1.5 nm to > 10 nm) spans the size range of biomacromolecules, providing surface area and size comparability for efficient interaction with target biomacromolecules.[6]
The ability to introduce a wide range of functionalities on the surface of NPs is one of the key attractions for the use of these materials in biology. Appropriate choice of functionality facilitates the creation of surface-specific receptors for a variety of targets.[7] These selective interactions also provide an adaptable scaffold for the self-assembly of nanomaterials using biomolecules[8] and have been used in biosensing and nanoelectronics.[9] Likewise, controlled interaction of NPs with cell membranes and lipid bilayers is crucial for the development of new strategies for photothermal therapy, imaging, and delivery applications,[10] as well as providing new tool for provoking differential cellular and sub-cellular responses for therapeutic applications.[11] The size and functionality available with NPs likewise make them excellent carriers for cytosolic delivery of drugs, genes, siRNA, and other macromolecular systems by overcoming the natural inability of these molecules to penetrate the cell membrane,[12] a process that can be further enhanced by introducing cell-penetrating peptides on the NP surface monolayer.[13]
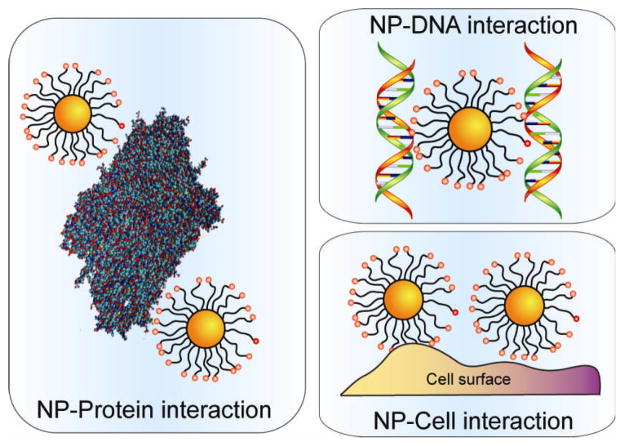
In this current review, we will focus on the recent advances in the use of the functional organic monolayers of NPs as multivalent recognition elements for protein, DNA and cell interactions, (Figure 1) focusing on non-covalent supramolecular strategies complementary to widely-used covalent conjugation methodologies.[14]
Figure 1.
Schematic presentation of NP interactions with proteins, DNA and cells.
2. Nanoparticle-protein interactions
2.1. Regulating the structure and function of proteins using nanoparticles
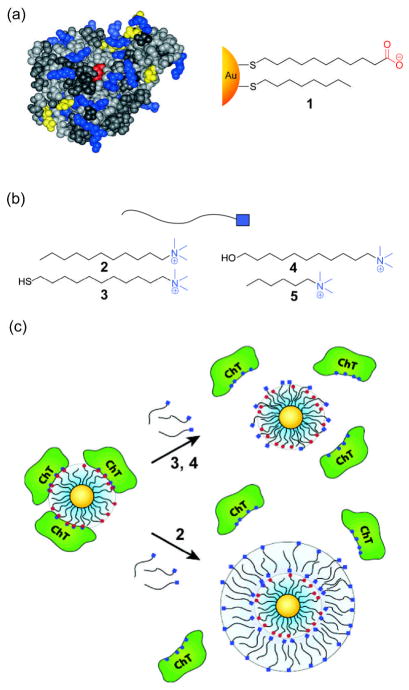
Selective binding of engineered NPs to enzymes provides a tool for modulating both their structure and activity. In early studies, mixed monolayer NPs bearing anionic ligands were used for surface recognition and activity inhibition of chymotrypsin (ChT),[15] targeting the cationic patch around the active site (Figure 2a).[16] Protein inhibition with these simple alkanethiol functionalized NPs was a two-step process featuring a fast reversible association followed by a slower irreversible denaturation process. The efficient complementary electrostatic interaction resulted in an apparent Ki=10 nM and was selective relative to other proteins. These electrostatic interactions can be tuned by ionic strength of aqueous media[17] and cationic surfactants.[18] For example, Rotello and coworkers have shown that cationic surfactants (Figure 2b) allow the release of ChT molecules from the surface of the anionic NPs with partial restoration in enzyme activity. The release of ChT from the NP-protein assembly was dependent upon the surfactant, as surfactant 2 liberated the bound proteins through the formation of bilayer structures, as evidenced by a 4.5 nm increase in the hydrodynamic radius of the complex (Figure 2c). In contrast, cationic thiol 3 and alcohol 4 incorporate themselves into the NP monolayer, with no increase in the hydrodynamic radius of the NP observed.
Figure 2.
(a) Space-filling model of ChT and structure of anionic gold NP 1 used in activity inhibition study (b) Structures of various cationic surfactants (c) Release mechanisms of ChT from the surface of gold NP 1 by addition of different surfactants (Ref [16] and [18]).
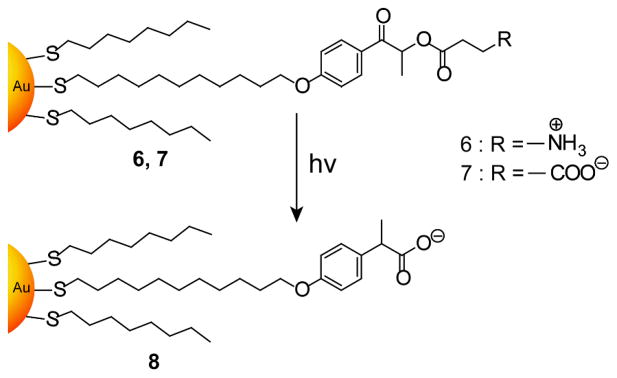
Photochemical stimuli provide an external trigger to modulate the activity of enzymes assembled on NPs,[19] a potentially useful tool for biomedical applications. Rotello and coworkers synthesized gold NPs bearing photoactive phenacyl ester linkages and explored their interactions with ChT (Figure 3).[20] Cationic gold NP 6 does not interact with ChT while anionic NP 7 can bind and inhibit ChT activity. Upon irradiation with UV light, both of these NPs are converted into anionic NP 8 that interacts strongly with ChT, inhibiting activity.
Figure 3.
Structures of photocleavable gold NPs. Reproduced with permission from Ref [20]. Copyright 2004 Royal Society of Chemistry.
Binding of enzymes to NP surfaces alters access to the active site in a selective fashion by virtue of substrates and products having to navigate the protein-NP interface. This modulation was demonstrated through changes in enzymatic activity of NP-bound ChT toward substrates with different charges.[21] Electrostatic interactions between cationic substrates and the anionic NPs facilitate the transport of substrate into the enzyme, accelerating catalysis. Electrostatic repulsion between anionic substrates and negatively charged NPs prevents accessibility to the active pocket of the surface-bound enzyme, with hydrolysis of these substrates being strongly inhibited by particle binding.
Binding studies of ChT with NPs bearing amino acid termini showed that both electrostatic and hydrophobic interactions contribute to the stability of the complex.[22] The binding constants (106–107 M−1) for these synthetic receptor-protein complexes are comparable with naturally occurring protein-ChT inhibitor interactions[23] and increase with the hydrophobicity of the amino acid chains. Protein stability is likewise altered in the non-covalent assemblies. Hydrophobic amino acids have little effect on the native structure of ChT, while hydrophilic amino acids destabilize the protein due to competitive hydrogen-bonding and breakage of salt bridges in the protein. Further control of protein stability can be achieved by changing the length of oligo(ethylene glycol) (OEG) tethers,[24] with shorter OEG segments increasing the rate of denaturation. In a related study using L-amino acid functionalized gold NPs, the cooperative role of electrostatics and hydrophobicity on the NP surface for complexation with proteins was explored. Detailed thermodynamic investigations using isothermal titration calorimetry (ITC) with three model proteins, ChT, cytochrome C (CC) and histone, revealed that enthalpy and entropy changes for the complex formation depend upon the NP structure and the surface distributions of charged and hydrophobic residues in the particular protein.[25] The linear relationship for ΔH-TΔS indicated significant conformational changes and substantial dehydration of the partners, providing an effective mimic for native protein-protein interactions.
Subtle structural effects can strongly alter NP-protein interactions. Rotello and coworkers investigated the effect of chirality of the NP surface towards the model proteins ChT and CC using a series of functional gold NPs bearing phenylalanine and/or leucine residues.[26] ITC studies revealed NPs bearing enantiomeric and diastereoisomeric groups can have markedly different interactions towards target proteins. Similarly, regiospecificity is also achieved between the gold NP-CC interactions by introducing anionic particles with different side chains.[27]
Preservation of protein structure upon binding is important for in vivo protein delivery and in vitro enzyme stabilization. Nanoparticles bearing tetra(ethylene glycol) chains improve the stability of ChT at the NP surface[28] owing to the fact that OEG minimizes the nonspecific interaction with biomacromolecules.[29] Recently, Ghosh et al. has shown highly efficient binding as well as intracellular protein delivery using gold NPs bearing a short peptide conjugated to a tetra(ethylene glycol) moiety. The engineered NPs were shown to translocate β-galactosidase, a large exogenous protein, into a variety of cells without exhibiting any cytotoxicity. These NP-protein complexes were able to escape from endosomes and retained their biological activity.[30]
2.2. Specificity in NP-protein interactions
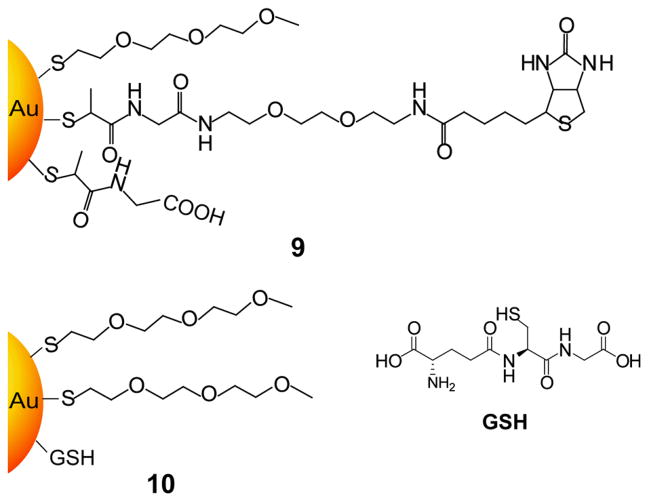
The above studies focus on selective protein binding; specific binding has generally been accomplished via conjugation of the NPs with biomolecular ligands. The biotin/streptavidin interaction has been extensively employed for the conjugation of NP systems[31] due to its strong interaction (Ks ~ 1014 dm3/mol) and stability over a broad range of pH values and temperatures.[32] For example, Zheng and Huang have synthesized biotin and glutathione moieties capped onto the surface of gold NPs protected by tri(ethylene glycol) thiols (Figure 4).[33] Biotin-containing gold NPs specifically bind to streptavidin while glutathione capped NPs are targeted to glutathione-S-transferase (GST). The binding of GSH by GST is far weaker than the biotin/streptavidin interaction; however, nonspecific binding with other proteins was diminished in this system.
Figure 4.
Chemical structures of biotin and glutathione capped gold NPs for specific interaction with streptavidin and glutathione-S-transferase respectively (Ref [33]).
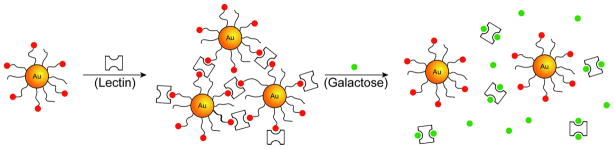
Another potential approach to gain specificity is through carbohydrate-lectin interactions.[34] Engineering of NP surfaces with carbohydrate ligands provides a means to couple carbohydrate-lectin interaction with increased specificity through the multivalent interactions.[35] For example, mannose-coated gold NPs bind with the plant lectin concanavalin A (Con A), with 10–100 fold higher affinity than monovalent mannose ligands.[36] This specific interaction between mannose functionalized gold NPs and Con A was explored to develop a competitive colorimetric assay to detect protein-protein interactions.[37] In another study, agglutinin, a bivalent lectin, induced the aggregation of lactose-functionalized gold NPs, leading to changes in its absorption spectrum. This NP-protein assembly can be dissociated by the addition of galactose, as agglutinin has β-D-galactose specificity (Figure 5). Moreover, the extent of aggregation was proportional to the lectin concentration leading to highly sensitive detection of the target molecule.[38]β-N-acetylglucosamine-conjugated quantum dots have been used to study specific and multivalent interactions with lectin and sperm by fluorometry.[39] Recently, Yan et al. have demonstrated a rapid and versatile way for coupling of unmodified sugars to gold NPs via a photochemical reaction with efficient lectin recognition observed.[40]
Figure 5.
Schematic representation of reversible lectin-induced association of lactose-conjugated gold NPs (Ref [38]).
Transition metal complexes have been used to target surface-exposed histidines of proteins via coordinative interactions,[41] providing specific receptors for proteins.[42] Nickel-terminated nitrilotriacetic acid (NTA) coated FePt NPs bind to proteins that have six consecutive histidine residues, allowing separation, transport and anchoring of recombinant proteins.[43] Using these magnetic NPs, pure proteins were separated from lysed cell mixtures within 10 min. Mattoussi et al. utilized the affinity of hexa-His peptides to semiconductor surfaces to directly conjugate fluorescent proteins onto quantum dot (QD) surfaces to develop caspase-3 sensors.[44] Recently, De et al. have shown that NTA functionalized gold NPs can exhibit high affinity for binding with His-tagged proteins,[45] with the protein-particle stoichiometry controlled by varying Ni2+ concentrations.
2.3. Protein sensing
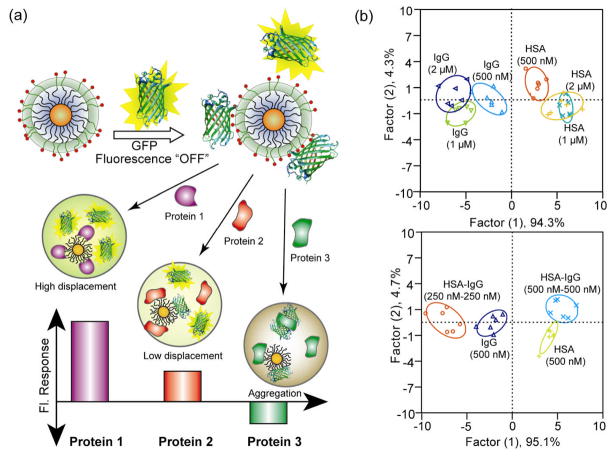
Differential affinities between NPs and proteins can generate distinct patterns of responses from sensor arrays, generating a fingerprint response for individual proteins. Using this strategy, Rotello and coworkers fabricated a sensor array composed of six cationic gold NPs and one anionic poly(p-phenylene ethynylene) (PPE) polymer that could properly identify seven proteins of different sizes and isoelectric points.[46] Linear discrimination analysis (LDA) was used to differentiate the response patterns with high accuracy (94.2%). Later studies focused on sensing proteins in human serum, a complex matrix with more than 20,000 proteins and an overall protein concentration of ~ 1mM. The same group achieved differentiation of proteins spiked into human serum using a green fluorescent protein (GFP)-NP sensor array (Figure 6a).[47] Positively charged NPs combined with anionic GFP generated a sensor that could detect 500 nM changes in the five most abundant human serum proteins in undiluted serum. Mixtures of proteins in different concentrations led to specific and reproducible changes in the LDA-based patterns (Figure 6b).
Figure 6.
a) Schematic illustration showing the competitive binding between proteins and quenched gold NP-GFP complexes. b) Discrimination of human serum albumin (HSA) and immunoglobulin G (IgG) at different concentrations and mixture of proteins. Top: canonical score plot for the fluorescence patterns as obtained from LDA for HSA and IgG at different concentrations (500 nM, 1 μM, and 2 μM) with 95% confidence ellipses. Bottom: HSA and IgG were mixed at 1:1 molar ratio with 250 nM each and 500 nM each, and added to five gold NP-GFP complexes. Reproduced with permission from Ref [47]. Copyright 2009 Nature Publishing Group.
The above fluorescence-based detection systems are restricted by the inherent emissivity of the fluorophore. To overcome this limitation, The Rotello group used enzymes to provide array-based sensors with enhanced sensitivity.[48] Cationic gold NPs electrostatically bind the anionic β-Galactosidase (β-gal), inhibiting the enzyme without denaturation. Displacement of the particles by analytes restores enzyme activity, providing an amplification of the recognition event that was used for sensing proteins in buffer and in urine with a limit of detection of 1 nM.
2.4. Peptide and protein assisted assembly of surface functionalized NPs
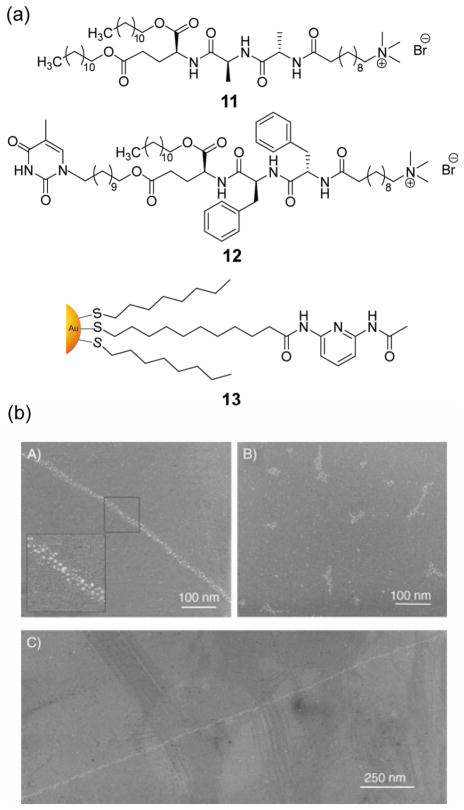
Biomolecule-mediated “bottom-up” self-assembly of nanobiocomposites provides a versatile strategy for the creation of functional materials. Stupp et al. used amphiphilic tripeptide based nanofibres 11 and 12 for one-dimensional assembly of lipophilic gold NPs (Figure 7a).[49] Co-assembly 11 and 12 into fibrils generates supramolecular structures in aprotic solvents. With the addition of gold NP 13, the diaminopyridine groups on the NP surface interact with thymine moieties, leading to a linear assembly of gold NPs (Figure 7b). In a related study, double-helical and single-chain arrays of NPs were obtained through peptide self-assembly.[50] These structures could be controlled by pH and the size of the NPs. Chmielewski et al. demonstrated an alternative non-covalent approach for gold NP assembly using coiled-coil peptides.[51] Polypeptide-functionalized gold NPs were also employed for controlled aggregation by induced folding of immobilized peptides.[52] Stulz et al. have shown a programmed assembly of gold NPs functionalized with DNA-binding peptides onto self-organized oligonucleotide templates,[53] integrating peptide and nucleic acid-based assembly strategies. Moreover, by controlling the amount of peptide on the NP surface and using specific oligonucleotide sequences, the assembly can be tuned to form dimers, trimers, and higher ordered structures.
Figure 7.
(a) Structures of amphiphilic tripeptide derivatives and diaminopyridine functionalized gold NPs (b) TEM images of linear arrays of surface modified gold NPs in the presence of surface modified peptide amphiphile nanofibers [A) & C)] and unmodified gold NPs and nanofibers [B)]. Reproduced with permission from Ref [49]. Copyright 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
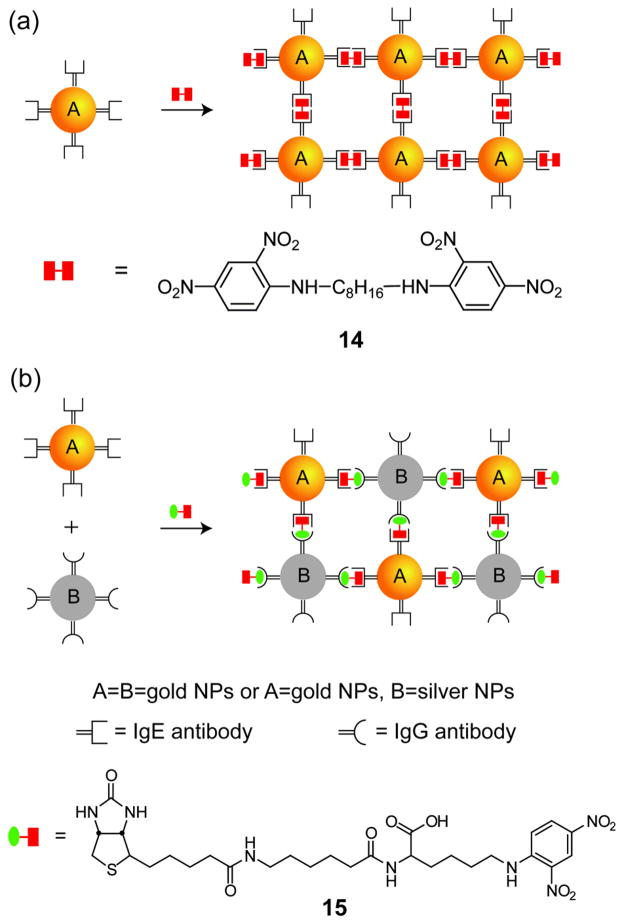
Proteins likewise provide a scaffold for NP assembly. Mukherjee et al. have illustrated a shape-selective assembly of gold NPs using antibodies as templates to provide a scaffold to direct the assembly of gold NPs into rod-shaped structures.[54] Mann and coworkers used antigen-antibody recognition for self-assembly of NPs into extended 3D networks such as wires and filaments using bivalent antigen 14 and DNP-biotin bivalent antigen 15 (Figure 8).[55] Whitesides et al. have demonstrated a related approach where monoclonal anti-fluorescein isothiocyanate (FITC) IgGs can spontaneously assemble into linear, triangular and square-planar nanostructures in the presence of divalent fluorescein antigens, serving as a template to grow gold NP clusters.[56]
Figure 8.
Schematic representation showing the directed self-assembly of NPs using antibody-antigen recognition (Ref [55]).
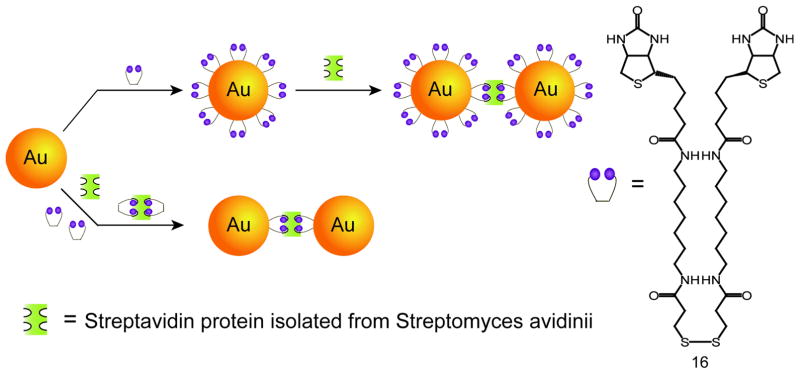
Streptavidin-biotin binding is a widely employed strategy for the self-assembly of nanostructures due to the high stability and specificity of the interaction.[57] Fitzmaurice and coworkers have shown two distinct routes to assemble gold NPs using a streptavidin/biotin analogue (Figure 9).[58] The first pathway involves the chemisorption of a disulfide biotin analogue 16 onto gold NPs followed by the subsequent addition of streptavidin. The second route involves the binding of 16 to the streptavidin before adding to gold NPs. In similar studies by Perez-Luna and colleagues, interactions of streptavidin with biotinylated gold NPs form aggregates with a shift in the plasmon resonance peak and broadening of the absorption spectrum.[59]
Figure 9.
Schematic illustration demonstrating two distinct pathways of self-assembly of gold NPs using streptavidin-disulfide biotin analogue (Ref [58]).
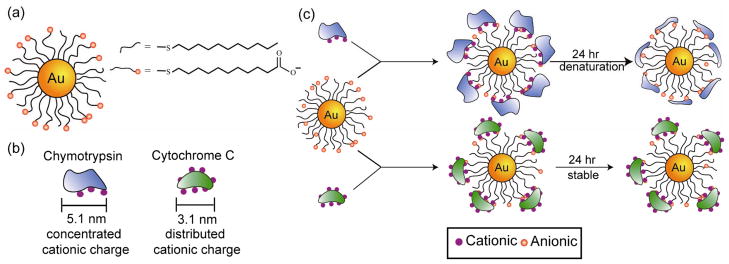
Assembly of NPs provides access to spacer-dependent modulation of optical, electronic, and magnetic properties of NP assemblies for materials applications. NP-protein assembly can be used to control interparticle spacing[60] and morphology[61] through appropriate choice of protein size, shape and charge.[62] Srivastava et al. has used two proteins, CC and ChT to assemble NPs into organized composites (Figure 10a & b).[60] These proteins have different stability behaviors on the surface of carboxylate-functionalized gold NPs: ChT unfolds onto the particle surface and acts as a linear polymer, whereas CC retains its native conformation on the NP (Figure 10c). This behavior was observed in the solid state as well: ChT-mediated NP assemblies showed increases in interparticle spacing consistent with uncoiled peptide, while CC generated NP assemblies with larger interparticle spacing consistent with the retention of the native protein structure. Later studies showed that lysozyme could be used to generate a wide range of interparticle spacings using an analogous strategy.[63] In recent studies, FePt and gold NPs were used to assemble ferritin protein, with ferritin providing added magnetic volume fraction to the magnetic bionanocomposite.[64] Samanta et al. has also demonstrated NP-protein assembly at oil-water interfaces. Through proper choice of cationic gold NPs, β-gal has been assembled onto oil-water interfaces with high retention of β-gal activity (>70%).[65]
Figure 10.
(a) Mixed monolayer gold NPs featuring a hydrophobic interior with carboxylate end groups. (b) Schematic depiction of protein electrostatic surfaces: positive residues on ChT are concentrated on one face, while they are distributed over the entire CC surface. (c) Protein-particle assembly of gold NPs with ChT and CC. Reproduced with permission from Ref [60]. Copyright 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
3. Nanoparticle-DNA interactions
3.1 Diagnostic and therapeutic applications
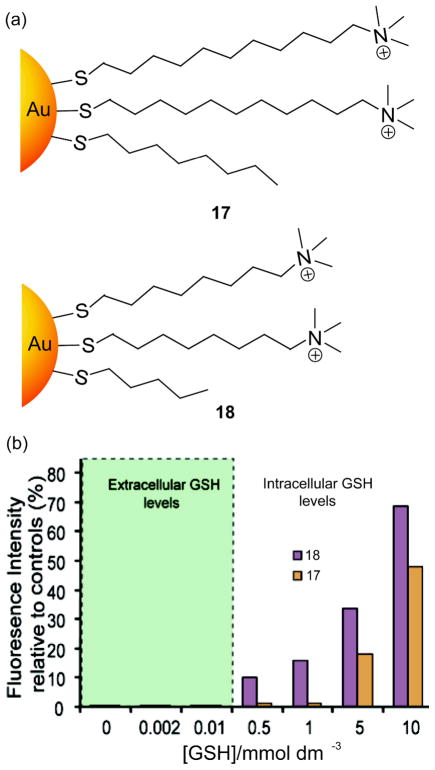
Inhibition of DNA transcription provides a direct strategy to modulate disease states and/or cellular behavior through gene regulation. Interaction with DNA can occur through three primary modes of association: surface binding, groove binding and intercalation.[66] DNA binding and consequent transcription regulation has been achieved with sequence-selective substituted polyamides,[67] nuclear binding peptides,[68] and small molecule binders.[69] Monolayer protected NPs provide an attractive alternative scaffold for binding nucleic acids, mimicking naturally occurring protein-nucleic acid interactions.[70,71] NPs bearing cationic ligands provide highly efficient DNA binding via electrostatic interactions. McIntosh et al. have shown that gold NP 17 bearing cationic headgroups (Figure 11a) binds with 37-mer DNA at a binding stoichiometry of 4:1, preventing transcription.[72] Moreover, NP 17 can protect DNA from enzymatic digestion,[73] with binding efficiency of analogs tuned by varying the surface hydrophobicity.[74] Analogous protective behavior was reported by Wang et al. where cationic silica NPs were shown to protect DNA from enzymatic cleavage.[75] The monolayer structure of NPs can be tuned to regulate NP stability for controlled delivery or release applications. For example, Han et al. used the intracellular glutathione (GSH) concentration as a trigger to restore the transcription of DNA bound to cationic gold NPs 17 and 18 (Figure 11a).[76] These NP-DNA complexes were stable at extracellular glutathione concentrations and inhibit DNA transcription, whereas at increased GSH concentrations, these complexes showed GSH-dependent recovery of DNA transcription (Figure 11b).
Figure 11.
(a) Structures of cationic gold NPs (b) The amount of 20-mer runoff RNA detected relative to that produced in the absence of gold NPs 17 and 18 at varying concentrations of GSH. Reproduced with permission from Ref [76]. Copyright 2005 American Chemical Society.
Cationic NPs provide a synthetic platform for DNA delivery applications due to strong interactions with both the DNA backbone and cell membranes. A series of gold NP 17 analogues with various positive charges and different tethers of the unfunctionalized alkane thiols were used for gene delivery into cells,[77] demonstrating that DNA-NP complexes with an overall positive charge are required for effective cellular uptake of NPs.[78] In a later study, polyethylenimine (PEI) coated gold NPs were used by Klibanov as gene delivery vectors,[79] with increased transfection observed for higher N/P (PEI nitrogen/DNA phosphate) ratios. Moreover, NPs bearing dodecyl-PEI showed higher DNA delivery than PEI-coated NPs, confirming the hydrophobicity on the monolayer can be a key attribute for superior transfection efficacy. Gold NPs functionalized with lysine and lysine dendrons were demonstrated to be highly efficient gene delivery vectors without any observed cytotoxicity, and were shown to be modulated by GSH levels consistent with a GSH-mediated release process.[80]
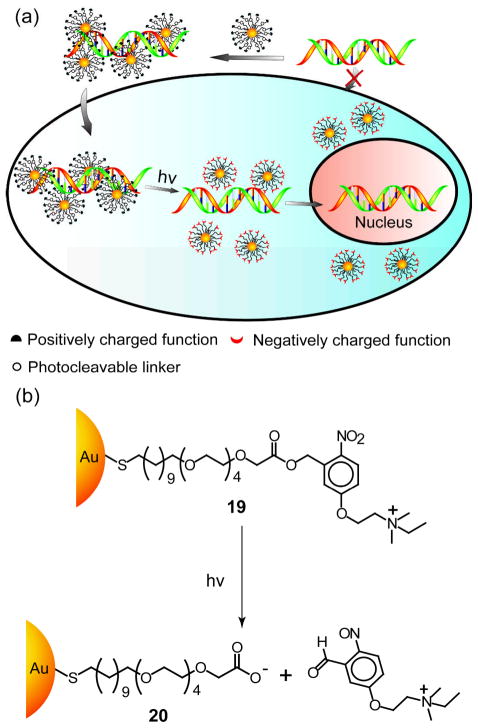
In addition to GSH-mediated DNA release, the DNA delivery can be controlled by photochemical means using photolabile linkages on the surface of NPs. Han et al. fabricated a cationic gold NP 19 that can be photo-switched to anionic gold NP 20 upon light irradiation to release DNA intracellularly (Figure 12a & b).[81]
Figure 12.
(a) Schematic illustration of delivery and photo-triggered release of DNA from gold NPs 19-DNA complex inside living cells, (b) Schematic of light-induced charge switching from cationic gold NP 19 to anionic gold NP 20. Reproduced with permission from Ref [81]. Copyright 2006 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
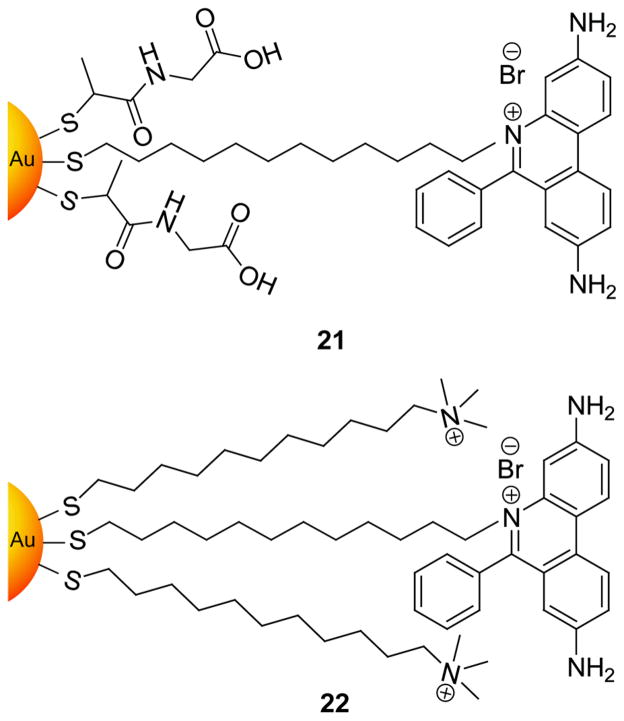
Intercalating ligands provide an alternative to electrostatic binding for DNA recognition. Murray et al. have synthesized anionic gold NP 21 and cationic gold NP 22 (Figure 13) with ethidium bromide (EtBr) headgroups.[82] Binding of cationic trimethylammonium gold NP 22 was efficient and rapid due to the cooperative nature of intercalation and electrostatics. The binding of anionic tiopronin gold NP 21, however, did not occur until NaCl concentrations were greater than 0.1 M to screen charge repulsion.
Figure 13.
Structures of ethidium-incorporated gold NPs for intercalation with DNA (Ref 82).
3.2. NP-DNA composite materials
The structural and functional diversity of DNA has made it a versatile building block for nanocomposite assembly.[83] Sequence-specific assembly motifs involving the hybridization of complementary DNA strands tethered to NPs have been widely employed to produce NP chains or networks[84] with a high level of sequence specificity.[85] Nanostructured assemblies can likewise be generated through non-covalent NP-DNA interactions. For example, cationic gold NPs were used for DNA-NP assembly by Murray et al.,[86] generating multi-micron one-dimensional chains. Similarly, cetyltrimethylammonium bromide (CTAB) functionalized gold NPs[87] and silver NPs[88] have also been used for programmed assembly with DNA. Srivastava et al. have demonstrated a ‘bricks and mortar’ based self-assembly approach to construct hybrid magnetic bionanocomposites using cationic FePt NPs and 37-mer duplex DNA.[89]
DNA-mediated NP assembly on solid surfaces provides a ‘bottom up’ approach for nanoscale electronics. For example, cationic NPs were assembled with DNA on solid surfaces to generate NP wires and threads,[90] with network structures generated by tuning the NP-DNA ratios.[91] NPs bearing labile ligands can self-fuse in situ into metal wire-like structures upon DNA binding via electrostatic interactions.[92] Although cationic NPs provide complementary electrostatic interaction for DNA assembly, Wessels and coworkers have reported negatively charged tris(hydroxymethyl)phosphine-capped gold NPs bind with calf thymus DNA immobilized on silicon, forming 30–40 nm narrow nanowires.[93]
In a route exploiting both supramolecular and covalent approaches, Willner et al. have shown the wire-like assembly of psoralen-coated NPs with DNA.[94] Psoralen was intercalated into double stranded polyA/polyT duplex forming a wire-like assembly (Figure 14), then irradiated with UV light, forming 2π + 2π cycloaddition adducts with thymine residues, consequently forming a dense covalent attachment of the NPs to the DNA. Use of λ-DNA also formed gold NP wires, however, with a substantially lower density of particles.
Figure 14.
(a) Chemical structure of psolaren-coated gold NP, (b) AFM image of wire-like assembly of psoralen-coated gold NPs in poly A/poly T template. Reproduced with permission from Ref [94]. Copyright 2002 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
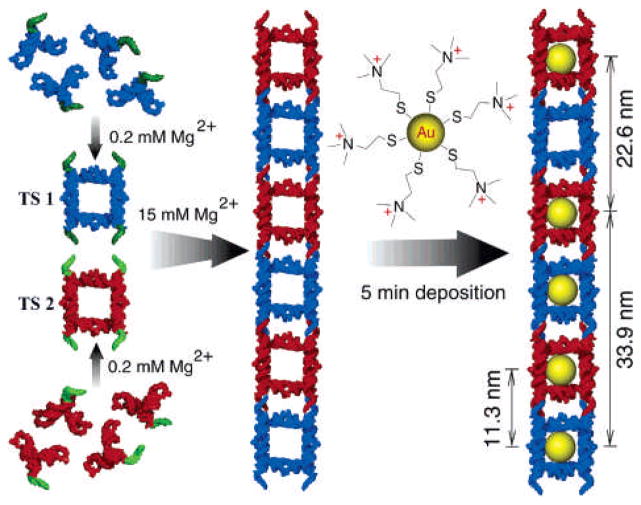
Precise positioning and controlled spacing of NP hybrid nanostructures can be used to generate effective structures for nanoelectronics. Jaeger et al. have examined the assembly of NPs using a nanocrown RNA scaffold.[95] They have generated two small tecto squares (TS) in the presence of 0.2 mM of Mg2+ by self-assembly of four different RNA subunits via loop-loop interactions[96] that can further self assemble to form ladders via complementary tail-tail connectors (Figure 15).
Figure 15.
Schematic illustration of hierarchical supramolecular assembly of TS ladder decorated with cationic gold NPs. Reproduced with permission from Ref [95]. Copyright 2005 American Chemical Society.
4. Nanoparticle-cell interactions
4.1. Effect of NP surface charge on cellular internalization
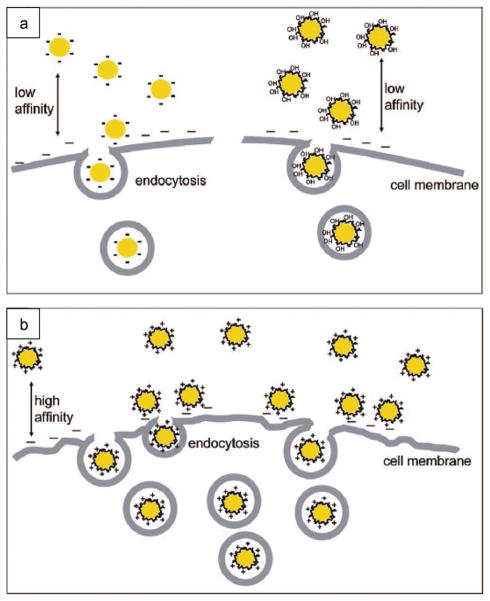
Cellular uptake of nanomaterials can occur through a variety of mechanisms.[97] Xia et al. recently examined the role of surface charge on the internalization of gold NPs (Figure 16).[98] In their findings, positively charged NPs were adsorbed more efficiently on the negatively charged cell surface and consequently showed greater internalization than that of neutral and negatively charged NPs.[98] Surprisingly, negatively charged iron oxide NPs showed cellular uptake and toxicity in HeLa cells.[99] The internalization of negatively charged NPs may occur through nonspecific binding and accumulating of the particles on cationic sites of the plasma membrane followed by endocytosis. Similarly, carboxyl-functionalized poly(amidoamine) (PAMAM) dendrimer-stabilized iron oxide NPs have shown to be uptaken into human epithelial carcinoma cells either via pinocytosis or through a membrane diffusion mechanism.[100]
Figure 16.
Schematic representation of the interaction between gold NPs bearing different surface charge and SK-BR-3 breast cancer cells. (a) Citrate-coated (negative) and PVA-coated (neutral) NPs, (b) poly(allyamine hydrochloride) –coated (positive) NPs. Reproduced with permission from Ref. [98]. Copyright 2009 American Chemical Society.
Recently, Arvizo et al. have demonstrated how surface charge of gold NPs can modulate membrane potential of malignant and normal cell types and their subsequent downstream intracellular events.[101] In their findings, positively charged particles depolarized the cell membrane and increased intracellular Ca2+ concentration. This membrane potential perturbation induced apoptosis in normal cells but cancerous cells remained unaffected. This differential behavior of positively charged gold NPs on normal versus malignant cells presents a potential lead for the creation of therapeutics.
Cationic NPs tend to be toxic, immunogenic,[102] and prone to interactions with serum proteins present in blood, thereby altering delivery profiles of NPs.[103] Therefore the proper choice of surface coating of NPs is crucial to lower toxicity and immunogenicity while increasing transport and delivery efficiency.[104] Proteins provide promising agents for NP functionalization due to their biocompatibility and low toxicity. Samanta et al. have reported the synthesis and hyperthermic effect of BSA-coated Fe3O4 NPs[105] as well as the differential delivery of these NPs into cancerous and isogenic cell types.[106]
4.2. Effect of surface-ligand arrangement on cell-membrane penetration
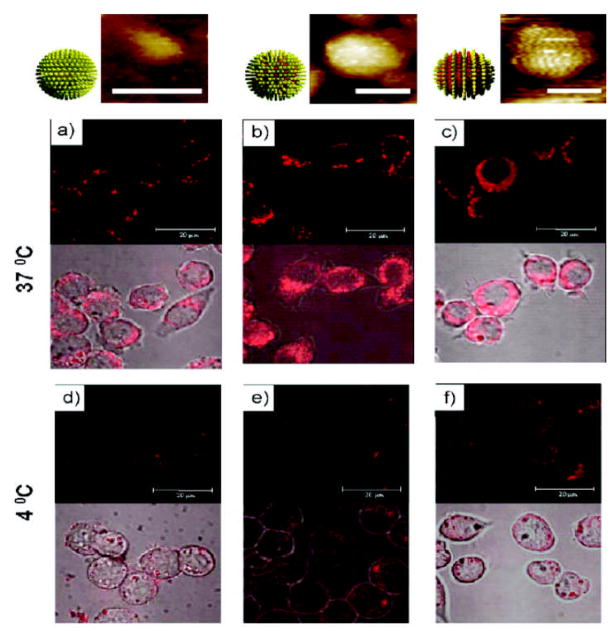
Stellacci et al. have explored the role of ligand arrangement on the NP surface for particle interactions with the cell membrane using amphiphilic gold NPs. The only observed difference between the particles resides in the arrangement of the ligands that coat the gold core.[107] They have shown NPs coated in an ordered alternating ribbon-like arrangement penetrated the cell membrane at 4°C and in the presence of an endocytosis inhibitor. However, NPs coated with the same molecules but in a random arrangement on the surface were inefficient at permeating the cell membrane barriers and were instead trapped in endosomes (Figure 17).
Figure 17.
The role of NP surface ligand arrangement in cell-membrane penetration. Top: Scanning tunneling microscopy (STM) images of NPs with homoligand, and unstructured and structured ligand shells (with scale bar 5 nm). Bottom: Confocal images of mouse dendritic cells incubated with the nanoparticles at a–c) 37 °C and d–f) 4 °C in serum-free condition. Reproduced with permission from Ref [107]. Copyright 2008 Nature Publishing Group.
4.3 Effect of cell-penetrating motifs on NP surface
Cell penetrating peptides (CPP) help in the fusion of NPs with cell membranes and their uptake into the cytoplasm.[108] Different peptides including RGD,[109] allatostatin 1,[110] PLL,[111] and arginine-rich peptides[112] have been used for effective cellular delivery of NPs. Medintz et al. have engineered quantum dots functionalized with cell penetrating peptides for intracellular delivery of fluorescent proteins.[113] They have shown the QD complexes remain endosomally entrapped. Quantum dots coated with cell penetrating TAT peptides[114] were also explored for cellular uptake studies via macropinocytosis.[115] Brust and coworkers have shown that NPs conjugated with CPPs enable cell penetration, whereas NPs coated with CPP and nuclear localizing sequences (NLS) become localized inside the nucleus.[116] These NPs have been used for delivery of drugs and DNA to target cells.[117] Gillies et al. have shown that coating of super paramagnetic iron oxide NPs with dendritic guanidines results in cell-penetration similar to human immunodeficiency virus-1 transactivator (HIV-TAT) peptide. They found that these NPs exhibited greater cell penetration than amine-coated particles and therefore are more toxic.[118]
4. Biosensors based on NP-cell interactions
NPs coated with synthetic ligands can bind with cell surfaces using electrostatic and hydrophobic interactions. These interactions are selective rather than specific, and can be used for identification and differentiation cells. Huang and coworkers used glycan functionalized NPs (MGNPs) to detect E. coli with a limit of 104 cells/mL. They have also achieved the removal of 88% of the E. coli from solutions with 103–104 cells/mL.[119] Phillips et al. have used NP-polymer arrays to identify bacteria.[120] In this study, cationic gold NPs and anionic fluorescent polymer were used to generate non-covalent complexes. In the presence of bacteria, the initially quenched fluorescent polymers recover their fluorescence. The sensor array was used to identify 12 bacteria including both Gram-positive (e.g. A. azurea, B. subtilis) and Gram-negative (e.g. E. coli, P. putida) species, as well as differentiate between three different strains of E. coli at 2×105 cells/mL.
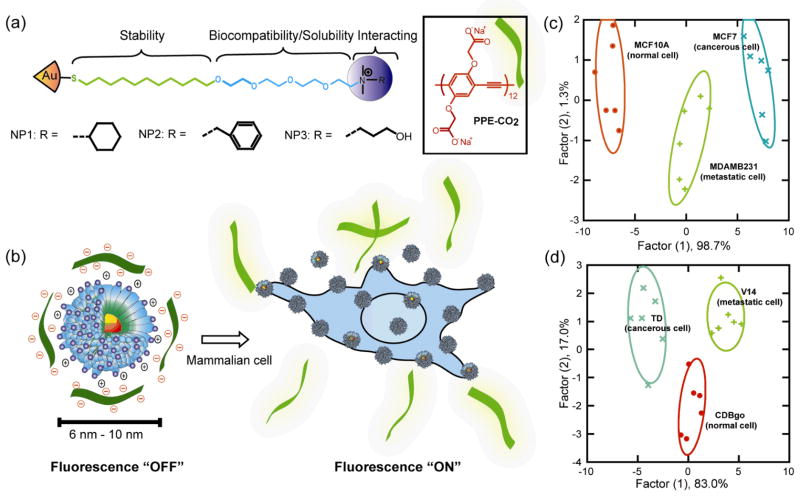
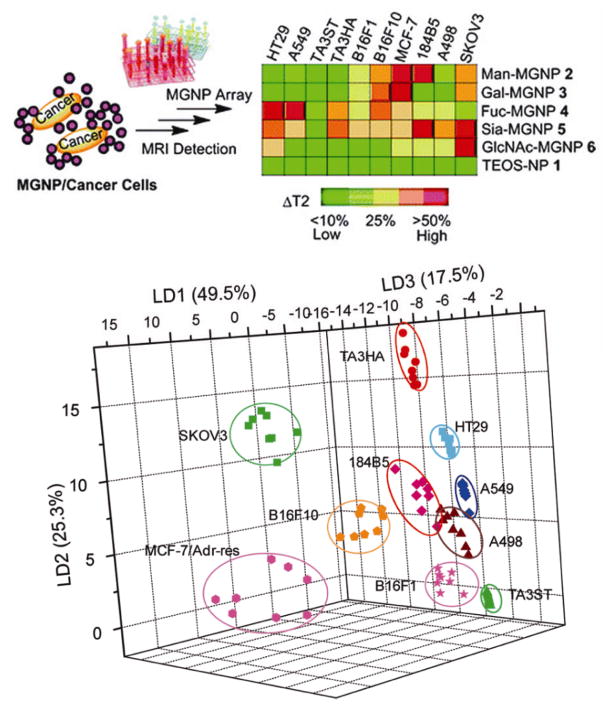
Extending this concept to mammalian cells, gold NP–fluorophore constructs were studied to differentiate normal human and murine cells from their cancerous and metastatic counterparts.[121] This detection system is based on conjugates between three structurally related cationic gold NPs (NP1–NP3) and the poly(para-phenyleneethynylene) (PPE) polymer PPE-CO2 (Figure 18a). Gold NPs quench the fluorescence upon binding with PPE, which was regenerated upon incubation with different cell types in a ‘turn on’ fashion (Figure 18b). With this sensor array construct, different normal, cancerous and metastatic cells from different genotypes (Figure 18c) as well as the same genotype (i.e. isogenic) were identified in a rapid fashion (Figure 18d). An analogous sensor using a gold NP and green fluorescence protein (GFP) complex[122] was capable of full differentiation of the mammalian cells with concentrations of ~5000 cells, a six-fold enhancement of sensitivity. Huang et al. reported a MGNP-based nanosensor array system utilizing carbohydrates as the ligands to detect and differentiate cancer cells and quantitatively profile their carbohydrate binding abilities by magnetic resonance imaging (MRI) signatures (Figure 19).[123]
Figure 18.
(a) Molecular structures of gold NPs and fluorescent polymer; (b) Schematic illustration of competitive binding between the quenched NP-polymer complexes and cell surface; canonical score plot of simplified fluorescence response patterns showing differentiation between normal, cancerous and metastatic cells from (c) different genotypes and (d) same genotype. Reproduced with permission from Ref [121]. Copyright 2009 National Academy of Sciences.
Figure 19.
Schematics of MGNP-based sensor array to differentiate cancer cells and LDA plots for the first three LDs of ΔT2 patterns showing 100% classification The ΔT2 was calculated by dividing the T2 differences between MGNP and MGNP/cancer cell by the corresponding highest ΔT2 from each MGNP category. Reproduced with permission from Ref [123]. Copyright 2010 American Chemical Society.
5. Summary & Outlook
NPs possess unique physical and chemical properties that make them versatile scaffolds for biomolecular recognition. The surface properties of NPs play a critical role in their interactions with biosystems. High affinity interaction of between NPs and biomolecules, can be achieved using biological ligand-receptor interaction, e.g. biotin-streptavidin, or multivalent interactions of simple organic ligands, in particular NPs bearing electrostatic and/or hydrophobic recognition elements. The examples presented in this review clearly demonstrate the use of NP monolayers as a platforms for biomoleculear recognition with applications in therapeutics and diagnostics. From a materials standpoint, DNA and proteins can be assembled onto NP surfaces to generate hybrid nanostructures with synergistic properties and functions. The surface properties of the NPs likewise.
As discussed above, a number of important fundamental and pragmatic advances have been made in the engineering of NP surfaces for controlled interactions with biomolecules or cells. Harnessing the complete capabilities of NPs for biological applications requires the integration of synthetic finesse with a true understanding of their interactions within biosystems. To date, substantial progress has been made in this direction, however much remains to be learned before we truly understand this complex interplay. We are currently at the early stages of the scientific method, where collection of empirical data is essential for formulating hypotheses. Generation of hypotheses through the integration of materials science, biology, and theory should provide us with the predictive capabilities required to access the full potential of NPs in biology and medicine.
Acknowledgments
Support from the NIH (GM077173, EB012246 and ES017871) is gratefully acknowledged.
Biographies

Krishnendu Saha
Krishnendu Saha received his B.Sc. in Chemistry from Jadavpur University, India in 2006 and M.Sc. in Chemistry from Indian Institute of Technology-Madras, India in 2008. He is currently pursuing his Ph.D. at the Department of Chemistry, University of Massachusetts at Amherst, USA under the supervision of Professor Vincent M. Rotello. His current research interest involves the use of gold nanoparticles for cell-surface recognition, cancer diagnosis and delivery applications.

Avinash Bajaj
Avinash Bajaj has obtained his PhD from Indian Institute of Science, Bangalore with Prof. Santanu Bhattacharya followed by a postdoctoral training with Prof. Vincent M. Rotello at University of Massachusetts, Amherst, USA. Currently, he is an assistant professor and Ramanujan Fellow at Regional Centre for Biotechnology. His research interests include engineering of nanomaterials for drug delivery, gene therapy, biosensors and tissue engineering.

Bradley Duncan
Bradley Duncan received his H.A.B. degree in Chemistry from Saint Anselm College in 2009. Currently, he is a Ph.D. student in the Department of Chemistry at the University of Massachusetts-Amherst under the guidance of Professor Vincent Rotello. His current research focuses on the self-assembly of nanoparticles at the liquid liquid interface and the fabrication of colloidal micro/nano capsules for delivery applications.

Vincent Rotello
Vincent Rotello is the Charles A. Goessmann Professor of Chemistry at the University of Massachusetts at Amherst, with an appointment in the Program in Molecular and Cellular Biology. He has been the recipient of the NSF CAREER and Cottrell Scholar awards, as well as the Camille Dreyfus Teacher-Scholar, and the Sloan Fellowships, and is a Fellow of the American Association for the Advancement of Science (AAAS) and of the Royal Society of Chemistry (U.K.). He is currently an Executive Editor for Advanced Drug Delivery Reviews and Associate Editor for North America for the Journal of Materials Chemistry, and is on the Editorial Board of nine other journals. His research program focuses on engineering the interface between hard and soft materials, and spans the areas of devices, polymers, and nanotechnology/bionanotechnology, with over 300 papers published to date.
Footnotes
Congratulating Chad Mirkin on twenty years (and counting!) of groundbreaking science.
References
- 1.(a) Stites WE. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]; (b) Jones S, Thornton JM. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Keskin O, Gursoy A, Ma B, Nussinov R. Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]; (d) de Rinaldis M, Ausiello G, Cesareni G, Helmer-Citterich M. J Mol Biol. 1998;284:1211–1221. doi: 10.1006/jmbi.1998.2248. [DOI] [PubMed] [Google Scholar]
- 2.(a) Spyrakis F, BidonChanal A, Barril X, Luque FJ. Curr Top Med Chem. 2011;11:192–210. doi: 10.2174/156802611794863571. [DOI] [PubMed] [Google Scholar]; (b) Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 3.(a) You CC, Verma A, Rotello VM. Soft Matter. 2006;2:190–204. doi: 10.1039/b517354j. [DOI] [PubMed] [Google Scholar]; (b) You CC, Chompoosor A, Rotello VM. Nano Today. 2007;2:34–43. [Google Scholar]; (c) Yin H, Hamilton AD. Angew Chem Int Ed. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 4.(a) De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225–4241. [Google Scholar]; (b) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 5.(a) Redl FX, Black CT, Papaefthymiou GC, Sandstrom RL, Yin M, Zeng H, Murray CB, O’Brien SP. J Am Chem Soc. 2004;126:14583–14599. doi: 10.1021/ja046808r. [DOI] [PubMed] [Google Scholar]; (b) Li Z, Sun Q, Gao MY. Angew Chem Int Ed. 2005;44:123–126. doi: 10.1002/anie.200460715. [DOI] [PubMed] [Google Scholar]; (c) Sun RWY, Chen R, Chung NPY, Ho CM, Lin CLS, Che CM. Chem Commun. 2005:5059–5061. doi: 10.1039/b510984a. [DOI] [PubMed] [Google Scholar]
- 6.(a) Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv Drug Delivery Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Daniel MC, Astruc D. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]; (c) Hostetler MJ, Wingate JE, Zhong CJ, Harris JE, Vachet RW, Clark MR, Londono JD, Green SJ, Stokes JJ, Wignall GD, Glish GL, Porter MD, Evans ND, Murray RW. Langmuir. 1998;14:17–30. [Google Scholar]; (d) Nam JM, Stoeva SI, Mirkin CA. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]; (e) Xiao Y, Patolsky F, Katz E, Hainfeld JF, Willner I. Science. 2003;299:1877–1881. doi: 10.1126/science.1080664. [DOI] [PubMed] [Google Scholar]; (f) Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]; (g) Boisselier E, Astruc D. Chem Soc Rev. 2009;38:1759–1782. doi: 10.1039/b806051g. [DOI] [PubMed] [Google Scholar]; (h) Hung A, Mwenifumbo S, Mager M, Kuna JJ, Stellacci F, Yarovsky I, Stevens MM. J Am Chem Soc. 2011;133:1438–1450. doi: 10.1021/ja108285u. [DOI] [PubMed] [Google Scholar]; (i) Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 7.(a) Thanh NTK, Green LAW. Nano Today. 2010;5:213–230. [Google Scholar]; (b) Hazarika P, Ceyhan B, Niemeyer CM. Small. 2005;1:844–848. doi: 10.1002/smll.200500063. [DOI] [PubMed] [Google Scholar]; (c) Agasti SS, Rana S, Park MH, Kim CK, You CC, Rotello VM. Adv Drug Delivery Rev. 2010;62:316–328. doi: 10.1016/j.addr.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Niemeyer CM. Angew Chem Int Ed. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]; (e) Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Nat Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 8.(a) Bayraktar H, Srivastava S, You CC, Rotello VM, Knapp MJ. Soft Matter. 2008;4:751–756. doi: 10.1039/b716386j. [DOI] [PubMed] [Google Scholar]; (b) Jones MR, Macfarlane RJ, Lee B, Zhang JA, Young KL, Senesi AJ, Mirkin CA. Nat Mater. 2010;9:913–917. doi: 10.1038/nmat2870. [DOI] [PubMed] [Google Scholar]; (c) Ofir Y, Samanta B, Rotello VM. Chem Soc Rev. 2008;37:1814–1823. doi: 10.1039/b712689c. [DOI] [PubMed] [Google Scholar]; (d) Fillon Y, Verma A, Ghosh P, Ernenwein D, Rotello VM, Chmielewski J. J Am Chem Soc. 2007;129:6676–6677. doi: 10.1021/ja070301+. [DOI] [PubMed] [Google Scholar]
- 9.(a) de la Rica R, Matsui H. Chem Soc Rev. 2010;39:3499–3509. doi: 10.1039/b917574c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wu ZC, Zhang B, Yan B. Int J Mol Sci. 2009;10:4198–4209. doi: 10.3390/ijms10104198. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dickerson MB, Sandhage KH, Naik RR. Chem Rev. 2008;108:4935–4978. doi: 10.1021/cr8002328. [DOI] [PubMed] [Google Scholar]
- 10.(a) Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]; (b) Berry CC. J Mater Chem. 2005;15:543–547. [Google Scholar]; (c) Mailander V, Landfester K. Biomacromolecules. 2009;10:2379–2400. doi: 10.1021/bm900266r. [DOI] [PubMed] [Google Scholar]; (d) Kumari A, Yadav SK. Expert Opin Drug Del. 2011;8:141–151. doi: 10.1517/17425247.2011.547934. [DOI] [PubMed] [Google Scholar]; (d) Vasir JK, Labhasetwar V. Biomaterials. 2008;29:4244–4252. doi: 10.1016/j.biomaterials.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Cho EC, Au L, Zhang Q, Xio Y. Small. 2010;6:517–522. doi: 10.1002/smll.200901622. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chompoosor A, Saha K, Ghosh PS, Macarthy DJ, Miranda OR, Zhu ZJ, Arcaro KF, Rotello VM. Small. 2010;6:2246–2249. doi: 10.1002/smll.201000463. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chung TH, Wu SH, Yao M, Lu CW, Lin YS, Hung Y, Mou CY, Chen YC, Huang DM. Biomaterials. 2007;28:2959–2966. doi: 10.1016/j.biomaterials.2007.03.006. [DOI] [PubMed] [Google Scholar]; (d) He CB, Hu YP, Yin LC, Tang C, Yin CH. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]; (e) El Badawy AM, Silva RG, Morris B, Scheckel KG, Suidan MT, Tolaymat TM. Environ Sci Technol. 2011;45:283–287. doi: 10.1021/es1034188. [DOI] [PubMed] [Google Scholar]
- 12.(a) Kim CK, Ghosh P, Rotello VM. Nanoscale. 2009;1:61–67. doi: 10.1039/b9nr00112c. [DOI] [PubMed] [Google Scholar]; (b) Ditto AJ, Shah PN, Yun YH. Expert Opin Drug Del. 2009;6:1149–1160. doi: 10.1517/17425240903241796. [DOI] [PubMed] [Google Scholar]; (c) Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Davis ME, Chen Z, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]; (e) Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]; (f) Peer D, Karp JM, Hong S, FaroKHzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]; (g) Xu ZP, Zeng QH, Lu GQ, Yu AB. Chem Eng Sci. 2006;61:1027–1040. [Google Scholar]; (h) Sokolova V, Epple M. Angew Chem Int Ed. 2008;47:1382–1395. doi: 10.1002/anie.200703039. [DOI] [PubMed] [Google Scholar]; (i) Faraji AH, Wipf P. Biorg Med Chem. 2009;17:2950–2962. doi: 10.1016/j.bmc.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 13.(a) Rao KS, Reddy MK, Horning JL, Labhasetwar V. Biomaterials. 2008;29:4429–4438. doi: 10.1016/j.biomaterials.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vives E, Schmidt J, Pelegrin A. BBA Rev Cancer. 2008;1786:126–138. doi: 10.1016/j.bbcan.2008.03.001. [DOI] [PubMed] [Google Scholar]; (c) Mandal D, Maran A, Yaszemski MJ, Bolander ME, Sarkar G. J Mater Sci Mater Med. 2009;20:347–350. doi: 10.1007/s10856-008-3588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Olson ES, Jiang T, Aguilera TA, Nguyen QT, Ellies LG, Scadeng M, Tsien RY. Proc Natl Acad Sci USA. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Liu JQ, Zhang Q, Remsen EE, Wooley KL. Biomacromolecules. 2001;2:362–368. doi: 10.1021/bm015515c. [DOI] [PubMed] [Google Scholar]; (f) Desai P, Patlolla RR, Singh M. Mol Membr Biol. 2010;27:247–259. doi: 10.3109/09687688.2010.522203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Katz E, Willner I. Angew Chem Int Ed. 2004;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]; (b) Sastry M, Rao M, Ganesh KN. Acc Chem Res. 2002;35:847–855. doi: 10.1021/ar010094x. [DOI] [PubMed] [Google Scholar]; (c) Pellegrino T, Kudera S, Liedl T, Javier AM, Manna L, Parak WJ. Small. 2005;1:48–63. doi: 10.1002/smll.200400071. [DOI] [PubMed] [Google Scholar]; (d) Ackerson CJ, Sykes MT, Kornberg RD. Proc Natl Acad Sci USA. 2005;102:13383–13385. doi: 10.1073/pnas.0506290102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; (f) Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]; (g) Mitchell GP, Mirkin CA, Letsinger RL. J Am Chem Soc. 1999;121:8122–8123. [Google Scholar]; (h) Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]; (i) Dyadyusha L, Yin H, Jaiswal S, Brown T, Baumberg JJ, Booy FP, Melvin T. Chem Commun. 2005:3201–3203. doi: 10.1039/b500664c. [DOI] [PubMed] [Google Scholar]; (j) Aubin-Tam ME, Hamad-Schifferli K. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 15.Fischer NO, McIntosh CM, Simard JM, Rotello VM. Proc Natl Acad Sci USA. 2002;99:5018–5023. doi: 10.1073/pnas.082644099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Blow DM. Acc Chem Res. 1976;9:145–152. [Google Scholar]; (b) Tsukada H, Blow DM. J Mol Biol. 1985;184:703–711. doi: 10.1016/0022-2836(85)90314-6. [DOI] [PubMed] [Google Scholar]
- 17.Verma A, Simard JM, Rotello VM. Langmuir. 2004;20:4178–4181. doi: 10.1021/la036183v. [DOI] [PubMed] [Google Scholar]
- 18.Fischer NO, Verma A, Goodman CM, Simard JM, Rotello VM. J Am Chem Soc. 2003;125:13387–13391. doi: 10.1021/ja0352505. [DOI] [PubMed] [Google Scholar]
- 19.Plunkett KN, Mohraz A, Haasch RT, Lewis JA, Moore JS. J Am Chem Soc. 2005;127:14574–14575. doi: 10.1021/ja054666a. [DOI] [PubMed] [Google Scholar]
- 20.Fischer NO, Paulini R, Drechsler U, Rotello VM. Chem Commun. 2004:2866–2867. doi: 10.1039/b408972c. [DOI] [PubMed] [Google Scholar]
- 21.a) Hong R, Emrick T, Rotello VM. J Am Chem Soc. 2004;126:13572–13573. doi: 10.1021/ja0461163. [DOI] [PubMed] [Google Scholar]; b) Jordan BJ, Hong R, Gider B, Hill J, Emrick T, Rotello VM. Soft Matter. 2006;2:558–560. doi: 10.1039/b603980d. [DOI] [PubMed] [Google Scholar]
- 22.You CC, De M, Han G, Rotello VM. J Am Chem Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 23.(a) Fukada H, Takahashi K, Sturtevant JM. Biochemistry. 1985;24:5109–5115. doi: 10.1021/bi00340a023. [DOI] [PubMed] [Google Scholar]; (b) Chen CP, Hsu CH, Su NY, Lin YC, Chiou SH, Wu SH. J Biol Chem. 2001;276:45079–45087. doi: 10.1074/jbc.M106182200. [DOI] [PubMed] [Google Scholar]
- 24.You CC, De M, Rotello VM. Org Lett. 2005;7:5685–5688. doi: 10.1021/ol052367k. [DOI] [PubMed] [Google Scholar]
- 25.De M, You CC, Srivastava S, Rotello VM. J Am Chem Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 26.You CC, Agasti SS, Rotello VM. Chem Eur J. 2008;14:143–150. doi: 10.1002/chem.200701234. [DOI] [PubMed] [Google Scholar]
- 27.Bayraktar H, You CC, Rotello VM, Knapp MJ. J Am Chem Soc. 2007;129:2732–2733. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]
- 28.Hong R, Fischer NO, Verma A, Goodman CM, Emrick T, Rotello VM. J Am Chem Soc. 2004;126:739–743. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- 29.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh P, Yang XC, Arvizo R, Zhu ZJ, Agasti SS, Mo ZH, Rotello VM. J Am Chem Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Susumu K, Uyeda HT, Medintz IL, Pons T, Delehanty JB, Mattoussi H. J Am Chem Soc. 2007;129:13987–13996. doi: 10.1021/ja0749744. [DOI] [PubMed] [Google Scholar]; (b) Hildebrandt N, Charbonniere LJ, Beck M, Ziessel RF, Lohmannsroben HG. Angew Chem Int Ed. 2005;44:7612–7615. doi: 10.1002/anie.200501552. [DOI] [PubMed] [Google Scholar]
- 32.(a) Miyamoto S, Kollman PA. Proc Natl Acad Sci USA. 1993;90:8402–8406. doi: 10.1073/pnas.90.18.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 33.Zheng M, Huang XY. J Am Chem Soc. 2004;126:12047–12054. doi: 10.1021/ja047029d. [DOI] [PubMed] [Google Scholar]
- 34.Lee YC. FASEB J. 1992;6:3193–3200. doi: 10.1096/fasebj.6.13.1397841. [DOI] [PubMed] [Google Scholar]
- 35.Jiang XZ, Housni A, Gody G, Boullanger P, Charreyre MT, Delair T, Narain R. Bioconjugate Chem. 2010;21:521–530. doi: 10.1021/bc900431p. [DOI] [PubMed] [Google Scholar]
- 36.Lin CC, Yeh YC, Yang CY, Chen GF, Chen YC, Wu YC, Chen CC. Chem Commun. 2003:2920–2921. doi: 10.1039/b308995a. [DOI] [PubMed] [Google Scholar]
- 37.Tsai CS, Yu TB, Chen CT. Chem Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 38.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J Am Chem Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 39.Robinson A, Fang JM, Chou PT, Liao KW, Chu RM, Lee SJ. ChemBioChem. 2005;6:1899–1905. doi: 10.1002/cbic.200500112. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Ramstrom O, Yan MD. J Mater Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fazal MA, Roy BC, Sun SG, Mallik S, Rodgers KR. J Am Chem Soc. 2001;123:6283–6290. doi: 10.1021/ja003193z. [DOI] [PubMed] [Google Scholar]
- 42.(a) Abad JM, Mertens SFL, Pita M, Fernandez VM, Schiffrin DJ. J Am Chem Soc. 2005;127:5689–5694. doi: 10.1021/ja042717i. [DOI] [PubMed] [Google Scholar]; (b) Kim JS, Valencia CA, Liu RH, Lin WB. Bioconjugate Chem. 2007;18:333–341. doi: 10.1021/bc060195l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu CJ, Xu KM, Gu HW, Zhong XF, Guo ZH, Zheng RK, Zhang XX, Xu B. J Am Chem Soc. 2004;126:3392–3393. doi: 10.1021/ja031776d. [DOI] [PubMed] [Google Scholar]
- 44.Boeneman K, Mei BC, Dennis AM, Bao G, Deschamps JR, Mattoussi H, Medintz IL. J Am Chem Soc. 2009;131:3828–3829. doi: 10.1021/ja809721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De M, Rana S, Rotello VM. Macromol Biosci. 2009;9:174–178. doi: 10.1002/mabi.200800289. [DOI] [PubMed] [Google Scholar]
- 46.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 47.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li LS, Stupp SI. Angew Chem Int Ed. 2005;44:1833–1836. doi: 10.1002/anie.200462142. [DOI] [PubMed] [Google Scholar]
- 50.Fu XY, Wang Y, Huang LX, Sha YL, Gui LL, Lai LH, Tang YQ. Adv Mater. 2003;15:902–906. [Google Scholar]
- 51.Ernenwein D, Ghosh P, Rotello V, Chmielewski J. J Mater Chem. 2010;20:5608–5611. [Google Scholar]
- 52.(a) Aili D, Enander K, Rydberg J, Lundstrom I, Baltzer L, Liedberg B. J Am Chem Soc. 2006;128:2194–2195. doi: 10.1021/ja057056j. [DOI] [PubMed] [Google Scholar]; (b) Aili D, Enander K, Rydberg J, Nesterenko I, Bjorefors F, Baltzer L, Liedberg B. J Am Chem Soc. 2008;130:5780–5788. doi: 10.1021/ja711330f. [DOI] [PubMed] [Google Scholar]; (c) Stevens MM, Flynn NT, Wang C, Tirrell DA, Langer R. Adv Mater. 2004;16:915–918. [Google Scholar]
- 53.Coomber D, Bartczak D, Gerrard SR, Tyas S, Kanaras AG, Stulz E. Langmuir. 2010;26:13760–13762. doi: 10.1021/la1023554. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharya R, Patra CR, Wang SF, Lu LC, Yaszemski MJ, Mukhopadhyay D, Mukherjee P. Adv Funct Mater. 2006;16:395–400. [Google Scholar]
- 55.Shenton W, Davis SA, Mann S. Adv Mater. 1999;11:449–452. [Google Scholar]
- 56.Yang J, Mayer M, Kriebel JK, Garstecki P, Whitesides GM. Angew Chem Int Ed. 2004;43:1555–1558. doi: 10.1002/anie.200353161. [DOI] [PubMed] [Google Scholar]
- 57.(a) Cobbe S, Connolly S, Ryan D, Nagle L, Eritja R, Fitzmaurice D. J Phys Chem B. 2003;107:470–477. [Google Scholar]; (b) Li M, Wong KKW, Mann S. Chem Mater. 1999;11:23–26. [Google Scholar]; (c) Costanzo PJ, Patten TE, Seery TAP. Chem Mater. 2004;16:1775–1785. [Google Scholar]; (d) Smorodin T, Beierlein U, Kotthaus JP. Nanotechnology. 2005;16:1123–1125. [Google Scholar]; (e) Aslan K, Holley P, Davies L, Lakowicz JR, Geddes CD. J Am Chem Soc. 2005;127:12115–12121. doi: 10.1021/ja052739k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connolly S, Fitzmaurice D. Adv Mater. 1999;11:1202–1205. [Google Scholar]
- 59.Aslan K, Luhrs CC, Perez-Luna VH. J Phys Chem B. 2004;108:15631–15639. [Google Scholar]
- 60.Srivastava S, Verma A, Frankamp BL, Rotello VM. Adv Mater. 2005;17:617–621. [Google Scholar]
- 61.Wei G, Pan C, Reichert J, Jandt KD. Carbon. 2010;48:645–653. [Google Scholar]
- 62.(a) De M, Miranda OR, Rana S, Rotello VM. Chem Commun. 2009:2157–2159. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]; (b) Hu MH, Qian LP, Brinas RP, Lymar ES, Hainfeld JF. Angew Chem Int Ed. 2007;46:5111–5114. doi: 10.1002/anie.200701180. [DOI] [PubMed] [Google Scholar]
- 63.Verma A, Srivastava S, Rotello VM. Chem Mater. 2005;17:6317–6322. [Google Scholar]
- 64.Srivastava S, Samanta B, Jordan BJ, Hong R, Xiao Q, Tuominen MT, Rotello VM. J Am Chem Soc. 2007;129:11776–11780. doi: 10.1021/ja073163x. [DOI] [PubMed] [Google Scholar]
- 65.Samanta B, Yang XC, Ofir Y, Park MH, Patra D, Agasti SS, Miranda OR, Mo ZH, Rotello VM. Angew Chem Int Ed. 2009;48:5341–5344. doi: 10.1002/anie.200901590. [DOI] [PubMed] [Google Scholar]
- 66.Armitage BA. Top Curr Chem. 2005;253:55–76. [Google Scholar]
- 67.(a) Gopal YNV, Van Dyke MW. Biochemistry. 2003;42:6891–6903. doi: 10.1021/bi027373s. [DOI] [PubMed] [Google Scholar]; (b) Fechter EJ, Dervan PB. J Am Chem Soc. 2003;125:8476–8485. doi: 10.1021/ja030125e. [DOI] [PubMed] [Google Scholar]; (c) Oyoshi T, Kawakami W, Narita A, Bando T, Sugiyama H. J Am Chem Soc. 2003;125:4752–4754. doi: 10.1021/ja029196o. [DOI] [PubMed] [Google Scholar]
- 68.(a) Vazquez ME, Caamano AM, Mascarenas JL. Chem Soc Rev. 2003;32:338–349. doi: 10.1039/b206274g. [DOI] [PubMed] [Google Scholar]; (b) Kovacic RT, Welch JT, Franklin SJ. J Am Chem Soc. 2003;125:6656–6662. doi: 10.1021/ja0210998. [DOI] [PubMed] [Google Scholar]
- 69.(a) Koehler AN, Shamji AF, Schreiber SL. J Am Chem Soc. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]; (b) Fechter EJ, Olenyuk B, Dervan PB. Angew Chem Int Ed. 2004;43:3591–3594. doi: 10.1002/anie.200454231. [DOI] [PubMed] [Google Scholar]
- 70.(a) Mahtab R, Harden HH, Murphy CJ. J Am Chem Soc. 2000;122:14–17. [Google Scholar]; (b) Lakowicz JR, Gryczynski I, Gryczynski Z, Nowaczyk K, Murphy CJ. Anal Biochem. 2000;280:128–136. doi: 10.1006/abio.2000.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Capek I. Adv Colloid Interface Sci. 2011;163:123–143. doi: 10.1016/j.cis.2011.02.007. [DOI] [PubMed] [Google Scholar]; (d) Basu S, Jana S, Pande S, Pal T. J Colloid Interface Sci. 2008;321:288–293. doi: 10.1016/j.jcis.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 71.Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv Drug Delivery Rev. 2008;60:1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 72.McIntosh CM, Esposito EA, Boal AK, Simard JM, Martin CT, Rotello VM. J Am Chem Soc. 2001;123:7626–7629. doi: 10.1021/ja015556g. [DOI] [PubMed] [Google Scholar]
- 73.Han G, Martin CT, Rotello VM. Chem Biol Drug Des. 2006;67:78–82. doi: 10.1111/j.1747-0285.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 74.Goodman CM, Chari NS, Han G, Hong R, Ghosh P, Rotello VM. Chem Biol Drug Des. 2006;67:297–304. doi: 10.1111/j.1747-0285.2006.00372.x. [DOI] [PubMed] [Google Scholar]
- 75.He XX, Wang KM, Tan WH, Liu B, Lin X, He CM, Li D, Huang SS, Li J. J Am Chem Soc. 2003;125:7168–7169. doi: 10.1021/ja034450d. [DOI] [PubMed] [Google Scholar]
- 76.Han G, Chari NS, Verma A, Hong R, Martin CT, Rotello VM. Bioconjugate Chem. 2005;16:1356–1359. doi: 10.1021/bc050173j. [DOI] [PubMed] [Google Scholar]
- 77.Sandhu KK, McIntosh CM, Simard JM, Smith SW, Rotello VM. Bioconjugate Chem. 2002;13:3–6. doi: 10.1021/bc015545c. [DOI] [PubMed] [Google Scholar]
- 78.(a) Kneuer C, Sameti M, Bakowsky U, Schiestel T, Schirra H, Schmidt H, Lehr CM. Bioconjugate Chem. 2000;11:926–932. doi: 10.1021/bc0000637. [DOI] [PubMed] [Google Scholar]; (b) Song HT, Choi JS, Huh YM, Kim S, Jun YW, Suh JS, Cheon J. J Am Chem Soc. 2005;127:9992–9993. doi: 10.1021/ja051833y. [DOI] [PubMed] [Google Scholar]
- 79.Thomas M, Klibanov AM. Proc Natl Acad Sci USA. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghosh PS, Kim CK, Han G, Forbes NS, Rotello VM. ACS Nano. 2008;2:2213–2218. doi: 10.1021/nn800507t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han G, You CC, Kim BJ, Turingan RS, Forbes NS, Martin CT, Rotello VM. Angew Chem Int Ed. 2006;45:3165–3169. doi: 10.1002/anie.200600214. [DOI] [PubMed] [Google Scholar]
- 82.Wang GL, Zhang J, Murray RW. Anal Chem. 2002;74:4320–4327. doi: 10.1021/ac0257804. [DOI] [PubMed] [Google Scholar]
- 83.Storhoff JJ, Mirkin CA. Chem Rev. 1999;99:1849–1862. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]
- 84.(a) Storhoff JJ, Lazarides AA, Mucic RC, Mirkin CA, Letsinger RL, Schatz GC. J Am Chem Soc. 2000;122:4640–4650. [Google Scholar]; (b) Park SJ, Lazarides AA, Storhoff JJ, Pesce L, Mirkin CA. J Phys Chem B. 2004;108:12375–12380. [Google Scholar]; (c) Deng ZX, Tian Y, Lee SH, Ribbe AE, Mao CD. Angew Chem Int Ed. 2005;44:3582–3585. doi: 10.1002/anie.200463096. [DOI] [PubMed] [Google Scholar]
- 85.(a) Yun CS, Khitrov GA, Vergona DE, Reich NO, Strouse GF. J Am Chem Soc. 2002;124:7644–7645. doi: 10.1021/ja025971o. [DOI] [PubMed] [Google Scholar]; (b) Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]; (c) Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]; (d) McKenzie F, Faulds K, Graham D. Small. 2007;3:1866–1868. doi: 10.1002/smll.200700225. [DOI] [PubMed] [Google Scholar]; (e) Das J, Yang H. J Phys Chem C. 2009;113:6093–6099. [Google Scholar]
- 86.Wang GL, Murray RW. Nano Lett. 2004;4:95–101. [Google Scholar]
- 87.Wu AG, Cheng WL, Li ZA, Jiang JG, Wang EK. Talanta. 2006;68:693–699. doi: 10.1016/j.talanta.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 88.Wei G, Wang L, Zhou HL, Liu ZG, Song YH, Li ZA. Appl Surf Sci. 2005;252:1189–1196. [Google Scholar]
- 89.Srivastava S, Samanta B, Arumugam P, Han G, Rotello VM. J Mater Chem. 2007;17:52–55. [Google Scholar]
- 90.(a) Kumar A, Pattarkine M, Bhadbhade M, Mandale AB, Ganesh KN, Datar SS, Dharmadhikari CV, Sastry M. Adv Mater. 2001;13:341–344. [Google Scholar]; (b) Sastry M, Kumar A, Datar S, Dharmadhikari CV, Ganesh KN. Appl Phys Lett. 2001;78:2943–2945. [Google Scholar]; (c) Gourishankar A, Shukla S, Pasricha R, Sastry M, Ganesh KN. Curr Appl Phys. 2005;5:102–107. [Google Scholar]; (d) Warner MG, Hutchison JE. Nat Mater. 2003;2:272–277. doi: 10.1038/nmat853. [DOI] [PubMed] [Google Scholar]; (e) Woehrle GH, Warner MG, Hutchison JE. Langmuir. 2004;20:5982–5988. doi: 10.1021/la049491h. [DOI] [PubMed] [Google Scholar]; (f) Nakao H, Shiigi H, Yamamoto Y, Tokonami S, Nagaoka T, Sugiyama S, Ohtani T. Nano Lett. 2003;3:1391–1394. [Google Scholar]; (g) Kinsella JM, Ivanisevic A. J Am Chem Soc. 2005;127:3276–3277. doi: 10.1021/ja043865b. [DOI] [PubMed] [Google Scholar]; (h) Braun G, Inagaki K, Estabrook RA, Wood DK, Levy E, Cleland AN, Strouse GF, Reich NO. Langmuir. 2005;21:10699–10701. doi: 10.1021/la0515367. [DOI] [PubMed] [Google Scholar]
- 91.Ganguli M, Babu JV, Maiti S. Langmuir. 2004;20:5165–5170. doi: 10.1021/la036049a. [DOI] [PubMed] [Google Scholar]
- 92.Yonezawa T, Onoue SY, Kimizuka N. Chem Lett. 2002:1172–1173. [Google Scholar]
- 93.Harnack O, Ford WE, Yasuda A, Wessels JM. Nano Lett. 2002;2:919–923. [Google Scholar]
- 94.Patolsky F, Weizmann Y, Lioubashevski O, Willner I. Angew Chem Int Ed. 2002;41:2323–2327. doi: 10.1002/1521-3773(20020703)41:13<2323::AID-ANIE2323>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 95.Koyfman AY, Braun G, Magonov S, Chworos A, Reich NO, Jaeger L. J Am Chem Soc. 2005;127:11886–11887. doi: 10.1021/ja051144m. [DOI] [PubMed] [Google Scholar]
- 96.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 97.(a) Harush-Frenkel O, Debotton N, Benita S, Altschuler Y. Biochem Biophys Res Commun. 2007;353:26–32. doi: 10.1016/j.bbrc.2006.11.135. [DOI] [PubMed] [Google Scholar]; (b) Harush-Frenkel O, Rozentur E, Benita S, Altschuler Y. Biomacromolecules. 2008;9:435–443. doi: 10.1021/bm700535p. [DOI] [PubMed] [Google Scholar]; (c) Zaki NM, Tirelli N. Expert Opin Drug Del. 2010;7:895–913. doi: 10.1517/17425247.2010.501792. [DOI] [PubMed] [Google Scholar]; (d) Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhang LW, Monteiro-Riviere NA. Toxicol Sci. 2009;110:138–155. doi: 10.1093/toxsci/kfp087. [DOI] [PubMed] [Google Scholar]
- 98.Cho EC, Xie JW, Wurm PA, Xia YN. Nano Lett. 2009;9:1080–1084. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- 99.(a) Villanueva A, Canete M, Roca AG, Calero M, Veintemillas-Verdaguer S, Serna CJ, Morales MD, Miranda R. Nanotechnology. 2009;20:115103. doi: 10.1088/0957-4484/20/11/115103. [DOI] [PubMed] [Google Scholar]; (b) Wilhelm C, Billotey C, Roger J, Pons JN, Bacri JC, Gazeau F. Biomaterials. 2003;24:1001–1011. doi: 10.1016/s0142-9612(02)00440-4. [DOI] [PubMed] [Google Scholar]
- 100.Shi XY, Thomas TP, Myc LA, Kotlyar A, Baker JR. Phys Chem Chem Phys. 2007;9:5712–5720. doi: 10.1039/b709147h. [DOI] [PubMed] [Google Scholar]
- 101.Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, Rotello VM, Prakash YS, Mukherjee P. Nano Lett. 2010;10:2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.(a) Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Bioconjugate Chem. 2004;15:897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]; (b) Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Holl MMB. Nano Lett. 2008;8:420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]; (c) Lin JQ, Zhang HW, Chen Z, Zheng YG. ACS Nano. 2010;4:5421–5429. doi: 10.1021/nn1010792. [DOI] [PubMed] [Google Scholar]; (d) Chen JM, Hessler JA, Putchakayala K, Panama BK, Khan DP, Hong S, Mullen DG, DiMaggio SC, Som A, Tew GN, Lopatin AN, Baker JR, Holl MMB, Orr BG. J Phys Chem B. 2009;113:11179–11185. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Fadeel B, Garcia-Bennett AE. Adv Drug Delivery Rev. 2010;62:362–374. doi: 10.1016/j.addr.2009.11.008. [DOI] [PubMed] [Google Scholar]; (f) Kedmi R, Ben-Arie N, Peer D. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 103.(a) Bajaj A, Kondaiah P, Bhattacharya S. Biomacromolecules. 2008;9:991–999. doi: 10.1021/bm700930y. [DOI] [PubMed] [Google Scholar]; (b) Patil S, Sandberg A, Heckert E, Self W, Seal S. Biomaterials. 2007;28:4600–4607. doi: 10.1016/j.biomaterials.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Oupicky D, Ogris M, Howard KA, Dash PR, Ulbrich K, Seymour LW. Mol Ther. 2002;5:463. doi: 10.1006/mthe.2002.0568. [DOI] [PubMed] [Google Scholar]
- 104.Bajaj A, Paul B, Kondaiah P, Bhattacharya S. Bioconjugate Chem. 2008;19:1283–1300. doi: 10.1021/bc700474r. [DOI] [PubMed] [Google Scholar]
- 105.Samanta B, Yan H, Fischer NO, Shi J, Jerry DJ, Rotello VM. J Mater Chem. 2008;18:1204–1208. doi: 10.1039/b718745a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bajaj A, Samanta B, Yan HH, Jerry DJ, Rotello VM. J Mater Chem. 2009;19:6328–6331. [Google Scholar]
- 107.Verma A, Uzun O, Hu YH, Hu Y, Han HS, Watson N, Chen SL, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.(a) Luhmann T, Rimann M, Bitterman AG, Hall H. Bioconjugate Chem. 2008;19:1907–1916. doi: 10.1021/bc800206r. [DOI] [PubMed] [Google Scholar]; (b) Crombez L, Morris MC, Deshayes S, Heitz F, Divita G. Curr Pharm Design. 2008;14:3656–3665. doi: 10.2174/138161208786898842. [DOI] [PubMed] [Google Scholar]; (c) Olson ES, Jiang T, Aguilera TA, Nguyen QT, Ellies LG, Scadeng M, Tsien RY. Proc Natl Acad Sci USA. 2010;107:4311–4316. doi: 10.1073/pnas.0910283107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liu BR, Huang YW, Chiang HJ, Lee HJ. J Nanosci Nanotechnol. 2010;10:7897–7905. doi: 10.1166/jnn.2010.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Lett. 2007;7:874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 110.Biju V, Muraleedharan D, Nakayama K, Shinohara Y, Itoh T, Baba Y, Ishikawa M. Langmuir. 2007;23:10254–10261. doi: 10.1021/la7012705. [DOI] [PubMed] [Google Scholar]
- 111.Mok H, Park JW, Park TG. Bioconjugate Chem. 2008;19:797–801. doi: 10.1021/bc700464m. [DOI] [PubMed] [Google Scholar]
- 112.Sun LL, Liu DJ, Wang ZX. Langmuir. 2008;24:10293–10297. doi: 10.1021/la8015063. [DOI] [PubMed] [Google Scholar]
- 113.Medintz IL, Pons T, Delehanty JB, Susumu K, Brunel FM, Dawson PE, Mattoussi H. Bioconjugate Chem. 2008;19:1785–1795. doi: 10.1021/bc800089r. [DOI] [PubMed] [Google Scholar]
- 114.(a) Zhang K, Fang HF, Chen ZY, Taylor JSA, Wooley KL. Bioconjugate Chem. 2008;19:1880–1887. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xue FL, Chen JY, Guo J, Wang CC, Yang WL, Wang PN, Lu DR. J Fluoresc. 2007;17:149–154. doi: 10.1007/s10895-006-0152-2. [DOI] [PubMed] [Google Scholar]; (c) Xiong RL, Li Z, Mi L, Wang PN, Chen JY, Wang LX, Yang WL. J Fluoresc. 2010;20:551–556. doi: 10.1007/s10895-009-0579-3. [DOI] [PubMed] [Google Scholar]; (d) Ruan G, Agrawal A, Marcus AI, Nie S. J Am Chem Soc. 2007;129:14759–14766. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 115.Chen B, Liu QL, Zhang YL, Xu L, Fang XH. Langmuir. 2008;24:11866–11871. doi: 10.1021/la802048s. [DOI] [PubMed] [Google Scholar]
- 116.Nativo P, Prior IA, Brust M. ACS Nano. 2008;2:1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 117.Patel PC, Giljohann DA, Seferos DS, Mirkin CA. Proc Natl Acad Sci USA. 2008;105:17222–17226. doi: 10.1073/pnas.0801609105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin AL, Bernas LM, Rutt BK, Foster PJ, Gillies ER. Bioconjugate Chem. 2008;19:2375–2384. doi: 10.1021/bc800209u. [DOI] [PubMed] [Google Scholar]
- 119.El-Boubbou K, Gruden C, Huang X. J Am Chem Soc. 2007;129:13392–13393. doi: 10.1021/ja076086e. [DOI] [PubMed] [Google Scholar]
- 120.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Angew Chem Int Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 121.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Proc Natl Acad Sci USA. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem Sci. 2010;1:134–138. [Google Scholar]
- 123.El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, Huang XF. J Am Chem Soc. 2010;132:4490–4499. doi: 10.1021/ja100455c. [DOI] [PubMed] [Google Scholar]