Abstract
The human salivary proteome is extremely complex, including proteins from salivary glands, serum, and oral microbes. Much has been learned about the host component, but little is known about the microbial component. Here we report a metaproteomic analysis of salivary supernatant pooled from 6 healthy subjects. For deep interrogation of the salivary proteome, we combined protein dynamic range compression, multidimensional peptide fractionation, and high mass accuracy MS/MS with a novel two-step peptide identification method using a database of human proteins plus those translated from oral microbe genomes. Peptides were identified from 124 microbial species as well as uncultured phylotypes such as TM7. Streptococcus, Rothia, Actinomyces, Prevotella, Neisseria, Veilonella, Lactobacillus, Selenomonas, Pseudomonas, Staphylococcus, and Campylobacter were abundant among the 65 genera from 12 phyla represented. Taxonomic diversity in our study was broadly consistent with metagenomic studies of saliva. Proteins mapped to twenty KEGG pathways, with Carbohydrate Metabolism, Amino Acid Metabolism, Energy Metabolism, Translation, Membrane Transport, and Signal Transduction most represented. The communities sampled appear to be actively engaged in glycolysis and protein synthesis. This first deep metaproteomic catalog from human salivary supernatant provides a baseline for future studies of shifts in microbial diversity and protein activities potentially associated with oral disease.
Keywords: whole saliva, bacteria, microbiome, metaproteomics, phylogenetic analysis
INTRODUCTION
The proteome of human whole saliva is extremely complex, encompassing proteins from several types of salivary glands with distinct secretory profiles, surface and secreted proteins from oral epithelial cells, as well as serum and neutrophil proteins that enter the mouth through the gingival crevice [1]. Moreover, the mouth also is home to an extremely diverse microbial community. Recent metagenomic studies indicate that the microbiota of whole saliva samples may include over 10,000 distinct taxa, and the vast majority of them have never been cultured [2–7]. This suggests that oral microbes may contribute thousands of additional proteins to the salivary proteome.
Much has been learned about the human components of the salivary proteome [8, 9], but far less is known regarding salivary proteins of microbial origin. It has become clear in recent years that the members of the oral microbiota interact extensively, and function together as a polymicrobial community [10]. Thus approaches are needed which allow one to analyze the salivary microbial proteome in the context of its host. The community-based approach to microbial proteomics has become known as metaproteomics. Conceptually, metaproteomics is the proteomic counterpart to metagenomics. Metaproteomic analyses have been carried out on bacterial communities from a wide variety of environments, including human feces [11, 12], marine microbial ecosystems [13], acid mine drainage [14–16], activated sludge wastewater treatment systems [16, 17], and rhizosphere soil [18, 19].
However, several challenges hold back routine metaproteomic analyses in saliva, as well as other samples. First is the challenge of the wide dynamic range of protein abundance. In saliva, human proteins are most likely orders of magnitude more abundant than microbial proteins, thereby suppressing detection and identification of the microbial proteins. Therefore, advanced analytical techniques are needed to “dig deep” into the salivary proteome and confidently identify even the low abundance proteins of microbial origin. The second challenge is peptide sequence matching using sequence database searching. Potential expressed protein sequences can be translated from microbial genome sequences. However, the collective genomes of sequenced microbes exceed the size of the human genome by several orders of magnitude. Thus, database searches are extremely computing-intensive. Most significantly, peptide sequence matching against such very large databases suffers from the increased potential for false-positive matches which lowers the number of high confidence true matches [20].
Our group previously published a metaproteomic analysis of 357 microbial peptides obtained from the pellet fraction of saliva, pooled from four oral cancer patients. Those peptides were assignable to five bacterial phyla representing 26 genera, of which Streptococcus, Neisseria, and Haemophilus were the most abundant. The primary functions represented by the parent proteins were translation, carbohydrate metabolism, amino acid metabolism, and energy metabolism [21]. Although those findings were generally representative of the species composition and activities of known oral microbes, there also was concern that a substantial number of peptides were assigned to exotic taxa that were very unlikely to be actually present in the mouth. This was particularly surprising given that the overall number of microbial peptides obtained was relatively small.
Here, we improve on our previous study using dynamic range compression (DRC) via ProteoMiner™ hexapeptide libraries, and three-dimensional peptide fractionation prior to MS/MS analysis on an LTQ-Orbitrap instrument [8]. We also applied a novel two-step peptide identification method for more effective metaproteomic analysis. These collective improvements enabled the first deep cataloging of microbial proteins in human salivary supernatant.
MATERIALS AND METHODS
Salivary supernatant dataset
Salivary supernatant was collected and pooled from six healthy subjects who refrained from eating or drinking for 90 minutes. Proteins were analyzed using Proteominer™ (Bio-Rad Laboratories, Hercules, CA) for DRC, multidimensional peptide fractionation and a LTQ-Orbitrap mass spectrometer as described in Bandhakavi et al 2009 [8]. An additional 45 RAW files generated from ProteoMiner ™ Library-2-treated saliva were also analyzed.
Two-step method for peptide sequence matching and protein identification
RAW files generated (200 total) from the LTQ-Orbitrap salivary supernatant dataset were processed using the MaxQuant (v 1.0.13.13) "Quant" module to generate .MSM files [8, 22–24]. Individual Peak and iso MSM files corresponding to each .RAW file were converted to Mascot generic format (MGF) and searched using ProteinPilot v 4.0 (ProteinPilot Software 4.0; Revision: 148085; Paragon Algorithm: 4.0.0.0. 148083). Paragon searches [35] were conducted using LTQ-Orbitrap subppm instrument settings. Other parameters used for the search were as follows: Sample Type: Identification; Cys alkylation: None; Digestion: Trypsin; ID Focus: Biological Modifications; Search effort: Thorough.
In the first step, all 200 RAW files were searched against a database consisting of all the translated human oral microbial genomic sequences from the Human Oral Microbiome Database (HOMD) [25], along with the human IPI v3.52 database and contaminant proteins (1,687,426 total protein sequences) [26]. The ProteinPilot searches generated a .group file that was used to generate a Protein Report from the peptide sequence matches. All non-human protein sequences that were identified at the threshold of at least 66% Conf score (0.47 ProtScore) in the first step were merged with the Human IPI v3.52 database along with contaminant proteins to generate a “refined” FASTA database for the second step.
In the second step, all 200 RAW files were searched against a "Target-Decoy" version of the FASTA database mentioned above, by appending the reversed protein sequences to the forward sequences, resulting in a database containing 152,724 total protein sequences. Parameters for ProteinPilot were the same described for the first step with the exception of searching to generate a false discovery rate (FDR) report. These ProteinPilot searches generated a .group file and a Proteomics System Performance Evaluation Pipeline (PSPEP) FDR report [27]. The .group file was used to generate a Protein Summary and a Peptide Summary Report. The FDR report was used to estimate the number of proteins, distinct peptides and spectra at 5% local FDR. Spectra identified at 5% local FDR were used to filter for non-human (microbial) peptide identifications. A list of non-human distinct peptide sequences that were identified was generated from the Peptide Summary Report. Supplemental Table S1 contains information on all non-human peptides identified in this study. Supplemental Table S3 provides a list of human proteins identified in an independent search from the 3D-fractionated fractions.
Metaproteomic analysis
MEGAN4 software was used for metaproteomic analysis [28, 29]. MEGAN4 specifically requires BLAST searches of nucleotide or protein sequences as input. The software then parses the BLAST results, and generates phylogenetic assignments for input sequences using a homology-based algorithm. MEGAN4 also uses Refseq IDs in BLAST hits (when available), to assign peptides to KEGG pathways. The distinct peptide sequences retained after FDR analysis by ProteinPilot were used for BLASTP searches (v BLASTP 2.2.25+) against a database containing all non-redundant GenBank sequences (13,812,207 sequences). We used the online version of the BLASTP server, with default parameters adjusted for short sequences (except in the case of three peptides that were more than 30 amino acids long). The default maximum of 100 hits per peptide was specified. The compiled BLAST results were then input to MEGAN4, for phylogenetic and KEGG analysis.
RESULTS
Two-step database searching for microbial peptide and protein identification
At the outset of our studies, we first addressed the challenges of matching a large number of MS/MS spectra (988,974 total spectra for our current study) to peptide sequences within the very large database containing both human and translated oral microbial proteins (over 1.6 million total proteins). A large database increases the possibility for false-positive peptide sequence matches [20], thereby necessitating more stringent filtering thresholds and effectively lowering the number of high confidence matches.
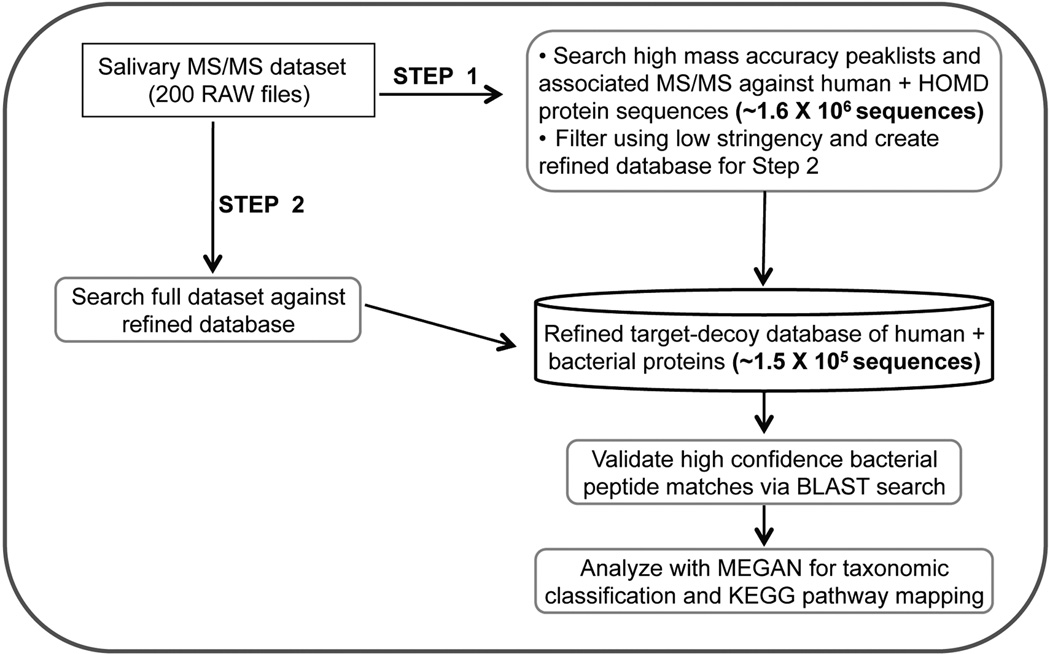
To address this challenge, we explored a novel two-step database searching method, detailed in Figure 1 and in Materials and Methods. In the first step we match MS/MS to peptides in the forward database using low-stringency scoring. A refined, much smaller database is created containing all identified proteins from the first step, and a target-decoy version of this database used to re-search all MS/MS data, followed by stringent filtering of peptide matches. We first evaluated the effectiveness of this two-step method using a representative set of 20 RAW files from our salivary dataset. We compared the numbers of human and microbial proteins identified using the two-step method to the ‘traditional’ one-step direct searching of the very large target-decoy version of the combined human and microbial protein sequence database. As shown in Supplemental Figure S1, the two step method increased by 63% the number of microbial proteins identified while having no discernable effect on identification of human proteins.
Figure 1. Two-step method for human salivary metaproteome analysis.
See text for details
Given this positive result, we analyzed the entire dataset of 200 RAW files using the two-step method. Table 1 summarizes the results. The database search in the first step resulted in identification of 2176 non-human proteins using a relatively low threshold for matching. For the second step, a refined target-decoy database containing all human proteins and the 2176 microbial protein sequences was constructed. This second search resulted in matching of 1927 distinct peptide sequences.
Table 1.
Summary of two-step database searching method for salivary metaproteomics
| Number of RAW files |
Total number of MS/MS spectra |
First stepa | Second stepb | ||
|---|---|---|---|---|---|
| Total sequences in initial database |
Non-human proteins identified |
Total sequences in refined target-decoy database |
Non- human, distinct peptides identified |
||
| 200 | 988,974 | 1,687,426 | 2176 | 152,724 | 1927 |
Database used contains human proteins + translated sequences from HOMD. Proteins identified at a low stringency 66% confidence value in ProteinPilot.
Database contains human proteins + microbial sequences identified from first step. Peptides were identified using a stringent 5% local FDR value in ProteinPilot.
Phylogenetic analysis
A BLAST output file containing results for 1,927 peptide sequences was input to MEGAN4. We initially used MEGAN4’s default minimum BLAST bit score criterion of 35.0. However, we obtained almost the same phylogenetic tree when the minimum bit score was reduced to zero, with the main difference being that more peptides were assigned to the different taxa. Accordingly, we present the results obtained when all 1,927 peptides were included.
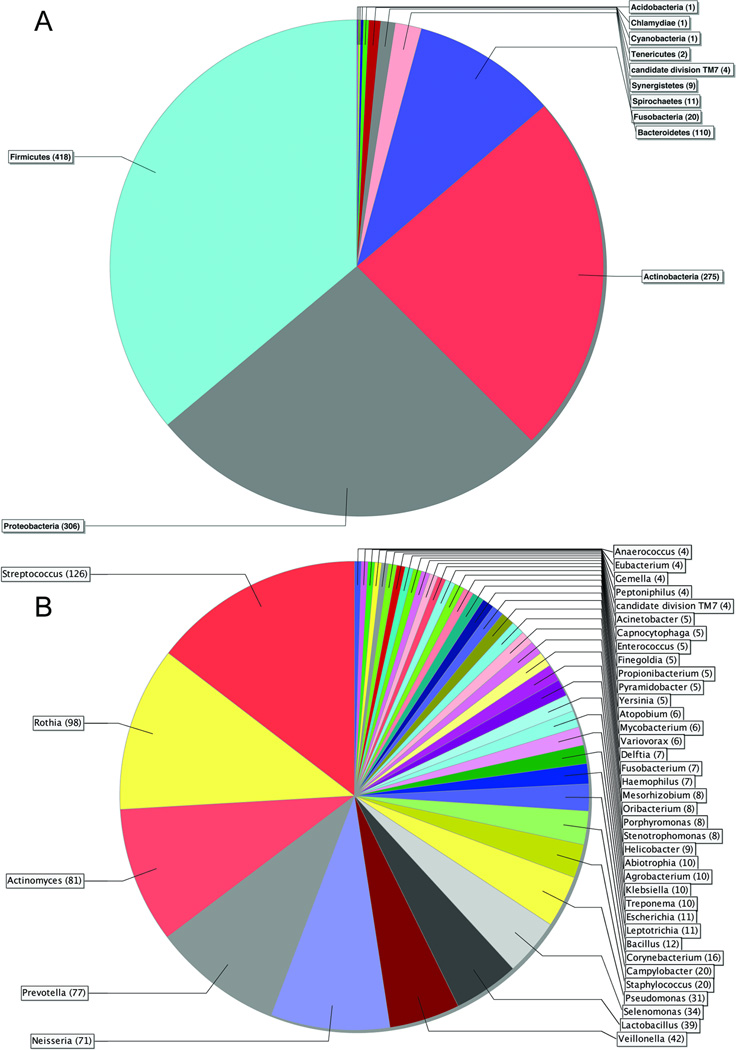
The complete phylogenetic tree is available as Supplemental Figure S2. Some peptides were so highly conserved that they could not be assigned at the kingdom level (539; 28%). The remaining peptides were all assigned to the kingdom Bacteria with the exception of 10 being assigned to Eukaryota, and 2 to viruses. Sixteen percent of the Bacteria peptides could not be assigned below the kingdom level. The remaining 1,156 peptides were distributed among twelve phyla (Figure 2A). The four most abundant phyla collectively accounted for 81% of bacterial peptides. Firmicutes were the most prevalent, followed by Proteobacteria, Actinobacteria, and Bacteroidetes. Sixty-six percent of bacterial peptides were assignable at the genus level, representing 65 genera (Figure 2B). The most abundant genera were Streptococcus, Rothia, Actinomyces, Prevotella, and Neisseria.
Figure 2. Phyla and genus level assignments for human salivary proteome.
A. Pie chart showing peptides assigned to bacterial phyla. The numbers in parentheses indicate the number of peptides assigned to each phylum. B. Pie chart showing peptides assigned to bacterial genera. The numbers in parentheses indicate the number of peptides assigned to each genus. For reasons of legibility, genera with fewer than four peptide assignments have been excluded from the chart. The excluded genera are shown in Supplemental Table S2.
Thirty-five percent of bacterial peptides were assignable at the species level, representing 124 species (Supplemental Figure S2 and Supplemental Table S2). The most common species were Rothia mucilaginosa, Prevotella melanogenica, Selenomonas sputigena, Pseudomonas aeruginosa, Cornybacterium matrochotii, Streptococcus salivarius, Actinomyces odontolyticus, and Abiotrophia defectiva. The remaining 115 species represented a broad range of oral bacterial diversity. They included species that are presently non-cultivable, most notably two members of candidate division TM7. All of those species were listed in the current version of the HOMD database, with the exception of six.
The six bacterial species not in HOMD included representatives of a variety of environments, including the human rectum, seawater, soil, plant pathogens, and legume root nodules (Table S2). There was only a single peptide assigned to each. Five of them showed mismatches with the target sequence for their assigned species in BLAST. Mismatches also applied to the five peptides in the complete phylogenetic tree that were assigned to non-human eukaryotic species, and one of two peptides assigned to viruses (Figure S2).
It is important to note that peptide counts for particular species were not always representative of the prevalence of their parent genus. A good example is Neisseria, where only 5 peptides were assignable at the species level, but 66 were assignable at the genus level (Table S2). Likewise, only 4 peptides were assignable to Veilonella at the species level, but 38 were assignable at the genus level. In the case of Streptococcus, 37 peptides were assigned at the species level, but 89 were assignable at the genus level. Thus, the genus level may provide a better representation of prevalence patterns within this dataset.
Ontology analysis
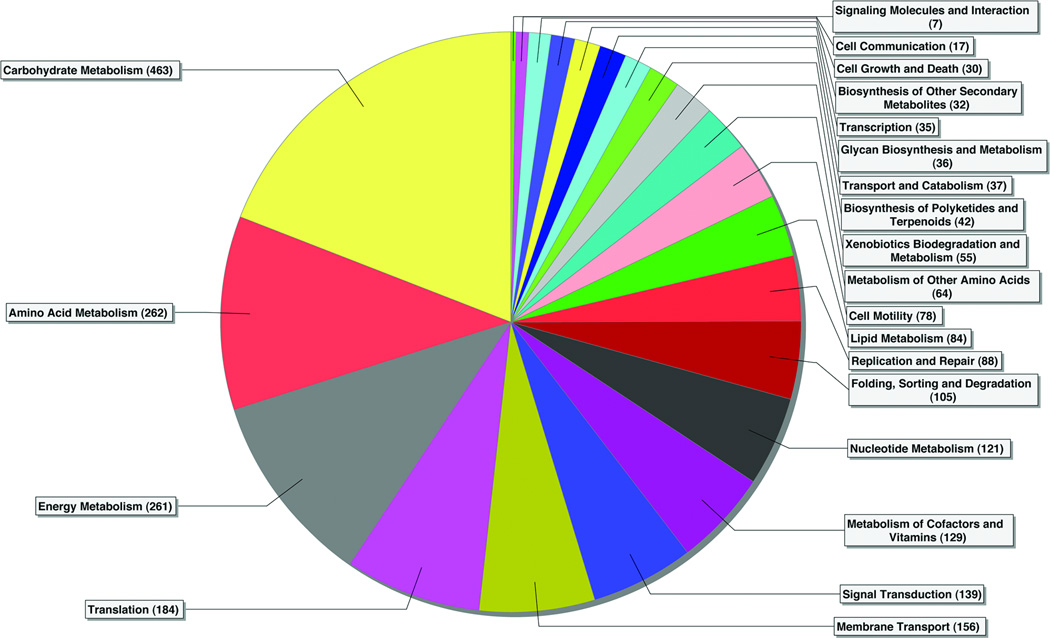
The KEGG analysis assigned 1,774 peptides to 20 pathways (Fig. 3). It is important to note that a number of key enzymes were represented in multiple KEGG pathways. Carbohydrate Metabolism was the most prevalent pathway, Proteins involved in glycolysis were among the most common on the basis of the number of peptides assigned to them (Table 2). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) had the highest number of sequence assignments in the entire dataset, and enolase, aldolase, phosphoglycerate mutase, triose-phosphate isomerase, pyruvate kinase, phosphoenolpyruvate carboxykinase, and phosphoglycerate kinase also were relatively abundant. Pyruvate Metabolism was represented by pyruvate formate lyase, and the Citrate Cycle by fumarate hydratase and succinyl-CoA synthetase. Levansucrase provided evidence for Sucrose Metabolism. Downstream from the Carbohydrate Metabolism pathway, Energy Metabolism was represented by ATP synthase.
Figure 3. KEGG analysis of human salivary metaproteome.
Pie chart showing bacterial peptides assigned to KEGG pathways. The numbers in parentheses indicate the number of peptides assigned to each pathway. Because many proteins are assigned to multiple KEGG pathways, the total number of peptide assignments is greater than the total number of peptides identified in our study.
Table 2.
KEGG analysis of abundant microbial proteins from the salivary metaproteome
| KEGG pathway |
Protein name (EC # or gene name) | Number of Peptides a |
|---|---|---|
|
Carbohydrate metabolism |
||
| glyceraldehyde-3-phosphate dehydrogenase (1.2.1.12) | 58 | |
| Enolase (4.2.1.11) | 27 | |
| fructose-bisphosphate aldolase (4.1.2.13) | 18 | |
| fumarate hydratase (4.2.1.2) | 14 | |
| pyruvate formate-lyase (2.3.1.54) | 14 | |
| phosphoglycerate mutase (5.4.2.1) | 13 | |
| triose-phosphate isomerase (5.3.1.1) | 11 | |
| pyruvate kinase (2.7.1.40) | 11 | |
| succinyl-CoA synthetase (6.2.1.5) | 9 | |
| phosphoenolpyruvate carboxykinase (4.1.1.32) | 8 | |
| levansucrase (2.4.1.10) | 8 | |
| phosphoglycerate kinase (2.7.2.3) | 8 | |
| Cell motility | ||
| flagellin (FliC) | 25 | |
| methyl-accepting chemotaxis protein (MCP) | 16 | |
|
DNA Replication and repair |
||
| DNA polymerase (2.7.7.7) | 36 | |
|
Energy metabolism |
||
| ATP synthase (3.6.3.14) | 12 | |
|
Folding, sorting, and degradation |
||
| molecular chaperone DnaK (DnaK) | 12 | |
| Transcription | ||
| RNA polymerase (2.7.7.6) | 13 | |
| Translation | ||
| Translation elongation factor G (EF-2) | 23 | |
| Ribosomal Protein L7/L12 (L7/L12) | 14 | |
| Ribosomal Protein S1 (S1) | 13 | |
| Ribosomal Protein S2 (S2) | 11 | |
Number of distinct peptides assigned to a given protein. Proteins listed in this table were represented by at least eight peptides.
Within the DNA Replication and Repair pathway, DNA polymerase was the second most abundant protein in the dataset. Molecular chaperone DnaK was the major component of Folding Sorting, and Degradation, while Transcription was primarily represented by RNA polymerase. Translation was represented by translation elongation factor G, as well as ribosomal protein L7/L12 and ribosomal protein S1. Other translation elongation factors and ribosomal proteins were present, but with fewer matching peptides.
The Cell Motility pathway was dominated by flagellin and methyl-accepting chemotaxis protein. The Membrane Transport group consisted mostly of diverse phosphotransferases associated with carbohydrate transport, although ABC transporters also were detected. The Signal Transduction pathway was represented by a variety of bacterial two-component systems. (not shown).
DISCUSSION
Our previous metaproteomic study identified 357 bacterial peptides in a pooled sample of salivary pellets from four oral squamous cell carcinoma patients [21]. The pellet fraction of saliva is a mixture consisting mostly of exfoliated epithelial cells, bacteria, and high molecular weight salivary proteins, while the salivary supernatant fraction is largely bacteria-free. Nevertheless, in this study, we achieved an almost six-fold increase in the number of bacterial peptides detected in a pooled sample of salivary supernatants from six healthy subjects.
Several factors likely increased the depth of information obtained in the current study. Protease inhibitors were used here to better preserve proteins prior to DRC [8]. The compression of high-abundance host salivary proteins via ProteomeMiner™ treatment likely enhanced the detection of bacterial proteins, which are at lower abundance compared to the dominant human proteins from the host. Interestingly, there was relatively little overlap between the bacterial peptides identified from the ProteomeMiner™-treated portion of the pool and those identified from the untreated portion (data not shown). Thus a parallel analysis of a non-treated sample may be warranted when using this approach for metaproteomic studies. We also used sensitive LTQ-Orbitrap mass spectrometry, with high peptide precursor mass accuracy, recommended for metaproteomics studies[30]. Mass accuracy can increase confident peptide identifications[31], especially when processing the data via the “Quant” module from MaxQuant [23], [32].
Our novel two-step database searching method also significantly increased bacterial protein identifications. We believe the increased bacterial protein identifications was due to the use of the refined database in the second-step, which was an order of magnitude smaller than the target-decoy database used for the one-step method. A smaller database has less potential for false-positive matches[20], thereby requiring lower stringency filtering and providing more protein identifications. Also beneficial to our study was the use of ProteinPilot database searching software designed for use with large sequence databases and capable of robust FDR estimation [33, 34], making it well suited for our two-step method.
One problem in metaproteomics is the choice of an appropriate database for peptide sequence matching. For our study in saliva, a database that encompasses a wide range of microbial environments runs the risk of producing a large number of “false positive” identifications of proteins from microbes that may share common sequences with oral taxa, even though they are quite unlikely to be actually present in the oral environment. Our previous metaproteomics study of the salivary cell pellet suffered from this problem, as we identified a number of microbes likely not found in the oral environment [21].
One solution to this problem is to perform a metaproteomic analysis on a sample for which corresponding metagenomic data already exist. That allows the peptides to be matched against genomic data that is specific to the same individuals. This was done in a recent metaproteomic analysis of the human gut microbiota, with considerable success [12]. We did not have a metagenomic dataset to work from, so we chose the translated protein sequences from the HOMD genomic dataset instead. HOMD is a curated database of species and uncultured phylotypes that have been identified from oral samples on the basis of 16S rRNA sequencing. It also incorporates genomic data for approximately 150 members of the oral microbiota that have been completely or partially sequenced [25, 35]. That allowed a search strategy focused on species of verified oral origin. Our strategy appears to have been successful, since only six peptides were assigned by MEGAN4 to species not present in HOMD, and five of those assignments were based on imperfect matches in BLAST.
Recent 16S rRNA-based metagenomic studies of microbial diversity in human saliva have shown that there is considerable variation between individuals, and possibly between different geographical populations as well. However, there also is consistent evidence for a “core oral microbiome” consisting of a more limited number of abundant taxa at the phylum, genus, and species level. Thus, although there is great diversity in saliva, that diversity is very unevenly distributed with respect to prevalence [2–7]. Because of the large amounts of protein required for DRC and subsequent peptide fractionation, it was necessary to pool saliva samples for this study. That pool included six individuals of varied ethnicity. Our data can be considered analogous to population summary data from a metagenomic study, although the data represent a relatively small number of people.
From that perspective, our data shows patterns that are consistent with existing metagenomic data. At the phylum level, our study is consistent with others showing Firmicutes to be the most prevalent phylum, with Proteobacteria, Actinobacteria, and Bacteroidetes also being abundant [2–7]. Our data showed a relatively higher prevalence of Actinobacteria than other studies [2–7], but this is more likely to be due to individual variation than to any bias towards Actinobacteria in our proteomic approach. The same broad patterns of similarity existed at the genus level, although we observed relatively lower prevalence of Haemophilus than has been seen by others [2–7]. Again, we believe that is most likely due to variation between individuals.
It has been suggested that the salivary microbiota corresponds most closely to that of the tongue [5, 36]. Our findings are consistent with that hypothesis, since we observed a high relative abundance for species common on the tongue, notably R. mucilaginosa, S. salivarius, and P. melaninogenica [5, 36, 37]. However, metaproteomic species classifications have to be interpreted cautiously, since important bacterial proteins may include regions, such as enzyme active sites, that are highly conserved. Thus, some genera that were poorly represented at the species level were in fact more abundant when viewed at the genus level.
The same caveat applies to metaproteomic data at any taxonomic level. There is presently no method for selectively removing conserved peptides. Thus, it is likely that there will be a consistent bias towards such sequences. Nevertheless, the high degree of consistency between our data and previous 16s rRNA metagenomic studies suggest that our approach for deep metaproteomic analysis can be used to provide comparable information about microbial diversity in saliva, although it may not provide the same depth of coverage of taxa that are extremely rare.
The diversity of proteins detected by our approach also facilitated ontological analysis of the activities carried out by the oral microbial community at large. The KEGG analysis suggested that the saliva supernatant community appeared to be actively engaged in growth and metabolism. Evidence for active DNA replication, transcription, and translation was provided by the relatively large number of peptides derived from DNA polymerase, RNA polymerase, ribosomal proteins, and translation elongation factors, while numerous peptides from ATP synthase suggested that activated energy carriers were being produced. Glycolysis is the most likely mechanism for ATP production, since peptides for key enzymes in that pathway were among the most abundant. Moreover, the Membrane Transport pathway included peptides for various components of sugar phosphotransferase systems, consistent with active glycolysis. The overall pattern is consistent with metabolism of dietary carbohydrates and salivary glycoproteins by oral bacteria.
KEGG pathway maps may incorporate data from both prokaryotes and eukaryotes [38]. As a consequence, some microbial enzymes in our dataset were also cross-referenced to eukaryote-specific KEGG pathways. Obvious misclassifications such as that were easy to eliminate. A subtler issue was presented by microbial proteins that have functions additional to their defined roles in KEGG pathways. GAPDH provides an excellent case in point. GAPDH is known to occur on streptococcal surfaces [39], and oral streptococci appear to release GAPDH when grown in batch culture under certain conditions [40], The extracellular form of streptococcal GAPDH is enzymatically active [39, 40], but GAPDH has also been shown to act as a bacterial adhesin for host cell proteins, such as fibronectin [39], and other oral bacterial species, such as Porphyromonas gingivalis [41]. There is only a single copy of the GAPDH gene in streptococci [39, 40], so the same gene product appears to be responsible for both intracellular and extracellular activities.
More recent proteomic studies of oral streptococci grown in monoculture have established that other members of the glycolytic pathway are surface proteins on streptococci. Those include proteins that are relatively abundant in our dataset, such as enolase, fructose-bisphosphate aldolase, phosphoglycerate mutase, triose-phosphate isomerase, pyruvate kinase, and phosphoglycerate kinase. Adhesin functions have been demonstrated for many of those proteins [42–47]. Moreover, streptococcal surfaces also incorporate proteins from other pathways that are relatively abundant in our dataset, including ATP synthase, molecular chaperone DnaK, RNA polymerase, Translation elongation factor G, Ribosomal Protein L7/L12, and Ribosomal Protein S1. Those proteins also are capable of functioning as adhesins [42–47]. Taken together, those reports suggest that many of the most abundant proteins in our dataset may function as extracellular adhesins, in addition to their established roles in intracellular pathways.
To conclude, our metaproteomic analysis has provided the first in-depth catalog of bacterial proteins in human saliva supernatant. We believe this could serve as a basis for future studies relevant to oral disease. Common oral diseases such as dental caries and periodontal disease are associated with major shifts in microbial ecology [48, 49]. Moreover, changes in the salivary microbiota also have been documented for rare but very serious conditions such as oral cancer [50]. Most studies have emphasized taxonomic changes in the composition of the oral microbiota in disease, but it is reasonable to suggest that such changes are likely to be accompanied by changes in the expression of microbial proteins. Our data now can be used as a basis for comparison in future metaproteomic studies of oral diseases. Our findings suggest that the salivary metaproteome is likely to be closely correlated with that of the tongue. Since the tongue is a frequent site of occurrence for oral cancer [51], comparisons of the salivary metaproteome in health, dysplasia, and disease may be particularly useful for testing the hypothesis that oral microbes are directly or indirectly involved in the pathogenesis of oral cancer. We are actively engaged in comparative studies to address that question.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded in part by NIH grant 1R01DE017734. We also thank the Center for Mass Spectrometry and Proteomics for instrumental support and maintenance and the Minnesota Supercomputing Institute for computational proteomics support.
ABBREVIATIONS
- ATP
Adenosine triphosphate
- BLAST
Basic Local Alignment Search Tool
- DRC
Dynamic range compression
- FDR
False discovery rate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HOMD
Human Oral Microbiome Database
- ID
identification
- IPI
International Protein Index
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LTQ
linear trap quadrupole
- MEGAN
MEtaGenome Analyzer
- MGF
Mascot generic format
- PSPEP
Proteomics System Performance Evaluation Pipeline
- SCX
Strong cation exchange
Footnotes
CONFLICT OF INTEREST STATEMENT
All authors declare that there is no financial / commercial conflict of interest.
SUPPORTING INFORMATION
Additional tables, text and figures are described in the text are included in the supporting information. The data associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the passphrase ‘saliva’ along with the following hash: T6YUFF/s0PhO2HFJ0KEH/AXoAK/vlpPaYcgBFzpNOojQ0SZVoX9k2Ts2YOsuWi5IkdySa8bF0av4ZvN2cUvh8NO+SXYAAAAAAAA1tw==
REFERENCES
- 1.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J. Dent. Res. 2007;86:680–693. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 2.Keijser BJ, Zaura E, Huse SM, van der Vossen JM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 3.Nasidze I, Li J, Quinque D, Tang K, et al. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasidze I, Quinque D, Li J, Li M, et al. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal. Biochem. 2009;391:64–68. doi: 10.1016/j.ab.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 5.Zaura E, Keijser B, Huse S, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik EM, Long CD, Armitage GC, Loomer P, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling Z, Kong J, Jia P, Wei C, et al. Analysis of Oral Microbiota in Children with Dental Caries by PCR-DGGE and Barcoded Pyrosequencing. Microbial Ecol. 2010;60:677–690. doi: 10.1007/s00248-010-9712-8. [DOI] [PubMed] [Google Scholar]
- 8.Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A Dynamic Range Compression and Three-Dimensional Peptide Fractionation Analysis Platform Expands Proteome Coverage and the Diagnostic Potential of Whole Saliva. J. Proteome Res. 2009;8:5590–5600. doi: 10.1021/pr900675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny P, Hagen FK, Hardt M, Liao L, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh PD, Moter A, Devine DA. Dental plaque biofilms: communities, conflict and control. Periodontol 2000. 2011;55:16–35. doi: 10.1111/j.1600-0757.2009.00339.x. [DOI] [PubMed] [Google Scholar]
- 11.Klaassens ES, de Vos WM, Vaughan EE. Metaproteomics approach to study the functionality of the microbiota in the human infant gastrointestinal tract. Appl Environ Microbiol. 2007;73:1388–1392. doi: 10.1128/AEM.01921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verberkmoes NC, Russell AL, Shah M, Godzik A, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 13.Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, et al. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009;3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 14.Mueller RS, Dill BD, Pan C, Belnap CP, et al. Proteome changes in the initial bacterial colonist during ecological succession in an acid mine drainage biofilm community. Environ Microbiol. 2011;8:2279–2292. doi: 10.1111/j.1462-2920.2011.02486.x. [DOI] [PubMed] [Google Scholar]
- 15.Jiao Y, D'Haeseleer P, Dill BD, Shah M, et al. Identification of biofilm matrix-associated proteins from an Acid mine drainage microbial community. Appl Environ Microbiol. 2011;77:5230–5237. doi: 10.1128/AEM.03005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham C, McMullan G, Graham RL. Proteomics in the microbial sciences. Bioeng Bugs. 2011;2:17–30. doi: 10.4161/bbug.2.1.14413. [DOI] [PubMed] [Google Scholar]
- 17.Abram F, Enright AM, O'Reilly J, Botting CH, et al. A metaproteomic approach gives functional insights into anaerobic digestion. J Appl Microbiol. 2011;110:1550–1560. doi: 10.1111/j.1365-2672.2011.05011.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang HB, Zhang ZX, Li H, He HB, et al. Characterization of metaproteomics in crop rhizospheric soil. J Proteome Res. 2011;10:932–940. doi: 10.1021/pr100981r. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Wang H, Zhang Z, Lin R, Lin W. Comparative metaproteomic analysis on consecutively Rehmannia glutinosa-monocultured rhizosphere soil. PLoS One. 2011;6:e20611. doi: 10.1371/journal.pone.0020611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cargile BJ, Bundy JL, Stephenson JL., Jr Potential for false positive identifications from large databases through tandem mass spectrometry. J Proteome Res. 2004;3:1082–1085. doi: 10.1021/pr049946o. [DOI] [PubMed] [Google Scholar]
- 21.Rudney JD, Xie H, Rhodus NL, Ondrey FG, Griffin TJ. A metaproteomic analysis of the human salivary microbiota by three-dimensional peptide fractionation and tandem mass spectrometry. Mol. Oral Microbiol. 2010;25:38–49. doi: 10.1111/j.2041-1014.2009.00558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J, Mann M. Computational principles of determining and improving mass precision and accuracy for proteome measurements in an Orbitrap. J Am Soc Mass Spectrom. 2009;20:1477–1485. doi: 10.1016/j.jasms.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 24.Olsen JV, de Godoy LM, Li G, Macek B, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Chen T, Yu W-H, Izard J, Baranova OV, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010 doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 27.Tang WH, Shilov IV, Seymour SL. Nonlinear Fitting Method for Determining Local False Discovery Rates from Decoy Database Searches. J. Proteome Res. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- 28.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2011;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 31.Boyne MT, Garcia BA, Li M, Zamdborg L, et al. Tandem mass spectrometry with ultrahigh mass accuracy clarifies peptide identification by database retrieval. J Proteome Res. 2009;8:374–379. doi: 10.1021/pr800635m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renard BY, Kirchner M, Monigatti F, Ivanov AR, et al. When less can yield more - Computational preprocessing of MS/MS spectra for peptide identification. Proteomics. 2009;9:4978–4984. doi: 10.1002/pmic.200900326. [DOI] [PubMed] [Google Scholar]
- 33.Shilov IV, Seymour SL, Patel AA, Loboda A, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- 35.Dewhirst FE, Chen T, Izard J, Paster BJ, et al. The Human Oral Microbiome. J. Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 37.Kazor CE, Mitchell PM, Lee AM, Stokes LN, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exper. Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson D, Goldstein JM, Boatright K, Harty DWS, et al. pH-regulated Secretion of a Glyceraldehyde-3-Phosphate Dehydrogenase from Streptococcus gordonii FSS2: Purification, Characterization, and Cloning of the Gene Encoding this Enzyme. J. Dent. Res. 2001;80:371–377. doi: 10.1177/00220345010800011301. [DOI] [PubMed] [Google Scholar]
- 41.Maeda K, Nagata H, Yamamoto Y, Tanaka M, et al. Glyceraldehyde-3-Phosphate Dehydrogenase of Streptococcus oralis Functions as a Coadhesin for Porphyromonas gingivalis Major Fimbriae. Infect. Immun. 2004;72:1341–1348. doi: 10.1128/IAI.72.3.1341-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkins JC, Beighton D, Homer KA. Effect of Acidic pH on Expression of Surface-Associated Proteins of Streptococcus oralis. Appl. Environ. Microbiol. 2003;69:5290–5296. doi: 10.1128/AEM.69.9.5290-5296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole JN, Ramirez RD, Currie BJ, Cordwell SJ, et al. Surface Analyses and Immune Reactivities of Major Cell Wall-Associated Proteins of Group A Streptococcus. Infect. Immun. 2005;73:3137–3146. doi: 10.1128/IAI.73.5.3137-3146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blau K, Portnoi M, Shagan M, Kaganovich A, et al. Flamingo cadherin: a putative host receptor for Streptococcus pneumoniae. J. Infect. Dis. 2007;195:1828–1837. doi: 10.1086/518038. [DOI] [PubMed] [Google Scholar]
- 45.Severin A, Nickbarg E, Wooters J, Quazi SA, et al. Proteomic Analysis and Identification of Streptococcus pyogenes Surface-Associated Proteins. J. Bacteriol. 2007;189:1514–1522. doi: 10.1128/JB.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kesimer M, Kilic N, Mehrotra R, Thornton DJ, Sheehan JK. Identification of salivary mucin MUC7 binding proteins from Streptococcus gordonii. BMC Microbiol. 2009;9:163. doi: 10.1186/1471-2180-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkins JC, Homer KA, Beighton D. Altered Protein Expression of Streptococcus oralis Cultured at Low pH Revealed by Two-Dimensional Gel Electrophoresis. Appl. Environ. Microbiol. 2001;67:3396–3405. doi: 10.1128/AEM.67.8.3396-3405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aas JA, Griffen AL, Dardis SR, Lee AM, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colombo APV, Boches SK, Cotton SL, Goodson JM, et al. Comparisons of Subgingival Microbial Profiles of Refractory Periodontitis, Severe Periodontitis, and Periodontal Health Using the Human Oral Microbe Identification Microarray. J. Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mager DL, Haffajee AD, Devlin PM, Norris CM, et al. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiboski CH, Shiboski SC, Silverman S. Trends in oral cancer rates in the United States, 1973-1996. Community Dent. Oral Epidemiol. 2000;28:249–256. doi: 10.1034/j.1600-0528.2000.280402.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.