Copper is a transition metal essential for life. At elevated concentrations, however, it is highly toxic to organisms such as algae, fungi and many bacteria, and in humans, may adversely affect the gastrointestinal, hepatic and renal systems.[1, 2] As such, the detection and measurement of copper ions in water has become increasingly important, especially in point-of-use formats. Several methods exist for the detection of Cu2+ ions, including those based on organic dyes,[3–6] semiconductor nanocrystals,[7] and spectroscopy.[8–10] The read out mechanisms for these methods, however, often require sophisticated instrumentation. In contrast, colorimetric methods are extremely attractive for point-of-use applications, since they can be easily interpreted with the naked eye or low-cost portable instruments.
DNA gold nanoparticle (Au NP) conjugates[11–13] have been used in a variety of detection formats for DNA,[14–18] proteins,[19–21] metal ions,[22–25] enzyme inhibitors,[26] small molecules,[27–29] and intracellular mRNA.[30] Carboxylate[31] and peptide[32] modified Au NPs have also been used for colorimetric metal ion sensing. DNA Au NPs have high extinction coefficients (about 4 orders of magnitude greater than typical organic dyes) and unique distance-dependent plasmonic properties. When hybridized to complementary particles, DNA Au NPs turn from red to purple, and their aggregates exhibit extremely sharp melting transitions which make them useful for the colorimetric detection of oligonucleotides.[12] These optical and melting properties have also been utilized in mercuric ion detection,[24] where deliberately designed T – T mismatches in the interparticle oligonucleotide duplexes selectively complex Hg2+ via coordination chemistry. The formation of the T – Hg2+ – T coordination complexes raises the Tm of the polymeric aggregates, so the solution color measured as a function temperature can be used to determine Hg2+ concentration.[23, 24]
Recently, gold nanoparticle-based colorimetric detection approaches for copper ions have been developed based upon: 1) aptamers and Cu2+ catalyzed DNA ligation, which induces subsequent nanoparticle aggregation,[33] and 2) the use of Cu2+ catalyzed click chemistry off the surfaces of alkyne and azide terminated nanoparticles without DNA to aggregate Au NPs.[34] However, these systems suffer from limitations inhibiting their utility for on-site use. In the first example, the aptamers used a labile imidazole leaving group for oligonucleotide ligation, which requires special storage and handling. In the latter example, the assay requires an overnight incubation to facilitate aggregation and was not quantitative. Herein, we report a colorimetric method for the detection of Cu2+ ions based on densely functionalized DNA Au NP conjugates and click chemistry. This new approach relies on the ligation of two 15 base pair (bp) oligonucleotide strands within polymeric aggregates of the DNA Au NPs. The oligonucleotide strands were terminated in a hexynyl or azide group, allowing them to be ligated into a 30 bp strand with click chemistry. We hypothesized that this ligation would raise the Tm of the aggregate in proportion to the amount of copper ion present after a given time (a kinetic endpoint), which could be monitored by UV Vis spectroscopy or the naked eye. The use of click chemistry and DNA Au NPs confers three specific advantages that the previous Au NP-based systems lacked in whole or in part. First, this approach uses the oligonucleotides as a template to align the alkyne and azide groups for optimal reactivity.[35, 36] Second, the sharp melting properties of the DNA Au NPs allow one to distinguish subtle difference in Tm, allowing for Cu2+ quantification.[37] Finally, the alkyne and azide groups are robust, and their cyclization can be catalyzed specifically (almost exclusively) by copper ions.[38, 39]
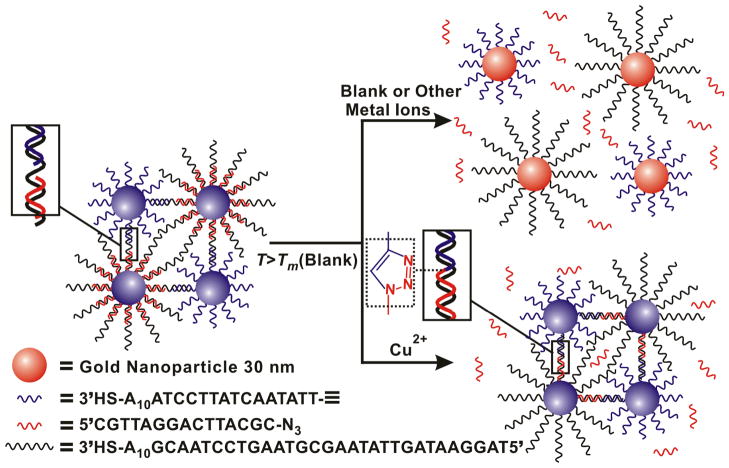
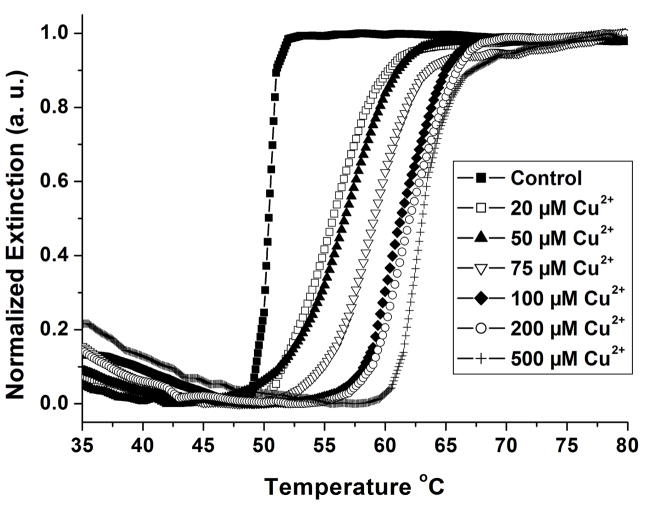
In a typical experiment, two sets of DNA Au NP conjugates were prepared by functionalizing two batches of 30 nm Au NPs with different alkylthiol-modified oligonucleotide strands. The first type of particle, called the template particle, was modified with 3′-propylthiol-terminated 40-mer oligonucleotides (5′ TAG GAA TAG TTA TAA GCG TAA GTC CTA ACG A10 SH 3′). The second particle, the alkyne Au NP, was functionalized with 3′-propylthiolated and 5′-alkylated 25-mer oligonucleotides that are complementary to half of the DNA on the template particles (5′ hexynyl TTA TAA CTA TTC CTA A10 SH 3′). These functionalized Au NPs formed polymeric networks upon mixing due to their complementary DNA. A third oligonucleotide, the azide strand, was added. These strands are 3′ azidobutyrate labeled 15-mer oligonucleotides (5′ CGT TAG GAC TTA CGC azidobutyrate 3′) complementary to the other half of the template strand. Together, these oligonucleotides formed three stranded aggregates with a concomitant red-to-purple color change (Scheme 1). These aggregates serve as the Cu2+ probes in our novel assay. In a proof-of-concept detection experiment, the Cu2+ concentration was increased to 200 μM, and excess sodium ascorbate and a water soluble tris-triazolylamine Cu+ binding ligand[35, 40] were added to the solution of aggregates and incubated for 2 hours to allow for the click-chemistry ligation to occur. The melting temperature of these aggregates, which was monitored at the Au NP surface plasmon resonance maximum of 525 nm, was 62.6 °C. In a control experiment performed in the absence of copper ion, the aggregates melted at 50.4 °C, approximately 12°C below the Tm after ligation (Figure 1). This increase in Tm was due to the conversion of the three-strand structure to a ligated two-strand 30 bp duplex. The presence of the ligated strand was confirmed with mass spectrometry (Figure S1).
Scheme 1.
Schematic illustration representing the aggregation and dissociation of the gold nanoparticle probes used in the colorimetric detection of copper ions.
Figure 1.
Normalized melting curves of the aggregates in the presence of Cu2+ concentrations ranging from 0 μM to 500 μM. The change in the extinction was monitored at 525 nm.
The sharp melting properties of DNA Au NPs allowed the measurement of small differences in Tm, which was proportional to the concentration of the Cu2+ added in the range of 20 to 100 μM (Figure 1). Greater concentrations of the ion increase the number of ligated 30 bp interstrand duplexes, increasing the Tm of the aggregates. The limit of detection of the assay is 20 μM and its dynamic range is 20 to 100 μM (Figure S2). The Environmental Protection Agency-defined maximum contaminant level for copper in drinking water is 20 μM, making it relevant for testing drinking water.
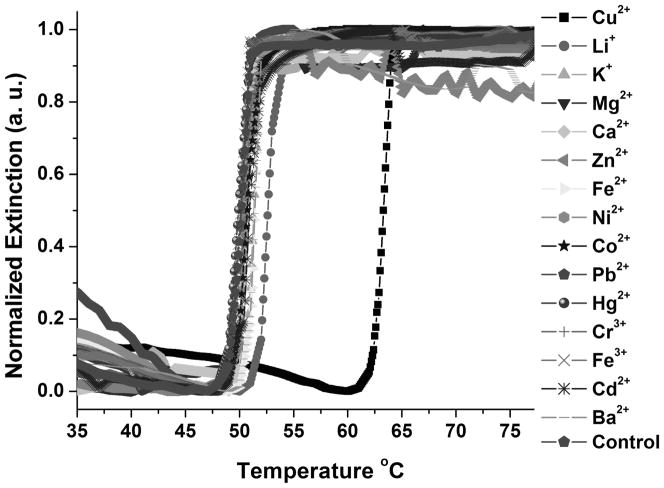
To evaluate the selectivity of this assay, it was challenged with other environmentally relevant metal ions, including Li+, K+, Mg2+, Ca2+, Fe2+, Mn2+, Co2+, Zn2+, Ni2+, Ba2+, Cd2+, Pb2+, Hg2+, Cr3+ and Fe3+. In a typical experiment, one of these metal ions was added to a solution of the DNA Au NP aggregates at a final concentration of 200 μM and incubated for 2 hours in the presence of the tris-triazolylamine Cu+-binding ligand and sodium ascorbate. Only the Cu2+ sample showed an increased melting temperature relative to the blank (Figure 2). No obvious melting temperature increase was observed for other metal ions. Importantly, the large extinction coefficient of the DNA Au NPs allowed for specific copper ion detection through visual inspection, which is particularly attractive for on-site use (Figure 3).
Figure 2.
Normalized melting curves of the aggregates in the presence of 200 μM of various metal ions. The change in extinction was monitored at 525 nm.
Figure 3.
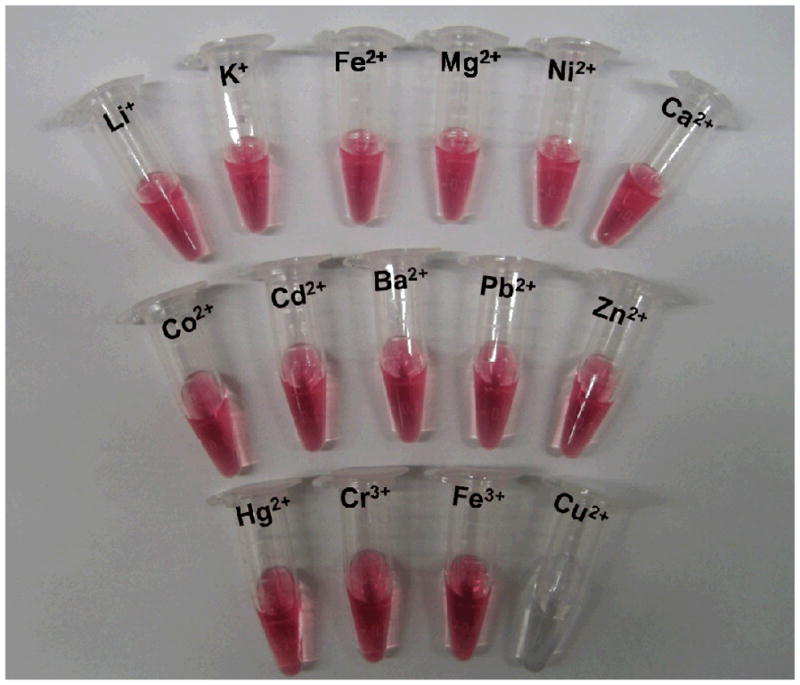
A photograph of the DNA Au NPs at 55 °C in the presence of different metal ions after ligation.
In conclusion, we have developed a colorimetric copper ion detection system with high selectivity, and high sensitivity for a colorimetric assay. The concentration of Cu2+ can be determined by the change in solution color at a given temperature, or through a measurement of the melting temperature of the DNA Au NP aggregates. In contrast to other Au NP based detection systems, this method does not require labile reactive groups or long incubation times due to the robustness of the alkyne and azide functionalities and the fact that those groups are templated together for optimal reactivity via oligonucleotide hybridization, respectively. Taken together, these advantages make this assay simple and robust, and therefore promising for on-site water testing. As such, the strategy presented in this work represents a promising new addition to the growing library of NP-based probes for protein, nucleic acid, metal ion, and small molecule detection.[13, 41–48]
Experimental Section
Materials
All reagents and solvents were purchased from Sigma-Aldrich unless otherwise noted. 30 nm diameter Au NPs were purchased from Ted Pella. Oligonucleotides were synthesized on an Expedite 8909 Nucleotide Synthesis System by standard solid-phase phosphoramidite synthesis techniques. All bases and reagents were purchased from Glen Research. The oligonucleotides were purified using reverse-phase high-performance liquid chromatography (RP-HPLC). After purification, the oligonucleotides were lyophilized and stored at −78 °C until use. Before nanoparticle conjugation, the 3′ disulfide functionality was reduced with dithiothreitol (DTT) following published procedures.[37]
Synthesis of alkyne-modified oligonucleotides
Alkyne-modified oligonucleotide (5′ hexynyl-TTA TAA CTA TTC CTA A10 SH- 3′) were synthesized using 3′-thiol-modifier C3 S-S CPG and 5′-hexynyl phosphoramidite. The oligonucleotides were characterized by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS, supporting information).
Synthesis of azido-modified oligonucleotides
Azido-modified oligonucleotides were obtained by conjugating an amino-modified oligonucleotide with an azide N-hydroxysuccinimide (NHS) ester (azidobutyrate NHS ester, Glen Research). Lyophilized amino-modified oligonucleotides (1 μmol) were dissolved in 0.5 mL of 0.1M Na2CO3/NaHCO3 buffer (pH 8.5). To this solution, 5 mg of azide NHS ester in 100 μL of DMSO was added. The resulting mixture was incubated overnight at room temperature, purified by RP-HPLC and characterized by MALDI-MS (supporting information).
Copper ion detection assay
Au NP aggregates were formed by mixing the template particles (modified with 5′ TAG GAA TAG TTA TAA GCG TAA GTC CTA ACG A10 SH 3′) (0.5 ml, 1 nM), the alkyne Au NPs (modified with 5′ hexynyl TTA TAA CTA TTC CTA A10 SH 3′) (0.5 ml, 1 nM) and 3′ azidobutyrate labeled oligonucleotides (5′ CGT TAG GAC TTA CGC azidobutyrate 3′). These oligonucleotides formed three stranded aggregates with a concomitant red-to-purple color change. The Au NP aggregates were washed (3 x) with PBS buffer (10 mM sodium phosphate, 0.1 M NaCl, pH 7.0). They were then resuspended in 950 μL of PBS buffer. Tris-hydroxypropyl triazolyl ligand (2.0 μmol), sodium ascorbic acid (2.0 μmol) and copper (II) sulphate pentahydrate (at predetermined concentrations) were then added to a solution of the aggregates, respectively, and incubated for 2 h. The melting transitions were monitored by UV-vis spectroscopy (Cary 5000, Varian) and the solution was continuously stirred with a magnetic stir bar to keep the aggregates suspended.
Supplementary Material
Footnotes
This work was supported by the AFOSR, ARO, DDRE, and NSF. CAM is grateful for a NIH Director’s Pioneer Award.
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
References
- 1.Georgopoulos PG, Roy A, Yonone-Lioy MJ, Opiekun RE, Lioy PJ. J Toxicol Env Health-Pt b-Crit Rev. 2001;4:341. doi: 10.1080/109374001753146207. [DOI] [PubMed] [Google Scholar]
- 2.Zietz BP, Dieter HH, Lakomek M, Schneider H, Kessler-Gaedtke B, Dunkelberg H. Sci Total Environ. 2003;302:127. doi: 10.1016/s0048-9697(02)00399-6. [DOI] [PubMed] [Google Scholar]
- 3.Viguier RFH, Hulme AN. J Am Chem Soc. 2006;128:11370. doi: 10.1021/ja064232v. [DOI] [PubMed] [Google Scholar]
- 4.Banthia S, Samanta A. New J Chem. 2005;29:1007. [Google Scholar]
- 5.Zheng YJ, Gattas-Asfura KM, Konka V, Leblanc RM. Chem Commun. 2002:2350. doi: 10.1039/b208012e. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Lu Y. J Am Chem Soc. 2007;129:9838. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- 7.Gattas-Asfura KA, Leblanc RM. Chem Commun. 2003:2684. doi: 10.1039/b308991f. [DOI] [PubMed] [Google Scholar]
- 8.Chan MS, Huang SD. Talanta. 2000;51:373. doi: 10.1016/s0039-9140(99)00283-0. [DOI] [PubMed] [Google Scholar]
- 9.Wu JF, Boyle EA. Anal Chem. 1997;69:2464. doi: 10.1021/ac961204u. [DOI] [PubMed] [Google Scholar]
- 10.Callahan MR, Rose JB, Byrne RH. Talanta. 2002;58:891. doi: 10.1016/s0039-9140(02)00403-4. [DOI] [PubMed] [Google Scholar]
- 11.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 12.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 13.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 14.Xu XY, Georganopoulou DG, Hill HD, Mirkin CA. Anal Chem. 2007;79:6650. doi: 10.1021/ac070867g. [DOI] [PubMed] [Google Scholar]
- 15.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Nature Biotech. 2004;22:883. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds RA, Mirkin CA, Letsinger RL. J Am Chem Soc. 2000;122:3795. [Google Scholar]
- 17.Cao YWC, Jin RC, Mirkin CA. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 18.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 19.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 20.Zheng GF, Daniel WL, Mirkin CA. J Am Chem Soc. 2008;130:9644. doi: 10.1021/ja803035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Proc Natl Acad Sci USA. 2005;102:2273. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JW, Lu Y. Chem Mat. 2004;16:3231. [Google Scholar]
- 23.Xue XJ, Wang F, Liu XG. J Am Chem Soc. 2008;130:3244. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 24.Lee JS, Han MS, Mirkin CA. Angew Chem Int Ed. 2007;46:4093. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Wieckowska A, Willner I. Angew Chem Int Ed. 2008;47:3927. doi: 10.1002/anie.200705991. [DOI] [PubMed] [Google Scholar]
- 26.Xu XY, Han MS, Mirkin CA. Angew Chem Int Ed. 2007;46:3468. doi: 10.1002/anie.200605249. [DOI] [PubMed] [Google Scholar]
- 27.Han MS, Lytton-Jean AKR, Mirkin CA. J Am Chem Soc. 2006;128:4954. doi: 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JS, Ulmann PA, Han MS, Mirkin CA. Nano Lett. 2008;8:529. doi: 10.1021/nl0727563. [DOI] [PubMed] [Google Scholar]
- 29.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Nano Lett. 2009;9:3258. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. J Am Chem Soc. 2007;129:15477. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y, Johnson RC, Hupp JT. Nano Lett. 2001;1:165. [Google Scholar]
- 32.Yang WR, Gooding JJ, He ZC, Li Q, Chen GN. J Nanosci Nanotech. 2007;7:712. [PubMed] [Google Scholar]
- 33.Liu J, Lu Y. Chem Commun. 2007:4872. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Wang SX, Zhang K, Jiang XY. Angew Chem Int Ed. 2008;47:7454. doi: 10.1002/anie.200802317. [DOI] [PubMed] [Google Scholar]
- 35.Kumar R, El-Sagheer A, Tumpane J, Lincoln P, Wilhelmsson LM, Brown T. J Am Chem Soc. 2007;129:6859. doi: 10.1021/ja070273v. [DOI] [PubMed] [Google Scholar]
- 36.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. Science. 2004;305:1601. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin RC, Wu GS, Li Z, Mirkin CA, Schatz GC. J Am Chem Soc. 2003;125:1643. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]
- 38.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 40.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 41.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J Am Chem Soc. 2000;122:9071. [Google Scholar]
- 42.Maxwell DJ, Taylor JR, Nie SM. J Am Chem Soc. 2002;124:9606. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 43.Chen XL, Huang YF, Tan WH. J of Biomed Nanotech. 2008;4:400. [Google Scholar]
- 44.Li D, Shlyahovsky B, Elbaz J, Willner I. J Am Chem Soc. 2007;129:5804. doi: 10.1021/ja070180d. [DOI] [PubMed] [Google Scholar]
- 45.Katz E, Willner I. Angew Chem Int Ed. 2004;43:6042. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 46.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Curr Opin Biotech. 2002;13:40. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 47.Niemeyer CM, Simon U. Eur J Inorg Chem. 2005:3641. [Google Scholar]
- 48.Daniel WL, Han MS, Lee JS, Mirkin CA. J Am Chem Soc. 2009;131:6362. doi: 10.1021/ja901609k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.