Abstract
Standard fluorescence microscopy approaches rely on measurements at single excitation and emission bands to identify specific fluorophores and the setting of thresholds to quantify fluorophore intensity. This is often insufficient to reliably resolve and quantify fluorescent labels in tissues due to high autofluorescence. Here we describe the use of hyperspectral analysis techniques to resolve and quantify fluorescently labeled cells in highly autofluorescent lung tissue. This approach allowed accurate detection of green fluorescent protein (GFP) emission spectra, even when GFP intensity was as little as 15% of the autofluorescence intensity. GFP-expressing cells were readily quantified with zero false positives detected. In contrast, when the same images were analyzed using standard (single-band) thresholding approaches, either few GFP cells (high thresholds) or substantial false positives (intermediate and low thresholds) were detected. These results demonstrate that hyperspectral analysis approaches uniquely offer accurate and precise detection and quantification of fluorescence signals in highly autofluorescent tissues.
Keywords: Hyperspectral analysis, lung autofluorescence, pulmonary microvascular endothelial cells
1. Introduction
Resolving, tracking, and quantifying proteins and cells using fluorescence microscopy have become hallmark techniques for visualizing the inner workings of complex biological systems. However, it is often daunting to reliably identify fluorescent signals in tissues, due to high levels of autofluorescence and low signal-to-noise ratios. One tissue that exhibits a high level of autofluorescence is lung tissue, likely due to the high content of collagen and elastin which have fluorescence emission peaks in the 450-550 nm range.[1], [2] Quantifying fluorescent signals in the presence of high autofluorescence has been even more problematic. Fluorescent protein assays present additional challenges, as there are limits to protein expression levels while maintaining cellular viability. In specific, it is particularly difficult to discriminate between lung autofluorescence (peak wavelength ≈ 505 nm) and fluorescent labels in the green portion of the spectrum, such as green fluorescent protein (GFP).[3] This has been prohibitive for long-term proliferation and migration studies of endothelial cells, as many of these studies use GFP-expression as the primary labeling method.[4], [5] To compensate, image contrast and linearity (gamma) are often adjusted to display only regions of high fluorescence emission. This intensity thresholding assumes that high intensity fluorescence is due to GFP emission and that low-intensity fluorescence is due to autofluorescence (or other unwanted signals). Hence, while fluorescence imaging of GFP-labeled cells has been used in lung tissue studies,[6] it remains difficult to accurately differentiate between lung autofluorescence and fluorescent protein signals using standard (single-band) fluorescence microscopy techniques. This uncertainty results in a low specificity and sensitivity for detecting GFP, which subsequently results in a high incidence of false-positive identification and/or occurrence of type II errors. Thus, accurate, quantitative detection of GFP-expressing cells in lung tissues is prohibitively difficult using single-band (a.k.a. single-wavelength) fluorescence microscopy.
To overcome these limitations we have utilized hyperspectral imaging and quantitative image-analysis approaches. Hyperspectral imaging was originally developed by NASA and the DOD to solve specific remote sensing problems.[7] From a sampling standpoint, hyperspectral imaging refers to the acquisition of spectral bands contiguously (often at constant wavelength spacing) over a specified wavelength range.[8] From an analysis standpoint, hyperspectral imaging often carries with it the assumption that many more spectral bands are collected than there are constituent species in the sample (which allows for band-reduction and orthogonal sub-space projection methods to be applied with little loss in the ability to identify these species). The approach described herein meets both of these requirements.
Hyperspectral image analysis can identify unique emission spectra of specific features or fluorophores within complex image datasets. While hyperspectral imaging techniques have been demonstrated in wide-field fluorescence microscopy,[9-11] confocal microscopy,[12-15] and in in vivo fluorescence imaging,[16-21] a thorough and quantitative comparison between single-band and hyperspectral fluorescence imaging has not been performed. Despite this, hyperspectral systems are now available from major microscope manufacturers.[13], [22] Hence, there is a significant need to demonstrate a definitive approach for designing hyperspectral assays. This necessitates performing spectral calibration, understanding hyperspectral image analysis, and quantitatively comparing results from single-band and hyperspectral microscopy assays.
Hyperspectral instruments typically use a large array of optical filters,[23] a dispersive element,[9] a tunable filter,[16], [17], [24], [25] or interferometry[26] to select wavelengths from either the excitation[16], [27] or emission lightpath (common in most instruments). This makes hyperspectral imaging systems inherently more complex – and typically more expensive – than their single-band counterparts. The analysis of hyperspectral image data is correspondingly complex, and while steps have been made in commercial software to streamline this process, it is critical to understand the basic principles of spectral image analysis before planning a hyperspectral assay. In specific, accurate definition of the spectral library and flat-field spectral correction are steps that directly affect the sensitivity and specificity of the spectral image analysis, as we demonstrate herein. For a further introduction into hyperspectral imaging, Garini et al. provide a broad overview of hyperspectral filtering configurations for life sciences applications,[12] while McNamara et al. provide a summary of spectral microscopy data and sources.[28] Although applications of hyperspectral analysis have been demonstrated for separating multiple fluorescence signals[7], [14], [16], [22] and separating autofluorescence and fluorescence signals,[17] a definitive approach to hyperspectral imaging, and comparison between hyperspectral and single-band microscopy is lacking.
The goal of this work is to develop a definitive approach for hyperspectral microscopy, and to compare this approach to traditional single band (single-wavelength) microscopy. This is demonstrated through identifying and quantifying GFP-expressing pulmonary microvascular endothelial cells (PMVECs) in highly autofluorescent tissues – specifically lung tissue. The results clearly indicate that this hyperspectral imaging approach results in improved sensitivity and specificity, while outlining standardized methods for experimental design and image analysis. This approach is applicable to a wide range of conditions and samples, whether separating tissue autofluorescence from fluorescence (as demonstrated here) or separating signals from multiple fluorescent labels.
2. Methods
Cell, animal, and sample preparation
Pulmonary microvascular endothelial cells (PMVECs) were isolated from CD rats as described elsewhere.[29] Briefly, distal lung parenchyma was dissected from each lobe, minced in 1 mm pieces and digested with collagenase II. Cells were filtered, by using a 40 mm BD strainer and seeded in tissue culture coated dishes. Cells were suspended in DMEM media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. After a month, endothelial cell colonies, characterized by a panel of surface markers, were expanded and sorted using a BD Aria II sorter. Two weeks later, cells that exhibited high proliferative behavior (e.g. those that form colonies of more than 10,000 cells) were further expanded. We have previously documented the endothelial progenitor capacity of these cells.[30]
Highly proliferative PMVECs were then transfected for 48 hours with a lentivirus encoding green fluorescent protein (GFP) or an empty vector control, both under a CMV promoter. GFP positive PMVECs were selected one week post-transfection by a cell sorter (BD Aria II sorter). The day GFP positive- and GFP negative-PMVECs were infused, cells were trypsinized, counted using a trypan blue exclusion approach (countess, Invitrogen) and suspended at a concentration of 3 millions cells per ml of clinical grade saline solution. Samples of GFP positive- and GFP negative-PMVECs were prepared as a confluent monolayer on 25mm round glass coverslips, for use as control samples. Prior to imaging, cells were incubated with Hoechst 33342 (Invitrogen Corporation, Carlsbad, CA) at a concentration of 5 μg/mL for 30 minutes. Coverslips with confluent cells were then fixed in a circular holder and bathed in 2 mL of a buffer containing (mM): 145 NaCl, 4 KCl, 10 HEPES, 10 D-glucose, 1 MgCl2, 1 CaCl2, pH 7.35.
Animal care and procedures conformed to the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals and The Institutional Animal Care and Use Committee (IACUC). Adult male CD rats were anesthetized with a blend of ketamine (75 mg/kg, BW) and xylazine (10 mg/kg, BW) intraperitoneally. Mechanical ventilation was initiated following a tracheostomy at 6 mL/kg, BW with room air. A thoracotomy was performed and the pulmonary artery and left atrium cannulated with PE-240 tubing. Blood from the pulmonary circulation was rinsed with 50 mL saline solution using a roller pump. After that, 3 million GFP positive PMVECs, 3 million GFP negative PMVECs, or saline solution containing 5% Bovine serum albumin were perfused into the lungs of 3 different animals by using the same constant volume perfusion approach described. Perfusion was maintained for 5 minutes, after which the pulmonary circulation was tightened using umbilical tape and the trachea tightened during maximal inspiration to achieve adequate lung inflation. Perfused lungs were then fixed in optimal cutting temperature (OCT) compound and stored in a light protected container at −80°C for at least 24 hours. Tissues were processed for cryosections of 10 μm thickness. Sections were mounted on poly-L-lysine coated slides and stained with 10 μL of 0.225 μM Hoechst 33342. Fluorescent Mounting Medium (Dako Cytomation, Carpinteria, CA) was used to apply the coverslips.
Instrument configuration, calibration, and image acquisition
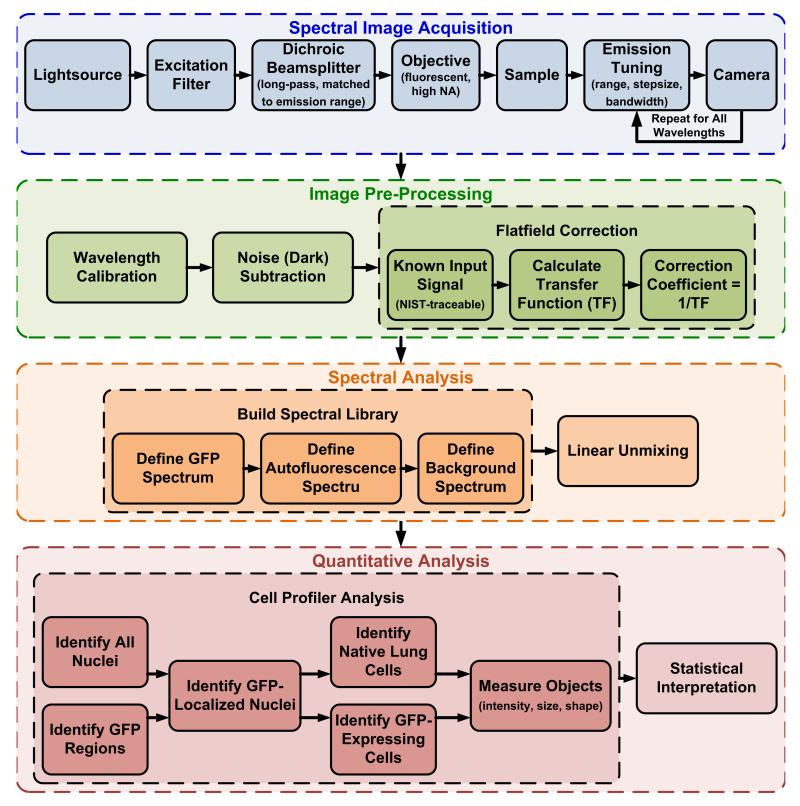
The series of steps in this spectral imaging approach have been designed to maximize the sensitivity for detection of GFP-expressing cells while minimizing (and in many cases, eliminating) false-positive errors. These steps are outlined in Figure 1.
Figure 1.
Schematic depicting the steps required for hyperspectral image acquisition, spectral flat-field correction, and spectral analysis.
Spectral microscope
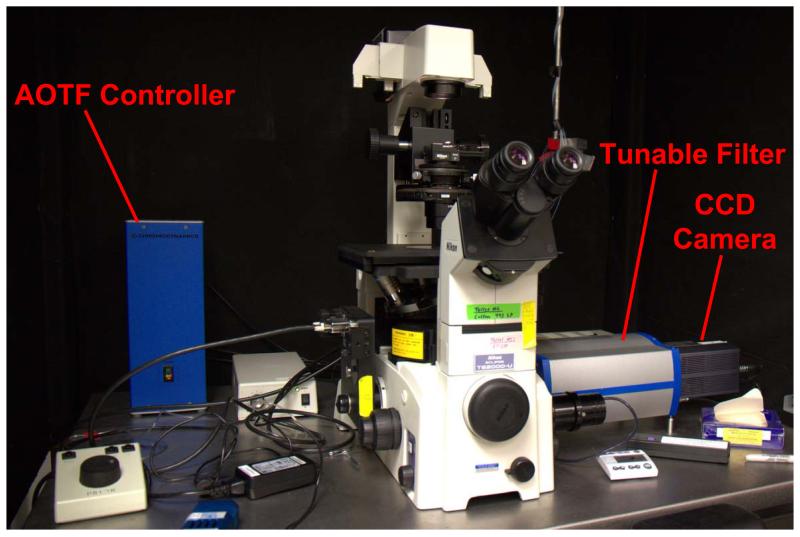
Fluorescence microscopy (Figure 2) was performed using an inverted fluorescence microscope (TE2000-U, Nikon Instruments, Melville, NY), equipped with a 40X-oil immersion objective (S Fluor, 40X/1.30 Oil, DIC H/N2, Nikon Instruments). Although chromatic correction information is not available from the manufacturer, we have observed little change in focal length between 450 and 650 nm. It should be noted that high degree of chromatic aberration, were it present, would result in a distorted spectral profile that would affect the transfer function of the microscope and could result in nonlinear spectral mixing. Fluorescence excitation was provided by a liquid light guide-coupled Xe arc lamp (Lambda DG-4, Sutter Instrument Company, Novato, CA). Spectral filtering of the fluorescence emission was accomplished using a variable-bandwidth acousto-optic tunable filter (AOTF), specially configured for microscopic use (HSi-300 Hyperspectral Imaging System, ChromoDynamics, Orlando, FL). For Hoechst 33342 imaging, a 360/40 nm excitation was used with a 400 nm long-pass dichroic beamsplitter and the emission filter tuned to 450 nm. For spectral imaging of GFP and lung autofluorescence, a 430 nm excitation was used with a 450 nm long-pass dichroic beamsplitter (and a variable wavelength emission). Images were detected using a cooled, back-illuminated, charge-coupled device (CCD) camera (Cascade 512B, Photometrics, San Diego, CA). The tunable filter allows selection of a bandwidth from 1.7-47 nm FWHM and tuning over a range of 450-800 nm. The transmission band of the HSi-300 is roughly Gaussian in shape. Hence, with appropriate excitation and beam-splitting optics (provided through additional filter cubes), a wide variety of fluorophores may be imaged using this system. The tunable filter and CCD camera were both controlled using μManager software (Vale Lab, UCSF).[31]
Figure 2.
Photograph of the spectral widefield fluorescence microscope showing the tunable filter (AOTF), AOTF controller, and charge-coupled device camera.
Instrument calibration
The factory wavelength calibration of the HSi-300 was verified using a multi-ion discharge lamp (MIDL, LightForm, Inc., Asheville, NC) and methods outlined by Lerner and Zucker[32]. In summary, the MIDL emission spectrum was measured using a fiber-coupled, high-sensitivity CCD spectrometer (QE65000, Ocean Optics, Inc., Dunedin, FL). The MIDL was then placed on the microscope stage and a spectral image stack was collected. The spectrum from the image stack was extracted and compared with the spectrometer-measured spectrum to verify that emission peaks occurred at correct wavelengths. As the spectral peaks were aligned within the wavelength step-size of the HSi-300, no further wavelength-calibration was performed.
A modified version of a spectrofluorimeter correction procedure was used to account for wavelength-dependent response of the imaging system[33]. A sample blank was made for each type of sample imaged – a blank slide and coverslip mounted using antifade fluorescence mounting medium was used as the tissue blank, and a 25 mm round coverslip in a round holder bathed in Tyrodes solution was used as the cell-suspension blank. For a given measurement, the corresponding sample blank was placed on the stage and background (dark) and control (bright) spectral image sets were acquired using identical exposure settings as the tissue and cell samples. The dark image set was acquired with the excitation shutter open. The dark image set is a measure of background fluorescence, stray light, noise, and instrument offset that is present in all image sets for a particular acquisition configuration. The bright image set was acquired similarly to the dark image set, with the addition of a NIST-traceable light source (LS-1-CAL-INT, Ocean Optics, Inc., Dunedin, FL) that was adjusted to fill the dynamic range of the CCD camera at the highest output wavelength. The bright image set is a measure of how the spectral imaging system attenuates a known light source in a wavelength-dependent manner, for a particular acquisition configuration. The average dark and bright spectra were extracted from each of these image sets. The spectral transfer function was calculated as:
| (1) |
Where IBright is the bright spectrum, IDark is the dark spectrum, and ILamp is the real (NIST-traceable) lamp spectrum. A correction coefficient was also calculated as:
| (2) |
All images in this study were corrected by subtracting a dark spectrum and then multiplying by the correction coefficient, as follows:
| (3) |
Where IRaw is the raw spectral image stack and ICorrected is the corrected spectral image stack. Image correction and conversion to band-sequential image files was performed using MATLAB software (The MathWorks, Inc., Natick, MA).
Image acquisition
Spectral images of fluorescence emission were acquired at 5 nm increments, from 450-800 nm, using a 8.5 nm bandwidth (FWHM). The overlap in adjacent wavelengths results in a slightly decreased spectral resolution, but an increased signal strength, allowing spectral image stacks to be acquired with a shorter acquisition time and less photobleaching. For adherent cells on 25 mm round coverslips, an exposure time of 100 ms per wavelength was used. Five fields of view were acquired for each cell type (non-expressing and GFP-expressing PMVECs).
For lung cryoslices, an exposure time of 2000 ms was used. 16 fields of view were acquired from each animal (control and PMVEC-perfused lung). Fields of view were selected sequentially in a 4 × 4 field-of-view region to avoid operator bias. Background (dark) subtraction and flat-field correction were applied to all samples using a corresponding blank sample and identical acquisition settings.
For all samples, a nuclear (Hoechst 33342) and brightfield image was also collected for each field of view. Nuclear images were used in image processing to identify the center of cells while brightfield images were used to verify the structure of tissue cryoslices.
Single-band analysis
Single-band (a.k.a. single wavelength) analysis of the identical image sets was performed by summing fluorescence emission bands corresponding to a standard GFP emission filter, (ET525/50m, Chroma Technology Corp., Bellows Falls, VT). Single-band images were then thresholded to identify GFP-positive and negative pixels.
Theoretical sensitivity analysis
The sensitivity of single-band thresholding was assessed by artificially adding known amounts of GFP signal to spectral images. The GFP signal that was added was taken from a control sample of GFP-expressing cells grown on coverslip (as described above). Hence, this signal includes both the reference GFP spectrum, and some elements of instrument noise. This GFP signal was added to a 30 × 30 pixel square in the image. These images were summed to produce a single-band image, as described above, and subsequently thresholded, using three thresholds (corresponding to low, medium, and high sensitivities for GFP). The number of GFP-positive pixels was measured and plotted as a function of the amount of GFP added to the original spectral image.
Cell quantitation analysis
Measurement of GFP-expressing PMVECs was accomplished using Cell Profiler software.[34] An image analysis pipeline was created, using the nuclei and thresholded image as inputs. Thresholding and primary object detection was performed to locate all nuclei and GFP regions (defined as positive pixels in the single-band thresholded image). Nuclei contained within GFP regions were then identified through masking and categorized as GFP nuclei. Partially masked nuclei were excluded. All other nuclei were categorized as native (lung tissue) nuclei. The number, size, and shape of GFP-expressing cells per field-of-view was counted and output to an excel file.
Spectral analysis
The first step in supervised analysis of spectral image data is to construct a spectral library. ENVI software (ITT Visual Information Solutions, Boulder, CO) was used for spectral analysis and unmixing, and Excel (Microsoft Corporation, Redmond, WA) was used for manipulation, normalization, and plotting of extracted spectra. Spectral image stacks were visualized as false-colored RGB composites (red = 610 nm, green = 515 nm, blue = 480 nm).
GFP-expressing PMVECs were used as the pure GFP signal in the spectral library. For each field of view (FOV), cellular and background ROIs were defined through thresholding. Intensity-thresholding was performed at the peak bandwidth for GFP (500 nm). Pixels at or below the background threshold were identified as background, while pixels at or above the cellular threshold were identified as the cell spectrum. The intensity threshold was set arbitrarily high, to ensure that non-fluorescent or weakly fluorescent regions were not selected. The same intensity threshold was applied to all cellular FOV. For each FOV, the pixel-averaged background spectrum was subtracted from the pixel-averaged cell spectrum, producing a background-corrected fluorescent protein spectrum. This spectrum was averaged over all FOV and the resulting spectrum saved in the spectral library.
The tissue autofluorescence spectrum was selected through thresholding negative control samples to select regions of tissue autofluorescence, using the same method as described for cellular ROIs. As with the control cell images, intensity-thresholding was performed using the 500 nm band (the peak emission wavelength of autofluorescence). The intensity threshold was set arbitrarily high, to ensure that non-fluorescent or weakly autofluorescent regions were not selected. The same intensity threshold value was applied to the 16 fields of view acquired for the control lung. The pixel-averaged spectra from these ROIs were extracted from all 16 fields of view. These spectra were averaged, resulting in a FOV-averaged spectrum for tissue autofluorescence and background. The final spectral library contained three spectra (end-members): GFP, tissue autofluorescence, and tissue background. These spectra were normalized to unity and were used as the basis for linear unmixing and the theoretical sensitivity analysis.
Linear unmixing was performed using ENVI software. ENVI uses a linear least squares approach to calculate the abundance of each component (end-member) in the spectral library, for each pixel in the image. Because linear unmixing estimates the amount, or abundance, of each signal source in a pixel, it is often referred to as a sub-pixel analysis technique. Unmixed images were saved as tiff files and used as the input images for quantitative analysis.
Theoretical sensitivity analysis
The theoretical assay sensitivity was tested by artificially adding known amounts of GFP signal to negative control images, using a custom MATLAB script. For each negative control image, a 30 × 30 pixel square area was selected. Fluorescent protein signal was artificially added to this square area in fixed amounts, chosen as a percentage of the maximum autofluorescence intensity, at 500 nm, found across all of the control samples.
Spectral image stacks with artificially added GFP signal were unmixed using the linear unmixing procedure described. To determine the minimum detection limit of GFP, the unmixed GFP image was normalized by dividing by the intensity of the spectral image (at 500 nm – the peak wavelength for GFP and autofluorescence in this study). The normalized unmixed GFP intensity of pixels within the 30 × 30 pixel square was then plotted as a function of the amount of GFP added. The minimum detection limit was defined as the unmixed GFP intensity that was 3 standard deviations above a value of 0. Hence a pixel with GFP would have to be 3 standard deviations from the mean in order to not be detected as GFP-positive (Type II error).
All unmixed GFP images were then thresholded using this minimum detection limit to define GFP-positive pixels. The number of GFP-positive pixels was plotted as a function of the amount of GFP added. While spectral unmixing is not typically used to make a positive/negative decision such as this, this analysis was performed to compare the sensitivity of this spectral approach to single-band fluorescence microscopy (described above).
Cell quantitation analysis
Measurement of GFP-expressing PMVECs was accomplished using Cell Profiler software[34], similar to the process described above for single-band images. An image analysis pipeline was created, using the nuclei image and unmixed GFP, autofluorescence, and background images as inputs. Thresholding and primary object detection were performed to locate all nuclei and GFP regions. Nuclei contained within GFP regions were then identified through masking and categorized as GFP nuclei. All other nuclei were categorized as native (lung tissue) nuclei. Secondary object detection was subsequently performed to identify GFP-expressing cells and native (lung) cells. The number, size, and shape of each type of cell per field-of-view was counted and output to an excel file.
3. Results
We have modified an inverted fluorescence microscope with a tunable filter for hyperspectral image acquisition. We have used this microscope to acquire hyperspectral images of lung cryoslices containing GFP-expressing PMVECs. We have also developed analysis methods using spectral flat-field correction and linear unmixing that allow separation of GFP signals from autofluorescence and quantitation of PMVEC count. We have compared this hyperspectral microscopy approach to standard, single-band (a.k.a. single-wavelength) fluorescence microscopy, using both experimental and theoretical methods for assessing sensitivity and specificity. This approach holds great promise for cell-tracking and proliferation studies in both ex vivo and in vivo samples, especially for cases where traditional fluorescence microscopy fails to delineate between a desired fluorescence signal and background autofluorescence. The steps described herein, as well as measures for performing instrument calibration and estimating assay sensitivity, form a basic understanding from which many variants of biological hyperspectral imaging assays may be developed.
Limitations of single-band fluorescence microscopy
The standard approach for fluorescence imaging (single-band fluorescence microscopy) was evaluated for the case of GFP expressing cells in the pulmonary vasculature. The equivalent of a single-band fluorescence image was calculated by summing spectral bands within the range of a standard GFP-emission filter (ET525/50m, Chroma Technology Corp., Bellows Falls, VT). Single-band images were then intensity-thresholded to identify GFP emission.
Determining the theoretical sensitivity of single-band fluorescence microscopy
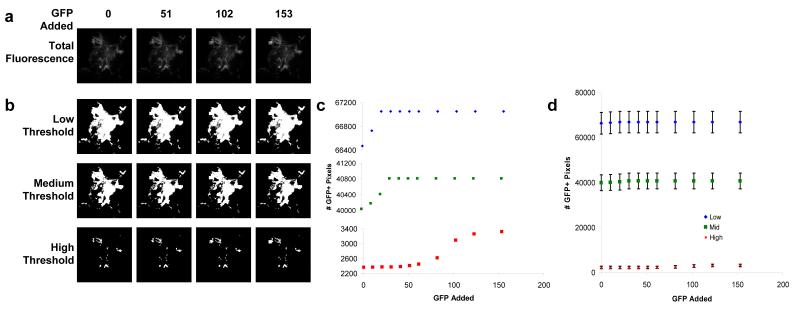
The sensitivity of single-band thresholding was assessed by artificially adding a 30 × 30 pixel region of known amounts of GFP signal to spectral images of lung autofluorescence. Spectral images were summed as described above to generate single-band images (Figure 3, A), which were intensity-thresholded to measure the sensitivity for detecting GFP (Figure 3, B). A high threshold resulted in a lower number of false-positives, but also a low sensitivity for detecting GFP with significant type II errors (Figure 3, C). By contrast, a low threshold resulted in a high number of false-positives, but also a higher sensitivity for GFP detection. It may be possible to slightly decrease the number of false-positives by using a narrower emission filter. However, we believe this improvement would be minimal, as the lung autofluorescence in these studies has the same peak fluorescence wavelength as GFP. Regardless of the threshold level, the number of false-positive events was far higher than that achieved through the hyperspectral approach, as described below. In addition, the variation in autofluorescence levels across different fields-of-view of the lung was much higher than the 30 × 30 pixel region, as evidenced by the standard error of the mean associated with this measurement (Figure 3, D, error bars).
Figure 3.
Theoretical sensitivity analysis revealed that single-band fluorescence imaging assays can be designed to have either high sensitivity with low specificity or low sensitivity with slightly improved specificity. A 30 × 30 pixel square of GFP signal was added to control hyperspectral images (A). Wavelengths between 500-550 nm were summed to approximate a standard GFP emission filter and thresholded to detect GFP signal based on intensity differences (B). Thresholded data are shown without error bars (C) to visualize the dependence of sensitivity (the minimum amount of GFP added that can be detected) and specificity (type I errors are represented by the number of GFP-positive pixels when 0 GFP was added); and with error bars (± SEM, n = 16 fields-of-view) to illustrate the high variance that is inherent in using single-band thresholding for GFP detection when there is a high level of autofluorescence (D).
Quantifying GFP-expressing cells using single-band intensity thresholds
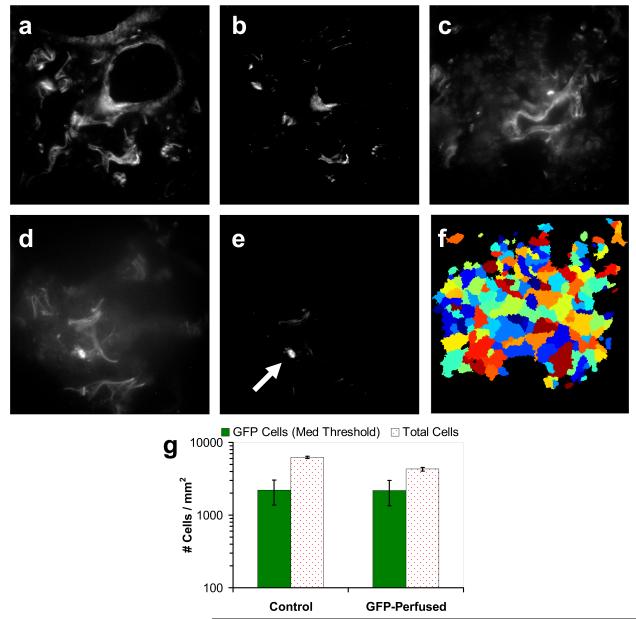
Experimental images of GFP-expressing cells in lung cryoslices were summed as described above and analyzed to determine whether a single-band intensity threshold could accurately detect these cells. The analysis results were nonspecific, identifying regions of high autofluorescence as well as GFP-expressing cells (Figure 4, A,B & C,D). Thresholded single-band images were analyzed using Cell Profiler software[34] to identify GFP regions based on fluorescence intensity (Figure 4, F). It is uncertain whether GFP-expressing cells were accurately identified using this technique (Figure 4, G), as there was an equivalent number of GFP-expressing cells detected in the control tissue (high incidence of type I errors). Although a high threshold decreased the incidence of false-positives, it also decreased the sensitivity for GFP detection (Figure 3, C), with no significant increase in specificity (Figure 4, B). Given the limitations of single-band fluorescence microscopy for detecting a fluorophore in highly autofluorescent tissue, we have developed the hyperspectral imaging approach (below) for differentiating GFP-expressing PMVECs from lung autofluorescence using linear spectral unmixing and quantitative image analysis.
Figure 4.
Single-band analysis of GFP-expressing pulmonary microvascular endothelial cells in lung cryosections reveals poor sensitivity and specificity for GFP-expressing cells. Using single-band (500 nm, 50 nm bandwidth) fluorescence emission, both control lung (A & B) and lung perfused with GFP-expressing endothelial cells (D & E) show high levels of fluorescence. Thresholding may be used in an attempt to identify GFP emission (arrow indicates a possible GFP-expressing cell), but also results in a significant number of false-positive regions, as evident in the control. For quantitative analysis, single-band images (C) were analyzed using Cell Profiler software[34] and intensity-thresholding techniques. Nuclei and GFP regions (F) were identified, and the total number of nuclei and the number of nuclei within GFP regions were measured (G). Results show a poor sensitivity and high incidence of type I errors. This threshold corresponds to the medium threshold in the theoretical sensitivity analysis (Figure 3) and the threshold used for spectral analysis (Figure 6).
Advantages of hyperspectral fluorescence microscopy and quantitative image analysis
The hyperspectral imaging approach described here consists of a combination of calibration, correction, sampling, and analysis steps. These steps ensure that: (1) the imaging system is wavelength-calibrated, (2) the measured signal is corrected for a flat spectral response, (3) non-specific system background is removed, (4) appropriate controls enable an accurate sampling of each spectral component, and (5) the spectra are accurately represented and quantitative analysis is possible.
Spectral calibration and flat-field correction
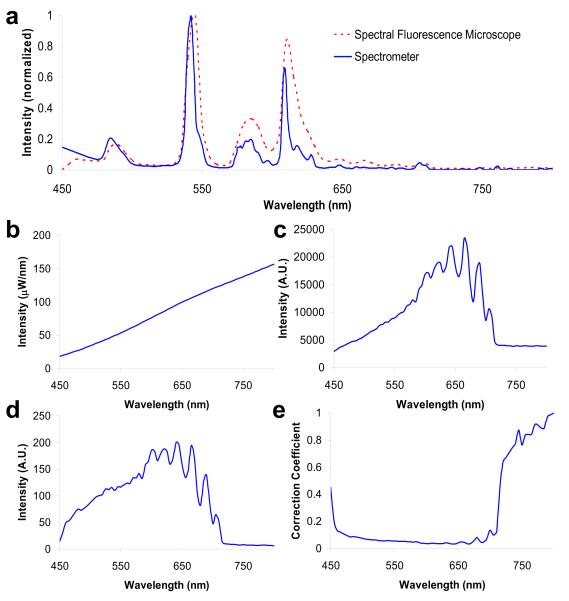
The first step in the hyperspectral microscopy approach was to ensure that the hyperspectral imaging system provided an accurate measurement of the fluorescence emission spectrum. Hence, prior to performing experimental measurements, the hyperspectral microscope was calibrated and corrected to produce a flat spectral response. These steps are important for insuring reproducible results and for comparing spectra across multiple systems. Wavelength calibration was performed using a multi-ion discharge lamp (MIDL). Measuring the MIDL spectrum using the spectral fluorescence microscope revealed that the factory calibration of the tunable filter was within the 8.5 nm bandwidth used for these experiments (Figure 5, A). Hence, no further wavelength calibration was performed.
Figure 5.
Spectral flat-field correction was used to compensate for wavelength-dependent attenuation of the fluorescence emission. Wavelength (A) and spectral flat-field calibration (B-E) were performed for the hyperspectral microscope. (A) Multi-ion discharge lamp peaks measured with the hyperspectral microscope aligned with peaks measured using a spectrometer to within the 5 nm spectral step size of the tunable filter; (B) the spectrum of the NIST-traceable lamp used to measure flat spectral response; (C) the NIST-traceable lamp spectrum as measured through the hyperspectral microscope; (D) the transfer function of the hyperspectral microscope; (E) the correction coefficient needed to achieve a flat spectral response.
Spectral flat-field correction was performed using a NIST-traceable light source (Figure 5, B). These measurements revealed significant wavelength-dependent attenuation, as evidenced by the spectral transfer function (Figure 5, D). This attenuation was primarily due to the transmittance spectrum of the long-pass dichroic beamsplitter, the transmission properties (at each tuning wavelength) of the tunable filter, and the quantum efficiency of the CCD detector. The oscillations in the transfer function that are visible above 650 nm are artifacts of the dichroic beamsplitter, as is the abrupt cut-off at 710 nm. Wavelength-dependent effects of the tunable filter and the CCD detector quantum efficiency are less obvious, but also contribute to the spectral transfer function. The inverse of the transfer function (Figure 5, E) was used as a correction coefficient that was applied to each image acquired to restore the system to a flat spectral response.
Determining the theoretical sensitivity of hyperspectral microscopy
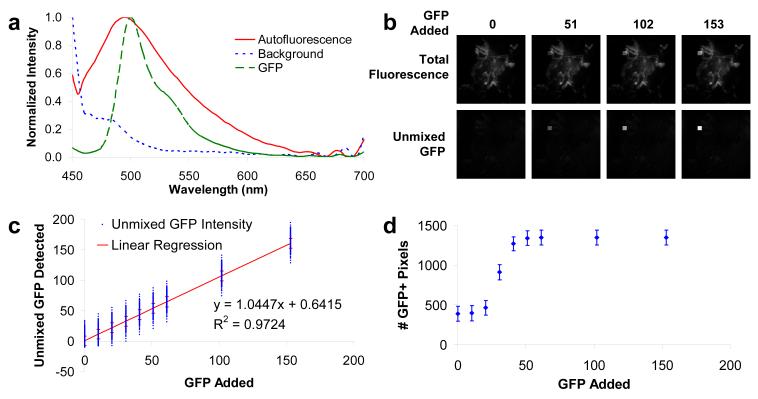
Once the hyperspectral microscope was calibrated and corrected for flat-field response, the sensitivity for detecting GFP in the presence of lung autofluorescence was measured. A theoretical sensitivity analysis was performed by artificially adding a 30 × 30 pixel square of GFP spectrum to control lung images (Figure 6, B). Linearly unmixed images accurately identified the 30 × 30 square, even at low levels of GFP. Results from this analysis revealed that GFP may be detected when the GFP signal contribution is as little as 15% of the peak autofluorescence intensity. Sensitivity results were recorded in terms of absolute GFP signal added and unmixed GFP signal detected – a standard method for assessing the static sensitivity of a detection system (Figure 6, C).[35] For reference, the peak autofluorescence signal across all fields-of-view was 204 intensity units. This curve represents an overall measure of the sensitivity that is achievable with hyperspectral microscopy for this specific assay, and includes experimental and equipment characteristics. Hence, this is a lumped sensitivity measurement, and should be repeated for changes in experimental or equipment conditions (different probes, tissues, equipment configurations, etc.).
Figure 6.
The theoretical sensitivity of the hyperspectral microscope for detecting GFP in lung cryoslices was tested by adding a 30 × 30 pixel square of GFP signal to control tissue hyperspectral images and performing linear unmixing. (A) The spectral library used for linear unmixing; (B) total fluorescence emission (top) and unmixed GFP (bottom) of a negative control hyperspectral image showing increasing amounts of added GFP signal; (C) the sensitivity response of the hyperspectral microscopy assay for GFP in lung tissues was used to select a GFP-detection threshold (in this case, when the unmixed signal was 3 std. dev. above the baseline), which was then used to determine whether a pixel was GFP-positive or GFP-negative; (D) the total number of GFP-positive pixels increases when the GFP signal is higher than the threshold determined from (C). Using this threshold, the mean occurrence of type I errors across all fields-of-view is 386 pixels (out of 262,144 pixels, or 0.15%). By contrast, the mean occurrence of type I errors for the highest threshold using single-band microscopy was 2367 pixels (0.9 %). Both specificity and sensitivity were significantly increased compared to single-band thresholding (Figure 3).
Once the minimum detection limit was identified, it was then used as the cut-off threshold for identifying GFP-positive pixels. The GFP detection threshold was defined as the value when the unmixed GFP signal was 3 standard deviations above 0. Hence, at this spectral threshold a pixel with GFP would have to be 3 standard deviations from the mean in order to be detected as containing no GFP (type II error). This threshold was applied across all unmixed control images with varying amounts of GFP signal added. Results show that GFP is detectable across a range of fields-of-view with a low incidence of type I errors and a high sensitivity (Figure 6, D), when compared to the single-band analysis (Figure 3, C). There was also more than an order-of-magnitude lower variation in unmixed hyperspectral images than in single-band images (as seen by comparing Figure 6, D and Figure 3, D). These results suggest this hyperspectral microscopy approach decreases the number of false-positive pixels while greatly improving the GFP sensitivity (decreasing the number of type II errors).
Spectral image analysis allows separation of GFP from autofluorescence signal
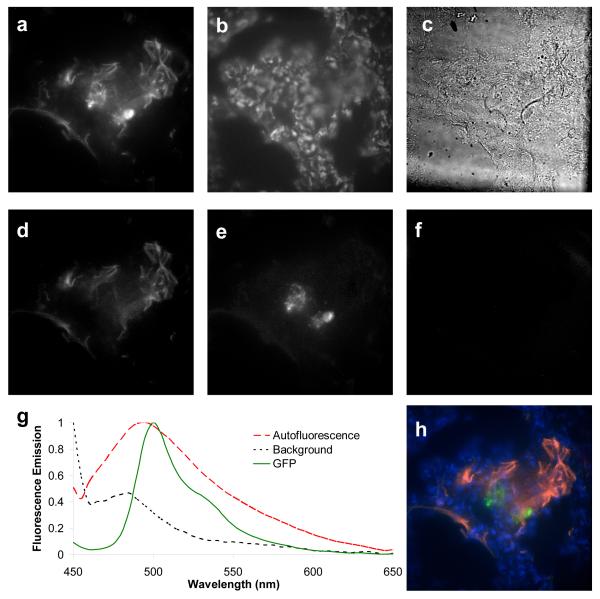
Accurate identification of GFP-expressing PMVECs is the critical outcome of the approach presented in this article. In contrast to single-band fluorescence microscopy, hyperspectral microscopy and subsequent spectral image analysis of PMVEC-perfused lung tissues revealed that GFP emission could be adequately separated from lung autofluorescence using linear unmixing. GFP-expressing PMVECs contributed to the total fluorescence emission, but were of no greater intensity than surrounding lung autofluorescence (Figure 7, A). However, linear unmixing of hyperspectral images effectively separated autofluorescence from GFP emission (Figure 7, D-F). False-colored image overlays allowed visualization of GFP and autofluorescence, within the context of Hoechst-labeled nuclei (Figure 7, H), while grayscale unmixed images were retained for quantitative analysis of PMVECs. Visual inspection of merged images revealed that perfused PMVECs accumulated in the pulmonary septal network (Figure 8, C). In this initial study, PMVECs were present as clusters, probably resulting from an inability to pass through narrow portions of the microvasculature or from possible rupturing of the microvasculature under perfusion conditions. Inspection of Hematoxylin and Eosin – stained cryoslices revealed relatively little disruption of the alveoli or septal network (Figure 8, A-B), confirming that the microvasculature was generally intact for these studies.
Figure 7.
Linear unmixing was applied to flat-field corrected spectral images. (A) Total fluorescence intensity of a 10 μm-thick lung cryoslice containing perfused GFP-expressing pulmonary microvascular endothelial cells (PMVECs); (B) image of Nuclei (Hoechst 33342 staining) with 360/40 nm excitation and 450 nm emission; (C) brightfield image at 500 nm; unmixed images of autofluorescence (D), GFP (E), and background (F); the spectra used for unmixing (G) were acquired from GFP-expressing PMVECs and negative control tissue; (H) false-colored overlay of unmixed image showing autofluorescence (red), GFP (green), nuclei (blue), and background (orange). Control lung cryoslices showed minimal false-positive GFP signal with identical imaging and linear unmixing (data not shown).
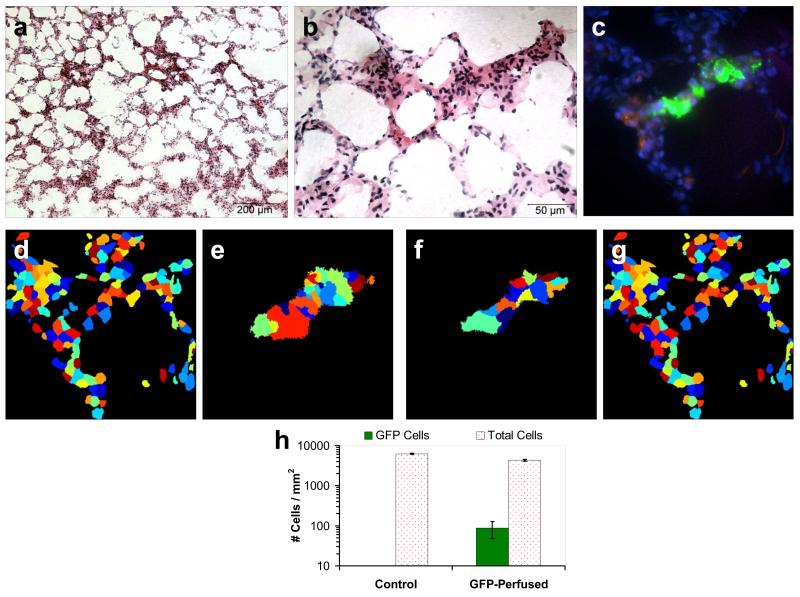
Figure 8.
Perfused pulmonary microvascular endothelial cells (PMVECs) accumulated within the alveolar septal network of rat lungs. Hematoxylin and Eosin stained 10 μm cryoslices at 10X (A) and 40X (B) magnification showed that the alveoli and surrounding septal network were intact. The linear-unmixed spectral fluorescence image of the subsequent 10 μm cryoslice shows GFP-expressing PMVECs (green), nuclei (blue), and autofluorescence (red). Unmixed images were quantitatively analyzed using Cell Profiler software[34]. Nuclei (D) and GFP-positive (E) regions were identified using thresholding algorithms – nuclei contained within GFP-positive regions were classified as PMVEC nuclei. The threshold for detecting GFP in spectral images was equivalent to the threshold used for single-band analysis (Figure 4, F). GFP-expressing PMVECs (F) and native tissue cells (G) were identified using secondary object detection. Using this approach, a mean value of 88 GFP-expressing cells was detected for the PMVEC-perfused tissue (H), with 0 type I errors detected within this sample size (n = 16 fields-of-view).
Quantifying GFP-expressing cells using hyperspectral microscopy
Quantitative image analysis was performed on unmixed images using Cell Profiler software[34]. Nuclei were defined as features in the image of Hoechst emission (360/40 nm excitation, 450 nm emission) with a diameter between 1.6-6 μm (Figure 8, D). Similarly, GFP cells were defined as having a diameter between 4-39 μm (Figure 8, E). Analysis results revealed an average of 88 ± 40 (standard error of the mean) GFP-expressing cells per field-of-view (Figure 8, H). By contrast, control images produced an average of 0 ± 0 GFP-expressing cells per field-of-view. These results indicate that the combination of hyperspectral imaging, linear unmixing, and quantitative image analysis can result in an approach with high sensitivity for GFP and the capability to detect individual GFP-expressing cells with little-to-no type I errors. By contrast, single-band fluorescence microscopy could only distinguish between changes in intensity, and for cases where GFP emission is equivalent to autofluorescence emission, resulted in a poor GFP detection sensitivity and a high level of false-positives.
4. Discussion
In this study, we have developed a hyperspectral imaging approach for tracking GFP-expressing PMVECs in the lung vasculature. This hyperspectral imaging approach was compared to traditional single-band (a.k.a. single-wavelength) fluorescence microscopy to assess its validity, and improvements in sensitivity and specificity, with corresponding abolishment of type I and II errors. While GFP emission was detectable using single-band fluorescence imaging, it was indistinguishable from surrounding autofluorescence (Figure 4, B & D). Consequently, intensity thresholding and image analysis using single-band fluorescence microscopy resulted in a much higher cell count than hyperspectral microscopy techniques (Figure 4, G and Figure 8, H). This higher cell count was the result of false-positive detection of GFP-expressing cells, as evidenced by the positive count in control tissue samples.
A theoretical sensitivity analysis was performed to measure the static sensitivity of the each approach to incremental increases in GFP signal. Results revealed that, for single-band microscopy, the number of false-positive pixels can be reduced by increasing the detection threshold (Figure 3, C). However, in doing so, the sensitivity for GFP detection was decreased to the point that very few true GFP-labeled pixels were detected (increased type II errors). More importantly, the image-to-image variations in autofluorescence intensity and the high number of autofluorescent pixels made it impossible to accurately identify GFP-labeled structures, whether they were the 30 × 30 pixel square or a GFP-labeled PMVEC (the occurrence of type I errors can vary significantly from one field-of-view to another). Hence, although the GFP region is in theory detectable, there is no way to discriminate this region from other highly autofluorescent pixels present in many of the fields-of-view. This implies that, in single-band microscopy, GFP-expressing cells will only be detectable when they are located in regions surrounded by relatively low autofluorescence. Conversely, regions of high autofluorescence will be inaccurately identified as GFP. These limitations of single-band fluorescence microscopy make it prohibitive for accurate and sensitive detection of GFP-expressing cells in highly autofluorescent tissues.
To address these limitations, the custom hyperspectral widefield fluorescence microscope and corresponding spectral calibration, correction, and analysis methods were developed. Linear unmixing of hyperspectral image sets revealed that GFP could be adequately separated from lung autofluorescence. A theoretical sensitivity study confirmed that GFP could be detected within lung tissues when the GFP intensity was only 15% of the peak autofluorescence signal. Visualization of overlayed unmixed images revealed that PMVECs aggregated in narrow portions of the pulmonary septal network (Figure 8, C). These results are in agreement with the expected distribution of cells in pressurized perfusion studies. Quantitative analysis of unmixed hyperspectral image sets found an average of 88 cells per mm2. This hyperspectral imaging and quantitative image analysis approach should be applicable in experiments in which single-band fluorescence microscopy fails to adequately delineate a given fluorophore from tissue autofluorescence. These experiments also highlight the need to determine the accuracy with which a label can be identified within a specific sample prior to performing quantitative analyses, such as cell counting or tracking.
Wavelength calibration and flat-field correction were critical components of this approach, and should be applied to correct for wavelength-dependent response in hyperspectral imaging systems. It should be noted that while flat-field correction restores a measured spectrum to its actual characteristics, photon-dependent effects, such as signal-to-noise, are not compensated for. For example, if equal intensity fluorescence emission were present at 600 and at 700 nm, the flat-field corrected image would display this spectrum as having equal intensities at both wavelengths. However, given the transfer function of Figure 5, D, the noise characteristics of 700 nm emission would be much poorer than those at 600 nm, as an order-of-magnitude fewer photons would be detected at 700 nm. Hence, specifying that each of the optical elements have as flat of a spectral response as possible is critical in a hyperspectral microscopy system. In the case of dichroic beam splitters, there are relatively few off-the-shelf solutions that present a sharp edge and also maintain a high percent transmission over a large wavelength range (450-800 nm, for example). Hence, although the dichroic beamsplitter used in this study is more than sufficient for GFP (with a wavelength range of 480-550 nm), a custom dichroic filter would be desirable if fluorophores in the far-red (>700 nm) portion of the spectrum were present.
To verify the accuracy of the hyperspectral system, the GFP-emission from in vitro culture (Figure 6, A) was also measured using a 32-channel spectral confocal microscope (A1, Nikon Instruments) and a spectrofluorimeter (data not shown). All three spectra were in agreement, although it should be noted that the GFP peak emission wavelength measured (500 nm) is blueshifted by about 10 nm from spectra that are often supplied in the default library of several spectral analysis software packages. This shift can cause significant effects when performing linear unmixing. Hence, when possible we recommend that the spectral library should always be collected from negative and positive test samples, rather than from manufacturer-measured spectra (often measured under different experimental conditions). As a final note, the flat-field correction performed here is a lumped-parameter correction. It is also possible to test each component in the fluorescence emission light path independently. While these optical testing methods are beyond the scope of this discussion, they are similar to those used in flat-field correction and may be helpful in diagnosing the source of unexpected spectral attenuation in a system.
The theoretical sensitivity of this hyperspectral microscopy approach was assessed by adding GFP signal to control (autofluorescence) images. This method maintains the variability of image information while adding known contributions of a specific signal. To determine a minimum sensitivity for GFP detection, the unmixed GFP signal was normalized to the total pixel intensity at the peak emission wavelength (500 nm, for both GFP and autofluorescence). Because the spectral library was also normalized (to unity), this represents a method for calculating the percentage of the total signal contributed by GFP. Using this method, a minimum detectible level of GFP was selected to be when the unmixed GFP signal was 3 standard deviations above 0 (as measured on a per-pixel basis). Detecting GFP-positive pixels based on this unmixed threshold revealed a large increase in both sensitivity and specificity, when compared to single-band microscopy (Figure 3, C and Figure 6, D). These results demonstrate that hyperspectral fluorescence microscopy is theoretically much more sensitive for detecting GFP emission than single-band microscopy and display a lower incidence of false-positive pixels. Although this sensitivity study applies to a specific experimental and equipment configuration, the improvements in sensitivity and specificity calculated here clearly portray the power of hyperspectral microscopy for separating fluorescence signals with similar – in this case identical – peak emission wavelengths. This theoretical sensitivity analysis is applicable for many other assay types, and could be modified to measure sensitivities of multiple fluorophores in complex hyperspectral microscopy assays.
The accuracy of spectral imaging analysis is highly influenced by the signal strength and noise characteristics of the detector, as well as perturbations due to the system as a whole (fluctuations in illumination intensity, ambient light, temperature, etc.). Hyperspectral measurements of the fluorescence emission spectrum are inherently noisy, when compared to broad-band measurements. This is because the narrow-band filtering required to image the fluorescence emission at many discrete wavelengths naturally results in decreased signal strength. We have estimated the SNR in selected hyperspectral images by using the method of Amer and colleagues [36], which has been previously demonstrated as applicable to fluorescence microscopy images [37]. For these images, the SNR of dominant spectral bands ranged from 6-20, while the SNR of lesser bands ranged from 0.3-0.7 (Supplemental Figure 2). As would be expected, the wavelength distribution of the SNR is similar in shape to the main spectral components that comprise an image, as these components represent the main signal source. One advantage to hyperspectral imaging – in addition to its discriminatory abilities – is that linear unmixing analysis often results in improved SNR. For example, prior to unmixing the SNR at 500 nm ranged from 10-15 for selected cryoslices containing both lung tissue and GFP-expressing cells. After unmixing, the SNR for the GFP-unmixed images ranged from 14-40, while the SNR for AF ranged from 0.3-10 (highly variable due to the varying levels of autofluorescence in different fields-of-view). Hence, there is commonly a 2-3 fold increase in SNR after unmixing, when compared to single-channel images. This is expected, as most linear unmixing algorithms take into account all of the spectral bands, and hence, are somewhat insensitive to noise or fluctuations in one single band. In this manner, hyperspectral imaging with linear unmixing could be thought of as a dimensionality-reduction technique that improves SNR characteristics.
While linear unmixing often increases the SNR ratio for components in the spectral library, the SNR of individual wavelength bands (prior to unmixing) is often relatively low, due to narrow bandwidth filtering. However, the SNR of individual wavelength bands can usually be improved through several methods: increasing the bandwidth of the spectral filter, increasing the image acquisition time, and increasing the detector gain. Increasing the bandwidth of the spectral filter can be achieved post-acquisition by binning spectral bands. Spectral band binning has the effect of improving the SNR, but decreasing the number of available bands for analysis. A more desirable alternative is to widen the bandwidth of the filter during image acquisition. In the HSi-300 tunable filter we have used for these studies, it is possible to vary the bandwidth of the filtered light, which allows for spectral oversampling. In these studies, a 5 nm step size was used between each band, while an 8.5 nm bandwidth was specified. This has the advantage of increasing the signal strength while decreasing the spectral resolution to a lesser extent than spectral band binning. More importantly, using a wider spectral band during acquisition (as opposed to post-acquisition spectral band binning) allows a decreased acquisition time to be used, minimizing spectral photobleaching artifacts (discussed below).
Another method for improving the SNR in hyperspectral imaging is to increase the image acquisition time. However, there are limits to the maximum acquisition time that can be tolerated before significant photobleaching has occurred. These limits should be more stringent in hyperspectral imaging instruments that acquire spectral bands sequentially – such as in this study – as time-dependent photobleaching will alter the measured spectral signature (spectral artifact). Because photobleaching rates vary between fluorophores, spectral artifact may affect different spectral components in an image to varying degrees, which is very difficult to compensate for. Hence, there is a trade-off between achieving a high SNR through increased image acquisition time and minimizing spectral photobleaching artifacts. An alternative to increasing the acquisition time is to increase the detector gain. These studies were performed using a cooled, electron-multiplying CCD (EMCCD) camera, in which it was possible to achieve a very high gain, although at the expense of increased noise characteristics (as demonstrated by the single-channel SNR values mentioned above).
Reproducible sample preparation played a key role in these studies, due to the fact that subtle variations in fluorescence spectra can have a large impact on unmixing results. It has been commonly reported that tissue fixation techniques can affect the tissue autofluorescence properties. Prior to beginning these studies, we compared lung tissues fixed through paraffin-embedding and cryofixation using optimal cutting temperature (OCT) compound. We found that paraffin-embedding resulted in much higher autofluorescence levels that OCT compound. Because of this, OCT-embedding and cryoslicing were selected as the method of fixation for these studies. To reduce the sample-to-sample variability, a background (dark) spectrum was subtracted from each hyperspectral image set prior to multiplying by the flat-field correction coefficient. However, if there is significant variability in tissue processing or imaging conditions, this background spectrum may not be uniform across cryosections, or even consecutive fields-of-view. Hence, it is important to maintain a stringent experimental protocol and to reduce stray (room) light during the imaging process. For these studies, the hyperspectral fluorescence microscope was installed on a vibration-isolated (floating) optical bench and electronically and optically shielded using a Faraday cage enclosure with photographic black-out cloth. Room lights were turned off during all imaging sessions.
During tissue preparation, nuclei were labeled with what would normally be considered a very low concentration of Hoechst stain. This was done to minimize the contribution of Hoechst emission to the tissue autofluorescence spectrum. Stained and unstained tissues were further compared spectrally to confirm that Hoechst emission contributed little to the autofluorescence properties of the tissue.
In addition to tissue processing, the optical scattering nature of tissues may also affect the accuracy of spectral analysis. The present analysis makes use of a linear mixing model, which assumes that each spectral component, or end-member, contributes independently to the total signal measured for any given pixel. The presence of optical scattering molecules, however, may remove this independence, as the optical pathlength will vary depending on the scatter coefficient, which is in turn dependent on the molecular composition of the tissue at any point. For example, a location composed primarily of collagen may present a longer effective pathlength than a location containing GFP. If linear unmixing were used to analyze this image, the abundance values for the pixel corresponding to the collagen location would deviate from the actual abundances that would be expected if the nonlinear nature of the mixing were taken into account. While we believe these effects are relatively small for the 10 μm cryoslices used in this study, comparing the differences in abundances that are produced with a linear versus a nonlinear mixing model remains a topic for further investigation. For an introduction into linear and nonlinear mixing models used in remote sensing, the reader is referred to Keshava and Mustard.[38]
Tissue autofluorescence emission spectra can vary widely with excitation wavelength.[2] Autofluorescence emission in the lung was found to be associated primarily with vascular, microvascular, and bronchiolar structures (Supplemental Figure 1, A). These structures have a complex cellular and extracellular composition. When multiple fluorophores in these structures contribute to a bulk autofluorescence spectrum, the rule of thumb of independent excitation and emission spectra cannot be applied (i.e., the “bulk” autofluorescence spectrum for a structure will be dependent on the excitation wavelength). In addition, micro-environmental changes (pH, ionic gradients, etc.) may affect the spectrum and quantum yield of fluorescent proteins. Because of this, we reiterate that spectral libraries, especially those containing tissue autofluorescence, should be generated from negative and positive controls using the exact settings as those used to image actual test samples. It is possible to store or compile large spectral libraries for future analysis. While this is a useful tool for saving and organizing spectra from many different experiments, extreme care should be taken when using spectra acquired under different conditions (because of the factors described above). The tissue autofluorescence spectrum from a previously saved library may not be the same autofluorescence spectrum presented by a new sample. At best, these differences will produce inaccuracies and higher levels of error in the spectral analysis. At worst, these differences will produce misleading results.
The broad emission spectrum of lung autofluorescence – between 450 and 600 nm – is likely due to combinations of native fluorophores, in varying environmental conditions. Emission peak wavelengths of elastin have been identified at 420 nm[1], 500 nm, and 520 nm[39]. This variance may indicate the dependence of fluorescence excitation and emission spectra on the cellular microenvironment and state of the fluorophore (e.g., changes in pH, elastin bundles, collagen cross-linking). Therefore, the broad fluorescence emission we have observed may very well be due to elastin, as well as other fluorophores native to the lung. In our studies we have observed a wide variation in tissue autofluorescence, with clearly identifiable changes in autofluorescence associated with different structures of the lung. For example, bronchiolar smooth muscle produces a predominantly 500-550 nm fluorescence emission while columnar epithelium has clear contributions in the 650-750 nm range (supplemental data). Thus, the analysis of spectral properties of native tissues may be a promising area for extracting additional information during a fluorescent protein assay. However, it should be noted that the addition of multiple variants of autofluorescence to the spectral library will result in more possible combinations of fluorophores that minimize the least-squares error solution that is often implemented in linear unmixing. Hence, for autofluorescence spectra that have small amounts of natural variation an appropriate number of controls should be performed to accurately sample the mean spectrum, while spectra that are clearly different in nature should be accounted for with different components in the spectral library. In this research, the autofluorescence spectrum represents a mean sampled of 16 fields-of-view, and should be widely applicable as a representative autofluorescence spectrum.
5. Conclusion
Hyperspectral microscopy and corresponding spectral analysis are powerful methods for detecting weak fluorescent signals within a high autofluorescence background. Our results indicate that hyperspectral microscopy has both increased sensitivity and specificity, compared to single-band microscopy methods. When combined with quantitative image processing, this approach has allowed us to detect single GFP-labeled cells in highly autofluroescent tissues with zero type I errors (false-positives). Our results also reinforce the need to accurately correct for the transfer function of the imager, and to include appropriate controls and training-set samples for building the spectral library. There are many opportunities for further advancing the effectiveness of hyperspectral microscopy. Other spectral analysis methods (such as mixture tuned matched filtering[40]) may better account for multiple, similar signals present in a sample and should be further investigated as potential methods for analyzing the molecular composition of cells and tissues. A combination of several excitation wavelengths while scanning the fluorescence emission spectrum may yield improved ability to discriminate amongst similar spectra. These advances will further increase the sensitivity and specificity of hyperspectral microscopy, as well as allow facilitate the translation of this technology to clinical devices.
Supplementary Material
Supplemental Figure 2 (A) Unmixed field-of-view from a lung cryoslice showing nuclei (blue), green fluorescent protein (GFP)-expressing pulmonary microvascular endothelial cells (green), autofluorescence (red), and nonspecific background (purple). (B) the corresponding signal-to-noise ratio (SNR) for: each spectral band prior to unmixing (black solid line), the unmixed GFP image (green dotted line, SNR = 39.6), the unmixed autofluorescence (AF) image (red dotted line, SNR = 4.2), and the unmixed background (BG) image (blue dotted line, SNR = 0.4).
Supplemental Figure 1 The native autofluorescence spectrum of lung tissue may vary greatly within a field of view. (A) Spectral image of a negative control lung (10 μm thick cryoslice, false-colored: 610 nm = red, 515 nm = green, 480 nm = blue); (B) normalized spectra from bronchiolar smooth muscle presents a predominantly green autofluorescence (510 nm peak) while bronchiolar epithelium is shifted towards the red and contains a significantly higher spectral contribution in the 650-700 nm range. Fluctuations between 650-700 nm are a result of imperfections in the dichroic beamsplitter.
Acknowledgements
Support for this work was provided by AHA0835134N (D.F.A.) and NIH R01HL094455 and S10RR027535 (T.C.R.). Student stipends were provided by the Alabama Space Grant Consortium and the University of South Alabama Undergraduate Research Program. The authors would also like to acknowledge support from the International Society for the Advancement of Cytometry Scholar’s Program and to thank Nikon Instruments for providing the HSi-300 tunable filter for evaluation.
Biography
Silas J. Leavesley received a B.Sc. in Chemical Engineering in 2003 from Florida State University and a Ph.D. in Biomedical Engineering in 2008 from Purdue University. He is currently an assistant professor of Chemical and Biomolecular Engineering and Pharmacology at the University of South Alabama. His research interests lie in the development of spectral imaging technologies for medical sciences research and clinical applications.
Naga Annamdevula received a B.Tech. in Biotechnology from Jawaharlal Nehru Technological (JNT) University, India in 2008. She is currently completing a M.Sc. in Chemical and Biomolecular Engineering at University of South Alabama, and pursuing a Ph.D. in Basic Medical Sciences at University of South Alabama. Her research interests lie in modeling and optimizing the detector characteristics of hyperspectral imaging microscopy systems for biomedical assays.
John Boni received a B.Sc. in Chemical Engineering (with departmental honors) from the University of South Alabama in May 2010. In August 2010, he began pursuing a Master’s degree in Chemical Engineering from the University of South Alabama. In his first year as a Master’s student, he assisted in teaching, and led experiments, for the Unit Operations Laboratory class for senior-level students. He began his professional career in June 2011 as a process engineer co-op at the Monsanto Company in Luling, LA.
Samantha Stocker received a B.Sc. in Chemical and Biomolecular Engineering from the University of South Alabama. She is currently employed at Chevron Corporation.
Kristin Grant received her B.S. in Chemical Engineering and Paper Science and Engineering in 2011 from North Carolina State University. She currently is working with DuPont.
Boris Troyanovsky received his Doctoral Degree from the Institute of Biochemistry & Physiology of Microorganisms in Pushino, Moscow in 1985. After three years as Posdoctoral fellow at the Institute of Molecular Biology & Genetic Ukranian Academy of Sciences, he joined for eight years the Cancer and Tumor Biology departments at the Karolinska Institute in Sweden. Since 2009 he has been working in Dr. Alvarez’s laboratory as a Research Associate at the University of South Alabama, College of Medicine, USA.
Thomas C. Rich received his B.AE. and M.Sc. in aerospace engineering from the Georgia Institute of Technology in 1988 and 1990, and his Ph.D. in biomedical engineering from Vanderbilt University in 1996. He is currently an Associate Professor in The Center of Lung Biology and Department of Pharmacology at the University of South Alabama College of Medicine. His research interests include the development of novel approaches to measure second messenger signals in real time, and the roles of intracellular signaling pathways in pulmonary physiology and pathophysiology.
Diego F. Alvarez received his Medical Degree in 1994 from La Universidad Libre de Colombia, Cali, Colombia, and his Ph.D. Degree in Basic Medical Sciences in 2005 from The University of South Alabama, Mobile, Alabama, USA. He is currently an Assistant Professor of Internal Medicine and Pharmacology. In conjunction with an enthusiastic group of collaborators, he has developed a model of acute lung injury and has pioneering work using endothelial progenitor cells to repair the lung. An strength of his research, is provide by integrating reductionist approaches using molecular and cellular biology, experimental animal models, and bedside work all of which aims to address the reparative capacity after acute lung injury.
Footnotes
Supporting information for this article is available free of charge under http://dx.doi.org/10.1002/jbio.[DOI]
References
- [1].Wagnieres GA, Star WM, Wilson BC. Photochem. Photobiol. 1998;68:603–632. [PubMed] [Google Scholar]
- [2].DaCosta R, Andersson H, Wilson B. Photochem. Photobiol. 2003;78:384–392. doi: 10.1562/0031-8655(2003)078<0384:mfemrt>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [3].Tsien R. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- [4].Ingram D, et al. Blood. 2004;104:2752–2760. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- [5].Ingram D, Mead L, Moore D, Woodard W, Fenoglio A, Yoder M. Blood. 2005;105:2783–2786. doi: 10.1182/blood-2004-08-3057. [DOI] [PubMed] [Google Scholar]
- [6].Serikov D, Popov B, Kropotov A, Tomilin N. Cytotherapy. 2005;7:483–493. doi: 10.1080/14653240500361012. [DOI] [PubMed] [Google Scholar]
- [7].Harris AT. Cytometry A. 2006;69:872–879. doi: 10.1002/cyto.a.20309. [DOI] [PubMed] [Google Scholar]
- [8].Lerner J, Gat N, Wachman E. Curr. Protoc. Cytom. doi: 10.1002/0471142956.cy1220s53. [DOI] [PubMed] [Google Scholar]
- [9].Tsagkatakis I, Peper S, Bakker E. Anal. Chem. 2001 Jan;73:315–320. doi: 10.1021/ac000832f. [DOI] [PubMed] [Google Scholar]
- [10].Forest S, Grothaus J. Microsc. Microanal. 2004;10:1304–1305. [Google Scholar]
- [11].Liyanage M, et al. Nat. Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- [12].Garini Y, Young I, McNamara G. Cytometry A. 2006;69:735–747. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- [13].Larson J. Cytometry A. 2006;69:825–834. doi: 10.1002/cyto.a.20305. [DOI] [PubMed] [Google Scholar]
- [14].Zimmermann T, Rietdorf J, Pepperkok R. FEBS Lett. 2003 Jul;546:87–92. doi: 10.1016/s0014-5793(03)00521-0. [DOI] [PubMed] [Google Scholar]
- [15].Rizzo M, Springer G, Granada B, Piston D. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- [16].Leavesley S, Jiang Y, Patsekin V, Rajwa B, Robinson J. Rev. Sci. Instrum. 2008 Feb;79:023707–023710. doi: 10.1063/1.2885043. [DOI] [PubMed] [Google Scholar]
- [17].Mansfield JR, Gossage K, Hoyt C, Levenson R. J. Biomed. Opt. 2005 Jul;10:41207. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- [18].Chaudhari A, et al. Phys. Med. Biol. 2005 Dec;50:5421–5441. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- [19].Zhao M, et al. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9814–9818. doi: 10.1073/pnas.161275798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gao X, Cui Y, Levenson R, Chung L, Nie S. Nat. Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- [21].So M, Xu C, Loening A, Gambhir S, Rao J. Nat. Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- [22].Thaler C, Vogel S. Cytometry A. 2006;69:904–911. doi: 10.1002/cyto.a.20267. [DOI] [PubMed] [Google Scholar]
- [23].Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. J. Biomed. Opt. 2005 Jul;10:041210–9. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- [24].Romier J, Selves J, Gastellu-Etchegorry J. Rev. Sci. Instrum. 1998;69:2859. [Google Scholar]
- [25].Vila-Francés J, et al. Rev. Sci. Instrum. 2006;77:073108. [Google Scholar]
- [26].Junttila M. Meas. Sci. Technol. 1993 Mar;4:740. [Google Scholar]
- [27].Frank J, Elder A, Swartling J, Venkitaraman A, Jeyasekharan A, Kaminski C. J. Microsc. 2007;227:203–215. doi: 10.1111/j.1365-2818.2007.01803.x. [DOI] [PubMed] [Google Scholar]
- [28].McNamara G, Gupta A, Reynaert J, Coates T, Boswell C. Cytometry A. 2006;69:863–871. doi: 10.1002/cyto.a.20304. [DOI] [PubMed] [Google Scholar]
- [29].King J, et al. Microvasc. Res. 2004;67:139–151. doi: 10.1016/j.mvr.2003.11.006. [DOI] [PubMed] [Google Scholar]
- [30].Alvarez D, Huang L, King J, ElZarrad M, Yoder M, Stevens T. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008;294:L419. doi: 10.1152/ajplung.00314.2007. [DOI] [PubMed] [Google Scholar]
- [31].Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Curr. Protoc. Mol. Biol. 2010:14.20.1–14.20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lerner J, Zucker R. Cytometry A. 2004 Nov;62:8–34. doi: 10.1002/cyto.a.20087. [DOI] [PubMed] [Google Scholar]
- [33].Lakowicz JR. Principles of fluorescence spectroscopy. Springer; 2006. [Google Scholar]
- [34].Carpenter A, et al. Genome. Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Webster JG. Medical instrumentation: application and design. John Wiley & Sons; 2010. [Google Scholar]
- [36].Amer A, Mitiche A, Dubois E. Reliable and fast structure-oriented video noise estimation; presented at the Proceedings of the International Conference on Image Processing, I-840; 2002. [Google Scholar]
- [37].Bernas T, Robinson JP, Asem EK, Rajwa B. J. Biomed. Opt. 2005 Nov;10:064015–9. doi: 10.1117/1.2136313. [DOI] [PubMed] [Google Scholar]
- [38].Keshava N, Mustard J. IEEE Signal. Process. Mag. 2002;19:44–57. [Google Scholar]
- [39].Richards-Kortum R, Sevick-Muraca E. Annu. Rev. Phys. Chem. 1996;47:555–606. doi: 10.1146/annurev.physchem.47.1.555. [DOI] [PubMed] [Google Scholar]
- [40].Williams P, Hunt A, Remote Sens ER., Jr. Environ. 2002;82:446–456. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 2 (A) Unmixed field-of-view from a lung cryoslice showing nuclei (blue), green fluorescent protein (GFP)-expressing pulmonary microvascular endothelial cells (green), autofluorescence (red), and nonspecific background (purple). (B) the corresponding signal-to-noise ratio (SNR) for: each spectral band prior to unmixing (black solid line), the unmixed GFP image (green dotted line, SNR = 39.6), the unmixed autofluorescence (AF) image (red dotted line, SNR = 4.2), and the unmixed background (BG) image (blue dotted line, SNR = 0.4).
Supplemental Figure 1 The native autofluorescence spectrum of lung tissue may vary greatly within a field of view. (A) Spectral image of a negative control lung (10 μm thick cryoslice, false-colored: 610 nm = red, 515 nm = green, 480 nm = blue); (B) normalized spectra from bronchiolar smooth muscle presents a predominantly green autofluorescence (510 nm peak) while bronchiolar epithelium is shifted towards the red and contains a significantly higher spectral contribution in the 650-700 nm range. Fluctuations between 650-700 nm are a result of imperfections in the dichroic beamsplitter.