Abstract
Succinate has been recognized as an important platform chemical that can be produced from biomass. While a number of organisms are capable of succinate production naturally, this review focuses on the engineering of Escherichia coli for production of the four-carbon dicarboxylic acid. Important features of a succinate production system are to achieve optimal balance of reducing equivalents generated by consumption of the feedstock, while maximizing the amount of carbon that is channeled to the product. Aerobic and anaerobic production strains have been developed and applied to production from glucose as well as other abundant carbon sources. Metabolic engineering methods and strain evolution have been used and supplemented by the recent application of systems biology and in silico modeling tools to construct optimal production strains. The metabolic capacity of the production strain, as well as the requirement for efficient recovery of succinate and the reliability of the performance under scale-up are important in the overall process. The costs of the overall biorefinery compatible process will determine the economical commercialization of succinate and its impact in larger chemical markets.
Keywords: Metabolic engineering, Industrial biotechnology, Succinate, Genetic engineering, Biomass feedstock
Introduction
The role of microbial production of useful chemicals from biomass has become of considerable interest to companies, governments, and downstream users as environmental concerns and high petroleum prices provoke assessment of long-term strategies. The higher value of chemicals vs biofuel molecules has also led to heightened interest on the part of biofuel companies as production of such chemicals in a biorefinery could contribute significantly to the overall value generated by the operation. This review will focus on succinate and strategies for engineering its economic production.
Succinic acid has been considered a valuable molecule that could be produced by microorganisms, however the placement of this molecule on the DOE list of twelve platform chemicals from biomass [1] spurred widespread attention to its potential [2]. Among the molecules that can be derived from succinic acid by known chemical processes are 1,4-butanediol, maleic anhydride, succinimide, 2-pyrrolidinone and tetrahydrofuran, all of which have large markets and can be converted to a wide range of other valuable molecules for use in polymers [3], industrial solvents, and specialty chemicals e.g. biodegradable succinate esters [4, 5]. The use of biodegradable polymers in tissue engineering applications is also of interest in the biomedical community [6].
Succinate is used in foods as a flavoring agent as it contributes a somewhat sour and astringent component to umami taste (e.g. in shellfish) and is often added to foods [7]. It is widely available as the mono or disodium salt for adding flavor to meats, soups etc. Succinate is used as a counter ion in many pharmaceutical formulations. Various derivatives of succinate such have found uses for their innocuous properties in food. Esterification with various small alcohols produces additives for the food processing industry. Octenyl succinate modified starch is used as an emulsifier and thickener in many foods requiring an appropriate texture or consistency in the product. Sodium dioctyl sulphosuccinate is used as a surfactant and wetting agent in textile & printing products and was a component of the Corexit dispersant used in the recent Gulf oil spill cleanup operations.
Zeikus et al. [1999] reported a relatively small succinic acid market at 15,000 ton/year worldwide. Recent estimates of the market potential for succinic acid and its immediate derivatives were projected to be as much as 245 × 103 ton/year, with a market size for succinic acid-derived polymers up to 25 × 106 ton/year [8]. While succinate is produced petrochemically from butane through maleic anhydride, fermentation process costs have become more competitive at about $0.55–1.10 per kg [9], and further reductions in cost are anticipated. Several companies and affiliates such as Mitsubishi Chemical, Japan; Myriant Technologies, USA; Diversified Natural Products, USA; Michigan Biotechnology Institute International, USA; BASF, Germany; DSM, The Netherlands and Roquette, France; BioAmber, France are setting up commercial biobased succinate production plants and planning for additional production in Asia.
Recent reviews of succinate production [10] and recovery of succinate from fermentation broth [11] provide background of general interest. The recent development of a succinate production process with E. coli via engineering and strain evolution has been covered in recent articles with attention to genetic methods and culture optimization [12–14] and some earlier articles have discussed the strategies and efforts in succinate production in engineered E. coli [15–17].
While many organisms produce succinate at low levels the analysis of natural isolates that produce a high proportion of succinate as a final metabolite gives a broader context for the engineering efforts. The anaerobic environment of cattle rumen has provided a rich source of such organisms with several widely studied microorganisms capable of high natural production of succinic acid being isolated from rumen. These anaerobic organisms have been studied extensively and key aspects of their specialized metabolism relating to redox and formation of key intermediates such as oxaloacetate have been analyzed. Work with Actinobacillus succinogenes [18] has been recently reviewed [19–21] and it worthwhile to note its early use in electrochemical stimulation of succinate production [22]. Methods to obtain variants of this organisms have also been reported [23]. Recent work with this organism has analyzed the metabolic fluxes [24, 25] and has also focused on the organisms ability to use a number of low cost carbon sources for succinate production [26–29]. Another rumen organism used for succinate production has been Anaerobiospirillum succiniciproducens and examined in terms of its use of various carbon feedstocks and high yield production [30–32] and culture innovations [33]. The organism Mannheimia succiniciproducens was isolated [34], sequenced [35] and genetic tools developed were rapidly generated for the organism [36, 37]. Workers on the organism have studied the metabolism and its novel features have provided an illustration of the application of systems biology and computational approaches to engineering a new organism for production [37–40]. A genome-scale in silico metabolic model [41, 42] and “omics” has given better understanding of cellular physiology and metabolism [39, 43] and development of genetic tools [36, 39, 44] allowed genome-based metabolic engineering [39, 40]. Corynebacterium glutamicum strains [45, 46] and Zymomonas mobilis ZM4 [47], have also been studied. BASF in collaboration with CSM announced production in a bio-succinate facility located in Spain utilizing a newly isolated strain of rumen bacteria, Basfia succiniproducens [48]. A continuous cultivation culture of this bacterium has been studied by Scholten et al. [49] to produce 5.21 g/L succinate from 5.1 g/L glycerol with productivities and yields of 0.094 g/L/h and 1.02 g/g, respectively. Yeasts have also been explored for succinate production as the highly acid and osmotolerance are advantages for succinic acid production [50]. In glucose-grown shake flask cultures, the quadruple deletion strain Δsdh1Δsdh2Δidh1Δidp1 produced succinic acid at a titer of 3.62 g/L at a yield of 0.11 mol/mol glucose. A new strategy employed Yarrowia lipolytica with a deletion in the gene coding one of succinate dehydrogenase subunits was reported [51] and accumulated succinate at the level of 45 g/L in shaking flasks with CaCO3 buffering. DSM and Roquette have jointly developed a process using engineered S. cerevisiae SUC-297 that produced 43 g/L succinic acid in 95 h along with 16.4 g/L ethanol and 14.9 g/L glycerol [52]. The major advantage is that it operates at low pH which prevents bacterial contamination and aids the downstream purification process. These and other organisms have provided a context for the engineering of E. coli for succinate production and this will be the main focus for subsequent sections of the short review. E. coli remains a preferred organism for testing new succinate technologies due to the extensive knowledge of its genome, proteome, availability of genetic tools, simple nutrient requirements and facile cultivation.
The ability to grow easily on a variety of abundant feedstocks is an important practical advantage in developing a commercially viable process. The above organisms as well as some engineered E. coli strains have been examined for their ability to use waste agricultural material and increasingly a focus has been on the use of lignocellulosic derived biomass as a feedstock [53–55]. The processing of such material will not be covered here, only some features needed for efficient use of the mixture of hexoses and pentoses without severe inhibition due to residual acids. Several articles have focused on the use of xylose from hemicellulose found in baggasse [26], straw [56] or corn cobs [29] since this sugar is more readily released upon limited acid hydrolysis. These studies have shown promise in generating useful yields from the pentose available from such biomass sources.
Metabolic engineering strategies
Many microorganisms are natural producers of a wide variety of compounds of industrial interest, e.g. antioxidants, polymers, amino acids, hydroxyacids and chiral alcohols, among others. However, in many cases the production processes are not economically feasible due to a low product yield, low productivity, and/or difficulties on cultivating the native producers. Product yield and productivity are affected by a variety of factors. Reducing equivalents, and the metabolic pathway used by the cells for product synthesis is critical, different pathways leading to the same product could require different precursors and, have different reducing equivalents and energy requirements and theoretical yields [57].
Metabolic engineering (ME) is generally defined as the rational redesign of biological system using genetic engineering techniques by modifying existing or introducing new metabolic pathways to improve production of certain valuable compounds. Therefore, ME principles are applied to the design and construction of more efficient metabolic pathways to increase product yield and productivity, either in the native producer organism or a in a more suitable host [58]. Genes can be deleted to eliminate competing pathways [59–62], overexpressed to increase enzyme pool [61, 63] or introduced from a different organism into a suitable host for industrial purposes [64, 65]. The current availability of genomic information for many organisms has allowed a rapid increase in ME research.
The selected substrate together with the metabolic pathway used by the microorganism will determine the maximum theoretical yield of the product of interest according to carbon and redox balances as well as energy requirements. Substrates with higher reduction states (e.g. glycerol) favor the synthesis of the more reduced products [66–69]. Industrially, the relative cost of the feedstock is an important component of production and this as well as environmental factors need to be considered in the commercial context. The increasing genetic information available together with the development of computational tools has led to the development of different methodologies and models to explain or describe cell metabolic networks attempting to interconnect cell genotype to the corresponding phenotype [70].
An important tool used in ME is the metabolic flux analysis (MFA); this technique studies the effect of genetic changes in the distribution of pathway fluxes. Unknown metabolic fluxes are calculated using experimentally measured metabolite concentrations together with a stoichoimetric model that includes all the important reactions in the network, where the metabolic fluxes are determined for a specific culture condition. In recent years, 13C labeled substrates together with NMR or GC-MS techniques and complex computational schemes have been applied to solve the unknown metabolic flux vector [71]. Information obtained from MFA can serve to identify potential “bottleneck” (rate-limiting) reactions and to understand the effect of the genetic alterations in cell metabolism [72]. Further genetic modifications designed based on the MFA results have demonstrated whole-cell biocatalysis improvement [73, 74].
Other important methodologies to analyze cell metabolic fluxes are the flux balance analysis (FBA) and the metabolic pathway analysis (MPA). FBA uses linear programming which main difference with MFA is the use of an objective function, e.g. maximize cell growth or product formation, under defined constrains such as a determined substrate uptake rate and thermodynamic constrains. It can be used when not enough information is available for MFA. The result of this analysis is one optimal set of fluxes for a specific growth condition, but there are potentially many sets that would satisfy the system. Some examples of application of FBA have included the identification of the gene products essential for E. coli aerobic or anaerobic growth [75]. On the other hand, the MPA determines all the metabolic flux vectors possible in a specific metabolic network without the need of an objective function or previously defined flux rates [70]. All three methods, MFA, FBA and MPA assume no changes in internal metabolites concentrations at steady state.
The elementary mode analysis, a type of MPA, where the elementary modes are defined as a the minimum sub-set of reactions that allows the metabolic network to operate at steady state [76], has been applied to identify potential gene deletions or overexpression that would increase the synthesis of a desired product. This analysis has been applied to evaluate the potential use of glycerol for succinate production under either aerobic or anaerobic conditions [77]. Several other computational tools have been developed that include the methodologies mentioned above or a combination of them [78–80].
Engineering E. coli for succinate production
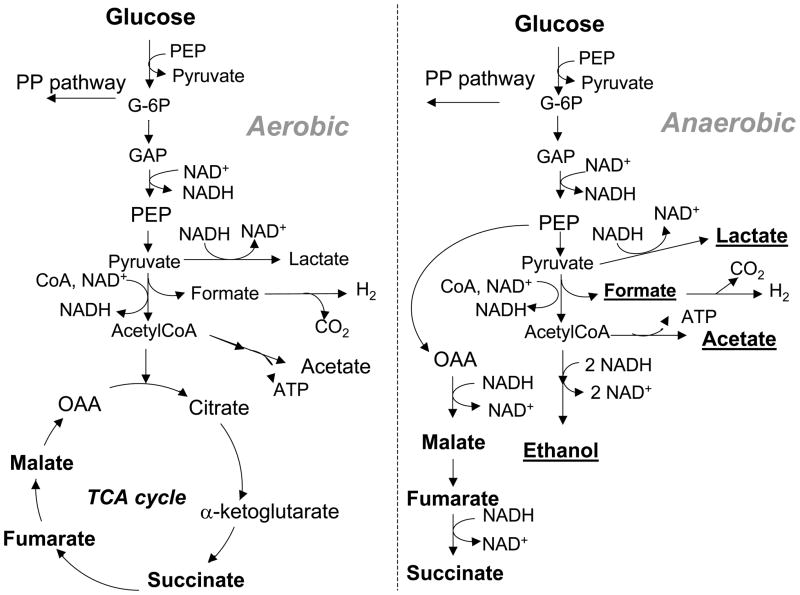
Under anaerobic conditions, wild type E. coli produces a mixture of fermentation products including acetate, formate, ethanol and minor amounts of succinate, while under aerobic conditions succinate is formed only as an intermediate of the TCA cycle unless the glyoxylate bypass is operating (Figure 1). In wild type E. coli the maximum theoretical yield of succinate is 1 mol/mol glucose in anaerobic conditions; this yield is limited by reducing equivalents availability (NADH). Strategies to improve succinate production in E. coli have included the elimination of competing pathways; the activation of pathways with lower reducing equivalents requirement, and the evolution of strains with improved succinate producing capabilities. Several mutant and recombinant strains have been constructed and tested under aerobic [81–83] anaerobic [12, 84] or two-stage fermentation, where the first stage is aerobic for biomass accumulation and the second stage is anaerobic for succinate production [85, 86]. A summary of metabolic engineered strains and conditions for succinate production was presented recently by Jantama and collaborators [84]. Some of the approaches are described below and the general pathway strategy is shown in Figure 2.
Figure 1.
Normal metabolic routes to succinate in wild type E. coli. The aerobic metabolism does not produce succinate as a final product unless the glyoxylate bypass is in operation. The anaerobic metabolism produces the mixed acid fermentation products, ethanol, acetate, formate, lactate and succinate, while the TCA cycle does not function.
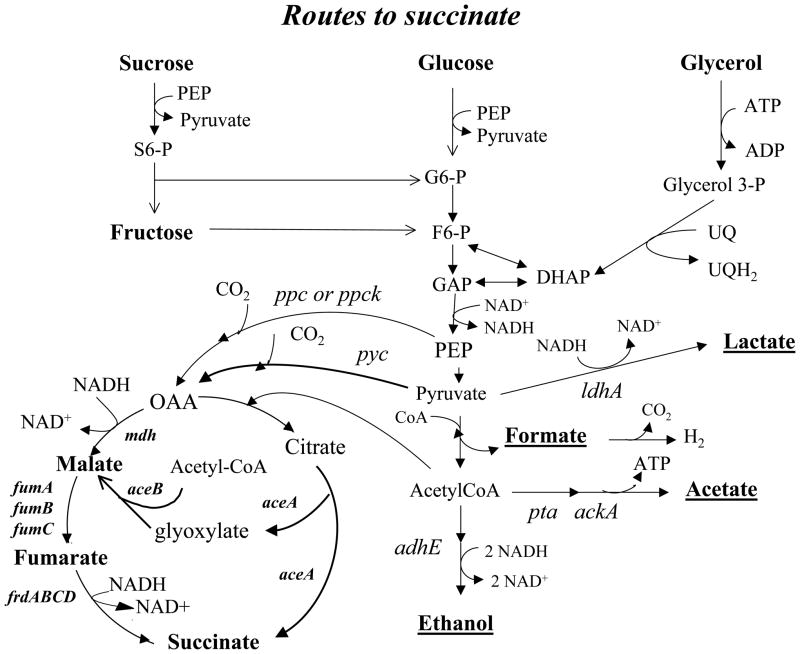
Figure 2.
Routes for overproduction of succinate in engineered E. coli. The routes from glucose, glycerol and sucrose to succinate are shown. The glycolytic reactions form PEP and pyruvate. Typically the routes to the other fermentation products, ethanol, lactate and acetate would be inactivated by disruption of genes ldhA, pta and/or ackA, and adhE. The glyoxylate bypass enzymes encoded by aceA and aceB would be expressed to convert the citrate via isocitrate to glyoxylate and succinate without requiring reductant. In the glyoxylate bypass, malate is formed by condensation of glyoxylate with acetyl-CoA formed from pyruvate. OAA is formed from PEP by either phosphoenolpyruvate carboxylase (ppc) or by phosphoenolpyruvate carboxykinase (ppck). OAA is reduced to malate by malate dehydrogenase (mdh) and the fumarase genes fumA, fumB, and fumC encode isoenzymes, with FumB appearing to play the major role. Fumarate is then reduced by fumarate reductase encoded by the frdABCD operon.
One of the first approaches to produce succinate in E. coli was to eliminate competing pathways by inactivating pyruvate-formate lyase gene (pflB) and lactate dehydrogenase (ldhA). This strain, named NZN111, only produced minor amounts of succinate and showed growth impairment on glucose in anaerobic conditions [87, 88]. This result was most likely due to a redox imbalance and intermediate metabolite accumulation, especially pyruvate [86]. A spontaneous NZN111 mutant strain, named AFP111, generated succinate as the major product; where succinate, acetate and ethanol were produced in a 2:1:1 ratio and molar yields of 1.0, 0.5 and 0.5 mol/mol glucose, respectively [88]. The spontaneous mutation was in the ptsG gene, encoding a glucose specific phosphotransferase system. Later, inactivation of ptsG in several E. coli mutant strains was shown to change product profile, favoring succinate production with various yields [89]. On the other hand, the overexpression of the native NAD+-dependent malate dehydrogenase or the NAD+-dependent malic enzyme in NZN111 resulted in a restoration of the strain ability to grow anaerobically on glucose and the strain produced succinate, with a yield of 1.19 and 1.04 mol/mol glucose, respectively [61, 90]. In addition, native genes whose overexpression can mitigate growth impairment under microaerobic conditions in NZN111 have been identified. Most of these genes were involved in oxidation-reduction reactions or cofactor synthesis related to electron transport [91].
From strictly a carbon balance arising from three-carbon precursors, two moles of succinate could be produced from one mole of glucose consumed, upon appropriate carboxylation and redox availability. To increase succinate yield, competing pathways have been inactivated, the pyruvate carboxylase (pyc) from Lactococcus lactis has been overexpressed and the glyoxylate shunt has been activated. The activation of the glyoxylate shunt contributes an alternative pathway for succinate production with a lower reducing equivalent requirement, leading to a dual pathway to produce succinate. This led to a maximum theoretical succinate yield of 1.6 mol/mol glucose [92] achieved experimentally with a metabolic engineered strain, SBS550MG, carrying ldhA adhE ackpta::Cm iclR inactivations and overexpressing the pyc from L. lactis in a two-stage system [85]. Related strains yielded also formed succinate (58.3 g/L) under aerobic conditions with an overall yield of 0.85 mol/mol glucose [83]. E. coli C strains, KJ060 and KJ073, have been reported to produce 622–733 mM of succinate with molar yields of 1.2–1.6 per mol of metabolized glucose [84]. On the other hand, it has been hypothesized that other strains showing yields over 1 mol/mol glucose have a mutation that activates pyruvate dehydrogenase (PDH) complex in anaerobic conditions, with this being the source of the extra NADH available for succinate production [12].
To produce succinate, CO2 fixation is required, one molecule of CO2 is incorporated to phosphoenolpyruvate (PEP) to form oxaloacetate (OAA) catalyzed normally by PEP carboxylase (PPC). In succinate production processes, CO2 is usually provided in the form of carbonate (e.g. MgCO3, NaHCO3) or directly by CO2 sparging in a bioreactor. CO2 sparging serves a double purpose, maintaining anaerobic conditions for succinate production and supplying the required CO2 for OAA formation from PEP. The effect of CO2 concentration in the gas phase has been evaluated on succinate production by AFP111. Going from 0 to 50% CO2 showed a dramatic increase in succinate specific productivity, however, the increase of CO2 concentration above 50% did not show a significant improvement in succinate production [93].
To increase metabolic flux to succinate, several enzymes involved in CO2 fixation have been overexpressed, including PPC and PEP carboxykinase (PEPCK, gene ppck), which transform PEP into OAA with the incorporation of one molecule of CO2 and the generation of one Pi or ATP, respectively [57, 83, 94, 95]. In addition, pyruvate carboxylase (PYC) and malic enzyme, that incorporates one molecule of CO2 into pyruvate to form OAA and malate, respectively, have also been overexpressed [12, 95]. The overexpression of PEPCK from A. succinogenes has an advantage over the native PPC in that it generates one molecule of ATP for each PEP converted to OAA. The ability to favor the carboxylation direction is important and specific enzymes derived from natural succinate producers or mutant PEPCK enzymes have been beneficial [57]. To increase availability of carbon dioxide for fixation, a cyanobacterial carbonic anhydrase has been cloned in a succinate producing host and improved the yield of succinate from glucose [96].
Several strains have been constructed where genes involved in succinate competing pathways, among others, were inactivated and no foreign genes were expressed. This approach together with several rounds of metabolic evolution, selecting for strains with improved growth in minimal medium, using glucose as sole carbon source, have led to strains with enhanced capability to produce succinate. Engineered E. coli strains KJ060 (ΔldhA, ΔadhE, ΔackA, ΔfocA, ΔpflB) and KJ073 (ΔldhA, ΔadhE, ΔackA, ΔfocA, ΔpflB, ΔmgsA, ΔpoxB) showed succinate yields of 1.2–1.6 mol/mol glucose and succinate concentrations of about 70–80 g/L. However, significant amounts of acetate and malate were also produced [84]. Further improvements, based on E. coli strain KJ073, were achieved by inactivating threonine decarboxylase (tdcD), 2-ketobutyrate formate-lyase (tdcE) and phosphotransacetylase (pta) genes, to conserve pyruvate for succinate production and decrease acetate synthesis; and inactivating aspartate aminotrasferase (aspC) and the NAD+-malic enzyme (sfcA) to conserve oxaloacetate and fumarate for succinate production. The resulting strain, KJ134, showed a succinate yield of 1.5 mol/mol glucose and a decrease in acetate production of about 80%, compared to the parent strain KJ073 [12].
An important approach to increase succinate production has been to manipulate glucose transport and the metabolic steps related to ATP generation [84]. In these strains PEPCK (ppck) was found to catalyze the carboxylation of PEP into OAA, increasing ATP generation. This change was related to a promoter mutation that increased gene expression [13]. In addition, a mutation in pstI resulting in the inactivation of the PEP-phosphotransferase system (PTS) was identified. The PTS function was spontaneously replaced by the D-galactose transporter (galP) and glucokinase (glk) by increasing their expression. These strains have higher PEP availability for succinate production and higher ATP level, showing increased succinate production and improved growth [97]. The resulting pathway was closely related to the native pathway for succinate production in A. succinogenes [13].
With each engineered strain the optimal growth and production conditions need to be examined and the optimal production window and the effect of parameters such as temperature, pH, cation, feedstock concentration, level of aeration if any, needs to be defined for appropriate scale-up.
Production of succinate using different carbon sources
Various carbon sources such as glucose, sucrose, xylose, galactose, and glycerol have been examined by different research groups. Recently, efforts are being made to use renewable agriculture resources with pretreatment to release sugar molecules for succinate production. Two of the most interesting feedstocks are sucrose and glycerol due to their abundance and low cost. See Figure 2 for how these feedstocks can fit into the engineered pathways discussed above.
Recently, Wang et al. [98] have engineered an E. coli strain capable of fermenting fructose, sucrose, and glucose mixtures. Xylose has been examined as sole carbon source for succinate production using E. coli. In a dual-phase batch fermentation containing 100 g/L of total initial xylose, E. coli strain AFP184 produced succinate with a yield of 0.50 g/g [99].
Researchers have demonstrated the use of various agricultural wastes and byproducts to produce succinate. Production of succinate from orange peel and wheat straw by consolidated bioprocessing using a cellulolytic bacterium, Fibrobacter succinogenes S85, was studied [100] giving succinate titers of 1.9 and 2.0 g/L respectively. Wheat flour milling byproducts have been used as the sole medium for A. succinogenes fermentations, which led to the production of 50.6 g/L succinic acid [101]. Corncob hydrolysate [29], crop stalk wastes, [28] corn stover [102], sugarcane bagasse hemicellulose hydrolysate, corn fiber hydrolysate [27] and galactose were studied for succinate production [30]. E. coli engineered to use various sugars in lignocellulose hydrolysates have been discussed [55].
Glycerol has attracted attention as a feedstock for production of bio-based chemicals due to its abundance, low cost, and high degree of reduction. Lee et al. [103] showed that A. succiniciproducens can efficiently convert glycerol to succinate. Succinic acid production from glycerol yields low levels of acetate which is advantageous for downstream process recovery of succinic acid [104]. By integrating the restriction of oxygen and redox sensing/regulatory system, elementary mode analysis was used to predict the metabolic potential of glycerol for succinate production by E. coli under either anaerobic or aerobic conditions [77]. Pathways and mechanisms for the utilization of glycerol by E. coli in minimal salts medium under microaerobic conditions have been reported and E. coli has been engineered for the production of succinate from glycerol [66]. Zhang et al. [105] reengineered fermentative metabolism of E. coli to convert glycerol to succinate under anaerobic conditions without the use of foreign genes. In mineral salt medium, an E. coli strain XZ721 (pck* ΔptsI ΔpflB) fermented 128 mM glycerol to 102 mM succinate with a molar yield of 0.8.
Downstream processing factors
In order to develop competitive biotechnological process for succinate production with petrochemical process, it is necessary to minimize the production costs. A recent review provides an overview of this topic [106]. Downstream processing can contribute about 60% of the total production costs, e.g. the isolation and the purification of the product from the fermentation broth [107]. Various methods such as precipitation, sorption and ion exchange, electrodialysis, and liquid–liquid extraction have been investigated for the recovery of succinic acid from fermentation broth. The first step is separation of cell debris which is performed by ultrafiltration through a bypass crossflow, hollow-fiber ultrafiltration unit [108].
The classical industrial method for the isolation of carboxylic acids from fermentation broth is precipitation with calcium hydroxide or calcium oxide [109–111] followed by filtration. The calcium salt is then treated with sulfuric acid which results in byproduct calcium sulfate after recovery of the succinic acid. Precipitation with ammonia has been described [112, 113]. With ammonia precipitation, succinic acid can be refined with a yield of 93.3% [113]. For some uses the ammonium salt is desirable for a specific downstream purpose such as the production of pyrrolidones [114].
The application of electrodialysis for downstream processing of succinic acid has been studied on a laboratory scale and is useful for industry. A desalting electrodialysis combined with a water-splitting electrodialysis was proposed by Glassner et al. [115]. A yield of 77% of succinic acid was achieved after electrodialysis. To yield a higher purity (>99%) of succinic acid the aqueous solution is subjected to anionic and cationic ion exchangers [111, 116, 117]. Disadvantages of electrodialysis are the high energy consumption, the material costs of the membranes and the low selectivity for succinic acid if other substances are present at high concentrations [106].
Liquid-liquid extraction is a commonly used method in the chemical industry. Most conventional extraction agents show very unfavorable distribution coefficients for organic acids. Reactive extraction is considered to be an effective primary separation step for the recovery of carboxylic acids [118] and long-chain aliphatic primary, secondary and tertiary amines have been proposed [119–122]. Reactive extraction with aliphatic amines depends on many factors [123]. If the separation process enables the recycling of the costly amines efficiently, then an optimized reactive extraction processes might serve a role in industrial production of bio-succinic acid although it is better suited to the processing of higher value compounds.
Summary
Metabolic engineering of E. coli to produce succinate has been a fruitful challenge, not only in producing higher yields of the product but as an example of the strategies and new techniques used for engineering high flux to a desired compound of medium value. The need to coordinate redox and carbon metabolism as well as consider the uptake of feedstock carbon and its partitioning at key metabolic nodes has illustrated key features that need to be addressed in other metabolic engineering efforts. Recently, the placement of such engineered strains into an appropriate industrial or biorefinery setting for implementation, has brought up the more traditional process engineering aspects of microbial production of chemicals and work on succinate has added to this broader, applied field. For the development of a highly efficient technology certain challenges such as developing high succinate tolerant strain with maximum theoretical yield and optimizing fed-batch cultivation method to increase and maintain the volumetric succinic acid productivity needs to be considered. Succinate production with no by-product formation would be desirable for the most efficient use of substrate and for less expensive downstream methods.
Acknowledgments
The authors acknowledge funding from grants from the National Institutes of Health (NIH GM090152) and NSF (CBET-0828516).
Abbreviations
- FBA
flux balance analysis
- ME
metabolic engineering
- MFA
metabolic flux analysis
- MPA
metabolic pathway analysis
- OAA
oxaloacetate
- PEPCK
PEP carboxykinase
- PEP
phosphoenol pyruvate
- PPC
PEP carboxylase
- PTS
PEP-phosphotransferase system
- PYC
pyruvate carboxylase
- PDH
pyruvate dehydrogenase
Footnotes
The authors have declared no conflict of interest.
References
- 1.Werpy T, Petersen G, editors. Top Value Added Chemicals from Biomass. USDOE; Washington DC: 2004. [Google Scholar]
- 2.McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 3.Xu J, Guo BH. Poly (butylene succinate) and its copolymers: research, development and industrialization. Biotechnol J. 2010;5:1149–1163. doi: 10.1002/biot.201000136. [DOI] [PubMed] [Google Scholar]
- 4.Budarin V, Luque R, Macquarrie DJ, Clark JH. Towards a bio-based industry: benign catalytic esterifications of succinic acid in the presence of water. Chemistry. 2007;13:6914–6919. doi: 10.1002/chem.200700037. [DOI] [PubMed] [Google Scholar]
- 5.Zeikus JG, Jain MK, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Environ Microbiol. 1999;51:545–552. [Google Scholar]
- 6.Barrett DG, Yousaf MN. Design and applications of biodegradable polyester tissue scaffolds based on endogenous monomers found in human metabolism. Molecules. 2009;14:4022–4050. doi: 10.3390/molecules14104022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukatani T, Matsumoto K. Flow-injection fluorometric quantification of succinate in foodstuffs based on the use of an immobilized enzyme reactor. Anal Chim Acta. 2000;416:197–203. [Google Scholar]
- 8.Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydrates - the US Department of Energy’s “Top 10” revisited. Green Chem. 2010;12:539–554. [Google Scholar]
- 9.Paster M, Pellegrino JL, Carole TM. Industrial Bioproducts: Today and Tomorrow. USDOE, Office of Energy Efficiency and Renewable Energy & Office of the Biomass Program; Washington DC: 2003. [Google Scholar]
- 10.Yu C, Cao Y, Zou H, Xian M. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl Microbiol Biotechnol. 2011;89:573–583. doi: 10.1007/s00253-010-2970-z. [DOI] [PubMed] [Google Scholar]
- 11.Kurzrock T, Weuster-Botz D. Recovery of succinic acid from fermentationbroth. Biotechnol Lett. 2011;32:331–339. doi: 10.1007/s10529-009-0163-6. [DOI] [PubMed] [Google Scholar]
- 12.Jantama K, Zhang X, Moore JC, Shanmugam KT, et al. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Jantama K, Shanmugam KT, Ingram LO. Reengineering Escherichia coli for succinate production in mineral salts medium. Appl Environ Microbiol. 2009;75:7807–7813. doi: 10.1128/AEM.01758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarboe LR, Zhang X, Wang X, Moore JC, et al. Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J Biomed Biotechnol. 2010;2010:761042. doi: 10.1155/2010/761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potera C. Making succinate more successful. Environ Health Persp. 2005;113:A832–835. doi: 10.1289/ehp.113-a832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Shanks BH. Unleashing biocatalysis/chemical catalysis synergies for efficient biomass conversion. ACS Chem Biol. 2007;2:533–535. doi: 10.1021/cb7001522. [DOI] [PubMed] [Google Scholar]
- 18.Guettler MV, Rumler D, Jain MK. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol. 1999;49(Pt 1):207–216. doi: 10.1099/00207713-49-1-207. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Zheng XY, Fang XJ, Liu SW, et al. A complete industrial system for economical succinic acid production by Actinobacillus succinogenes. Bioresour Technol. 2011;102:6147–6152. doi: 10.1016/j.biortech.2011.02.093. [DOI] [PubMed] [Google Scholar]
- 20.Liu YP, Zheng P, Sun ZH, Ni Y, et al. Economical succinic acid production from cane molasses by Actinobacillus succinogenes. Bioresour Technol. 2008;99:1736–1742. doi: 10.1016/j.biortech.2007.03.044. [DOI] [PubMed] [Google Scholar]
- 21.McKinlay JB, Laivenieks M, Schindler BD, McKinlay AA, et al. A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genomics. 2010;11:680. doi: 10.1186/1471-2164-11-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park DH, Zeikus JG. Utilization of electrically reduced neutral red by Actinobacillus succinogenes: physiological function of neutral red in membrane-driven fumarate reduction and energy conservation. J Bacteriol. 1999;181:2403–2410. doi: 10.1128/jb.181.8.2403-2410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye GZ, Jiang M, Li J, Chen KQ, et al. Isolation of NH4+-tolerant mutants of Actinobacillus succinogenes for succinic acid production by continuous selection. J Microbiol Biotechnol. 2010;20:1219–1225. doi: 10.4014/jmb.1003.03030. [DOI] [PubMed] [Google Scholar]
- 24.McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C. Determining Actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab Eng. 2007;9:177–192. doi: 10.1016/j.ymben.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 25.McKinlay JB, Vieille C. 13C-metabolic flux analysis of Actinobacillus succinogenes fermentative metabolism at different NaHCO3 and H2 concentrations. Metab Eng. 2008;10:55–68. doi: 10.1016/j.ymben.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Borges ER, Pereira N., Jr Succinic acid production from sugarcane bagasse hemicellulose hydrolysate by Actinobacillus succinogenes . J Ind Microbiol Biotechnol. 2010 doi: 10.1007/s10295-010-0874-7. [DOI] [PubMed] [Google Scholar]
- 27.Chen KQ, Li J, Ma JF, Jiang M, et al. Succinic acid production by Actinobacillus succinogenes using hydrolysates of spent yeast cells and corn fiber. Bioresour Technol. 2011;102:1704–1708. doi: 10.1016/j.biortech.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Yang M, Wang D, Li W, et al. Efficient conversion of crop stalk wastes into succinic acid production by Actinobacillus succinogenes. Bioresour Technol. 2010;101:3292–3294. doi: 10.1016/j.biortech.2009.12.064. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Li Z, Ye Q, Yang Y, Chen S. Development of succinic acid production from corncob hydrolysate by Actinobacillus succinogenes. J Ind Microbiol Biotechnol. 2010;37:1033–1040. doi: 10.1007/s10295-010-0750-5. [DOI] [PubMed] [Google Scholar]
- 30.Lee PC, Lee SY, Chang HN. Succinic acid production by Anaerobiospirillum succiniciproducens ATCC 29305 growing on galactose, galactose/glucose, and galactose/lactose. J Microbiol Biotechnol. 2008;18:1792–1796. doi: 10.4014/jmb.0800.129. [DOI] [PubMed] [Google Scholar]
- 31.Lee PC, Lee SY, Chang HN. Kinetic study on succinic acid and acetic acid formation during continuous cultures of Anaerobiospirillum succiniciproducens grown on glycerol. Bioprocess Biosyst Eng. 2010;33:465–471. doi: 10.1007/s00449-009-0355-4. [DOI] [PubMed] [Google Scholar]
- 32.Meynial-Salles I, Dorotyn S, Soucaille P. A new process for the continuous production of succinic acid from glucose at high yield, titer, and productivity. Biotechnol Bioeng. 2008;99:129–135. doi: 10.1002/bit.21521. [DOI] [PubMed] [Google Scholar]
- 33.Lee PC, Lee SY, Chang HN. Cell recycled culture of succinic acid-producing Anaerobiospirillum succiniciproducens using an internal membrane filtration system. J Microbiol Biotechnol. 2008;18:1252–1256. [PubMed] [Google Scholar]
- 34.Lee PC, Lee SY, Hong SH, Chang HN. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl Microbiol Biotechnol. 2002;58:663–668. doi: 10.1007/s00253-002-0935-6. [DOI] [PubMed] [Google Scholar]
- 35.Hong SH, Kim JS, Lee SY, In YH, et al. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat Biotechnol. 2004;22:1275–1281. doi: 10.1038/nbt1010. [DOI] [PubMed] [Google Scholar]
- 36.Kim JM, Lee KH, Lee SY. Development of a markerless gene knock-out system for Mannheimia succiniciproducens using a temperature-sensitive plasmid. FEMS Microbiol Lett. 2008;278:78–85. doi: 10.1111/j.1574-6968.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee SJ, Lee DY, Kim TY, Kim BH, et al. Metabolic engineering of Escherichia coli for enhanced production of succinic acid, based on genome comparison and in silico gene knockout simulation. Appl Environ Microbiol. 2005;71:7880–7887. doi: 10.1128/AEM.71.12.7880-7887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JW, Lee SY. Proteome-based physiological analysis of the metabolically engineered succinic acid producer Mannheimia succiniciproducensLPK7. Bioprocess Biosyst Eng. 2011;33:97–107. doi: 10.1007/s00449-009-0339-4. [DOI] [PubMed] [Google Scholar]
- 39.Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl Environ Microbiol. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SY, Kim JM, Song H, Lee JW, et al. From genome sequence to integrated bioprocess for succinic acid production by Mannheimia succiniciproducens. Appl Microbiol Biotechnol. 2008;79:11–22. doi: 10.1007/s00253-008-1424-3. [DOI] [PubMed] [Google Scholar]
- 41.Kim TY, Kim HU, Park JM, Song H, et al. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol Bioeng. 2007;97:657–671. doi: 10.1002/bit.21433. [DOI] [PubMed] [Google Scholar]
- 42.Kim TY, Kim HU, Song H, Lee SY. In silico analysis of the effects of H2 and CO2 on the metabolism of a capnophilic bacterium Mannheimia succiniciproducens. J Biotechnol. 2009;144:184–189. doi: 10.1016/j.jbiotec.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Lee JW, Lee SY, Song H, Yoo JS. The proteome of Mannheimia succiniciproducens, a capnophilic rumen bacterium. Proteomics. 2006;6:3550–3566. doi: 10.1002/pmic.200500837. [DOI] [PubMed] [Google Scholar]
- 44.Jang YS, Jung YR, Lee SY, Kim JM, et al. Construction and characterization of shuttle vectors for succinic acid-producing rumen bacteria. Appl Environ Microbiol. 2007;73:5411–5420. doi: 10.1128/AEM.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okino S, Noburyu R, Suda M, Jojima T, et al. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- 46.Huhn S, Jolkver E, Kramer R, Marin K. Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;89:327–335. doi: 10.1007/s00253-010-2855-1. [DOI] [PubMed] [Google Scholar]
- 47.Lee KY, Park JM, Kim TY, Yun H, Lee SY. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb Cell Fact. 2010;9:94. doi: 10.1186/1475-2859-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhnert P, Scholten E, Haefner S, Mayor D, Frey J. Basfia succiniciproducens gen. nov., sp. nov., a new member of the family Pasteurellaceae isolated from bovine rumen. Int J Syst Evol Microbiol. 2010;60:44–50. doi: 10.1099/ijs.0.011809-0. [DOI] [PubMed] [Google Scholar]
- 49.Scholten E, Renz T, Thomas J. Continuous cultivation approach for fermentative succinic acid production from crude glycerol by Basfia succiniciproducens DD1. Biotechnol Lett. 2009;31:1947–1951. doi: 10.1007/s10529-009-0104-4. [DOI] [PubMed] [Google Scholar]
- 50.Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng. 2010;12:518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Yuzbashev TV, Yuzbasheva EY, Sobolevskaya TI, Laptev IA, et al. Production of succinic acid at low pH by a recombinant strain of the aerobic yeast Yarrowia lipolytica. Biotechnol Bioeng. 2010;107:673–682. doi: 10.1002/bit.22859. [DOI] [PubMed] [Google Scholar]
- 52.van de Graaf MJ, Valianpoer F, Fiey G, Delattre L, Schulten EAM. Process for crystallization of succinic acid. WO 2011/064151. World Intellectual Property Organization. 2011:A064151.
- 53.Luo L, van der Voet E, Huppes G. Biorefining of lignocellulosic feedstock -Technical, economic and environmental considerations. Bioresour Technol. 2010;101:5023–5032. doi: 10.1016/j.biortech.2009.12.109. [DOI] [PubMed] [Google Scholar]
- 54.Shanmugam KT, Ingram LO. Engineering biocatalysts for production of commodity chemicals. J Mol Microbiol Biotechnol. 2008;15:8–15. doi: 10.1159/000111988. [DOI] [PubMed] [Google Scholar]
- 55.Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO. Development of ethanologenic bacteria. Adv Biochem Eng Biotechnol. 2007;108:237–261. doi: 10.1007/10_2007_068. [DOI] [PubMed] [Google Scholar]
- 56.Zheng P, Dong JJ, Sun ZH, Ni Y, Fang L. Fermentative production of succinic acid from straw hydrolysate by Actinobacillus succinogenes. Bioresour Technol. 2009;100:2425–2429. doi: 10.1016/j.biortech.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 57.Singh A, Cher Soh K, Hatzimanikatis V, Gill RT. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab Eng. 2011;13:76–81. doi: 10.1016/j.ymben.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 58.Stephanopoulos G. Metabolic fluxes and metabolic engineering. Metab Eng. 1999;1:1–11. doi: 10.1006/mben.1998.0101. [DOI] [PubMed] [Google Scholar]
- 59.Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO. Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol. 2003;69:399–407. doi: 10.1128/AEM.69.1.399-407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vadali RV, Horton CE, Rudolph FB, Bennett GN, San KY. Production of isoamyl acetate in ackA-pta and/or ldh mutants of Escherichia coli with overexpression of yeast ATF2. Appl Microbiol Biotechnol. 2004;63:698–704. doi: 10.1007/s00253-003-1452-y. [DOI] [PubMed] [Google Scholar]
- 61.Wang W, Li Z, Xie J, Ye Q. Production of succinate by a pflB ldhA double mutant of Escherichia coli overexpressing malate dehydrogenase. Bioprocess Biosyst Eng. 2009;32:737–745. doi: 10.1007/s00449-009-0298-9. [DOI] [PubMed] [Google Scholar]
- 62.Liu H, Kang J, Qi Q, Chen G. Production of lactate in Escherichia coli by redox regulation genetically and physiologically. Appl Biochem Biotechnol. 2011;164:162–169. doi: 10.1007/s12010-010-9123-9. [DOI] [PubMed] [Google Scholar]
- 63.Kim SW, Keasling JD. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng. 2001;72:408–415. doi: 10.1002/1097-0290(20000220)72:4<408::aid-bit1003>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 64.Choi JI, Lee SY, Han K. Cloning of the Alcaligenes latus polyhydroxyalkanoate biosynthesis genes and use of these genes for enhanced production of Poly(3-hydroxybutyrate) in Escherichia coli. Appl Environ Microbiol. 1998;64:4897–4903. doi: 10.1128/aem.64.12.4897-4903.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caetano T, Krawczyk JM, Mosker E, Sussmuth RD, Mendo S. Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol. 2011;18:90–100. doi: 10.1016/j.chembiol.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 66.Blankschien MD, Clomburg JM, Gonzalez R. Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng. 2010;12:409–419. doi: 10.1016/j.ymben.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 67.Berrios-Rivera SJ, Sanchez AM, Bennett GN, San KY. Effect of different levels of NADH availability on metabolite distribution in Escherichia coli fermentation in minimal and complex media. Appl Microbiol Biotechnol. 2004;65:426–432. doi: 10.1007/s00253-004-1609-3. [DOI] [PubMed] [Google Scholar]
- 68.Berrios-Rivera SJ, San KY, Bennett GN. The effect of carbon sources and lactate dehydrogenase deletion on 1,2-propanediol production in Escherichia coli. J Ind Microbiol Biotechnol. 2003;30:34–40. doi: 10.1007/s10295-002-0006-0. [DOI] [PubMed] [Google Scholar]
- 69.Sanchez AM, Bennett GN, San KY. Effect of different levels of NADH availability on metabolic fluxes of Escherichia coli chemostat cultures in defined medium. J Biotechnol. 2005;117:395–405. doi: 10.1016/j.jbiotec.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 70.Trinh CT, Wlaschin A, Srienc F. Elementary mode analysis: a useful metabolic pathway analysis tool for characterizing cellular metabolism. Appl Microbiol Biotechnol. 2009;81:813–826. doi: 10.1007/s00253-008-1770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 72.Stephanopoulos G. Metabolic engineering. Biotechnol Bioeng. 1998;58:119–120. [PubMed] [Google Scholar]
- 73.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Wang Q, Chen X, Yang Y, Zhao X. Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Appl Microbiol Biotechnol. 2006;73:887–894. doi: 10.1007/s00253-006-0535-y. [DOI] [PubMed] [Google Scholar]
- 75.Edwards JS, Palsson BO. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics. 2000;1:1. doi: 10.1186/1471-2105-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuster S, Dandekar T, Fell DA. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999;17:53–60. doi: 10.1016/s0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- 77.Chen Z, Liu H, Zhang J, Liu D. Elementary mode analysis for the rational design of efficient succinate conversion from glycerol by Escherichia coli. J Biomed Biotechnol. 2010;2010:518743. doi: 10.1155/2010/518743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranganathan S, Suthers PF, Maranas CD. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput Biol. 2010;6:e1000744. doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boghigian BA, Shi H, Lee K, Pfeifer BA. Utilizing elementary mode analysis, pathway thermodynamics, and a genetic algorithm for metabolic flux determination and optimal metabolic network design. BMC Syst Biol. 2010;4:49. doi: 10.1186/1752-0509-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JI, Varner JD, Ramkrishna D. A hybrid model of anaerobic E. coli GJT001: combination of elementary flux modes and cybernetic variables. Biotechnol Prog. 2008;24:993–1006. doi: 10.1002/btpr.73. [DOI] [PubMed] [Google Scholar]
- 81.Lin H, Bennett GN, San KY. Fed-batch culture of a metabolically engineered Escherichia coli strain designed for high-level succinate production and yield under aerobic conditions. Biotechnol Bioeng. 2005;90:775–779. doi: 10.1002/bit.20458. [DOI] [PubMed] [Google Scholar]
- 82.Lin H, Bennett GN, San KY. Genetic reconstruction of the aerobic central metabolism in Escherichia coli for the absolute aerobic production of succinate. Biotechnol Bioeng. 2005;89:148–156. doi: 10.1002/bit.20298. [DOI] [PubMed] [Google Scholar]
- 83.Lin H, Bennett GN, San KY. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab Eng. 2005;7:116–127. doi: 10.1016/j.ymben.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 84.Jantama K, Haupt MJ, Svoronos SA, Zhang X, et al. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng. 2008;99:1140–1153. doi: 10.1002/bit.21694. [DOI] [PubMed] [Google Scholar]
- 85.Sanchez AM, Bennett GN, San KY. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng. 2005;7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 86.Vemuri GN, Eiteman MA, Altman E. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2002;68:1715–1727. doi: 10.1128/AEM.68.4.1715-1727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bunch PK, Mat-Jan F, Lee N, Clark DP. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143(part1):187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 88.Donnelly MI, Millard CS, Clark DP, Chen MJ, Rathke JW. A novel fermentation pathway in an Escherichia coli mutant producing succinic acid, acetic acid, and ethanol. Appl Biochem Biotechnol. 1998;70–72:187–198. doi: 10.1007/BF02920135. [DOI] [PubMed] [Google Scholar]
- 89.Chatterjee R, Millard CS, Champion K, Clark DP, Donnelly MI. Mutation of the ptsG gene results in increased production of succinate in fermentation of glucose by Escherichia coli. Appl Environ Microbiol. 2001;67:148–154. doi: 10.1128/AEM.67.1.148-154.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stols L, Donnelly MI. Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl Environ Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh A, Lynch MD, Gill RT. Genes restoring redox balance in fermentation-deficient E. coli NZN111. Metab Eng. 2009;11:347–354. doi: 10.1016/j.ymben.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 92.Cox SJ, Shalel Levanon S, Sanchez A, Lin H, et al. Development of a metabolic network design and optimization framework incorporating implementation constraints: a succinate production case study. Metab Eng. 2006;8:46–57. doi: 10.1016/j.ymben.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 93.Lu S, Eiteman MA, Altman E. Effect of CO2 on succinate production in dual-phase Escherichia coli fermentations. J Biotechnol. 2009;143:213–223. doi: 10.1016/j.jbiotec.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 94.Kim P, Laivenieks M, Vieille C, Zeikus JG. Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl Environ Microbiol. 2004;70:1238–1241. doi: 10.1128/AEM.70.2.1238-1241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin H, San KY, Bennett GN. Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli. Appl Microbiol Biotechnol. 2005;67:515–523. doi: 10.1007/s00253-004-1789-x. [DOI] [PubMed] [Google Scholar]
- 96.Wang D, Li Q, Li W, Xing J, Su Z. Improvement of succinate production by overexpression of a cyanobacterial carbonic anhydrase in Escherichia coli. Enzyme Microb Tech. 2009;45:491–497. [Google Scholar]
- 97.Zhang X, Jantama K, Moore JC, Jarboe LR, et al. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc Natl Acad Sci U S A. 2009;106:20180–20185. doi: 10.1073/pnas.0905396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, Zhu J, Bennett GN, San KY. Succinate production from different carbon sources under anaerobic conditions by metabolic engineered Escherichia coli strains. Metab Eng. 2011;13:328–335. doi: 10.1016/j.ymben.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 99.Andersson C, Hodge D, Berglund KA, Rova U. Effect of different carbon sources on the production of succinic acid using metabolically engineered Escherichia coli. Biotechnol Prog. 2007;23:381–388. doi: 10.1021/bp060301y. [DOI] [PubMed] [Google Scholar]
- 100.Li Q, Siles JA, Thompson IP. Succinic acid production from orange peel and wheat straw by batch fermentations of Fibrobacter succinogenes S85. Appl Microbiol Biotechnol. 2010;88:671–678. doi: 10.1007/s00253-010-2726-9. [DOI] [PubMed] [Google Scholar]
- 101.Dorado MP, Lin SK, Koutinas A, Du C, et al. Cereal-based biorefinery development: utilisation of wheat milling by-products for the production of succinic acid. J Biotechnol. 2009;143:51–59. doi: 10.1016/j.jbiotec.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Zheng P, Fang L, Xu Y, Dong JJ, et al. Succinic acid production from corn stover by simultaneous saccharification and fermentation using Actinobacillus succinogenes. Bioresour Technol. 2010;101:7889–7894. doi: 10.1016/j.biortech.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 103.Lee PC, Lee WG, Lee SY, Chang HN. Succinic acid production with reduced by-product formation in the fermentation of Anaerobiospirillum succiniciproducens using glycerol as a carbon source. Biotechnol Bioeng. 2001;72:41–48. [PubMed] [Google Scholar]
- 104.Song H, Lee SY. Production of succinic acid by bacterial fermentation. Enzyme Microb Tech. 2006;39:352–361. [Google Scholar]
- 105.Zhang X, Shanmugam KT, Ingram LO. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kurzrock T, Weuster-Botz D. Recovery of succinic acid from fermentation broth. Biotechnol Lett. 2010;32:331–339. doi: 10.1007/s10529-009-0163-6. [DOI] [PubMed] [Google Scholar]
- 107.Bechthold I, Bretz K, Kabasci S, Kopitzky R, Springer A. Succinic acid: a new platform chemical for biobased polymers from renewable resources. Chem Eng Technol. 2008;5:647–654. [Google Scholar]
- 108.Ruffer N, Heidersdorf U, Kretzers I, Sprenger GA, et al. Fully integrated L-phenylalanine separation and concentration using reactive-extraction with liquid-liquid centrifuges in a fed-batch process with E. coli. Bioprocess Biosyst Eng. 2004;26:239–248. doi: 10.1007/s00449-004-0354-4. [DOI] [PubMed] [Google Scholar]
- 109.Datta R. Process for the production of succinic acid by anaerobic fermentation. 5:143,833. US Patent. 1992
- 110.Datta R, Glassner DA, Jain MK, Vickyroy JR. Fermentation and purification process for succinic acid. 5:168,055. US Patent. 1992
- 111.Berglund KA, Elankovan P, Glassner DA. Carboxylic acid purification and crystallization process. 5:034,105. US Patent. 1991
- 112.Berglund KA, Yedur S, Dunuwila D. Succinic acid production and purification. 5:958,744. US Patent. 1999
- 113.Yedur S, Berglung KS, Dunuwila DD. Succinic acid production and purification. 6:265,190. US Patent. 2001
- 114.Werpy T, Frye JG, Zacher AH, Wang Y. Chapter 17 Catalytic Preparation of Pyrrolidones from Renewable Resources. In: Sowa JGJ, editor. Catalysis of Organic Reactions. CRC Press/Taylor & Francis; Boca Raton, FL: 2005. pp. 145–154. [Google Scholar]
- 115.Glassner DA, Satinwood RD, Datta R. Process for the production and purification of succinic acid. 5:143,834. US Patent. 1992
- 116.Jun YS, Huh YS, Park HS, Thomas A, et al. Adsorption of pyruvic and succinic acid by amine-functionalized SBA-15 for the purification of succinic acid from fermentation broth. J Phys Chem C. 2007;200:13076–13086. [Google Scholar]
- 117.Li Q, Li WL, Wang D, Liu BB, et al. pH neutralization while succinic acid adsorption onto anion-exchange resins. Appl Biochem Biotechnol. 2010;160:438–445. doi: 10.1007/s12010-008-8355-4. [DOI] [PubMed] [Google Scholar]
- 118.Cascaval D, Balaction AI. New separation techniques on bioseparations 1. Reactive extraction. Chem Ind. 2004;58:375–386. [Google Scholar]
- 119.Tung LA, King CJ. Sorption and extraction of lactic and succinic acids at pH > pKa1 1. Factors governing equilibria. Ind Eng Chem Res. 1994;33:3217–3223. [Google Scholar]
- 120.Hong YK, Hong WH. Equilibrium studies on reactive extraction of succinic acid from aqueous solutions with tertiary amines. Bioproc Biosyst Eng. 2000;22:477–481. [Google Scholar]
- 121.Song H, Huh YS, Lee SY, Hong WH, Hong YK. Recovery of succinic acid produced by fermentation of a metabolically engineered Mannheimia succiniciproducens strain. J Biotechnol. 2007;132:445–452. doi: 10.1016/j.jbiotec.2007.07.496. [DOI] [PubMed] [Google Scholar]
- 122.Jun YS, Huh YS, Hong WH, Hong YK. Kinetics of the extraction of succinic acid with tri-n-octylamine in 1-octanol solution. Biotechnol Prog. 2005;21:1673–1679. doi: 10.1021/bp050083t. [DOI] [PubMed] [Google Scholar]
- 123.Kahya E, Bayraktar E, Mehmetoglu U. Optimization of process parameters for reactive lactic acid extraction. Turk J Chem. 2001;25:223–230. [Google Scholar]