Abstract
Tuning photoinduced electron transfer: Subtle differences in local sequence and conformation can produce diversity and specificity in electron transfer (ET) in proteins, despite high structural conservation of redox partners. For individual ET steps, distance is not necessarily the decisive parameter; orientation and solvent accessibility of ET partners, and therefore, stabilization of charge-separated states contribute substantially.
Keywords: electron transfer, EPR spectroscopy, flavins, radical reactions, tryptophan
Nature has developed amazing mechanisms for rapid long-range electron transfer (ET) within biological macromolecules.[1] These could in principle serve as templates for technological applications ranging from nano-electronics based on molecular wires (e.g., DNA filaments) to renewable energy conversion devices. Adaptation of the fundamentals of such ET motifs to organic solar cells, for example, could help optimize charge transport thereby decreasing charge recombination and increasing overall energy conversion efficiency.[2] Hence, dissecting and controlling motifs for rapid ET in bio-macromolecules is prerequisite to application in bionics. The purpose of this communication is to examine, in homologous proteins from different organisms, the robustness and conservation of long-range ET via a cascade of short steps. We discover unexpected variability and identify alternative ET pathways in an important member of a blue-light photoreceptor/DNA-repair protein class.
Multistep ET plays a key role in biological systems. For example, in complex I of the respiratory chain on the inner membrane of mitochondria, electrons are transferred over a cascade of iron-sulfur clusters from a flavin adenine mononucleotide cofactor to a quinone-binding site.[3] The overall pathway is nearly 10 nm long[4] and most likely involves aromatic amino-acid residues as ‘stepping stones’ to bridge the gaps between the iron-sulfur clusters.[5] Another paradigmatic example for long-range ET is charge transport in DNA.[6] The π-stacked nucleobases of double-helical DNA can serve as redox intermediates to transfer charges over long distances.[7] The nucleobases stack nearly perfectly parallel at distances of about 3.3 Å in B-DNA and allow for rapid photo-induced ET (PET). The mechanism of charge transfer in DNA depends on the structure and dynamics of the DNA assembly.
Some photoreactive proteins, including those responsive to UV light, utilize characteristic aligned tryptophan (Trp) residues for ET.[8] Very fast PET is feasible along the conserved linear Trp pathway[9] found in flavoproteins of the photolyase/cryptochrome family (Figures S1 and S2 of SI), including Arabidopsis UVR2 and UVR3. In these systems, three Trps are aligned between the active-site flavin adenine dinucleotide (FAD) cofactor and the enzyme surface, with edge-to-edge distances of about 5 and 4 Å between the proximal Trp (TrpA) and the middle Trp (TrpB), and between TrpB and the distal Trp (TrpC), respectively.[10, 11, 12] Upon photo-excitation, FAD, in the fully oxidized or semi-reduced (radical) form, abstracts an electron from nearby TrpA. In a subsequent ET step, the thus generated radical state on TrpA is transferred to TrpB of the chain. Further ET moves the electron hole to the solvent-exposed TrpC, where the resultant radical can be stabilized by interaction with water molecules.[13–15] Starting from the singly reduced (semiquinoid) state of FAD, the overall photoreduction process is completed within 30 ps, as has been shown recently by time-resolved optical absorption spectroscopy on Escherichia coli DNA photolyase (PHR).[16]
For FAD in the fully oxidized state, PET generates a sequence of radical-pair (RP) species that can be probed by time-resolved electron paramagnetic resonance (TREPR) spectroscopy.[17–20] When applied with sufficiently high temporal resolution, TREPR exploits the spin-polarized nature of the early transient paramagnetic species.[21] A quantitative spectral analysis of the spin polarization of TREPR signals reveals information on the chemical nature of RPs as well as the spin multiplicity of the precursor state of RP formation.[22] Here we use RP detection by TREPR to unravel ET pathways in homologous proteins of the PHR/CRY family.
In PHRs, the fully reduced FAD cofactor, FADH−, supplies the electron required for light-dependent catalytic repair of photo-products in UV-damaged DNA.[19] If the FAD cofactor in PHR is fully or partially oxidized, catalytic DNA repair activity can be restored by light-induced photoreduction (also termed photoactivation) via ET along the Trp cascade.[13] All PHRs examined to date functionally conserve this FAD photoactivation process,[23] despite sequence, structural and functional diversity. Although the relevance of this process for efficient DNA repair has been questioned,[24] it is likely critical for protein quality control during oxidative stress in vivo. PHRs, regardless of their FAD redox state, recognize and bind to damaged DNA; thus, DNA-bound PHRs inactivated by oxidation would inhibit essential repair, transcription and replication activities in cells.
In cryptochromes (CRYs), photolyase homologs without such DNA repair activities, the FAD redox state is proposed to play a key role in light-responsive activities.[25, 26] Both in vitro and in vivo experiments suggest that the FAD radical state is responsible for signaling[27] and magnetoreception.[28] By analogy to PHR, the conserved Trp triad has been proposed to function in FAD photoreduction,[25, 29] however, conclusive proof for these functions remains elusive. Nevertheless, CRYs do form photo-induced, spin-correlated RPs with lifetimes and spin-spin couplings appropriate for the central component of a biological magnetic compass.[20, 22, 30]
To characterize conservation and diversity of ET along the Trp pathway of the CRY/PHR family, we have conducted comparative TREPR experiments on DASH-type CRYs (CRYD) from the cyano-bacterium Synechocystis sp. PCC6803 and the frog Xenopus laevis. We recently showed that the Trp triad is active in Xenopus CRYD,[20] and previously determined the crystallographic structure of Synechocystis CRYD.[12] The two proteins have an overall amino acid sequence similarity of 67% and conserve the Trp triad (W396≡TrpA, W373≡TrpB, and W320≡TrpC in Synechocystis, and W400≡TrpA, W377≡TrpB, and W324≡TrpC in Xenopus) with similar geometry (Figure S1).[12]
PET in Xenopus CRYD starts out from a photoexcited singlet state of the protein’s redox-active FAD, as we recently showed by analyzing the electron-spin polarized spectral TREPR pattern for the generated RP state FAD•…TrpC•.[22] Replacement of the protein-surface exposed TrpC by phenylalanine (W324F) in Xenopus CRYD impedes RP formation on a timescale of about 10 ns (corresponding to the maximum temporal resolution of our instrumentation) and longer. PET to generate the RP state FAD•…TrpB•via FAD•…TrpA•could basically still occur. However, without the terminal TrpC, the photo-generated radicals on FAD and TrpA/TrpB are not sufficiently spatially separated for stable charge separation on a longer time scale. Thus, efficient backward ET and radical recombination would likely regenerate the diamagnetic ground-state reactants within less than 10 ns.
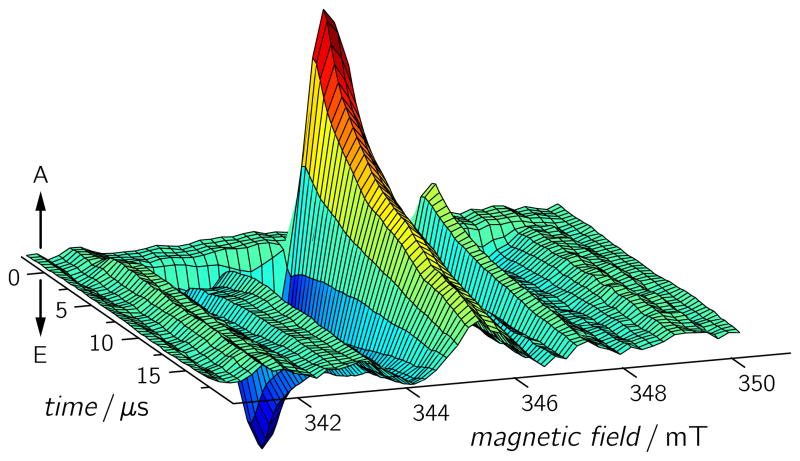
TREPR experiments on wild-type (WT) Synechocystis CRYD revealed a RP spectrum (Figure 1) that is roughly similar to that of WT Xenopus CRYD,[20, 22] in terms of electron-spin polarization and spectral width, implying an analogous RP generation scheme. Positive and negative TREPR amplitudes reflect the (enhanced) absorptive and emissive electron-spin polarizations of the EPR transitions, respectively. The spectral shape is determined by the magnetic interactions within (Zeeman and hyperfine interactions) and between (dipolar and exchange interactions) the two radicals of the RP. We recently presented details on spectral simulations of TREPR signals from RPs in Xenopus CRYD.[22] The time evolution of the TREPR signal revealed that the RP state lives for at least 6μs.[20]
Figure 1.
TREPR data set for Synechocystis CRYD measured at 274 K. Each time profile is the average of 120 acquisitions recorded with a laser pulse repetition rate of 1.25 Hz, a microwave frequency of 9.68 GHz, and a microwave power of 2 mW at a detection bandwidth of 100 MHz. A: enhanced absorption; E: emission.
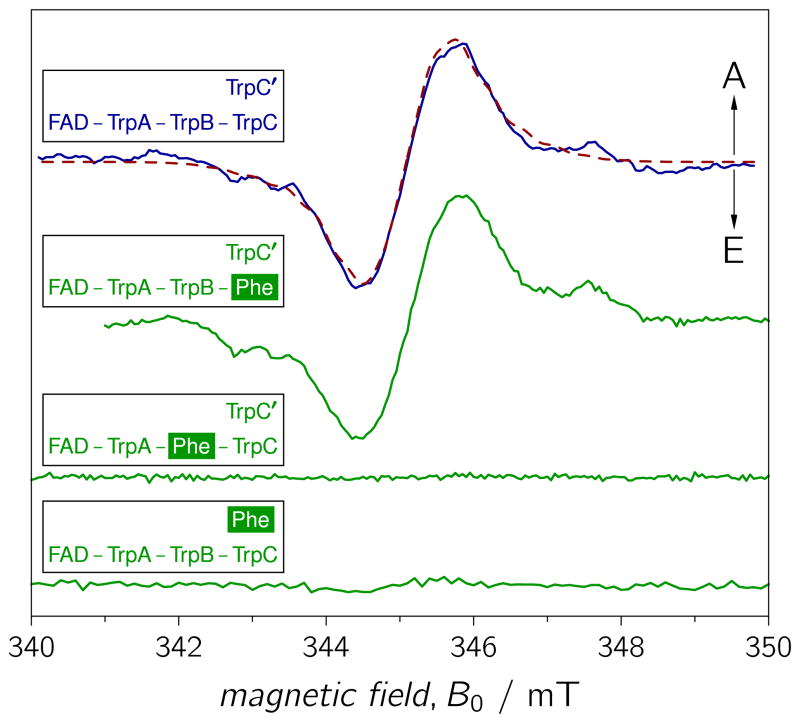
The Synechocystis CRYD W320F mutation, in which the terminal TrpC residue is replaced by phenylalanine, did not result in the disappearance of the TREPR signature (Figure 2), as was observed for the equivalent mutation of the frog protein. Furthermore, the Synechocystis mutant protein reveals a spectral pattern that is virtually unchanged from that of the WT protein. The Trp triad of Synechocystis CRYD is structurally well overlaid with that of E. coli PHR (Figure S1).[10, 12] Obviously, however, photo-excitation of both WT and W320F (TrpC) mutant proteins leads to RP formation, so we concluded that TrpC may be not the terminal electron donor of the FAD photoreduction cascade in Synechocystis CRYD.
Figure 2.
TREPR spectra of WT (solid blue curve) and different Trp mutant proteins (solid green curves) of Synechocystis CRYD recorded 500 ns after pulsed laser excitation. The spectra were scaled to comparable signal-to-noise ratio. TrpA≡W396, TrpB≡W373, TrpC≡W320, and TrpC’≡W375. From top to bottom: WT, W320F, W373F, and W375F. Experimental parameters as in Figure 1. The dashed red curve shows a spectral simulation (parameters, see SI) of the TREPR spectrum for the RP state FAD•…TrpC’•.
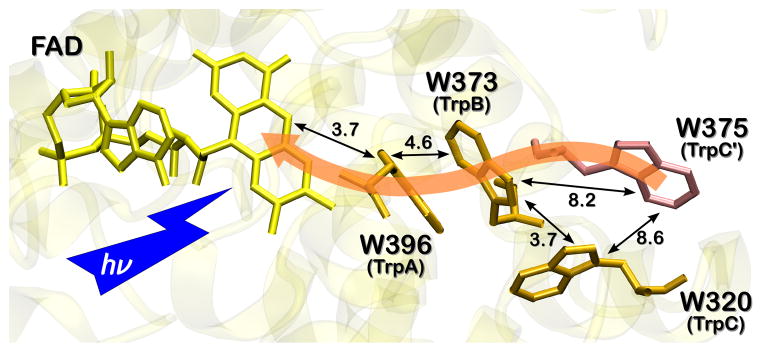
To elucidate the origin of the RP signature in Synechocystis CRYD, we examined two additional mutant proteins, W373F and W375F. The former was designed to test if the middle TrpB in the conserved Trp triad is required for ET in the cyanobacterial protein, and the latter for examination of potential PET beyond TrpC to W375≡TrpC’ (Figure 3). TrpC’ is relatively close to TrpC and is also exposed to the protein surface.[12] Hence, deprotonation and solvent–radical interactions could stabilize a photo-generated radical state on TrpC’, as on TrpC. The addition of TrpC’ to the classic Trp triad would then extend the ET pathway in Synechocystis CRYD to four Trps.
Figure 3.
The conserved Trp triad of Synechocystis CRYD (orange) and the alternative ET pathway including W375 (light red). The indole ring planes of TrpC and TrpB are nearly perpendicular to each other, whereas the ring planes of TrpB and TrpC’ assume a (for ET more favorable) nearly parallel orientation. The numbers at the black arrows are edge-to-edge distances in Å.
TREPR experiments on WT and Trp-to-Phe mutants of Synechocystis CRYD were conducted under identical experimental conditions (Figure 2). Remarkably, neither W373F (TrpB) nor W375F (TrpC’) revealed any significant RP signals (in the same time intervals), whereas both WT and W320F (TrpC) mutant proteins exhibited a signal. These results show that TrpC’ rather than TrpC is the more critical solvent-exposed distal Trp in the ET pathway in Synechocystis CRYD, unlike E. coli PHR and Xenopus CRYD, which both use TrpC as the terminal electron donor.[20, 31] Furthermore, the results on cyanobacterial CRYD unambiguously show that ET proceeds neither directly from TrpC’ (outer) to TrpA (inner), nor sequentially via TrpC, but – quite to our surprise – via TrpB in the middle of the conserved triad. This is despite the longer edge-to-edge distance between TrpB and TrpC’ (8.2 Å) as compared to TrpB and TrpC (3.7 Å), see Figure 3. Subtle differences between the TREPR spectra of Xenopus and Synechocystis WT cryptochrome (see Figure S3), as well as spectral simulations based on the theory of correlated coupled RPs corroborate these conclusions: The RP TREPR signature of the WT protein can be satisfactorily simulated with the magnetic interaction parameters (g-values, exchange and dipolar couplings) of the RP state FAD•…TrpC’• (Figure 2), but not with those of FAD•…TrpB•and FAD•…TrpC• (see SI). The alternative terminal electron donor, TrpC’, is generally not conserved throughout the PHR/CRY protein family; however, both CRY1 (W379) and CRY2 (W376) from A. thaliana do conserve this TrpC’ (Figure S1).
The novel ET pathway in Synechocystis CRYD, as compared to Xenopus CRYD or the related PHRs, triggers the question as to why ET in Synechocystis CRYD is preferred over the significantly longer distance from distal TrpC’ to TrpB than from TrpC to TrpB. We discuss our experimental observations on the basis of Marcus’ theory of charge transfer,[32] which predicts that the achievable ET rate kET depends, apart from the temperature T, on (i) the electronic coupling between the redox states, HAB, (ii) the change of total Gibbs free energy, ΔG, and (iii) the reorganization energy, λ:
| (1) |
In Eq. (1), kB is the Boltzmann constant.
In a previous contribution on the thermodynamics of ET along the Trp cascade in E. coli PHR, we computed the Marcus ET parameters, HAB, ΔG, and λ, by combining classical molecular dynamics based free-energy calculations with quantum-mechanical computations.[15] The resulting reorganization energies for each ET step, TrpB → TrpA (shorthand for TrpB + TrpA•→ TrpB• + TrpA) and TrpC → TrpB, had similar values. Because the three-dimensional structures of Synechocystis CRYD and E. coli PHR are comparable,[10, 12] we assume that λ-values are similar for the alternative TrpC’ → TrpB and (classic) TrpC → TrpB ET steps in Synechocystis CRYD. This leaves the electronic coupling elements, HAB(TrpC’→TrpB) and HAB(TrpC→TrpB), as well as the changes in free energy, ΔG, as the main sources for the observed differential ET behavior.
Electronic couplings between redox partners depend on their distance, provided that the intervening protein medium is similar, but also on their relative orientation.[32, 33] Relative orientation might compensate or even overcompensate for a more advantageous (i.e. shorter) ET distance.[34] In Synechocystis CRYD, the edge-to-edge distance between TrpC and TrpB is significantly shorter (3.7 Å) than that between TrpC’ and TrpB (8.2 Å). Thus, according to the phenomenological ‘Dutton ruler’,[33] an ~100-fold faster ET rate for TrpC → TrpB compared to TrpC’ → TrpB is expected, but clearly was not observed. Therefore, we expect that orientation effects between the aromatic rings of the Trps may favor the TrpC’ → TrpB route. Furthermore, the environment of the two Trps, W320 and W375 of Synechocystis CRYD, suggests that W375 (TrpC’) is more accessible to solvent molecules than W320 (TrpC), and thus, likely to be better stabilized by deprotonation and solvent–radical interactions (Figure 3).[12] Although we cannot rule out the possibility that TrpC’ is closer to TrpB in solution than in the crystal,[12] our molecular dynamics simulations of the Synechocystis CRYD protein in a water box gave no indication of significant spatial rearrangements of TrpC’. For the respective ET in E. coli PHR, the terminal TrpC (W306) was found to be the energetically favored radical, 30–45 kJ/mol below the other two Trps.[15] Hence, once populated, reverse charge transfer to the two buried TrpB and TrpA will be slow.
To address the thermodynamic stability at potential centers of charge localization from a theoretical perspective, we make use of a thermodynamic integration scheme[35] adapted for charge transfer reactions.[15] Without any loss of generality, we use the free energy of the presumed initial site of charge localization, TrpA, as the (zero) reference of the Gibbs free energy. We computed free energy changes for moving a positive charge from TrpA to TrpB (TrpB → TrpA) as ΔG = −49 kJ/mol, to TrpC (TrpC → TrpA) as −62 kJ/mol, and to TrpC’ (TrpC’ → TrpA) as −81 kJ/mol. This translates into an equilibrium Boltzmann population on TrpC’ in excess of 99% at room temperature. Computing the root mean square deviation of ΔG for charge transfer paths involving identical initial and terminal sites, but different intermediate sites, we estimate the statistical accuracy of the procedure as σ2(ΔG) ~5 kJ/mol. As a consequence, we are able to unambiguously identify TrpC’ (W375) in Synechocystis CRYD as a thermodynamic sink for hole transfer processes, relative to all amino acids considered here.
To conclude, we have demonstrated that tuning of local sequence and conformation can produce diversity in ET, despite a shared redox pathway in a highly conserved overall structural framework. Clearly, amino-acid sequence comparisons are not sufficient for predicting the photochemistry of a protein.[36] For the individual ET steps, distance is not necessarily the decisive parameter; orientation of ET partners[34] and their solvent accessibility, and therefore, stabilization of charge-separated states[13–15] contribute substantially.
The flexibility in ET pathways in CRY, as shown here, has a strong impact for understanding the photochemistry and functional diversity of the PHR/CRY family, including the proposed interactions of many CRYs with protein partners. Further spectroscopic investigations of this diversity will therefore lead to a much more detailed understanding of the reaction mechanism of these versatile proteins, and contribute to a deeper comprehension and better control of long-range ET in, but not restricted to, biological systems.
Experimental Section
Time-resolved detection of EPR following pulsed laser excitation was performed using a laboratory-built spectrometer.[18] Pulsed optical excitation of the Synechocystis CRYD samples (for details on protein synthesis and purification, see SI) was provided by a Nd:YAG laser (Spectra Physics GCR-11) pumping an optical parametric oscillator (Opta BBO-355-vis/IR, Opta GmbH, Bensheim, Germany) tuned to a wavelength of 460 nm (pulse with, 6 ns; pulse energy, 4 mJ).
Details on the thermodynamic integration scheme can be found in the Supporting Information to this communication.
Footnotes
This work was supported by the Deutsche Forschungs-gemeinschaft (FOR-526 to. S.W.), the U.S. National Institutes of Health (Grant R01 GM37684 to E.D.G.), and the Skaggs Institute for Chemical Biology (fellowship to K.H.). We thank R. Bittl (Free University Berlin) for use of instrumentation, M.E. Michel-Beyerle (Nanyang Technological University) for helpful discussions, and C. Hitomi for technical assistance.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Dr. Till Biskup, Department of Physics, Free University Berlin (Germany) and Department of Chemistry, University of Oxford (UK)
Dr. Kenichi Hitomi, Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA (USA)
Prof. Elizabeth D. Getzoff, Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA (USA)
Sebastian Krapf, Institute of Physical Chemistry, Albert-Ludwigs-University Freiburg, Albertstraße 21, 79104 Freiburg (Germany), Fax: (+49) 761 203-6222.
Prof. Dr. Thorsten Koslowski, Institute of Physical Chemistry, Albert-Ludwigs-University Freiburg, Albertstraße 21, 79104 Freiburg (Germany), Fax: (+49) 761 203-6222
Dr. Erik Schleicher, Institute of Physical Chemistry, Albert-Ludwigs-University Freiburg, Albertstraße 21, 79104 Freiburg (Germany), Fax: (+49) 761 203-6222
Prof. Dr. Stefan Weber, Email: Stefan.Weber@physchem.uni-freiburg.de, Institute of Physical Chemistry, Albert-Ludwigs-University Freiburg, Albertstraße 21, 79104 Freiburg (Germany), Fax: (+49) 761 203-6222.
References
- 1.Onuchic JN, Beratan DN, Winkler JR, Gray HB. Annu Rev Biophys Biomol Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]; Edwards PP, Gray HB, Lodge MTJ, Williams RJP. Angew Chem Int Ed. 2008;47:6758–6765. doi: 10.1002/anie.200703177. [DOI] [PubMed] [Google Scholar]; Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson BC, Fréchet JMJ. Angew Chem Int Ed. 2008;47:58–77. doi: 10.1002/anie.200702506. [DOI] [PubMed] [Google Scholar]
- 3.Hirst J. Biochem J. 2010;425:327–339. doi: 10.1042/BJ20091382. [DOI] [PubMed] [Google Scholar]
- 4.Efremov RG, Baradaran R, Sazanov LA. Nature. 2010;465:441–445. doi: 10.1038/nature09066. [DOI] [PubMed] [Google Scholar]
- 5.Wittekindt C, Schwarz M, Friedrich T, Koslowski T. J Am Chem Soc. 2009;131:8134–8140. doi: 10.1021/ja900352t. [DOI] [PubMed] [Google Scholar]
- 6.Genereux JC, Barton JK. Chem Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osakada Y, Kawai K, Fujitsuka M, Majima T. Proc Natl Acad Sci USA. 2006;103:18072–18076. doi: 10.1073/pnas.0607148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizzini L, et al. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 9.Byrdin M, Lukacs A, Thiagarajan V, Eker APM, Brettel K, Vos MH. J Phys Chem A. 2010;114:3207–3214. doi: 10.1021/jp9093589. [DOI] [PubMed] [Google Scholar]
- 10.Park HW, Kim ST, Sancar A, Deisenhofer J. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 11.Tamada T, Kitadokoro K, Higuchi Y, Inaka K, Yasui A, de Ruiter PE, Eker APM, Miki K. Nat Struct Biol. 1997;4:887–891. doi: 10.1038/nsb1197-887. [DOI] [PubMed] [Google Scholar]; Komori H, Masui R, Kuramitsu S, Yokoyama S, Shibata T, Inoue Y, Miki K. Proc Natl Acad Sci USA. 2001;98:13560–13565. doi: 10.1073/pnas.241371398. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klar T, Kaiser G, Hennecke U, Carell T, Batschauer A, Essen LO. ChemBioChem. 2006;7:1798–1806. doi: 10.1002/cbic.200600206. [DOI] [PubMed] [Google Scholar]; Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J. Proc Natl Acad Sci USA. 2006;103:17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Klar T, Pokorny R, Moldt J, Batschauer A, Essen LO. J Mol Biol. 2007;366:954–964. doi: 10.1016/j.jmb.2006.11.066. [DOI] [PubMed] [Google Scholar]
- 12.Brudler R, et al. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 13.Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 14.Popovic DM, Zmiric A, Zaric SD, Knapp EW. J Am Chem Soc. 2002;124:3775–3782. doi: 10.1021/ja016249d. [DOI] [PubMed] [Google Scholar]
- 15.Krapf S, Koslowski T, Steinbrecher T. Phys Chem Chem Phys. 2010;12:9516–9525. doi: 10.1039/c000876a. [DOI] [PubMed] [Google Scholar]
- 16.Lukacs A, Eker APM, Byrdin M, Brettel K, Vos MH. J Am Chem Soc. 2008;130:14394–14395. doi: 10.1021/ja805261m. [DOI] [PubMed] [Google Scholar]
- 17.Gindt YM, Vollenbroek E, Westphal K, Sackett H, Sancar A, Babcock GT. Biochemistry. 1999;38:3857–3866. doi: 10.1021/bi981191+. [DOI] [PubMed] [Google Scholar]
- 18.Weber S, Kay CWM, Mögling H, Möbius K, Hitomi K, Todo T. Proc Natl Acad Sci USA. 2002;99:1319–1322. doi: 10.1073/pnas.032469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber S. Biochim Biophys Acta. 2005;1707:1–23. doi: 10.1016/j.bbabio.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Biskup T, Schleicher E, Okafuji A, Link G, Hitomi K, Getzoff ED, Weber S. Angew Chem Int Ed. 2009;48:404–407. doi: 10.1002/anie.200803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittl R, Weber S. Biochim Biophys Acta. 2005;1707:117–126. doi: 10.1016/j.bbabio.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Weber S, Biskup T, Okafuji A, Marino AR, Berthold T, Link G, Hitomi K, Getzoff ED, Schleicher E, Norris JR. J Phys Chem B. 2010;114:14745–14754. doi: 10.1021/jp103401u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okafuji A, et al. DNA Repair. 2010;9:495–505. doi: 10.1016/j.dnarep.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Kavakli IH, Sancar A. Biochemistry. 2004;43:15103–15110. doi: 10.1021/bi0478796. [DOI] [PubMed] [Google Scholar]
- 25.Froy O, Chang DC, Reppert SM. Curr Biol. 2002;12:147–152. doi: 10.1016/s0960-9822(01)00656-x. [DOI] [PubMed] [Google Scholar]
- 26.Giovani B, Byrdin M, Ahmad M, Brettel K. Nat Struct Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]; Bouly JP, et al. J Biol Chem. 2007;282:9383–9391. doi: 10.1074/jbc.M609842200. [DOI] [PubMed] [Google Scholar]
- 27.Merrow M, Roenneberg T. Cell (Cambridge, Mass) 2001;106:141–143. doi: 10.1016/s0092-8674(01)00443-3. [DOI] [PubMed] [Google Scholar]; Banerjee R, Schleicher E, Meier S, Muñoz Viana R, Pokorny R, Ahmad M, Bittl R, Batschauer A. J Biol Chem. 2007;282:14916–14922. doi: 10.1074/jbc.M700616200. [DOI] [PubMed] [Google Scholar]
- 28.Henbest KB, Maeda K, Hore PJ, Joshi M, Bacher A, Bittl R, Weber S, Timmel CR, Schleicher E. Proc Natl Acad Sci USA. 2008;105:14395–14399. doi: 10.1073/pnas.0803620105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K, Ahmad M. J Biol Chem. 2005;280:19437–19440. doi: 10.1074/jbc.C500077200. [DOI] [PubMed] [Google Scholar]
- 30.Rodgers CT, Hore PJ. Proc Natl Acad Sci USA. 2009;106:353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YF, Heelis PF, Sancar A. Biochemistry. 1991;30:6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- 32.Marcus RA, Sutin N. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 33.Page CC, Moser CC, Chen X, Dutton L. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 34.Cave RJ, Siders P, Marcus RA. J Phys Chem. 1986;90:1436–1444. [Google Scholar]; Williams RM. Photochem Photobiol Sci. 2010;9:1018–1026. doi: 10.1039/c0pp00050g. [DOI] [PubMed] [Google Scholar]
- 35.Kirkwood JG. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 36.Whisstock JC, Lesk AM. Q Rev Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]