Abstract
Varying depth of high-throughput sequencing reads along a chromosome makes it possible to observe copy number variants (CNVs) in a sample relative to a reference. In exome and other targeted sequencing projects, technical factors increase variation in read depth while reducing the number of observed locations, adding difficulty to the problem of identifying CNVs. We present a hidden Markov model for detecting CNVs from raw read count data, using background read depth from a control set as well as other positional covariates such as GC-content. The model, exomeCopy, is applied to a large chromosome X exome sequencing project identifying a list of large unique CNVs. CNVs predicted by the model and experimentally validated are then recovered using a cross-platform control set from publicly available exome sequencing data. Simulations show high sensitivity for detecting heterozygous and homozygous CNVs, outperforming normalization and state-of-the-art segmentation methods.
Keywords: exorne sequencing, targeted sequencing, CNV, copy number variant, HMM, hidden Markov model
1 Introduction
Copy number variants (CNVs) are regions of a genome present in varying number in reference to another genome or population. CNVs are increasingly recognized as important components of genetic variation in the human genome and effective predictors of disease states. CNVs have been associated with a number of human diseases including cancer (Campbell et al., 2008), autism (Sebat et al., 2007, Glessner et al., 2009), schizophrenia (St Clair, 2009), HIV (susceptibility) (Gonzalez et al., 2005), and intellectual disability (Madrigal et al., 2007). These variants produce phenotypic changes through gene dosage effects, when the number of copies of a gene leads to more or less of a gene product, through gene disruption, when a CNV breakpoint falls within a gene, or through regulatory effects, when a CNV affects regulatory sequences such as enhancers and insulators (Kleinjan and van Heyningen, 1998). Recent studies report that 20–40 megabases, around 1% of the genome, are copy number variant in individual human genomes, making CNVs a larger source of basepair variation than single nucleotide polymorphisms (Conrad et al., 2010, Pang et al., 2010).
Two primary technologies for genome-wide detection of CNVs are array comparative genomic hybridization (arrayCGH) and high-throughput sequencing (HTS). ArrayCGH measures the fluorescence of two labeled DNA samples, which competitively bind to many probe sequences printed on an array. When the values from the probes are lined up according to genomic location, regions with variant copy number ratio can be observed as consecutive probes with higher or lower fluorescence ratio. CNVs exhibit a number of different signatures in resequencing data, where HTS reads from a sample are mapped to a reference genome, as reviewed by Medvedev et al. (2009). One kind of HTS signature is given by aberrant distances between the mapped positions of a paired end fragment overlapping a CNV, or between the ends of an unmappable read overlapping a CNV breakpoint. Another HTS signature, which this paper will focus on, is the amount of HTS reads mapping to regions along the chromosome, or “read depth”. The signature in this case is a region with higher or lower read depth compared to a control sequencing experiment, or compared to other regions within an experiment, assuming that HTS reads are distributed uniformly along the sample genome.
The read depth CNV signature is similar to the pattern seen in arrayCGH, so it is helpful to review the algorithms devised for this task. Popular algorithms for analyzing arrayCGH data include circular binary segmentation (Venkatraman and Olshen, 2007) and hidden Markov models (Fridlyand, 2004, Marioni et al., 2006). Hidden Markov models are useful for segmentation of many kinds of genomic data, as they represent linear sequences of observed data made up of homogeneous stretches associated with a hidden state. There are efficient algorithms for assessing the likelihood of an HMM with certain parameters given observed data and for estimating the most likely sequence of underlying states for a set of parameters (Rabiner, 1989). The HMMs designed for arrayCGH data take as input log ratios of measured fluorescence, a continuous variable, while read depth data consists of discrete counts of reads. We will therefore consider how to adjust the HMM framework to model read counts.
The main obstacle for CNV detection from read depth is the variance due to technical factors rather than copy number changes. HTS reads are subject to differential rates of amplification before sequencing and differential levels of errors during sequencing and mapping. For any HTS experiment, read depth in a genomic region can be related to local GC-content (Benjamini and Speed, 2011), as well as sequence complexity and sequence repetitiveness in the genome. In whole genome sequencing, it has been shown that normalizing read depth against GC-content can be sufficient to predict CNVs accurately (Campbell et al., 2008, Yoon et al., 2009, Alkan et al., 2009, Boeva et al., 2011, Miller et al., 2011). In paired sequencing experiments, such as in tumor/normal samples, position-specific effects can be eliminated through direct comparison, similarly to the elimination of probe-specific effects in arrayCGH (Chiang et al., 2008, Xie and Tammi, 2009, Ivakhno et al., 2010, Shen and Zhang, 2011, Sathirapongsasuti et al., 2011). However, HTS experiments do not always cover the whole genome and do not always include a reference sample sequenced using the same experimental protocol.
In targeted sequencing, such as exome sequencing, DNA fragments from regions of interest are enriched over other fragments and sequenced. Ideally, the sequenced reads map only to the targeted regions. Targeted sequencing therefore results in fewer positions at which to observe a change in read depth attributable to a CNV. Most target enrichment platforms use the following steps:
-
1.
DNA from a sample is fragmented and prepared for later sequencing.
-
2.
Prepared DNA fragments are hybridized to biotinylated RNA oligonucleotides and captured with magnetic beads or hybridized to probes on an array.
-
3.
The beads are washed, eluted and the RNA is digested or the array is washed and eluted.
-
4.
The remaining DNA sequences are amplified and sequenced.
Within the targeted regions, the enrichment steps lead to less uniform read depth than in whole genome sequencing, but the read depth pattern is consistent among samples using the same sequencing technology and enrichment platform. Sequencing with three different technologies using the same enrichment platform, Harismendy et al. (2009) find “a unique reproducible pattern of non-uniform sequence coverage” within each group and low correlation of read depth across different technologies. Testing three different target enrichment platforms with the same sequencing technology, Hedges et al. (2011) report high correlation within samples from the same platform and low correlation across different platforms. Taking advantage of the reproducibility of read depth, Herman et al. (2009) and Nord et al. (2011) are able to identify CNVs in targeted sequencing by normalizing read depth in individual samples against average depth over control samples, though thresholds must be set for calling a position as CNV.
We sought to extend the HMM framework for CNV detection in targeted sequencing data, modeling read counts in non-overlapping genomic windows as the observed variable generated from a distribution depending on the hidden copy number state. Similar to the usage of covariates by Marioni et al. (2006) in modulating transition probabilities, we outline a model which fits non-uniform read counts to positional covariates such as background read depth, GC-content and window width. Background read depth is generated similarly to the methods of Herman et al. (2009) and Nord et al. (2011) by taking the median of normalized read depth per window over a control set. By using a number of explanatory covariates, one can analyze samples which have positive but low correlation with background read depth and residual dependence on GC-content. Another benefit of the HMM framework is the forward algorithm, which allows for fitting the distributional parameters without knowing the underlying copy number state. The model formulation replaces preprocessing, thresholding, and window-merging steps with the optimization of a statistical likelihood over a parameter space.
We will present an HMM for predicting copy number state in exome and other targeted sequencing data using observed read counts and positional covariates. We show that this model can successfully detect private CNVs in an exome sequencing project using all samples to generate background read depth. We then evaluate the robustness of our method using a control set from publicly available exome sequencing data from an alternate enrichment platform. We simulate CNVs of various sizes and copy number in exome sequencing data and find that our model outperforms normalization and segmentation methods in recovering the simulated CNVs. Finally, we summarize the results and discuss possible extensions of the method.
2 Methods
2.1 Modeling resequencing read counts
As a measure of read depth, we count the number of start positions of reads with high mapping quality in non-overlapping windows along a chromosome. To examine the characteristics of targeted sequencing read depth, we will count reads from a whole exome sequencing project (Li et al. (2010), discussed later) in variable-size windows subdividing the consensus coding sequence (CCDS) (Pruitt et al., 2009). The distributions of counts per window for one sample often have positive skewness (Figure 1). Over all windows, the maximal count can be up to 20 times the mean count.
Figure 1: Distribution of read counts in windows covering the CCDS regions of chromosome 1 for one exome sequencing sample, cropped at 100 reads per window.
In this paper, we use two methods of generating windows from targeted regions. The method referred to above subdivides the CCDS regions such that a region of x basepairs (bp) is split evenly into max(1, ⎣x/100⎦) windows. This ensures that the individual windows covering smaller exons are of comparable size to the multiple windows dividing larger exons. Smaller windows could theoretically be chosen for higher resolution, but in exome sequencing data the resolution of CNV detection is inherently limited by the sparse distribution of exons in the genome. Furthermore, the windows must be large enough such that the read counts are sufficiently high (from sensitivity analysis ∼ 50 reads per window) for the samples with the least amount of sequencing. The windows generated from subdividing the CCDS regions of chromosome 1 are 112 bp on average. Another method of generating windows is to subdivide the targeted regions, which can increase the number of observed basepairs as the targeted regions in exome enrichment often overhang the CCDS regions. Both methods are comparable in terms of the qualitative signature of CNVs in read depth and the resulting predicted CNV breakpoints. Setting windows within the CCDS regions has two advantages though. First, the CCDS regions are more likely to be covered equally across different enrichment platforms, enabling cross-platform comparison or control sets. Second, we find that the extremes of the targeted regions have more variability than the centers. By starting with the CCDS regions we can avoid these variable flanking regions.
A suitable distribution for modeling the observed read counts in windows should have support on the non-negative integers. We could consider the Poisson distribution with a position-dependent mean parameter, representing the underlying rate of technical inflation of read counts. If the counts for a given window are distributed as a Poisson, then replicates should have equal mean and variance. We can check this assumption with read counts from a set of samples with similar amount of total sequencing. While these samples are not replicates, we expect that the private CNVs and SNPs which would alter read counts per sample should be rare in the coding regions. Plotting the variance over the mean for the read counts shows that most windows fall above the line y = x, and are therefore overdispersed for Poisson distributed data (Figure 2).
Robinson et al. (2010) and Anders and Huber (2010) suggest that the negative binomial is a more appropriate distribution for HTS read count data, having both a mean parameter µ and dispersion parameter ϕ. The density for a random variable X ∼ NB(µ, ϕ) is defined by
with mean and variance given by
The negative binomial is often used in ecological and biological contexts when the rate underlying a count statistic is variable and covariates cannot be found which would account for the variance. It can be derived as a mixture of Poisson distributions with the mean parameter following a gamma distribution, and it converges as ϕ → 0 to a Poisson with mean µ. We will use positional covariates to account for as much variance in read counts over windows as possible, but allow for the situation that unknown factors lead to overdispersed counts. We will first attempt to fit a single value of ϕ over all windows, then add model parameters to allow for ϕ to vary over windows.
Figure 2: Mean and variance of read count for 23,619 windows over 40 samples with similar amount of total mapped reads.
To obtain a measure of the positional non-uniformity in read depth, we calculate the median of sample-normalized read counts over a control set. Because samples vary in the total number of reads which map to the reference genome, we first need to normalize read counts per sample. Boxplots of read counts per window for 5 samples are shown in Figure 3. The distributions all exhibit positive skewness but the median and quartiles are shifted. Given a matrix C of counts of reads in T windows on a chromosome (rows) across N samples (columns), Cnorm is formed by dividing each column by its mean. Distributions of sample-normalized read counts per window (rows of Cnorm) indicate high variance in medians across consecutive windows (Figure 4). Some but not all of this variance of median read depth can be explained by GC-content (Figure 5). We calculate the background read depth by taking the median of the sample-normalized read count per window (median of rows of Cnorm), and the background variance similarly.
Figure 3: Boxplots of read counts for 5 samples over windows covering exons of chromosome 1.
Figure 4: Sample-normalized read counts for 15 consecutive windows over 200 samples.
2.2 Hidden Markov model to predict sample CNVs
HMMs are a natural framework to segment genomic data with a discrete number of states, and we can take advantage of the algorithms that have been developed to
Figure 5: Smooth scatterplot of median read depth over GC-content. Median read depth is the median of sample-normalized read counts from 200 samples.
evaluate these models. The observed variable is c*j, the j-th column of C, which represents the counts of HTS reads for sample j in T non-overlapping windows positioned linearly along a chromosome. Using the notation of Rabiner (1989) and Fridlyand (2004), we write c*j as O⃗ = {O1,...,OT}. We define exomeCopy, a homogeneous discrete-time HMM to generate O⃗, by the following:
-
1.
The number of states K. The set of states {S1,...,SK} represents the possible copy number states of the sample. Q⃗ = {q1,...,qT} represents the vector of underlying copy number states over T windows. qt = Si indicates that at window t, the sample has copy number Si.
-
2.
The initial state distribution π⃗ = {πi} where

-
3.
The state transition probability distribution A = {aij} where

-
4.
The emission distribution B = {bi(O⃗)} where


The choice of the number of underlying copy number states K must be fixed before fitting parameters, as well as the possible copy number values {Si} and expected copy number d. We tested the model for {Si} = {0,1,2,3,4} for the diploid genome (d = 2), and {Si} = {0,1,2} for the non-pseudoautosomal portion of the X chromosome in males (d = 1). Sets {Si} with higher possible copy number values can be used as well.
Two transition probabilities are fitted in the model: the probabilities of transitioning to a normal state and to a CNV state. These are depicted for a chromosome with expected copy count of 2 in Figure 6, with transitions going to the normal state as black lines and transitions going to a CNV state as gray dotted lines. The probability of staying in a state (grey solid lines) is set such that all transition probabilities from a state (rows of A) sum to 1. The initial distribution π is set equal to the transition probabilities from the normal state.
Figure 6: Transition probabilities for copy number states of the HMM with {Si} = {0,1,2,3,4} and expected copy number d = 2.
Consecutive windows in targeted sequencing can be adjacent on the chromosome if they subdivide the same targeted region or distant if they belong to different targeted regions. Therefore we might consider modifying the transition probabilities per window, because two positions that are close together on the chromosome should have a higher chance of being in the same copy number state than those which are distant. This is reflected in the heterogeneous HMM of Marioni et al. (2006) with transition probabilities that exponentially decay or grow to the stationary distribution as the distance grows. In testing we observed that a simple transition matrix results in similar CNV calls as the heterogeneous model without having to fit extra parameters.
While the HMMs of Fridlyand (2004) and Marioni et al. (2006) fit an unknown mean for the emission distribution of each hidden state, the emission distributions of exomeCopy for different states differ only by the discrete values {Si} associated with the hidden copy number state. Similar to the usage of positional covariates by Marioni et al. (2006) to modulate the transition probabilities, we use covariates to adjust the mean of the emission distribution, µti. We introduce the following variables: X, a matrix with leftmost column a vector of 1’s and remaining columns of median background read depth, window width and quadratic terms for GC-content; and β⃗ a column vector of coefficients with length equal to the number of columns of X. The mean parameter µti of the t-th window and the i-th state is calculated by the product of the sample to background copy number ratio and a linear combination of the covariates xt*, the t-th row of X. The mean parameter must be positive, so if the product is negative we take a small positive value ε.
The parameters of the HMM can be written compactly as λ⃗ = (π⃗,A,B). The underlying parameters necessary to fit are the transition probability to normal state, the transition probability to CNV state, β⃗ and ϕ. Parameters which are fixed are K, {Si} and d. The input data is O⃗ and X. The forward algorithm allows for efficient calculation of the likelihood of the parameters given the observed sequence of read counts, L(λ⃗|O⃗) (Rabiner, 1989). We use a slightly modified version of the likelihood function to deal with outlier positions. Some samples will occasionally have a very large count in window t such that bi(Ot) < ε for all states i and ε equal to the smallest positive number representable on the computer. In this case, the model likelihood is penalized and the previous column of normalized probabilities for the forward algorithm is duplicated.
To find an optimal λ⃗, we use Nelder-Mead optimization on the negative log likelihood function, with the optim function in the R package stats (R Development Core Team, 2011). A value of λ⃗ is chosen which decreases the negative log likelihood by an amount less than a specified relative tolerance. For this value of λ⃗, the Viterbi algorithm is used to evaluate the most likely sequence of copy number states at each window,
This most likely path is then reported as ranges of predicted constant copy number. The ranges extend from the starting position of window s with q̂s ≠ q̂s–1 to the ending position of window e, such that q̂e = q̂t, s ≤ t < e. For targeted sequencing, the nearest windows are not necessarily adjacent, so the breakpoints could occur anywhere in between the end of window s — 1 and the start of window s, for example. Ranges which correspond to CNVs can be intersected with gene annotations to build candidate lists of potentially pathogenic CNVs.
The optimization procedure requires that we set initial values for the various parameters to be fit. Initializing the probability to transition to a CNV state very low and the probability to transition to normal state high ensures that the Markov chain stays most often in the normal state. X is scaled to have non-intercept columns with zero mean and unit variance, as this was found to improve the results from numerical optimization. β⃗ is initialized to β̂ using linear regression of the raw counts O⃗ on the scaled matrix of covariates X. ϕ is initialized using the moment estimate for the dispersion parameter of a negative binomial random variable (Bliss and Fisher, 1953). Although each window is modeled with a different negative binomial distribution, we found a good initial estimate for ϕ uses the sample mean ō of O⃗ and the sample variance s2 of (O⃗–Xβ̂):
We extend exomeCopy to an alternate model, exomeCopyVar, where ϕ is replaced by ϕ⃗ which can vary across windows. The input data for modeling ϕ⃗ is the variance at each window of sample-normalized read depth, which can be seen in Figure 4. This modification could potentially improve CNV detection by accounting for highly variable windows using information from the background. We introduce Y, a matrix with leftmost column a vector of 1’s and other columns of background standard deviation and background variance. The emission distributions are then defined by
γ⃗ is a column vector of coefficients fitted similarly to β⃗ using numerical optimization of the likelihood. γ⃗ is initialized to [ϕ̂,0,0,...] with ϕ̂ defined in Equation 8.
| Table 1: Summary of exomeCopy notation | |
| Ot | observed count of reads in the t-th genomic window |
| ƒ | the emission distribution for read counts |
| µti | the mean parameter for ƒ at window t in copy state i |
| ϕ | the dispersion parameter for ƒ |
| Si | the copy number value for state i |
| d | the expected background copy number (2 for diploid, 1 for haploid) |
| X | the matrix of covariates for estimating µ |
| Y | the matrix of covariates for estimating ϕ |
| β | the coefficients for estimating µ |
| γ | the coefficients for estimating ϕ |
3 Results
3.1 XLID project: chromosome X exome resequencing
The accuracy with which a model can predict CNVs from read depth depends on many experimental factors, so we try to recover both experimentally validated and simulated CNVs using backgrounds from different enrichment platforms. First we run exomeCopy on data from a chromosome X exome sequencing project to find the potential genetic causes of disease in 248 male patients with X-linked Intellectual Disabilities (XLID) (Manuscript submitted). As males are haploid for the non-pseudoautosomal portion of chromosome X, detection of CNVs is easier than in the case of heterozygous CNVs, where read depth drops or increases by approximately one half. The high coverage of the targeted region in this experiment also facilitates discovery of CNVs from changes in read depth. Each patient’s chromosome X exons are targeted using a custom Agilent SureSelect platform and 76 bp single-end reads are generated using Illumina sequencing machines. Reads are mapped using RazerS software (Weese et al., 2009). Total sequencing varies from 1 to 20 million reads per patient over 3.8 Mb of targeted region. Reads are counted in 100 bp windows covering the targeted region, and only windows with positive median read depth across all samples are retained. The positional covariates used are background read depth from all patients and quadratic terms for GC-content.
exomeCopy predicts on average 0.3% of windows per patient to be CNV. This represents 11,581 CNV segments from all patients combined, with 60% being single windows with outlying read counts. For candidate CNV validation we retain 640 predicted CNVs covering 5 or more windows. The larger segments are stronger causal candidates and we suspect are less enriched with artifacts. The majority of the 640 predicted CNVs are common across many patients. There are 66 predicted CNVs present in 1-2 patients, 14 in 3-10 patients, 8 in 11-20 patients, and 7 in 21-75 patients, described further in Table 2. We retain 16 predicted novel CNVs, which are present in 1-2 patients, not in the Database of Genomic Variants (Zhang et al., 2006) and not already known to be associated with XLID.
As of writing, 10 predicted novel CNVs, 6 duplications and 4 deletions, have been tested and all were confirmed by arrayCGH or PCR. These CNVs are strong causal candidates based on segregation in the patients’ families and the genes which are contained in the CNVs. This estimated lower bound of patients with causal candidate CNVs, about 4%, is in agreement with results from a previous study suggesting that 5-10% of cases of XLID can be attributed to CNVs (Madrigal et al., 2007). Plots of experimentally validated CNVs found by our method are shown in Figure 7, with each point corresponding to the raw read count from a window covering the targeted region.
Table 2: Predicted XLID CNVs by type, frequency, genomic size and inclusion in the Database of Genomic Variants (DGV)
| Genomic size | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| [600bp-10kb] | (10-20kb] | (20-100kb] | (100kb-4Mb] | ||||||
| Type | Freq. | DGV+ | DGV- | DGV+ | DGV- | DGV+ | DGV- | DGV+ | DGV- |
| Dup. | 1-2 | 10 | 10 | 2 | 3 | 2 | 3 | 2 | 16 |
| 3-10 | 9 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | |
| 11-20 | 2 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | |
| 21-75 | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | |
| Del. | 1-2 | 6 | 6 | 0 | 1 | 1 | 2 | 0 | 2 |
| 3-10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 11-20 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 21-75 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
3.2 Recovering XLID CNVs with a cross-platform control set
To investigate the effect of background read depth on CNV detection, we attempt to recover the experimentally validated CNVs in the XLID patients, substituting the XLID read depth background used in the previous section with a read depth background from a whole exome sequencing project of 200 Danish male and female individuals published by Li et al. (2010) (referred to afterward as “Danish” or “Danish exomes”). We also run exomeCopy on the 9 XLID patients using no background read depth, but only GC-content and window width information. In contrast to the custom Agilent platform used in the XLID project, the Danish samples were enriched for exons using a NimbleGen array and the coverage is substantially lower, with a median of 15 reads per window compared to 326 per window in the XLID
Figure 7: Experimentally validated CNVs identified in the XLID read depth data. The y-axis corresponds to the raw read counts for windows along the targeted region. The x-axis corresponds to the index of the windows. The color is the predicted copy number with blue indicating a hemizygous duplication and red indicating a hemizygous deletion.
project. For comparison of background read depth between the XLID samples and the Danish samples, we restrict the analysis to 9,710 CCDS-based windows on chromosome X, excluding the pseudoautosomal regions and regions not covered by both enrichment platforms. The CCDS regions are split evenly into windows no larger than 200 bp.
Comparing median read depth for XLID samples with median read depth for Danish samples shows positive but not strong correlation across the different platforms (Figure 8). Comparing within groups shows that two randomly selected subsets of a group are highly correlated in both datasets. This is in agreement with the observations of Hedges et al. (2011) that read depth is highly correlated within enrichment platforms but only partially correlated across platforms.
As a robust measure of signal to noise, we calculate the median read depth divided by the median absolute deviation of read depth across windows on chromosome X covered by different enrichment platforms. We also provide read depth statistics from 16 high coverage paired-end exome sequencing samples and one whole genome sample from the 1000 Genomes Project (1000 Genomes Project Consortium, 2010). In the case of paired-end data, each sequenced read is counted in its respective window. The decreased signal to noise ratio displayed in Table 3 for the exon sequencing projects supports our assumption that exon enrichment leads to increased non-uniformity in read depth.
We run exomeCopy on 9 of the XLID patients with experimentally validated CNVs, once while substituting the XLID background with the Danish background,
Figure 8: XLID median normalized read depth and Danish exome median normalized read depth. Between groups there is positive but not strong Pearson correlation, while randomly dividing groups and comparing median read depth within groups gives very high correlation.
Table 3: Read depth statistics for experiments in CCDS-based windows on chr X
| study | submitted | Li et al. | 1000 Genomes | 1000 Genomes |
| population | XLID | Danish | PUR | NA12878 |
| sequencing target | chrX exons | exome | exome | whole genome |
| # samples used | 248 | 200 | 16 | 1 |
| median read count | 326 ± 96 | 15 ± 6 | 200 ± 102 | 105 |
| (mean ± sd) | ||||
| signal-noise ratio | 2.0 ± 0.2 | 1.3 ± .1 | 1.1 ± .03 | 2.7 |
| (mean ± sd) | ||||
| mean pairwise | .87 | .77 | .97 | – |
| correlation |
and again using no background read depth, only GC-content and window width as covariates. One experimentally validated duplication is removed from analysis, as it spans windows not targeted by the NimbleGen platform. The median read depth from the XLID dataset and the Danish exome dataset is only partially correlated (r = 0.58), so dividing one by the other would not necessarily help to recover CNV signal. However, exomeCopy is able to adapt to less correlated backgrounds by reducing the contribution of the background term and increasing the contribution of the other covariates, window width and quadratic terms for GC-content. The results in Table 4 demonstrate that with an independent control set for generating background, exomeCopy is frequently able to recover most of the windows contained within the experimentally validated CNVs. The sensitivity is measured as the percent of windows which are predicted as CNV out of the total number of windows contained within the validated CNV region, as the HMM does not always fit the entire span with the correct copy number state. The use of Danish exome background is always more sensitive in recovering CNVs than when exomeCopy is run without any read depth background. The average percent of windows predicted to be CNV is 5.4% and 1.9%, using Danish background and without background respectively. Also noteworthy in Table 4 is that CNVs with comparable genomic size can cover different numbers of windows, so methods for CNV detection in exome data should be sensitive to events covering only a few windows.
| Table 4: Recovery of experimentally validated XLID CNVs | ||||
|---|---|---|---|---|
| CNV type | # windows | genomic | % CNV windows recovered | |
| size in kb | Danish bg | without bg | ||
| duplication | 488 | 899 | 80 | 31 |
| duplication | 218 | 291 | 96 | 94 |
| duplication | 90 | 541 | 100 | 34 |
| duplication | 90 | 541 | 100 | 1 |
| duplication | 74 | 329 | 87 | 83 |
| deletion | 51 | 237 | 100 | 100 |
| deletion | 21 | 169 | 77 | 77 |
| deletion | 17 | 27 | 100 | 100 |
| deletion | 4 | 49 | 100 | 100 |
3.3 Sensitivity analysis on simulated autosomal CNVs
In order to further evaluate the performance of the model on CNVs in autosomes and in low coverage samples, we simulate heterozygous and homozygous CNVs of various size on chromosome 1 in the Danish exome data. Simulated heterozygous deletions and duplications are generated by randomly sampling 50% of reads in a specified region and either removing or doubling the counts respectively. Simulated homozygous deletions and duplications are generated by removing 95% of the reads or doubling the reads respectively.
For sensitivity analysis, we simulate CNVs overlapping varying numbers of CCDS-based windows on chromosome 1, and report the percent of windows within the simulated CNV with accurate predicted copy number, averaging over a number of simulation runs. We report the sensitivity in terms of windows rather than base-pairs, as the major factor influencing sensitivity is the amount of exonic (targeted) basepairs contained within the CNV. The number of windows is approximately the amount of targeted basepairs contained within the CNV divided by the average window size (112 bp for CCDS regions on chromosome 1). For reference, we include Table 5 which gives the estimated quartiles of genomic sizes in kilobases for varying number of CCDS-based windows on chromosome 1.
Table 5: Quartiles of genomic size (kb) by number of CCDS-based windows
| # CCDS-based windows | 1Q | 2Q | 3Q |
|---|---|---|---|
| 10 | 10 | 23 | 58 |
| 20 | 35 | 72 | 160 |
| 50 | 125 | 238 | 460 |
| 100 | 324 | 566 | 1043 |
| 200 | 684 | 1145 | 2037 |
| 400 | 1640 | 2656 | 4400 |
We test the recovery of simulated CNVs with or without background variance information using exomeCopy and exomeCopyVar respectively. The model incorporating background variance performs nearly the same, although it has increased calling outside of the simulated CNVs and longer running time (Figure 9). For both models we can calculate the variance-mean ratio of the emission distribution for the normal state, (1 + ϕµnormal), averaging over all windows. exomeCopy fits the dispersion parameter ϕ such that the variance of the emission distribution is on average 1.51 times the normal state mean. This supports the earlier analysis that read counts are overdispersed for Poisson. exomeCopyVar fits ϕ⃗ with a linear combination of columns in Y (Equation 10) such that the variance of the emission distribution is on average 1.32 times the normal state mean. ϕ⃗t is set to nearly zero for some windows, reducing the emission distributions to Poisson, but has higher ϕt than used by exomeCopy for windows with high background variance.
We further compare the sensitivity of exomeCopy against segmentation of normalized log ratios. We leave out exomeCopyVar as it uses background variance information in predicting copy number state which cannot be incorporated into normalization methods. We use two state-of-the-art segmentation algorithms for arrayCGH log ratios, the circular binary segmentation algorithm of Venkatraman and Olshen (2007) (referred to as “DNAcopy”) and the hidden Markov model of Marioni et al. (2006) (referred to as “BioHMM”), implemented in the R packages DNAcopy and snapCGH respectively. For comparing against normalization methods, we calculate the log2 ratio of sample counts plus a pseudocount of 0.1 over the median background. Log ratios are regressed on the remaining covariates (window
Figure 9: exomeCopy and exomeCopyVar perform similarly in recovering simulated CNVs of different type and size. Average percent of windows called CNV outside of the simulated CNVs is 0.5% and 0.8% and average run time is 7.6 s and 10.3 s for exomeCopy, exomeCopyVar respectively. Each point is the average over 100 simulations.
width, quadratic terms for GC-content, and an intercept term), and the residuals are used as input to the segmentation algorithms.
Segmentation algorithms on the normalized data are preferable to the many false positives that would result from using thresholds. DNAcopy and BioHMM are run using default settings, except the epsilon was lowered for BioHMM to 1e-4 to allow for sufficient number of simulations and var.fixed was set to TRUE. Predicted segment means are translated into estimates of discrete copy number by thresholding at intermediate values. For diploid genome sequences, normalized log ratio in (–∞,log2(0.25)] is recorded as homozygous deletion, normalized log ratio in (log2(0.25),log2(0.75)] is recorded as heterozygous deletion, etc. Relaxed evaluation allows any predicted value in (–∞,log2(0.75)] to be accepted for deletions and any predicted value in (log2(1.25), ∞) to be accepted for duplications.
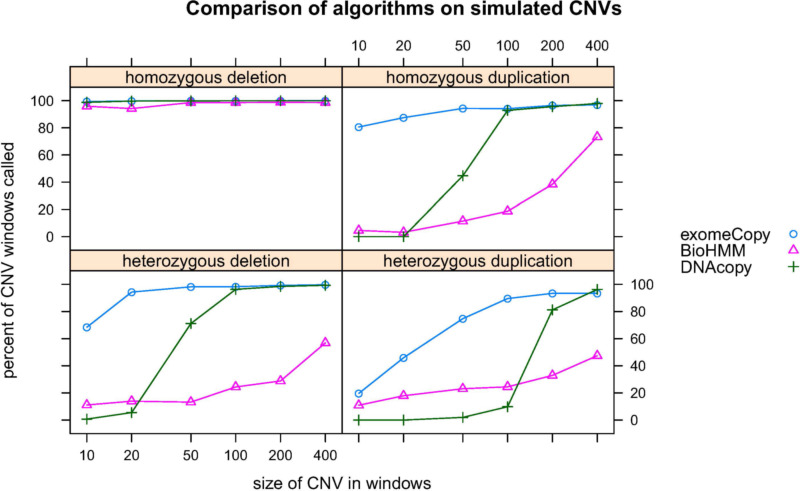
exomeCopy has equal or superior sensitivity to normalization and both segmentation methods for almost all types of CNVs (Figure 10). exomeCopy is often more sensitive for CNVs overlapping less than 100 windows, which is important as many of the experimentally validated CNVs from the XLID project overlapped 100 or fewer windows (Table 4). In the case of homozygous deletions, all methods can recover almost all windows of the simulated CNVs. In the relaxed evaluation,
Figure 10: Performance of algorithms in recovering simulated CNVs on chr 1 of the Danish exome samples. exomeCopy is equally or more sensitive for almost all types and sizes of CNVs. Average percent of windows called CNV outside of the simulated CNVs is 0.4%, 5.2%, 0.2% and average run time is 7.4 s, 111.9 s, 3.7 s for exomeCopy, BioHMM, and DNAcopy respectively. Each point is the average over 100 simulations.
the results are very similar, with improved recovery for BioHMM in homozygous duplications and heterozygous deletions (Figure 11).
As our method relies on the sample having increased read depth relative to the background, it can be expected that the presence of the identical CNV in the control set would reduce sensitivity. To estimate this effect on sensitivity, we simulate CNVs both in the test sample and at different minor allele frequencies (MAF) in the control population. 400 simulations are performed for both homozygous and heterozygous deletions/duplications covering 100 windows on chromosome 1 in the Danish exome data. We vary the MAF and the number of control samples used to make the background. The simulated CNV is inserted into control sample chromosomes with probability equal to the MAF. At MAF levels less than 10%, we find that exomeCopy has 86% sensitivity or greater, nearly equal to the sensitivity with an MAF of 0% (Table 6). The number of controls used does not seem to have a large effect on the sensitivity, however individual samples in small control sets might bias results. The average percent of windows called CNV outside of the simulated CNVs is less than 0.9% for all combinations.
Figure 11: Relaxed evaluation of algorithms in recovering simulated CNVs on chr 1 of the Danish exome samples. The same simulations as in Figure 10 are presented, but evaluation ignores the difference between heterozygous and homozygous predicted CNVs. BioHMM has improved recovery of small homozygous duplications and heterozygous deletions.
Table 6: Percent of simulated CNV windows recovered by minor allele frequency and number of controls
| MAF | |||||||
|---|---|---|---|---|---|---|---|
| CNV type | # controls | 0% | 1% | 5% | 10% | 25% | 50% |
| 10 | 100 | 100 | 100 | 100 | 98 | 68 | |
| homozygous deletion | 20 | 100 | 100 | 100 | 100 | 99 | 62 |
| 100 | 100 | 100 | 100 | 100 | 100 | 56 | |
| 10 | 97 | 96 | 92 | 88 | 59 | 16 | |
| homozygous duplication | 20 | 96 | 95 | 94 | 91 | 55 | 9 |
| 100 | 95 | 97 | 94 | 90 | 59 | 3 | |
| 10 | 99 | 99 | 98 | 96 | 51 | 0 | |
| heterozygous deletion | 20 | 99 | 99 | 98 | 96 | 48 | 0 |
| 100 | 99 | 98 | 99 | 97 | 42 | 0 | |
| 10 | 89 | 89 | 87 | 75 | 38 | 0 | |
| heterozygous duplication | 20 | 90 | 90 | 86 | 83 | 35 | 1 |
| 100 | 91 | 88 | 88 | 82 | 36 | 0 | |
Figure 12: Effect of background correlation on the absolute value of fitted coefficients. The x-axis shows the correlation of the simulated background with the original Danish background. Each point is the average over 100 simulations.
We demonstrate exomeCopy adjusting to less correlated or uncorrelated backgrounds in Figure 12. After adding increasing amounts of noise to the original Danish background, the absolute value of the coefficients for window width and quadratic terms for GC-content rise to replace the coefficient for noisy background. In the case that the sample is entirely uncorrelated with the background, the model will remove all contribution of the background in modeling the read counts.
Simulations on the Danish exome data demonstrates that exomeCopy can often recover CNVs in low coverage data if they overlap sufficient amount of targeted sequence. However, we expect that exomeCopy will have improved performance with higher coverage autosomal datasets. To assess the influence of total sequencing depth on recovery of different kinds of CNVs, we performed further simulations on 16 high coverage exome sequencing samples from the PUR population of the 1000 Genomes Project. (1000 Genomes Project Consortium, 2010) The library format is paired-end data, and we count both ends in their respective windows. Although this decision introduces dependency between the counts in nearby windows, it avoids the loss of sample coverage information at either or both positions.
To simulate experiments with different amounts of total sequencing, we sub-sample reads from the original PUR samples to achieve 10, 20, 50 and 100 average read counts in windows subdividing the CCDS regions of chromosome 1. At each level of read depth, we create a background across all 16 PUR samples, then simulate CNVs of varying length and type as before. As expected, increasing the
Figure 13: Performance of algorithms in recovering simulated CNVs on chr 1 after subsampling reads from the high coverage 1000 Genomes exome sequencing data. exomeCopy is increasingly sensitive with increasing average read counts. Average percent of windows called CNV outside of the simulated CNVs is always less than 0.7%. Each point is the average over 100 simulations.
read depth increases the sensitivity of exomeCopy, especially for the detection of the smallest heterozygous duplications, with 78% or more windows recovered at an average read count of 50. (Figure 13). This simulation suggests that average read counts of at least 50 per window will result in high sensitivity to detect both heterozygous and homozygous CNVs.
4 Discussion
Targeted sequencing is desirable for achieving high read coverage over regions of interest, while keeping costs and the size of generated data to manageable amounts. Exome sequencing prioritizes the discovery of variants in exons, as we expect these variants are more likely to be associated with a distinct phenotype than those which do not overlap exons. Nevertheless, methods for finding CNVs in targeted sequencing read depth data must overcome non-uniform patterns in read depth introduced by enrichment steps and a reduced number of genomic loci at which to observe changes.
We introduce a statistical model, exomeCopy, for detecting CNVs in targeted sequencing data which is robust across various enrichment platforms and different types and sizes of CNVs. In testing on exome sequencing data, our approach is more sensitive than normalization and state-of-the-art segmentation methods in finding duplications and heterozygous deletions which overlap few exons (Venkatraman and Olshen, 2007, Marioni et al., 2006). exomeCopy formulates the CNV detection problem as the optimization of a likelihood function over few parameters, and therefore requires no thresholds or preprocessing decisions which might affect downstream results. In modeling sample read count using a number of covariates in addition to background read depth, our method can find CNVs in samples which show low correlation with the background. This allows for targeted sequencing projects with few samples to use median read depth from another project as background. While intuitively exomeCopy could also be applied to detect amplifications in cancer sequencing using the healthy tissue read depth as background, we believe the paired tumor/normal sequencing setup deserves a different statistical treatment. We therefore recommend the use of methods specifically designed for segmentation of paired tumor/normal exome sequencing experiments. (Sathirapongsasuti et al., 2011)
Two limitations of CNV detection with targeted sequencing read depth are the effect of polymorphic CNVs in the control set and the inability to precisely localize CNV breakpoints. Although the median read depth method works well for finding CNVs which are rare in the control set, it might miss CNVs which are polymorphic. We formulate an HMM where the expected copy number d of the control set is constant over all windows. For genotyping polymorphic CNVs, one could locally cluster samples in the control set by read depth and attempt to assign absolute copy numbers to the samples in a given region (Alkan et al., 2009). Then the read depth for a copy number of d could be extrapolated from the clusters using their assigned copy numbers. Addressing the problem of localization, CNVs predicted from read depth in windows will not include exact breakpoints, and in the case of exome sequencing, the predicted breakpoints could fall anywhere between the outermost affected exons and the closest unaffected exons. Other sequencing based methods, such as partial mapping or anchored split mapping can be employed to recover breakpoints which fall within continuous targeted regions (Nord et al., 2011, O’Roak et al., 2011).
As sequence read counts are increasingly taken as quantitative measurements, statisticians and bioinformaticians must adapt methods to separate technical bias from biologically meaningful signal. From our investigations, we find increased sensitivity to the underlying CNV signal in statistical modeling of the raw count data compared to converting counts to normalized log ratios. We expect that similar methods of contrasting individual samples against a background capturing technical bias will be useful in other sequencing protocols such as RNA-Seq and ChIP-Seq.
5 Software
All calculations are performed in the R computing environment (R Development Core Team, 2011). exomeCopy is available as an R package through the Bioconductor project (http://www.bioconductor.org) (Gentleman et al., 2004).
Author Notes:
We thank our collaborators on the XLID project, Prof. Dr. H.-Hilger Ropers, Wei Chen, Hao Hu, Reinhard Ullmann and the EUROMRX consortium for providing the XLID data, validation of CNVs and for helpful discussion. We also thank Ho-Ryun Chung for suggestions. Part of this work was financed by the European Union’s Seventh Framework Program under grant agreement number 241995, project GENCODYS.
Contributor Information
Michael I. Love, Max Planck Institute for Molecular Genetics
Alena Myšičková, Max Planck Institute for Molecular Genetics .
Ruping Sun, Max Planck Institute for Molecular Genetics .
Vera Kalscheuer, Max Planck Institute for Molecular Genetics .
Martin Vingron, Max Planck Institute for Molecular Genetics .
Stefan A. Haas, Max Planck Institute for Molecular Genetics
References
- 1000 Genomes Project Consortium (2010): “A map of human genome variation from population-scale sequencing,” Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan, C., J. M. Kidd, T. Marques-Bonet, G. Aksay, F. Antonacci, F. Hormozdiari, J. O. Kitzman, C. Baker, M. Malig, O. Mutlu, S. C. Sahinalp, R. A. Gibbs, and E. E. Eichler (2009): “Personalized copy number and segmental duplication maps using next-generation sequencing,” Nature Genetics, 41, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. and W. Huber (2010): “Differential expression analysis for sequence count data.” Genome biology, 11, R106+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. and T. P. Speed (2011): “Estimation and correction for GC-content bias in high throughput sequencing,” Technical report, University of California at Berkeley. [Google Scholar]
- Bliss, C. I. and R. A. Fisher (1953): “Fitting the Negative Binomial Distribution to Biological Data,” Biometrics, 9. [Google Scholar]
- Boeva, V., A. Zinovyev, K. Bleakley, J.-P. Vert, I. Janoueix-Lerosey, O. Delattre, and E. Barillot (2011): “Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization,” Bioinformatics, 27, 268–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, P. J., P. J. Stephens, E. D. Pleasance, S. O’Meara, H. Li, T. Santarius, L. A. Stebbings, C. Leroy, S. Edkins, C. Hardy, J. W. Teague, A. Menzies, I. Goodhead, D. J. Turner, C. M. Clee, M. A. Quail, A. Cox, C. Brown, R. Durbin, M. E. Hurles, P. A. W. Edwards, G. R. Bignell, M. R. Stratton, and P. A. Futreal (2008): “Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing,” Nature Genetics, 40, 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, D. Y., G. Getz, D. B. Jaffe, M. J. T. O’Kelly, X. Zhao, S. L. Carter, C. Russ, C. Nusbaum, M. Meyerson, and E. S. Lander (2008): “High-resolution mapping of copy-number alterations with massively parallel sequencing,” Nature Methods, 6, 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, D. F., D. Pinto, R. Redon, L. Feuk, O. Gokcumen, Y. Zhang, J. Aerts, T. D. Andrews, C. Barnes, P. Campbell, T. Fitzgerald, M. Hu, C. H. Ihm, K. Kristiansson, D. G. MacArthur, J. R. MacDonald, I. Onyiah, A. W. Pang, S. Robson, K. Stirrups, A. Valsesia, K. Walter, J. Wei, C. Tyler-Smith, N. P. Carter, C. Lee, S. W. Scherer, and M. E. Hurles (2010): “Origins and functional impact of copy number variation in the human genome,” Nature, 464, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlyand, J. (2004): “Hidden Markov models approach to the analysis of array CGH data,” Journal of Multivariate Analysis, 90, 132–153. [Google Scholar]
- Gentleman, R., V. Carey, D. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Yang, and J. Zhang (2004): “Bioconductor: open software development for computational biology and bioinformatics,” Genome Biology, 5, R80+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner, J. T., K. Wang, G. Cai, O. Korvatska, C. E. Kim, S. Wood, H. Zhang, A. Estes, C. W. Brune, J. P. Bradfield, M. Imielinski, E. C. Frackelton, J. Reichert, E. L. Crawford, J. Munson, P. M. A. Sleiman, R. Chiavacci, K. Annaiah, K. Thomas, C. Hou, W. Glaberson, J. Flory, F. Otieno, M. Garris, L. Soorya, L. Klei, J. Piven, K. J. Meyer, E. Anagnostou, T. Sakurai, R. M. Game, D. S. Rudd, D. Zurawiecki, C. J. McDougle, L. K. Davis, J. Miller, D. J. Posey, S. Michaels, A. Kolevzon, J. M. Silverman, R. Bernier, S. E. Levy, R. T. Schultz, G. Dawson, T. Owley, W. M. McMahon, T. H. Wassink, J. A. Sweeney, J. I. Nurnberger, H. Coon, J. S. Sutcliffe, N. J. Minshew, S. F. A. Grant, M. Bucan, E. H. Cook, J. D. Buxbaum, B. Devlin, G. D. Schellenberg, and H. Hakonarson (2009): “Autism genome-wide copy number variation reveals ubiquitin and neuronal genes,” Nature, 459, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez, G. Catano, R. J. Nibbs, B. I. Freedman, M. P. Quinones, M. J. Bamshad, K. K. Murthy, B. H. Rovin, W. Bradley, R. A. Clark, S. A. Anderson, R. J. O’Connell, B. K. Agan, S. S. Ahuja, R. Bologna, L. Sen, M. J. Dolan, and S. K. Ahuja (2005): “The Influence of CCL3L1 Gene-Containing Segmental Duplications on HIV-1/AIDS Susceptibility,” Science, 307, 1434–1440. [DOI] [PubMed] [Google Scholar]
- Harismendy, O., P. Ng, R. Strausberg, X. Wang, T. Stockwell, K. Beeson, N. Schork, S. Murray, E. Topol, S. Levy, and K. Frazer (2009): “Evaluation of next generation sequencing platforms for population targeted sequencing studies,” Genome Biology, 10, R32+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, D. J., T. Guettouche, S. Yang, G. Bademci, A. Diaz, A. Andersen, W. F. Hulme, S. Linker, A. Mehta, Y. J. K. Edwards, G. W. Beecham, E. R. Martin, M. A. Pericak-Vance, S. Zuchner, J. M. Vance, and J. R. Gilbert (2011): “Comparison of Three Targeted Enrichment Strategies on the SOLiD Sequencing Platform,” PLoS ONE, 6, e18595+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, D. S., G. K. Hovingh, O. Iartchouk, H. L. Rehm, R. Kucherlapati, J. G. Seidman, and C. E. Seidman (2009): “Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection.” Nature methods, 6, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivakhno, S., T. Royce, A. J. Cox, D. J. Evers, R. K. Cheetham, and S. Tavaré (2010): “CNAsega novel framework for identification of copy number changes in cancer from second-generation sequencing data,” Bioinformatics, 26, 3051–3058. [DOI] [PubMed] [Google Scholar]
- Kleinjan, D.-J. and V. van Heyningen (1998): “Position Effect in Human Genetic Disease,” Human Molecular Genetics, 7, 1611–1618. [DOI] [PubMed] [Google Scholar]
- Li, Y., N. Vinckenbosch, G. Tian, E. Huerta-Sanchez, T. Jiang, H. Jiang, A. Albrechtsen, G. Andersen, H. Cao, T. Korneliussen, N. Grarup, Y. Guo, I. Hellman, X. Jin, Q. Li, J. Liu, X. Liu, T. Sparso, M. Tang, H. Wu, R. Wu, C. Yu, H. Zheng, A. Astrup, L. Bolund, J. Holmkvist, T. Jorgensen, K. Kristiansen, O. Schmitz, T. W. Schwartz, X. Zhang, R. Li, H. Yang, J. Wang, T. Hansen, O. Pedersen, R. Nielsen, and J. Wang (2010): “Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants,” Nature Genetics, 42, 969–972. [DOI] [PubMed] [Google Scholar]
- Madrigal, I., L. Rodríguez-Revenga, L. Armengol, E. González, B. Rodriguez, C. Badenas, A. Sánchez, F. Martínez, M. Guitart, I. Fernández, J. A. Arranz, M. Tejada, L. A. Pérez-Jurado, X. Estivill, and M. Milà (2007): “X-chromosome tiling path array detection of copy number variants in patients with chromosome X-linked mental retardation.” BMC genomics, 8, 443+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni, J. C., N. P. Thorne, and S. Tavaré (2006): “BioHMM: a heterogeneous hidden Markov model for segmenting array CGH data.” Bioinformatics, 22, 1144–1146. [DOI] [PubMed] [Google Scholar]
- Medvedev, P., M. Stanciu, and M. Brudno (2009): “Computational methods for discovering structural variation with next-generation sequencing,” Nature Methods, 6, S13–S20. [DOI] [PubMed] [Google Scholar]
- Miller, C. A., O. Hampton, C. Coarfa, and A. Milosavljevic (2011): “ReadDepth: A Parallel R Package for Detecting Copy Number Alterations from Short Sequencing Reads,” PLoS ONE, 6, e16327+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord, A., M. Lee, M. C. King, and T. Walsh (2011): “Accurate and exact CNV identification from targeted high-throughput sequence data,” BMC Genomics, 12, 184+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak, B. J., P. Deriziotis, C. Lee, L. Vives, J. J. Schwartz, S. Girirajan, E. Karakoc, A. P. MacKenzie, S. B. Ng, C. Baker, M. J. Rieder, D. A. Nickerson, R. Bernier, S. E. Fisher, J. Shendure, and E. E. Eichler (2011): “Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations,” Nature Genetics, 43, 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, A., J. MacDonald, D. Pinto, J. Wei, M. Rafiq, D. Conrad, H. Park, M. Hurles, C. Lee, J. C. Venter, E. Kirkness, S. Levy, L. Feuk, and S. Scherer (2010): “Towards a comprehensive structural variation map of an individual human genome,” Genome Biology, 11, R52+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K. D., J. Harrow, R. A. Harte, C. Wallin, M. Diekhans, D. R. Maglott, S. Searle, C. M. Farrell, J. E. Loveland, B. J. Ruef, E. Hart, M.-M. M. Suner, M. J. Landrum, B. Aken, S. Ayling, R. Baertsch, J. Fernandez-Banet, J. L. Cherry, V. Curwen, M. Dicuccio, M. Kellis, J. Lee, M. F. Lin, M. Schuster, A. Shkeda, C. Amid, G. Brown, O. Dukhanina, A. Frankish, J. Hart, B. L. Maidak, J. Mudge, M. R. Murphy, T. Murphy, J. Rajan, B. Rajput, L. D. Riddick, C. Snow, C. Steward, D. Webb, J. A. Weber, L. Wilming, W. Wu, E. Birney, D. Haussler, T. Hubbard, J. Ostell, R. Durbin, and D. Lipman (2009): “The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes.” Genome research, 19, 1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2011): R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rabiner, L. R. (1989): “A tutorial on hidden Markov models and selected applications in speech recognition,” Proceedings of the IEEE, 77, 257–286. [Google Scholar]
- Robinson, M. D., D. J. McCarthy, and G. K. Smyth (2010): “edgeR: a Bioconductor package for differential expression analysis of digital gene expression data.” Bioinformatics (Oxford, England), 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti, J. F., H. Lee, B. A. Horst, G. Brunner, A. J. Cochran, S. Binder, J. Quackenbush, and S. F. Nelson (2011): “Exome Sequencing-Based Copy-Number Variation and Loss of Heterozygosity Detection: ExomeCNV.” Bioinformatics (Oxford, England). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J., B. Lakshmi, D. Malhotra, J. Troge, C. Lese-Martin, T. Walsh, B. Yamrom, S. Yoon, A. Krasnitz, J. Kendall, A. Leotta, D. Pai, R. Zhang, Y.-H. H. Lee, J. Hicks, S. J. Spence, A. T. Lee, K. Puura, T. Lehtimäki, D. Ledbetter, P. K. Gregersen, J. Bregman, J. S. Sutcliffe, V. Jobanputra, W. Chung, D. Warburton, M.-C. C. King, D. Skuse, D. H. Geschwind, T. C. Gilliam, K. Ye, and M. Wigler (2007): “Strong association of de novo copy number mutations with autism.” Science (New York, N.Y.), 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J. J. and N. R. Zhang (2011): “Change-Point Model on Non-Homogeneous Poisson Processes with Application in Copy Number Profiling by Next-Generation DNA Sequencing,” Technical report, Division of Biostatistics, Stanford University. [Google Scholar]
- St Clair, D. (2009): “Copy number variation and schizophrenia.” Schizophrenia bulletin, 35, 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman, E. S. and A. B. Olshen (2007): “A faster circular binary segmentation algorithm for the analysis of array CGH data,” Bioinformatics, 23, 657–663. [DOI] [PubMed] [Google Scholar]
- Weese, D., A.-K. Emde, T. Rausch, A. Döring, and K. Reinert (2009): “RazerSfast read mapping with sensitivity control,” Genome Research, 19, 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, C. and M. Tammi (2009): “CNV-seq, a new method to detect copy number variation using high-throughput sequencing,” BMC Bioinformatics, 10, 80+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S., Z. Xuan, V. Makarov, K. Ye, and J. Sebat (2009): “Sensitive and accurate detection of copy number variants using read depth of coverage,” Genome Research, 19, 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., L. Feuk, G. E. Duggan, R. Khaja, and S. W. Scherer (2006): “Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome,” Cytogenetic and Genome Research, 115, 205–214. [DOI] [PubMed] [Google Scholar]